Abstract
Background:
Rotavirus is a major cause of severe diarrhea worldwide. It causes 453,000 deaths in children annually. In the Democratic Republic of the Congo, sentinel site surveillance of rotavirus gastroenteritis started in 2009 and aimed to document burden of rotavirus diarrhea and identify circulating rotavirus genotypes.
Methods:
Between August 2009 to June 2012, stool samples were collected in Kinshasa and Lubumbashi, from children <5 years of age who met the WHO case definition for rotavirus gastroenteritis. Rotavirus antigen detection was performed using an enzyme immunoassay technique and rotavirus strains were characterized using a multiplex reverse transcription polymerase chain reaction assay.
Results:
During the study period, 1614 stool samples were screened for rotavirus by enzyme immunoassay and 990 (61%) were positive. Of these, the genotype was determined in 330 (33%) samples. The most common genotypes found in the samples analyzed were G1P[8] in 2009 (28%) and 2012 (33%), G2P[4] (33%) in 2010 and G2P[6] (28%) in 2011. Uncommon strains like G8P[6] (5%), G6P[6] (5%), G12P[6] (3%), G12P[8] (3%) and G8P[8] (2%) were also detected.
Conclusions:
In Democratic Republic of the Congo, 61% of the diarrhea in children in <5 years of age was caused by rotavirus infection and a variety of rotavirus genotypes were detected. Implementation of rotavirus genotyping at the national level has improved the timely identification of rotavirus strains. These results will help decision makers in Democratic Republic of the Congo plan the implementation of a rotavirus vaccination program.
Keywords: pediatric, rotavirus, surveillance, Democratic Republic of the Congo
Rotavirus gastroenteritis is a major public health concern globally, estimated to cause 453,000 deaths among children <5 years of age in 2008.1 More than 85% of the mortality occurs in south Asia and sub-Saharan Africa with 5 countries [India, Nigeria, the Democratic Republic of the Congo (DRC), Ethiopia and Pakistan] accounting for more than half of all rotavirus deaths.1,2 In Africa, DRC has the second highest rotavirus mortality rate, after Nigeria, resulting in an estimated 32,653 deaths of children in 2008.2
The DRC is a developing country located in central Africa with a population of 73 million (2012 estimate) living within an area of 2,345,000 km2.3 Despite the fact that diarrhea is one of the leading causes of admission to pediatric hospitals, there is little information on the causes of diarrhea in DRC. To generate local knowledge on rotavirus disease burden and provide evidence for national health policy decisions, surveillance for rotavirus gastroenteritis was established in DRC during 2009 through the African Rotavirus Surveillance Network and the World Health Organization (WHO) Regional Office for Africa. In 2010, rotavirus gastroenteritis surveillance in DRC was enhanced to include laboratory surveillance for rotavirus strains at the national level by training laboratory staff for rotavirus genotyping techniques in their home laboratory through the Surveillance Épidémiologique en Afrique Centrale (SURVAC) project.4
Rotavirus is a nonenveloped virus with an 11-segment, double-stranded RNA genome encased within a triple layer capsid. Two structural proteins, VP7 (the glycoprotein or G protein) and VP4 (the protease-cleaved protein or P protein) make up the outer layer of the capsid. VP7 and VP4 are considered important for vaccine development as they are major antigens involved in virus neutralization.5 The genes encoding these proteins segregate independently of each other during reassortment. Although a recommendation for classification of group A rotavirus using all 11 segments does exist,6 a dual classification system based on both VP7 (G) and VP4 (P) is commonly used.7 The G and P genotype combinations commonly found around the world include G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8].8 New genotypes such as G12 have emerged and spread worldwide.9 Previous studies in DRC on rotavirus strains circulating from 2003 to 2005 showed that genotypes G8P[6] and G8P[8] predominated in 2003; while in 2004 and 2005, G1P[6] was the most common strain detected.10
This surveillance aims to document the burden of rotavirus diarrhea and identify prevalent rotavirus genotypes circulating in DRC. This report presents the results of surveillance of rotavirus in 3 sentinel hospitals in DRC from August 2009 to June 2012. These data provide baseline information on rotavirus strains circulating in DRC before the planned introduction of rotavirus vaccination in the national childhood immunization program. These data will be helpful to assess the future impact of the rotavirus vaccination program.
