ABSTRACT
Deformed wing virus (DWV) is the most prevalent Iflavirus that is infecting honey bees worldwide. However, the mechanisms of its infection and replication in host cells are poorly understood. In this study, we analyzed the structure and function of DWV 3C protease (3Cpro), which is necessary for the cleavage of the polyprotein to synthesize mature viral proteins. Thus, it is one of the nonstructural viral proteins essential for the replication. We found that the 3Cpros of DWV and picornaviruses share common enzymatic properties, including sensitivity to the same inhibitors, such as rupintrivir. The predicted structure of DWV 3Cpro by AlphaFold2, the predicted rupintrivir binding domain, and the protease activities of mutant proteins revealed that it has a Cys-His-Asn catalytic triad. Moreover, 3Cpros of other Iflaviruses and Dicistrovirus appear to contain Asn, Ser, Asp, or Glu as the third residue of the catalytic triad, suggesting diversity in insect RNA viruses. Both precursor 3Cpro with RNA-dependent RNA polymerase and mature 3Cpro are present in DWV-infected cells, suggesting that they may have different enzymatic properties and functions. DWV 3Cpro is the first 3Cpro characterized among insect RNA viruses, and our study uncovered both the common and unique characteristics among 3Cpros of Picornavirales. Furthermore, it would be possible to use the specific inhibitors of DWV 3Cpro to control DWV infection in honey bees in future.
IMPORTANCE The number of managed honey bee (Apis mellifera) colonies has considerably declined in many developed countries in the recent years. Deformed wing virus (DWV) vectored by the mites is the major threat to honey bee colonies and health. To give insight into the mechanism of DWV replication in the host cells, we studied the structure–function relationship of 3C protease (3Cpro), which is necessary to cleave a viral polyprotein at the specific sites to produce the mature proteins. We found that the overall structure, some inhibitors, and processing of 3Cpro are shared between Picornavirales; however, there is diversity in the catalytic triad. DWV 3Cpro is the first viral protease characterized among insect RNA viruses and reveals the evolutionary history of 3Cpro among Picornavirales. Furthermore, DWV 3Cpro inhibitors identified in our study could also be applied to control DWV in honey bees in future.
KEYWORDS: deformed wing virus, 3C protease, catalytic triad
INTRODUCTION
Winter colony loss in honey bees is strongly correlated with the presence of deformed wing virus (DWV) and the ectoparasitic mites, Varroa destructor and Tropilaelaps mercedesae (1–3). The mites transmit DWV to honey bee (3–6) and increase viral loads (6–10). With the spread of V. destructor, DWV has become the most prevalent virus that is infecting honey bees worldwide (11). Honey bees often show multiple symptoms due to high DWV levels, including the death of pupae, deformed wings, shortened abdomen, and reduced life span (4, 5, 12, 13).
DWV belongs to the genus Iflavirus in the order Picornavirales. It has a nonenveloped virion approximately 30 nm in diameter and contains a positive-strand RNA genome. The RNA genome is translated into a polyprotein that is processed by the 3C protease (3Cpro) to produce mature viral proteins (14). The crucial 3Cpros have been well characterized in picornaviruses infecting vertebrates and share similar structures and functions (15–21). Indeed, all of them have a Cys-His-Asp/Glu (Asp or Glu) catalytic triad. 3Cpro plays important roles in the viral life cycle and host-virus interactions. For example, 3Cpro not only processes a viral polyprotein but also cleaves the specific host proteins necessary for transcription, translation, and nucleocytoplasmic trafficking to modify the cell physiology for viral replication. Furthermore, 3Cpro and its precursor with RNA-dependent RNA polymerase (RdRP) bind to the 5’UTR region of the viral genome RNA to initiate replication (22, 23). Based on structural conservation studies, inhibitors of picornavirus 3Cpros have been identified and characterized to develop potential antiviral treatments (24).
DWV is the best characterized virus among honey bee viruses; however, very little is known about its mechanism of infection and replication in host cells. Because 3Cpro of insect RNA virus has never been studied, we expressed and purified DWV 3Cpro as a glutathione S-transferase (GST) fusion protein to characterize the protease activity and the inhibitors in this study. We also studied the structure–function relationship of DWV 3Cpro based on the structure predicted by AlphaFold2 as well as the protease activities of mutant proteins and compared to that of the 3Cpros of other Iflaviruses and Dicistrovirus. We focused on characterizing the catalytic triad since it represents the most critical domain of picornavirus 3Cpro. We further discussed the mechanism of synthesis and maturation of DWV 3Cpro in infected cells. Our study on DWV 3Cpro gives insight into the mechanism of viral replication and may help to control DWV infection in honey bees using the specific inhibitor.
RESULTS
Enzymatic properties of DWV 3Cpro.
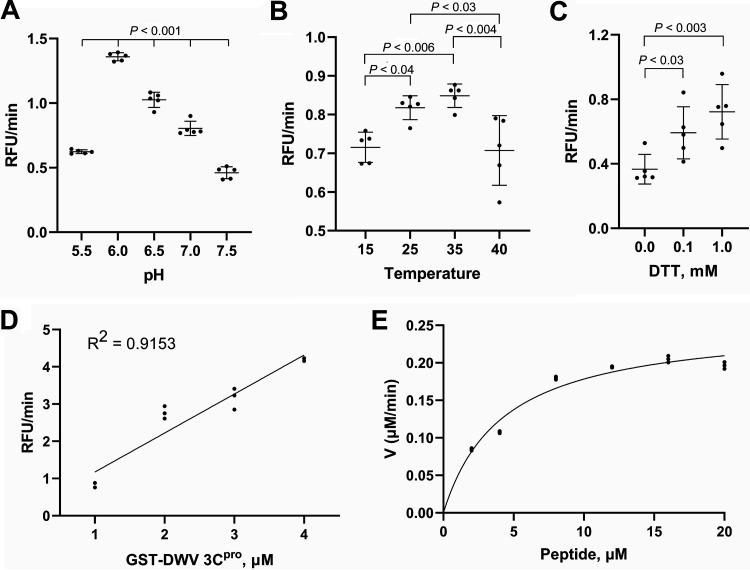
We expressed DWV 3Cpro as a GST-fusion protein and further included the potential VPg region (2094-2353 of DWV polyprotein) to make it soluble in E. coli. Protease activity of DWV 3Cpro was measured by a fluorescence resonance energy transfer (FRET) assay using a 15 amino acid peptide (PVQAKPEMDNPNPGE) with the cleavage site (25) as the substrate. Six amino acid sequence, AKPEMD, is present at three sites in the DWV polyprotein. To determine the optimum condition to measure the protease activity, we found that the optimum pH for protease activity is 6 (Fig. 1A), and the optimum temperature is between 25 and 35°C (Fig. 1B). Dithiothreitol was required for maximum activity as a cysteine protease (Fig. 1C) (26). The protease activity (RFU/min) was proportional to the amount of GST-DWV 3Cpro added in the reaction mixture (Fig. 1D). Based on the Michaelis-Menten equation, Km, Vmax, and Kcat/Km values were 4.233 ± 0.256 μM, 0.2538 ± 0.0067 μM min−1, and 59.96 ± 2.032 mM−1 min−1, respectively (Fig. 1E).
FIG 1.