MATERIALS AND METHODS
Sentinel Surveillance Hospitals
The surveillance was conducted at 3 sentinel hospitals located in the western province of Kinshasa (Centre Hospitalier Pédiatrique de Kalembelembe, CHPK; Centre Hospitalier de Kingasani, CHK) and the southern province of Katanga (Hôpital Général Sendwe, HGS) in the town of Lubumbashi (Fig. 1). These 3 hospitals are representative provincial hospitals, caring for most of the pediatric patients in the area. The surveillance of rotavirus started in DRC in 2009; August in CHPK, November in CHK and December in HGS.
FIGURE 1.
Geographical location of the 3 sentinel sites (black dots) conducting rotavirus surveillance in the Democratic Republic of the Congo. CHPK, Centre Hospitalier Pédiatrique de Kalembelembe; CHK, Centre Hospitalier de Kingasani; HGS, Hôpital Général Sendwe.
Case Definition and Data Collection
From August 2009 to June 2012, stool specimens were collected within 48 hours of hospital admission, from children <5 years of age with a diagnosis of gastroenteritis. The WHO case definition of gastroenteritis was the occurrence of at least 3 looser than normal or watery stools in a 24-hour period and/or 2 or more episodes of vomiting unexplained by other reasons.11
Laboratory Testing for Rotavirus
Samples collected from each sentinel site were first screened for group A rotavirus antigen by enzyme immunoassay (EIA), as described below and aliquots stored at −20°C before transportation to the Institut National de Recherche Biomedicale (INRB) where EIA results were confirmed by repeat EIA. Before 2011, the genotyping assays were performed using multiplex reverse transcription polymerase chain reaction (RT-PCR) at the rotavirus Regional Reference Laboratory in South Africa, Medical Research Council (MRC)/ Diarrhoeal Pathogens Research Unit, University of Limpopo, Medunsa Campus. Since 2011, the genotyping assays have been performed by the INRB laboratory, with the support of the SURVAC Project, using a multiplex RT-PCR technique. Samples subjected to genotyping were subsequently confirmed at the Centers for Disease Control and Prevention (CDC), Atlanta, GA, for quality control.
Enzyme Immunoassay
A 10% suspension of specimens was prepared and analyzed using a sandwich EIA for detection of group A rotavirus in fecal specimens (ProSpecT Rotavirus Microplate Assay (Oxoid, Ltd., Basingstoke, Hampshire, United Kingdom) according to manufacturer’s instructions.
Nucleic Acid Extraction
Viral RNA from rotavirus-positive samples was extracted from 140 μL of 10% stool suspension using a QIAamp viral RNA Mini kit (Qiagen, Inc., Valencia, CA) according to manufacturer’s instructions and extracted RNAs were stored at −80°C.
Genotyping by RT-PCR and Multiplexed PCR
The RNA extracts first were subjected to RT-PCR. Two gene regions, VP7 (896 bp) and VP4 (876 bp), were reverse transcribed and amplified with primers 9Con1-L/VP7-R and Con3/Con2, respectively.12,13 Reverse transcription of double strand RNA (dsRNA) was carried out with the OneStep RT-PCR Kit (Qiagen, Inc.). After 5 min denaturation at 97°C, the RNA was mixed with kit reagents and incubated at 42°C for 30 minutes to obtain complementary DNA (cDNA), immediately followed by the PCR reaction. These first round RT-PCR products then were used in a semi-nested, multiplexed PCR to identify G-types (G1, G2, G3, G4, G9 and G12) and P-types (P[4], P[6], P[8]).12,13 All PCR products were identified by electrophoresis in 2% agarose gels containing 10% Gel Red (Biotium, Hayward, CA) and visualized under ultraviolet illumination.
RESULTS
Seasonality and Temporal Distribution of Rotavirus Infection
Between August 2009 and June 2012, a total of 1614 children <5 years of age with diarrhea were enrolled at the 3 sentinel sites in DRC. They were distributed as follows: Kalembelembe Hospital 828/1614 (51%), Kingasani Hospital 628/1614 (39%) and Sendwe Hospital 159/1614 (10%). During the study period, rotavirus infection occurred year round but was more common from March through November, with a peak from April to August (Fig. 2). This corresponds to the dry season in these parts of the country.
FIGURE 2.
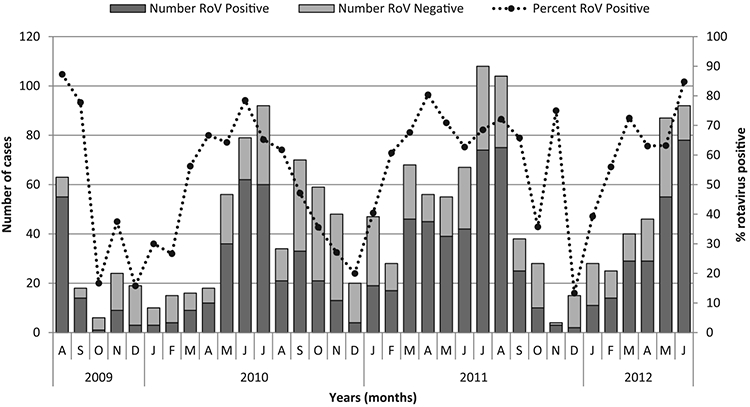
Seasonality of rotavirus diarrhea in DRC, from August 2009 to June 2012. Number of rotavirus positives (shaded dark bars), rotavirus negatives (shaded grey bars) and percentage of rotavirus positives (dotted line) are indicated.
Identification of Rotavirus and Age Distribution
During the study period, 1614 stool samples were tested for group A rotavirus antigen by EIA and rotavirus was identified in 990 (61%). The age distribution of rotavirus cases showed that 22% of rotavirus infections occurred during the first 6 months of life and peaked in the 6–11 month age group (54.3%; Table 1). Among children with rotavirus infection, 3 quarters of the patients (76.3%) were hospitalized before 12 months of age. Only 2% cases were observed in the 24–59 month age group.
TABLE 1.
Cumulative Age Distribution of Children <5 Years of Age Admitted With Gastroenteritis to 3 Sentinel Site Hospital in DRC, August 2009 to June 2012
| Age Groups |
Total | Rotavirus Negative |
Rotavirus Positive (%) |
Cumulative Rotavirus Positive Rate (%) |
|---|---|---|---|---|
| 0–5 | 329 | 111 | 218(22) | 22 |
| 6–11 | 863 | 325 | 538(54.3) | 76.3 |
| 12–23 | 376 | 161 | 215(21.7) | 98 |
| 24–59 | 46 | 27 | 19(2) | 100 |
| Total | 1614 | 624 | 990 | — |
Rotavirus Strain Characterization
Genotyping analyses were carried out to determine the type and distribution of rotavirus strains in the 3 sentinel sites. Of the 990 rotavirus-positive stool samples, 330 samples (33%) were characterized for both VP7 and VP4 types using multiplex RT-PCR. Among the G-types examined, 315 (95.5%) could be G-typed. The prevalence of G2, G1 and G8 strains was found to be 37%, 27% and 9%, respectively (Table 2). Among the P-types, 314 (95.2%) could be P-typed, the most common strains were P[6] (52%) followed by P[8] (22%) and P[4] (17%). Of all the strains that could be both G- and P-typed (299 strains, 90.6%), the most prevalent G-P combination was G2P[6] (18%) followed by G2P[4] (14%), G1P[8] (13%) and G1P[6] (11%). The uncommon G6P[6] strain was found and represented 5% of typed strains. Among the strains characterized in this study, 16 (5%) and 15 (5%) were P and G nontypeable, respectively (Table 2).