Enzymatic properties of the deformed wing virus (DWV) 3C protease (3Cpro). (A) Optimum pH for protease activity. Multiple comparisons were made by the Tukey-Kramer method and the P-value between pH 6 and other tested pH was < 0.001. (B) Optimum temperature for protease activity. Multiple comparisons were made by the Tukey-Kramer method. Pairs with statistically significant differences are indicated by the P-values. (C) Dithiothreitol (DTT) requirement for protease activity. Multiple comparisons were made by the Dunnett method (one-tailed). Mean values ± standard deviation (SD) (error bars) are shown (n = 5) for A-C. (D) Protease activity with the indicated concentrations of glutathione S-transferase (GST)-DWV 3Cpro protein. (E) Protease activity (V, μM/min) at the indicated peptide substrate concentrations (2-20 μM). Mean values ± SD (error bars) are shown (n = 3) for D and E.
Inhibitors of DWV 3Cpro.
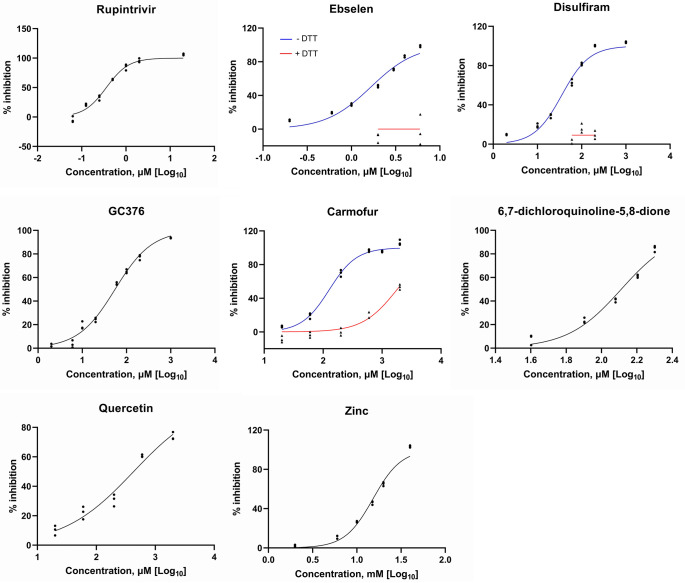
We next tested the effects of known protease inhibitors of picornaviruses and other viruses on DWV 3Cpro. Eight compounds inhibited enzyme activity with various IC50 values, as shown in Table 1. Fig. 2 shows the dose-response curves for rupintrivir (27), ebselen (28), disulfiram (29), GC376 (30), carmofur (29, 31), 6,7-dichloroquinoline-5,8-dione (32), quercetin (33), and zinc. Rupintrivir was the most effective compound with the IC50 value of 0.36 μM. Ebselen, disulfiram, and carmofur were only effective without dithiothreitol, as they modified cysteine in the catalytic triad (29). DWV 3Cpro is sensitive to some protease inhibitors of mammalian viruses, suggesting that their catalytic domains and substrate-binding domains share similar structures.
TABLE 1.
IC50 values of DWV 3Cpro inhibitors
| Compound | IC50 (μM) |
|---|---|
| Rupintrivir | 0.36 ± 0.03 |
| Ebselen | 1.66 ± 0.02 |
| Disulfiram | 35.6 ± 0.8 |
| GC376 | 55.2 ± 4.0 |
| Carmofur | 126.7 ± 6.2 |
| 6,7-dichloroquinoline-5,8-dione | 129.3 ± 1.8 |
| Quercetin | 430 ± 62.8 |
| Zinc | 15240 ± 422 |
FIG 2.
Compounds that inhibit the activity of the deformed wing virus (DWV) 3C protease (3Cpro). Protease activity of DWV 3Cpro in the presence of increasing concentrations of rupintrivir, ebselen, disulfiram, GC376, carmofur, 6,7-dichloroquinoline-5,8-dione, quercetin, and zinc. Ebselen, disulfiram, carmofur were tested in the absence (blue line) or presence (red line) of 2 mM dithiothreitol (DTT). Other compounds were tested in the presence of DTT. All data (n = 3) are shown at the indicated concentrations.
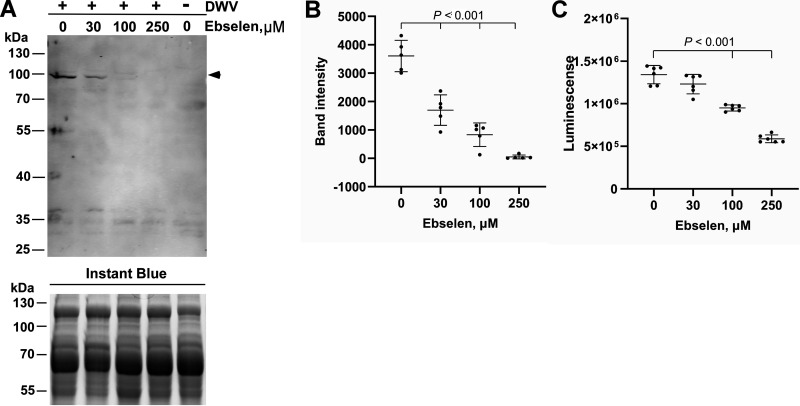
We previously showed that rupintrivir suppresses DWV replication in honey bee pupal cells based on the synthesis of RdRP (34). Thus, we tested the effect of ebselen on DWV replication. Ebselen decreased the synthesis of the 97 kDa precursor of RdRP and 3Cpro in a concentration-dependent manner (Fig. 3A and B). However, cell viability was reduced at concentrations of 100 and 250 μM (Fig. 3C), suggesting that ebselen affects the functions of host proteins as well.
FIG 3.
Ebselen suppresses RNA-dependent RNA polymerase (RdRP) synthesis in the deformed wing virus (DWV)-infected pupal head cells. (A) Effect of increasing concentrations of ebselen on RdRP synthesis in DWV-infected (+) pupal head cells. Dimethyl sulfoxide (DMSO) was used as the control. RdRP precursor with 3C protease (97 kDa) is indicated by a black arrowhead. This band was absent in DWV-uninfected (−) cells. The replicate gel was stained with Instant Blue to show that equal amount of protein was applied in each lane. Molecular weight (kDa) of the protein marker is indicated at the left. (B) Band intensity of the RdRP precursor was compared between DMSO and ebselen at the indicated concentrations by the Dunnett method (one-tailed). Mean values ± SD (error bars) are shown (n = 5). (C) Luminescence generated by the luciferase activity dependent on the intracellular ATP level was compared between the pupal head cells treated with DMSO or ebselen at the indicated concentrations by the Dunnett method (one-tailed). There was no significant difference between DMSO and 30 μM ebselen. Mean values ± SD (error bars) are shown (n = 5).
Predicted structure of DWV 3Cpro by AlphaFold2.