TABLE 2.
Distribution of Rotavirus G- and P-types in DRC from August 2009 to June 2012
| P-type | G-type | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gl(%) | G2 | G3 | G4 | G6 | G8 | G9 | G12 | GMIX | GNT | Total | |
| P[4] | 3(1) | 46(14) | — | 1(0) | — | 2(1) | — | 1(0) | 3(1) | 1(0) | 57(17) |
| P[6] | 36(11) | 60(18) | 1(0) | 1(0) | 16(5) | 18(5) | — | 11(3) | 20(6) | 9(3) | 172(52) |
| P[8] | 43(13) | 4(1) | — | — | 1(0) | 6(2) | 1(0) | 9(3) | 5(2) | 5(2) | 74(22) |
| P[MIX] | 1(0) | 6(2) | — | — | — | 1(0) | — | — | 3(1) | — | 11(3) |
| PINT] | 6(2) | 3(1) | — | — | — | 3(1) | — | — | 4(1) | — | 16(5) |
| Total | 89(27) | 119(37) | 1(0) | 2(1) | 17(5) | 30(9) | 1(0) | 21(6) | 35(11) | 15(5) | 330(100) |
MIX indicates at least 2 G- or P-types found in the same specimen. NT indicates nontypeable strains. The percentage of each type is indicated in bracket (%).
Temporal distribution of genotype combinations showed that G1P[8] and G1P[6] strains circulated during the entire study period, while G8P[6] and G12P[6] were not found in 2012 (Fig. 3). Mixed infection, defined as a sample containing multiple G and/or P-types, occurred in 44 cases (13.3%). Within these mixed infections, P[6] was involved in 25 cases (56%), of which 10 cases (40%) were P[6] in combination with G8 and G12 (Table 3). The age distribution for the “mixed infection” cases was proportional to the number of cases per age group and thus mixed genotype infections were detected at the same rate in all age groups.
FIGURE 3.
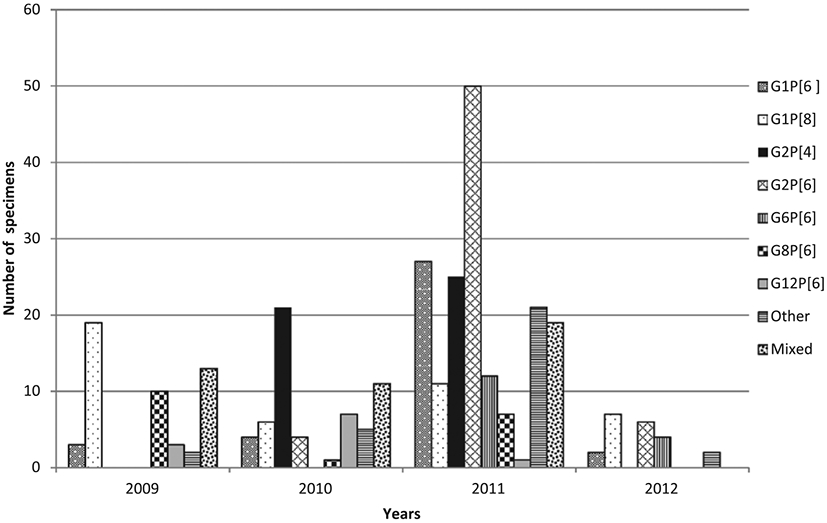
The distribution of the most common rotavirus genotypes by year from August 2009 through June 2012 in DRC. Other represents genotypes with a frequency over the study period with <10 cases. Mixed represents mixed infection.
TABLE 3.
P-G Type Combination in 44 Mixed Infections From Gastroenteritis Cases Admitted at 3 Sentinel Sites in DRC, 2009–2012
| Number of infections | ||||||
|---|---|---|---|---|---|---|
| P-type | G-type | 2009 | 2010 | 2011 | 2012 | Total(%) |
| P[4] | G2.12 | 0 | 0 | 1 | 0 | 1(2%) |
| P[4] | G6.2 | 0 | 0 | 1 | 1 | 2(4%) |
| P[4] | G12.1 | 0 | 0 | 1 | 0 | 1(2%) |
| P[4,8] | G2 | 0 | 2 | 3 | 0 | 5(11%) |
| P[4,8] | G1.2 | 0 | 1 | 0 | 0 | 1(2%) |
| P[6] | G1.2 | 0 | 0 | 3 | 0 | 3(7%) |
| P[6] | G1.12 | 0 | 0 | 1 | 0 | 1(2%) |
| P[6] | G2.1 | 0 | 0 | 1 | 0 | 1(2%) |
| P[6] | G3.8 | 1 | 0 | 0 | 0 | 1(2%) |
| P[6] | G8.12 | 7 | 0 | 0 | 0 | 7(16%) |
| P[6] | G12.1 | 0 | 1 | 2 | 0 | 3(7%) |
| P[6] | G12.8 | 3 | 0 | 0 | 0 | 3(7%) |
| P[6] | G12.3.9 | 0 | 1 | 0 | 0 | 1(2%) |
| P[6,4] | G2 | 0 | 1 | 0 | 0 | 1(2%) |
| P[6,4] | G12.1 | 0 | 1 | 0 | 0 | 1(2%) |
| P[6,8] | G1 | 0 | 1 | 0 | 0 | 1(2%) |
| P[6,8] | G12.1 | 0 | 0 | 1 | 0 | 1(2%) |
| PI6.4.8] | G8 | 0 | 1 | 0 | 0 | 1(2%) |
| P[8] | G1.8 | 0 | 0 | 1 | 0 | 1(2%) |
| P[8] | G1.9 | 0 | 1 | 0 | 0 | 1(2%) |
| P[8] | G2.1 | 0 | 0 | 1 | 0 | 1(2%) |
| P[8] | G8.12 | 1 | 0 | 0 | 0 | 1(2%) |
| P[8] | G12.1 | 0 | 0 | 1 | 0 | 1(2%) |
| PINT] | G1.2 | 0 | 1 | 0 | 0 | 1(2%) |
| PINT] | G2.1 | 0 | 0 | 2 | 0 | 2(2%) |
| PINT] | G9.12 | 1 | 0 | 0 | 0 | 1(2%) |
| Total | 13 | 11 | 19 | 2 | 44(100%! | |
Bold values indicate the most prominent strains with at least 2 cases per year. NT indicates nontypeable strains.
DISCUSSION
The objective of this study was to assess disease burden and baseline strain prevalence data in anticipation of vaccine introduction in DRC. A standardized protocol for rotavirus surveillance11 was used in 3 sentinel site hospitals in DRC from August 2009 to June 2012. Rotavirus was detected in 61% of stool specimens by antigen EIA from the 1614 hospitalized children enrolled in the study, varying from a low of 54% in 2010 to a high of 68% in the first 6 months of 2012. These rates of rotavirus gastroenteritis are higher than those reported from WHO global surveillance, estimated at 38%,14 and from the African Region, estimated at 40%.15 This high proportion of rotavirus gastroenteritis among hospitalized children is in concordance with previous studies from DRC during 2003–2005,10 where 70% of admitted children had diarrhea caused by rotavirus. The slight difference in rotavirus detection rate observed between the current study and the former may be due to the fact that for this study, samples were collected throughout the year whereas the former study focused on samples collected during rotavirus season only. In the current study, it was noted that rotavirus infection is predominant in children <12 months of age, in agreement with similar observations reported previously in studies from DRC and other parts of Africa.10,16,17
Significant temporal variation was observed in the prevalence of some genotypes. Genotype G1P[8], known as the most common human rotavirus strain globally, was detected throughout the study period and was the predominant strain in the samples analyzed in 2009 and 2012. Genotype G2P[4], in contrast, was detected only during 2 years, 2010 and 2011. Genotype G8P[6] was found circulating, albeit at lower levels, during the 2009–2011 rotavirus seasons and then disappeared during the first 6 months of 2012 rotavirus season. Kabue et al10 in 2010 reported the detection of G8P[6] in DRC accounting for 16% of rotavirus strains in circulation during the 2003–2005 rotavirus seasons. In this study, genotype G12P[6] emerged for the first time in DRC. Of note, the identification of mixed infections at high frequency among rotavirus cases is in agreement with previous studies from Africa and elsewhere and may account in part for the diversity of human-human rotavirus reassortant strains circulating in DRC. Some of these mixed infections, for example, G1P[6,8], were previously reported in DRC10 and Malawi,17 and G1,8P[8] strains were identified in Kenya.18 Strains like G12P[8] detected in this study have been found in Cameroon and Ethiopia.15