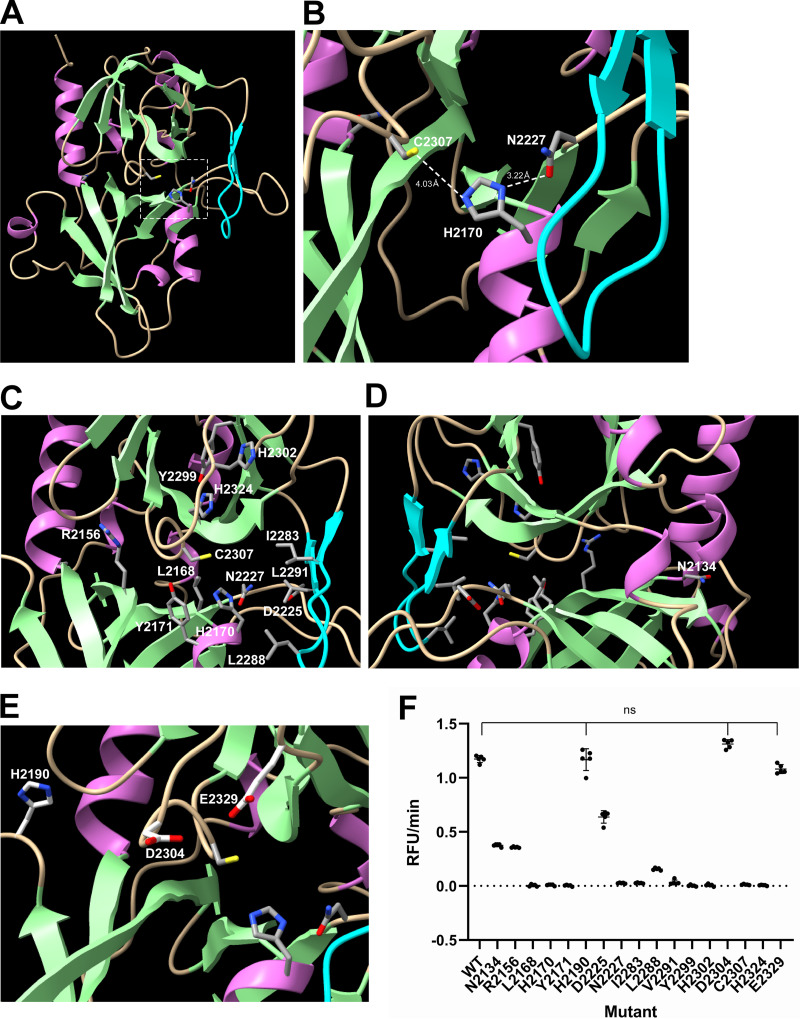
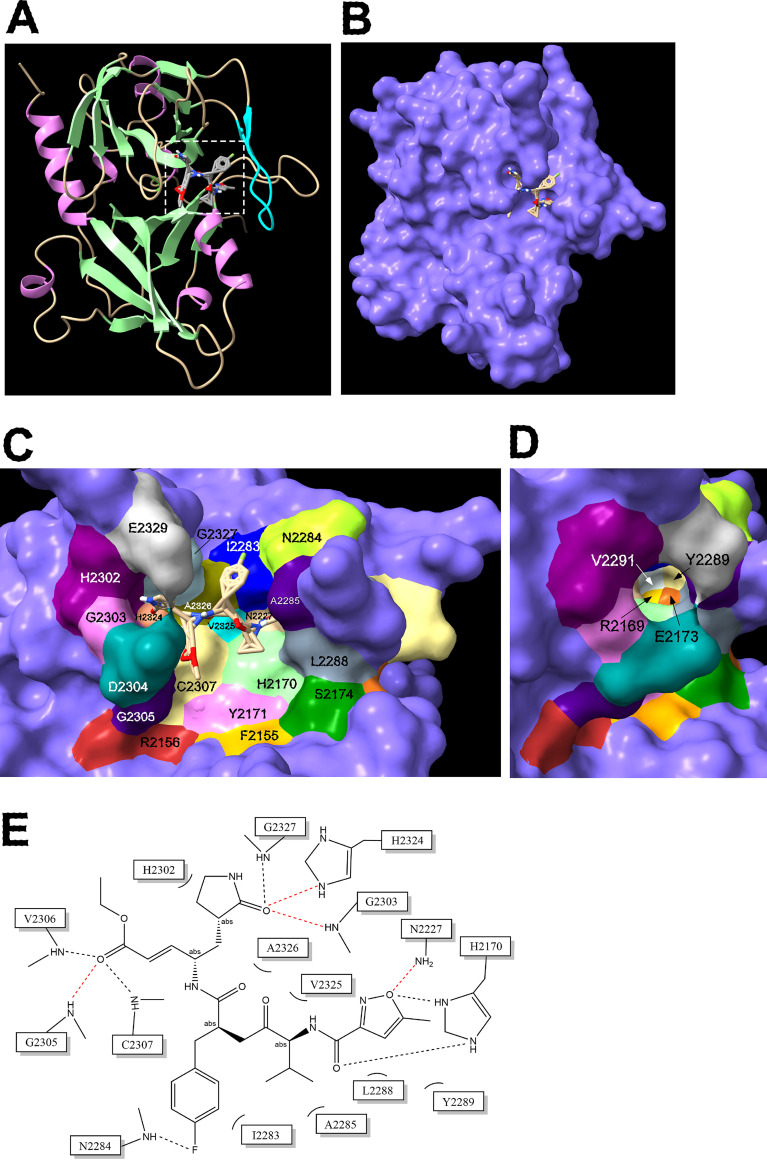
We used AlphaFold2 to predict the structure of DWV 3Cpro (polyprotein 2199–2353) (35, 36). We obtained multiple models and one of them with the overall structure is shown in Fig. 4A. Based on the IDDT values per position, the structure was predicted with high confidence, except for the N- and C termini as well as single loop (polyprotein 2330–2332) (Fig. S1 in the supplemental material). DWV 3Cpro has a trypsin-like structure consisting of six β-sheets, which are folded into two β-barrel domains packed perpendicularly to each other. The catalytic center appears to be present between the two domains. This is similar to those of other 3Cpros in addition to the position of β-ribbon (polyprotein 2281–2293) which constitutes the substrate-binding domain (15–21) (Fig. 4A). To identify the amino acids critical for protease activity, we aligned 3Cpro sequences of DWV and 23 other Iflaviruses (Fig. S2 in the supplemental material). Among the conserved 12 amino acids, the positions of C2307 and H2170 together with nonconserved N2227 in the DWV 3Cpro structure corresponded to those of the known 3Cpro catalytic triads (15–21). Although Y2171 was well conserved, its position relative to H2170 did not appear to be compatible with the third residue of the catalytic triad. There is also D2225, but the side chain with a carboxyl group was oriented in the opposite direction (Fig. 4C and D). The predicted distances between C2307 and H2170 as well as H2170 and N2227 were 4.03 Å and 3.22 Å, respectively in range of interaction (Fig. 4B). We also predicted the 3Cpro structures of four different Iflaviruses: Brevicoryne brassicae virus (BrBV), Laodelphax striatellus picorna-like virus 2 (LsPVI2), Sacbrood virus (SBV), and Spodoptera exigua iflavirus (SeIV-1), and one Dicistrovirus, Cricket paralysis virus (CrPV). For the catalytic triads, all of them had Cys and His residues; however, BrBV, LsPVI2, and SeIV-1 appeared to contain Asn, Ser, and Glu, respectively, as the third residue. SBV and CrPV appeared to have Asp (Fig. S3 in the supplemental material). Thus, 3Cpro of insect RNA virus is likely to have invariant Cys as well as His and various amino acid with side chain containing oxygen as the catalytic triad.
FIG 4.
Structure–function analysis of the deformed wing virus (DWV) 3C protease (3Cpro). (A) Structure of DWV 3Cpro predicted by AlphaFold2. The helix, strand, and coil structures are indicated by orchid, pale green, and wheat colors, respectively. β-ribbon is colored by cyan. The catalytic Cys-His-Asn triad is indicated by a square with a white dotted line. (B) Close-up view of the catalytic C2307-H2170-N2227 triad with distance information. (C) Positions of amino acid residues critical for the protease activity. (D) The position of N2134 that was well conserved between 3Cpros of Iflaviruses is shown by rotating the image (C) by 180°. (E) The positions of H2190, D2304, and E2329, which are not essential for the protease activity, are shown together with the catalytic C2307-H2170-N2227 triad. (F) Protease activities of wild type (WT) 3Cpro and alanine substituted mutants. Compared to WT, all mutants showed decreased activities, except H2190A, D2304A, and E2329A (ns), as indicated by the Dunnett test (one-tailed, n = 5).
To further examine the structure of DWV 3Cpro predicted by AlphaFold2, we prepared mutant proteins in which the potentially critical amino acid was substituted with alanine and measured the protease activities. The activities were dramatically reduced with the mutant proteins for the catalytic triad (C2307, H2170, and N2227) and substrate-binding domains, including conserved Y2299 and H2324 domains (Fig. 4C and F). N2134 of DWV is also well conserved between the 3Cpros of Iflaviruses, but it localizes far from the catalytic center (Fig. 4D). Nevertheless, we found that the protease activity of the N2134A mutant was reduced to approximately one-third of that of the wild-type protein (Fig. 4F). A previous study suggested that H2190 and D2225 constitute the catalytic triad together with C2307 (14). However, H2190 was far from C2307 (Fig. 4E), and the H2190A mutant and wild-type protein showed comparable protease activity (Fig. 4F). D2225 is close to N2227 as described above, but the protease activity of D2225A was reduced to only half of that of the wild-type protein (Fig. 4F). Thus, H2190 and D2225 are not part of the catalytic triad of DWV 3Cpro. D2304A and E2329A mutants have the same activity as the wild-type protein, indicating that they are not the third residue of the catalytic triad (Fig. 4E and F). The above results are consistent with the structure of DWV 3Cpro predicted by AlphaFold2.
We next determined how rupintrivir binds DWV 3Cpro to inhibit its activity using a molecular docking tool. Rupintrivir was predicted to interact with 16 amino acids in the catalytic triad (C2307, H2170, and N2227) as well as a substrate-binding domain. Nitrogen atoms in the side chains of N2227 and H2324 appeared to form hydrogen bonds with rupintrivir. Five amino acids (I2283, N2284, A2285, L2288, and Y2289) in β-ribbon also appeared to interact with rupintrivir (Fig. 5A, B, and E). Thus, 3Cpros of DWV and picornaviruses appear to bind rupintrivir in a similar manner (15, 37, 38). Analysis of rupintrivir binding with DWV 3Cpro revealed a precise map of the amino acids critical for protease activity at the protein surface. The catalytic triad (C2307, H2170, and N2227) are adjacent to each other; R2156 and Y2171 are adjacent to C2307 and H2170. The largely reduced protease activities of R2156A and Y2171A mutants were consistent with their localization (Fig. 4F). The substrate-binding domain is composed of two grooves on the left and right sides, and V2325 and A2326 are in the middle to connect them. The left groove was constructed using H2302, G2303, D2304, G2305, H2324, G2327, and E2329. Among them, H2302, G2303, D2304, and E2329 create a porous structure. The deep right groove is made by R2169, E2173, I2283, N2284, A2285, L2288, Y2289, and V2291. Most of the amino acids were in the β-ribbon structure (Fig. 5C and D).
FIG 5.
Amino acids interacting with rupintrivir in deformed wing virus (DWV) 3C protease (3Cpro). (A) Overall structure of rupintrivir-bound DWV 3Cpro. Rupintrivir binding site is indicated by a square with white dotted line. (B) Surface view of rupintrivir bound DWV 3Cpro. The binding site of rupintrivir was predicted by the molecular docking tool, AutoDock Vina. (C) Amino acids interacting with rupintrivir as well as the neighboring ones are indicated by different colors. Rupintrivir is also shown. (D) Amino acids in the deep right groove of the substrate-binding domain created by β-ribbon are indicated by rotating the image (C) 45° without rupintrivir. (E) Sixteen amino acids interacting with rupintrivir. Hydrogen bonds are indicated by red dotted lines.
Self-cleavage of GST-DWV 3Cpro.
When we purified the 56 kDa wild-type GST-DWV 3Cpro protein, a 30 kDa small band was co-purified. This band was absent in the mutant protein lacking protease activity, for example, C2307A (Fig. 6A). These results suggest that the 30 kDa band was generated by the self-cleavage of GST-DWV 3Cpro either by cis or trans and contained GST. Our DWV 3Cpro fusion protein contains two potential cleavage sites, Q2118 and E2180 (14). To determine the cleavage site, we prepared mutant proteins in which one of the above two amino acids was substituted with alanine. As shown in Fig. 6A, the 30 kDa band was absent only in the Q2118A mutant, suggesting that GST-DWV 3Cpro is self-cleaved at Q2118 in E. coli. We also tested whether the purified GST-DWV 3Cpro was self-cleaved during incubation at 33°C and found that it remained stable without self-cleavage for 18 h (Fig. 6B).
FIG 6.
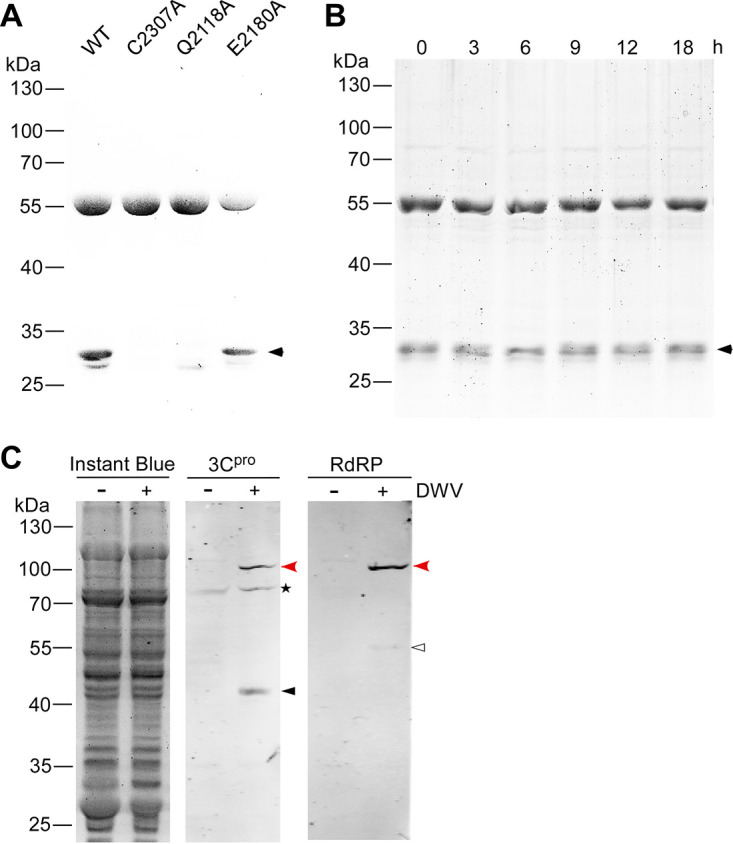
Self-cleavage of the deformed wing virus (DWV) 3C protease (3Cpro) and its synthesis in the virus-infected honey bee cells. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of purified wild type (WT) and three mutant (C2307A, Q2118A, and E2180A) glutathione S-transferase (GST)-DWV 3Cpro proteins. The 30 kDa band co-purified with 56 kDa GST-DWV 3Cpro is indicated by a black arrowhead. Molecular weights (kDa) of the protein marker are at the left. (B) SDS-PAGE of purified GST-DWV 3Cpro incubated at 33°C for the indicated time. (C) Western blotting of lysates of the control (−) or DWV-infected (+) honey bee pupal head cells by anti-3Cpro (3Cpro) or anti-RdRP (RdRP) antibody. The replicate gel was stained with Instant Blue to show that equal amount of protein was applied in each lane. Mature 42 kDa 3Cpro and 55 kDa RdRP are indicated by black and white arrowheads, respectively. The 97 kDa precursor of 3Cpro and RdRP (red arrowheads) was detected by both anti-3Cpro and anti-RdRP antibodies. The above-mentioned three bands were specifically present in DWV-infected cells. Asterisk represents the nonspecific band detected by the anti-3Cpro antibody.
Synthesis of 3Cpro in DWV-infected honey bee cells.
To characterize the 3Cpro synthesized in DWV-infected honey bee cells, we raised antibodies against DWV 3Cpro. We found that a 97 kDa protein specifically present in DWV-infected cells is recognized by both anti-3Cpro and anti-RdRP antibodies, confirming that this is a 3Cpro precursor with RdRP. There was also a 42 kDa band specifically recognized by the anti-3Cpro antibody, suggesting that this corresponds to the mature 3Cpro in DWV-infected cells (Fig. 6C). Thus, DWV 3Cpro is present as both a precursor and a mature protein in the infected cells.
DISCUSSION
Compared to the Km values of the best peptide substrate for 3Cpros of human rhinovirus (250 μM), poliovirus (7 μM), and enterovirus (30 and 43 μM) (21, 39–41), DWV 3Cpro has a higher affinity for the peptide substrate used in this study (Km: 4.233 μM). Nevertheless, DWV polyprotein has other potential cleavage sites by 3Cpro so that the efficiency of cleaving a peptide substrate containing these sites would be varied. The Kcat/Km value of DWV 3Cpro was 59.96 mM−1 min−1 and comparable to those of hepatitis A virus (126 mM−1 min−1), enterovirus (11.8 and 0.71 mM−1 min−1), and foot-and-mouth disease virus (59.4 mM−1 min−1) 3Cpros with their best substrates (21, 41–43). These results suggest that the presence of Asn instead of Asp/Glu in the catalytic triad of DWV 3Cpro does not dramatically affect protease activity. In fact, substituting Asp in the catalytic triad of hepatitis A virus 3Cpro with Asn only resulted in slower processing (44).
Rupintrivir was the most effective to inhibit DWV 3Cpro activity and the IC50 value (0.36 μM) was lower than that of enterovirus 3Cpro (1.65–7.3 μM) but higher than that of human rhinovirus 3Cpro (5 nM) (37, 45, 46). Ebselen, disulfiram, and carmofur have been shown to inhibit the main protease of SARS-CoV-2 by covalently modifying cysteine in the catalytic diad (28, 29, 31). Although we were not able to predict the binding site of ebselen in DWV 3Cpro using a molecular docking approach, it is likely to target C2307 in the catalytic triad to inhibit protease activity. Ebselen inhibited DWV replication in honey bee cells; however, it was also toxic to the cells (Fig. 3). Thus, ebselen (MW: 274.18) could be a potential lead compound for fragment-based drug discovery approaches to increase the specificity and potency to DWV 3Cpro. Rupintrivir and quercetin (34) as well as ebselen inhibited DWV replication in cultured honey bee cells. Thus, these compounds and the other 3Cpro inhibitors we identified in this study could be used to control DWV in honey bee colony.
In contrast to picornaviruses, DWV 3Cpro contains Asn (N2227) as the third residue of the catalytic triad instead of Asp/Glu (Fig. 4A and B). Moreover, Asn is not conserved among Iflaviruses, and in fact other Iflaviruses and Dicistrovirus appear to have Asn/Ser/Glu/Asp residue (Fig. S2 and S3 in the supplemental material). The third residue of the triad could be flexible as long as it contained an oxygen in the side chain. Oxygen is necessary to maintain the entire architecture of the active site and stabilize the positively charged imidazole side chain of His during proteolysis (37). Meanwhile, Cys and His in the catalytic triad are under strong purifying selection, suggesting that they are the most critical amino acids for protease reaction. This is consistent with the fact that the main protease of coronavirus has the Cys-His catalytic diad (47). Because the N2227A mutant completely lost its protease activity (Fig. 4F), and the same was observed with other 3Cpros (48–50), the catalytic triad seems to be necessary for Picornavirales. In many Iflaviruses, the Tyr residue next to the His residue of the catalytic triad (Y2171 for DWV) is conserved and is substituted with Phe in the Dicistrovirus, CrPV. Thus, Tyr appears to be important to position the imidazole side chain of His close to the side chain of Cys by stacking interactions.
The pores made by H2302, G2303, D2304, and E2329 in the left groove of the substrate-binding domain did not appear to be essential because the protease activities of the D2304A and E2329A mutants were not affected (Fig. 4F). β-ribbon is the major component of the deep right groove of the substrate-binding domain. In picornavirus 3Cpros, the β-ribbon adopts an open conformation to increase the substrate accessibility to its binding domain, and the interaction between the β-ribbon and the N-terminal end of the substrate stabilizes the closed conformation to form an ES complex (15–21). Comparing the surface views of the substrate-binding domains of DWV, BrBV, LsPVI2, SeIV-1, SBV, and CrPV 3Cpro (Fig. 5C and D; Fig. S4 in the supplemental material), the overall conformation of the catalytic triad and β-ribbon was similar, but the shapes of substrate-binding domains were quite different. Thus, the structure of the substrate-binding domain of each 3Cpro is shaped by a preferred cleavage site with a different amino acid sequence.
We found that GST-DWV 3Cpro was self-cleaved at Q2118. However, this seems to only occur during the synthesis in E. coli because the mature folded protein was not self-cleaved at 33°C for 18 h (Fig. 6B). These results suggest that only the fraction of unfolded GST-DWV 3Cpro is cleaved in trans at 15°C. It is likely that DWV polyprotein is cleaved at Q2118 to release the 97 kDa precursor of 3Cpro and RdRP. However, we need to assume that the 97 kDa precursor is active as a protease similar to picornavirus 3CD precursor (51) and that cleavage should occur in cis, at least during the early stage of polyprotein synthesis until a sufficient 97 kDa precursor and 42 kDa mature 3Cpros accumulate in DWV-infected cells. Because the 97 kDa precursor remains abundant in the virus-infected honey bee cells (Fig. 6C), processing between 3Cpro and RdRP appears to be slower than the cleavage at Q2118. These characteristics appear to be shared among picornaviruses as well (52). Considering the size of mature 3Cpro (42 kDa), the cleavage site should be beyond E2353 which was the C-terminal end of GST-DWV 3Cpro. It will be interesting to test whether the 97 kDa precursor and 42 kDa mature 3Cpros have different protease activities and functions for viral RNA replication in future studies.
MATERIALS AND METHODS
Expression and purification of GST-DWV 3Cpro.
The 3C protease cDNA corresponding to amino acid 2094–2353 of DWV polyprotein was amplified by PCR using two primers, GST-3Cpro-5-BamHI and GST-3Cpro-3-NotI (Table S1). The amplified PCR product was digested by BamHI (NEB) and NotI (NEB), and then subcloned to pGEX-4T-1vector (Cytiva) followed by transformation to BL21. The transformed BL21 was grown in 1 L of LB medium containing 1% glucose and 0.1 mg/mL Ampillicin at 37°C until A600 reached to 0.5. The cell suspension was cooled down, and then IPTG was added at 0.1 mM to induce the protein expression at 15°C for 16 h. E. coli was collected by centrifugation and resuspended in 100 mL of ice-cold TNED buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1 mM DTT) with 0.5% TX-100 and protease inhibitor cocktail (Beyotime). The cell lysate was prepared by sonication using Q700 Sonicator (Qsonica) at amplitude 100 on ice for 45 min (30 sec pulse with 3 min-interval). 1 mL of BeyoGold GST-tag Purification Resin (Beyotime) was added to the supernatant collected after centrifugation. After gently rotating at 4°C for 2 h, the resin was washed five times with 10 mL TNED buffer. The bound protein was eluted with 1. 5 mL TNED buffer containing 10 mM reduced glutathione. The two-thirds of eluted protein was dialyzed against 2L of TNED buffer twice at 4°C for 24 h. The rest of one-third was dialyzed against 2L of TNE buffer without DTT. We measured the protein concentration of purified protein using Enhanced BCA Protein assay kit (Beyotime).
GST-3Cpro mutant proteins were generated by fusion PCR of two PCR products amplified using pGEX 5′ primer as well as the reverse primer for each mutant and pGEX 3′ primer as well as the forward primer for each mutant (Table S1). The plasmid DNA to express GST-DWV 3Cpro was used as the template for the 1st PCR. The fusion PCR products were cloned in pGEX-4T-1vector and the mutant proteins were expressed and purified as above. All plasmid DNAs were sequenced to verify the intended mutations.
Protease assay of GST-DWV 3Cpro.
Dabcyl-PVQAKPEMDNPNPGE-Edans derived from the cleavage site between L-protein and VP2 (underlined, 205-218 of polyprotein) was used as a peptide substrate to measure the protease activity. E was added to the C-terminus to link Edans. FRET experiments were performed with Varioskan LUX multimode microplate reader (Thermo Fisher). We measured 100 μL of reaction mixture with 50 mM Citrate buffer, pH 6.0, 150 mM NaCl, 2 mM EDTA, 2 mM DTT containing 1 μM GST-DWV 3Cpro and 20 μM fluorogenic peptide substrate in a 96-well flat-bottom white microplate at 33°C. The relative fluorescence was measured using an excitation wavelength of 336 nm and by monitoring the emission at 490 nm every 3 min for 30 min. In order to determine the optimal reaction condition for GST-DWV 3Cpro, various pH values, temperatures, and DTT concentrations were used. To test the effect of DTT on the activity, the protein dialyzed against TNE buffer was used. To determine the velocity of product formation, we precalibrated the instrument with the free Edans standard in the presence of 20 μM Dabcyl to calculate the relationship between relative fluorescence and substrate concentration. We calculated the kinetic parameters, Km, Vmax, and catalytic efficiency (Kcat/Km) assuming Michaelis-Menten kinetics for the cleavage of peptide. No significant hydrolysis of the peptide substrates was observed in the absence of GST-DWV 3Cpro.
To identify the inhibitors for DWV 3Cpro, various concentrations of the tested compounds were added. To determine the inhibitory effects of the various compounds, we preincubated the compound and enzyme on ice for 10 min prior to addition of the peptide substrate. To analyze the effects of ebselen, disulfiram, and carmofur, we used the enzyme dialyzed against TNE buffer and carried out the reaction with or without DTT. We determined the initial velocities of the enzymatic reactions and fitted to a sigmoidal dose-response equation with nonlinear regression analysis using GraphPad Prism 9. The data from three independent assays were used to determine IC50 value of each compound.
Infection of honey bee pupal head cells with DWV.
Honey bee pupae with pale/pink eyes were collected from a mite-free colony. They were surface sterilized by washing with 10% bleach followed by sterile PBS three times (5 min for each wash). Heads from the pupae were dissected and homogenized seven times with 1 mL Grace culture medium using Dounce homogenizer (Loose fitting). The homogenate was then filtered through a cell strainer (Falcon) and the number of cells was counted. 106 cells were suspended in 100 μL of Grace medium containing 10% FBS, antibiotics (penicillin and streptomycin), and ebselen at the indicated concentration with or without DWV (MOI: 10) in 24-well plate at 33°C for 1 h. DWV used for this study was one of type A strains (VD-B7) we previously isolated from Varroa mite-infested pupa (8). Fresh culture medium (400 μL) with ebselen at the indicated concentration was then added (the final cell density at 2 × 106/mL) and incubated at 33°C for 16 h.
Western blot.
After DWV infection, the pupal head cells were collected by centrifugation, and then homogenized with 150 μL of RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitor cocktail. After centrifugation of the homogenate, the protein concentration of supernatant was measured using Enhanced BCA protein assay kit. The cell lysate with 20 μg of protein was analyzed by Western blot for each sample. The protein samples in SDS-PAGE sample buffer (2% SDS, 10% glycerol, 10% β-mercaptoethanol, 0.25% bromophenol blue, 50 mM Tris-HCl, pH 6.8) were heated at 95°C for 5 min. After centrifugation, the supernatants were applied to 10% SDS-PAGE and the proteins were transferred to a nitrocellulose membrane (Pall Life Sciences). Another 10% SDS-PAGE gel with the same samples was stained by Instant Blue. The membrane was then blocked with PBST (PBS with 0.1% Tween 20) containing 5% BSA at room temperature for 30 min followed by incubating with 1000-fold diluted anti-RdRP antibody (6) or 500-fold diluted anti-3Cpro antibody at 4°C overnight. The membrane was washed five times with PBST (5 min each), and then incubated with 10,000-fold diluted IRDye 680RD donkey anti-rabbit IgG (H+L) (LI-COR Biosciences) in PBST containing 5% skim milk at room temperature for 2 h. The membrane was washed as above, and then visualized using Odyssey Imaging System (LI-COR Biosciences). Band intensity of 97 kDa RdRP precursor was measured by image-J.
Testing cell viability.
Honey bee pupal head cells were prepared as described above and 100 μL cell suspension (2 × 106/mL) was inoculated to each well in 96-well plate. The cells were cultured in the presence of either DMSO or ebselen at the indicated concentrations at 33°C for 16 h. The cultured plate was first incubated at room temperature for 10 min followed by adding 100 μL of reagent solution (CellTiter-Lumi Luminescent Cell Viability assay kit, Beyotime). The plate was shaken at room temperature for 2 min to promote cell lysis, and then further incubated for 10 min to stabilize the chemiluminescence signal. The signal was detected using Varioskan LUX multimode microplate reader and depends on intracellular ATP level, thus the relative viability of cells.
Structural analysis of 3C proteases of DWV and other insect RNA viruses.
We determined the structures of 3C proteases of DWV, BrBV, LsPVI2, SeIV-1, SBV, and CrPV using ColabFold (36) based on AlphaFold v2.1.0 (35). The generated structures were analyzed and displayed using UCSF ChimeraX (53, 54). The molecular structure of rupintrivir was collected from PubChem (CID: 6440352). We used AutoDock Tools package (55) to prepare the ligand and receptor by removing water and other heteroatoms and assigning partial charges. The size of the grid box for docking was set at 20 Å in each direction. We provided explicit coordinate to find all possible binding sites. After preparing the ligand, receptor, definition of binding site, AutoDock Vina (version 1.1.2) (56) program was used for the molecular docking simulation.
Self-cleavage of GST-DWV 3Cpro.
Ten micrograms of wild type, C2307A, Q2118A, and E2180A GST-DWV 3Cpro protein was analyzed by 10% SDS-PAGE followed by Instant Blue staining. 20 μg of wild-type GST-DWV 3Cpro was incubated in the enzyme reaction buffer without the substrate at 33°C and the 4 μg protein was sampled every 3 h for 12 h and at 18 h. All samples including the one without the incubation (0 h) were analyzed by SDS-PAGE as above.
Preparation of anti-DWV 3Cpro antibody.
DWV 3Cpro insert in pGEX-4T-1 was transferred to pGEX-6P-3 followed by transformation to BL21. The protein was expressed and purified as above except 5 mL TNED buffer containing 100 units of PreScission Protease (Beyotime) was added to the resin after the final wash. The resin was incubated at 4°C overnight and the supernatant with the released DWV 3Cpro was collected. The remaining resin was eluted once with 5 mL TNED buffer, and the eluate was combined with the above supernatant. The total eluate was dialyzed against 2L of PBS with 1 mM EDTA, 0.5 mM DTT, and 0.1% sarcosyl twice at 4°C for 24 h. The purified protein was delivered to GeneScript-Nanjing to raise the anti-rabbit polyclonal antibody.
Alignment of Ifravirus 3C protease sequences.
3C protease sequences of various Iflaviruses were searched in NCBI by BLASTP using DWV 3Cpro sequence as a query. Twenty-three sequences were picked up and aligned together with DWV 3Cpro using MUSCLE program (EMBL-EBI).
Statistical analysis.
All data presented were from representative independent experiments. Statistical analyses were performed with Bell Curve for Excel (Social Survey Research Information Co., Ltd.) and no data point was excluded. The applied statistical tests and P-values are described in figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yixiang Tang, Shiqi Xu, Jiarui Li, Zhuoran Leng, and Shangqing Li for their contributions to conduct the experiments. We are grateful to David Ruiz-Carrillo for his comments on the paper.
This work was supported by Jinji Lake Double Hundred Talents Program and XJTLU Summer Undergraduate Research Fellowship (SURF 2021095) to T.K.
Footnotes
Supplemental material is available online only.
Contributor Information
Tatsuhiko Kadowaki, Email: Tatsuhiko.Kadowaki@xjtlu.edu.cn.
Frederick S. B. Kibenge, University of Prince Edward Island
REFERENCES
- 1.Highfield AC, El Nagar A, Mackinder LC, Noël LM, Hall MJ, Martin SJ, Schroeder DC. 2009. Deformed wing virus implicated in overwintering honeybee colony losses. Appl Environ Microbiol 75:7212–7220. doi: 10.1128/AEM.02227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nazzi F, Le Conte Y. 2016. Ecology of Varroa destructor, the major ectoparasite of the western honey bee, Apis mellifera. Annu Rev Entomol 61:417–432. doi: 10.1146/annurev-ento-010715-023731. [DOI] [PubMed] [Google Scholar]
- 3.Chantawannakul P, Ramsey S, vanEngelsdorp D, Khongphinitbunjong K, Phokasem P. 2018. Tropilaelaps mite: an emerging threat to European honey bee. Curr Opin Insect Sci 26:69–75. doi: 10.1016/j.cois.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 4.de Miranda JR, Genersch E. 2010. Deformed wing virus. J Invertebr Pathol 103(Suppl 1):S48–61. doi: 10.1016/j.jip.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Rosenkranz P, Aumeier P, Ziegelmann B. 2010. Biology and control of Varroa destructor. J Invertebr Pathol 103(Suppl 1):S96–119. doi: 10.1016/j.jip.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, Liu Q, Weiss B, Kaltenpoth M, Kadowaki T. 2020. Honey bee suppresses the parasitic mite vitellogenin by antimicrobial peptide. Front Microbiol 11:1037. doi: 10.3389/fmicb.2020.01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen M, Yang X, Cox-Foster D, Cui L. 2005. The role of varroa mites in infections of Kashmir bee virus (KBV) and deformed wing virus (DWV) in honey bees. Virology 342:141–149. doi: 10.1016/j.virol.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Dong X, Kadowaki T. 2017. Characterization of the copy number and variants of deformed wing virus (DWV) in the pairs of honey bee pupa and infesting. Front Microbiol 8:1558. doi: 10.3389/fmicb.2017.01558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsgren E, de Miranda JR, Isaksson M, Wei S, Fries I. 2009. Deformed wing virus associated with Tropilaelaps mercedesae infesting European honey bees (Apis mellifera). Exp Appl Acarol 47:87–97. doi: 10.1007/s10493-008-9204-4. [DOI] [PubMed] [Google Scholar]
- 10.Khongphinitbunjong K, Neumann P, Chantawannakul P, Williams G. 2016. The ectoparasitic mite Tropilaelaps mercedesae reduces western honey bee, Apis mellifera, longevity and emergence weight, and promotes Deformed wing virus infections. J Invertebr Pathol 137:38–42. doi: 10.1016/j.jip.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Wilfert L, Long G, Leggett HC, Schmid-Hempel P, Butlin R, Martin SJ, Boots M. 2016. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 351:594–597. doi: 10.1126/science.aac9976. [DOI] [PubMed] [Google Scholar]
- 12.Yue C, Schröder M, Gisder S, Genersch E. 2007. Vertical-transmission routes for deformed wing virus of honeybees (Apis mellifera). J Gen Virol 88:2329–2336. doi: 10.1099/vir.0.83101-0. [DOI] [PubMed] [Google Scholar]
- 13.Tentcheva D, Gauthier L, Zappulla N, Dainat B, Cousserans F, Colin ME, Bergoin M. 2004. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl Environ Microbiol 70:7185–7191. doi: 10.1128/AEM.70.12.7185-7191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanzi G, de Miranda JR, Boniotti MB, Cameron CE, Lavazza A, Capucci L, Camazine SM, Rossi C. 2006. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J Virol 80:4998–5009. doi: 10.1128/JVI.80.10.4998-5009.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthews DA, Smith WW, Ferre RA, Condon B, Budahazi G, Sisson W, Villafranca JE, Janson CA, McElroy HE, Gribskov CL. 1994. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 16.Bergmann EM, Mosimann SC, Chernaia MM, Malcolm BA, James MN. 1997. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J Virol 71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosimann SC, Cherney MM, Sia S, Plotch S, James MN. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol 273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 18.Birtley JR, Knox SR, Jaulent AM, Brick P, Leatherbarrow RJ, Curry S. 2005. Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J Biol Chem 280:11520–11527. doi: 10.1074/jbc.M413254200. [DOI] [PubMed] [Google Scholar]
- 19.Lee CC, Kuo CJ, Ko TP, Hsu MF, Tsui YC, Chang SC, Yang S, Chen SJ, Chen HC, Hsu MC, Shih SR, Liang PH, Wang AH. 2009. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J Biol Chem 284:7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohlenschläger O, Wöhnert J, Bucci E, Seitz S, Häfner S, Ramachandran R, Zell R, Görlach M. 2004. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Cui S, Wang J, Fan T, Qin B, Guo L, Lei X, Wang J, Wang M, Jin Q. 2011. Crystal structure of human enterovirus 71 3C protease. J Mol Biol 408:449–461. doi: 10.1016/j.jmb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi J, Peng J, Yang W, Zhu G, Ren J, Li D, Zheng H. 2021. Picornavirus 3C: a protease ensuring virus replication and subverting host responses. J Cell Sci 134. [DOI] [PubMed] [Google Scholar]
- 23.Sun D, Chen S, Cheng A, Wang M. 2016. Roles of the picornaviral 3C proteinase in the viral life cycle and host cells. Viruses 8:82. doi: 10.3390/v8030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lall MS, Jain RP, Vederas JC. 2004. Inhibitors of 3C cysteine proteinases from Picornaviridae. Curr Top Med Chem 4:1239–1253. doi: 10.2174/1568026043387836. [DOI] [PubMed] [Google Scholar]
- 25.Ryabov EV, Christmon K, Heerman MC, Posada-Florez F, Harrison RL, Chen Y, Evans JD. 2020. Development of a honey bee RNA virus vector based on the genome of a deformed wing virus. Viruses 12:374. doi: 10.3390/v12040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkesman J. 2017. Cysteine protease zymography: brief review. Methods Mol Biol 1626:25–31. doi: 10.1007/978-1-4939-7111-4_3. [DOI] [PubMed] [Google Scholar]
- 27.Patick AK, Binford SL, Brothers MA, Jackson RL, Ford CE, Diem MD, Maldonado F, Dragovich PS, Zhou R, Prins TJ, Fuhrman SA, Meador JW, Zalman LS, Matthews DA, Worland ST. 1999. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob Agents Chemother 43:2444–2450. doi: 10.1128/AAC.43.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amporndanai K, Meng X, Shang W, Jin Z, Rogers M, Zhao Y, Rao Z, Liu ZJ, Yang H, Zhang L, O'Neill PM, Samar Hasnain S. 2021. Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives. Nat Commun 12:3061. doi: 10.1038/s41467-021-23313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma C, Hu Y, Townsend JA, Lagarias PI, Marty MT, Kolocouris A, Wang J. 2020. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol Transl Sci 3:1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y, Lovell S, Tiew KC, Mandadapu SR, Alliston KR, Battaile KP, Groutas WC, Chang KO. 2012. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J Virol 86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Z, Zhao Y, Sun Y, Zhang B, Wang H, Wu Y, Zhu Y, Zhu C, Hu T, Du X, Duan Y, Yu J, Yang X, Yang X, Yang K, Liu X, Guddat LW, Xiao G, Zhang L, Yang H, Rao Z. 2020. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol 27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 32.Jung E, Lee JY, Kim HJ, Ryu CK, Lee KI, Kim M, Lee CK, Go YY. 2018. Identification of quinone analogues as potential inhibitors of picornavirus 3C protease in vitro. Bioorg Med Chem Lett 28:2533–2538. doi: 10.1016/j.bmcl.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 33.Yao C, Xi C, Hu K, Gao W, Cai X, Qin J, Lv S, Du C, Wei Y. 2018. Inhibition of enterovirus 71 replication and viral 3C protease by quercetin. Virol J 15:116. doi: 10.1186/s12985-018-1023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Yuan X, Li J, Kadowaki T. 2021. DWV Infection in vitro using honey bee pupal tissue. Front Microbiol 12:631889. doi: 10.3389/fmicb.2021.631889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2021. ColabFold: making protein folding accessible to all. bioRxiv doi: 10.1101/2021.08.15.456425... [DOI] [PMC free article] [PubMed]
- 37.Wang J, Fan T, Yao X, Wu Z, Guo L, Lei X, Wang J, Wang M, Jin Q, Cui S. 2011. Crystal structures of enterovirus 71 3C protease complexed with rupintrivir reveal the roles of catalytically important residues. J Virol 85:10021–10030. doi: 10.1128/JVI.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binford SL, Maldonado F, Brothers MA, Weady PT, Zalman LS, Meador JW, Matthews DA, Patick AK. 2005. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob Agents Chemother 49:619–626. doi: 10.1128/AAC.49.2.619-626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordingley MG, Register RB, Callahan PL, Garsky VM, Colonno RJ. 1989. Cleavage of small peptides in vitro by human rhinovirus 14 3C protease expressed in Escherichia coli. J Virol 63:5037–5045. doi: 10.1128/jvi.63.12.5037-5045.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weidner JR, Dunn BM. 1991. Development of synthetic peptide substrates for the poliovirus 3C proteinase. Arch Biochem Biophys 286:402–408. doi: 10.1016/0003-9861(91)90058-Q. [DOI] [PubMed] [Google Scholar]
- 41.Shang L, Zhang S, Yang X, Sun J, Li L, Cui Z, He Q, Guo Y, Sun Y, Yin Z. 2015. Biochemical characterization of recombinant Enterovirus 71 3C protease with fluorogenic model peptide substrates and development of a biochemical assay. Antimicrob Agents Chemother 59:1827–1836. doi: 10.1128/AAC.04698-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jewell DA, Swietnicki W, Dunn BM, Malcolm BA. 1992. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry 31:7862–7869. doi: 10.1021/bi00149a017. [DOI] [PubMed] [Google Scholar]
- 43.Sweeney TR, Roqué-Rosell N, Birtley JR, Leatherbarrow RJ, Curry S. 2007. Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J Virol 81:115–124. doi: 10.1128/JVI.01587-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gosert R, Dollenmaier G, Weitz M. 1997. Identification of active-site residues in protease 3C of hepatitis A virus by site-directed mutagenesis. J Virol 71:3062–3068. doi: 10.1128/jvi.71.4.3062-3068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan YW, Ang MJ, Lau QY, Poulsen A, Ng FM, Then SW, Peng J, Hill J, Hong WJ, Chia CS, Chu JJ. 2016. Antiviral activities of peptide-based covalent inhibitors of the Enterovirus 71 3C protease. Sci Rep 6:33663. doi: 10.1038/srep33663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dragovich PS, Prins TJ, Zhou R, Webber SE, Marakovits JT, Fuhrman SA, Patick AK, Matthews DA, Lee CA, Ford CE, Burke BJ, Rejto PA, Hendrickson TF, Tuntland T, Brown EL, Meador JW, Ferre RA, Harr JE, Kosa MB, Worland ST. 1999. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements. J Med Chem 42:1213–1224. doi: 10.1021/jm9805384. [DOI] [PubMed] [Google Scholar]
- 47.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. 2020. Structure of M. Nature 582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 48.Cheah KC, Leong LE, Porter AG. 1990. Site-directed mutagenesis suggests close functional relationship between a human rhinovirus 3C cysteine protease and cellular trypsin-like serine proteases. J Biol Chem 265:7180–7187. doi: 10.1016/S0021-9258(19)39096-9. [DOI] [PubMed] [Google Scholar]
- 49.Hämmerle T, Hellen CU, Wimmer E. 1991. Site-directed mutagenesis of the putative catalytic triad of poliovirus 3C proteinase. J Biol Chem 266:5412–5416. doi: 10.1016/S0021-9258(19)67610-6. [DOI] [PubMed] [Google Scholar]
- 50.Miyashita K, Kusumi M, Utsumi R, Katayama S, Noda M, Komano T, Satoh N. 1993. Site-directed mutagenesis of the putative active site residues of 3C proteinase of coxsackievirus B3: evidence of a functional relationship with trypsin-like serine proteinases. Protein Eng 6:189–193. doi: 10.1093/protein/6.2.189. [DOI] [PubMed] [Google Scholar]
- 51.Winston DS, Boehr DD. 2021. The picornavirus precursor 3CD has different conformational dynamics compared to 3C. Viruses 13:442. doi: 10.3390/v13030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang P, Liu Y, Ma HC, Paul AV, Wimmer E. 2014. Picornavirus morphogenesis. Microbiol Mol Biol Rev 78:418–437. doi: 10.1128/MMBR.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00068-22-s001.pdf, PDF file, 0.6 MB (668.2KB, pdf)