While the predominance of rotavirus strains G2P[6] was remarkable in 2011 and 2012, It is worth mentioning that G2P[4] strain reached a peak detection of 21/65 samples (32%) in a single season in 2010. These results suggested that a shift in predominant rotavirus strains can occur suddenly, raising the question of the impact of such strain changes on the vaccination program that is to be introduced in the country. The ability of currently available rotavirus vaccines to protect against emerging strains such as G12 cannot be predicted.19 However, the evidence that the majority (72%) of strains that caused rotavirus gastroenteritis in DRC contained at least 1 of the G or P genotypes present in both currently available rotavirus vaccines is very important and reassuring.
This multisite study in DRC has some limitations because it was geographically restricted to 2 provinces and thus, does not necessarily reflect what is happening in other provinces of the country. Within the 25 G-P combinations identified in this study, only 3, G1P[6], G1P[8] and G12P[6], and the single strain G3P[6] were found in Lubumbashi. Therefore, it is important to initiate rotavirus surveillance in other provinces to have a comprehensive picture of strain distribution in this large country. Specimens were randomly selected for genotyping assays and this could have biased the results. The fact that only a limited number of specimens were genotyped could have also resulted in skewed prevalence values. Genotyping were performed at different laboratory institutes and therefore, genotyping results may not be completely consistent among institutes.
In conclusion, this study shows additional evidence of a high burden of rotavirus hospitalizations in DRC and considerable variation in the number of rotavirus strains circulating each year. This study of the surveillance of rotavirus strains in DRC has played a key role in defining the G- and P-types circulating in the country. This information will be very important to decision makers for the introduction of rotavirus vaccine in DRC. It will be essential to continue to monitor changes in genotype prevalence in the future to determine whether the introduction of rotavirus vaccine has an impact on rotavirus strain evolution and diversity.
ACKNOWLEDGMENTS
The authors are grateful to the staff of the Centre Hospitalier Pédiatrique de Kalembelembe, the Centre Hospitalier de Kingasani and Hôpital Général Sendwe for their assistance. The authors also want to thank Dr Jason Mwenda, WHO Regional Office for Africa and the WHO DRC office for logistical help in the country. The authors would like to thank Sr Josée Bende, Micheline Katembo, Yvonne Lay Mowele, Naomie Mitonga and Frida Nkawa, Jean-Pierre Lonyema Wandje, Mazarin Nlandu Phezo and Jean Mukwela Mungulu for technical assistance. The authors are grateful to Albert Mbule, Jean-Claude Changa Changa and Médard Ngoy Kiluba for their help on data management. The authors are grateful to Olen Kew and Paul Rota for guidance, helpful discussions and constructive criticisms leading to the completion of this work.
The SURVAC working group included: Veronique Tubijinga, MS, Mediatrice Biamungu, MS, and Francis Ngoy, BS, Centre Hospitalier Pédiatrique de Kalembelembe, Kinshasa, DRC; Emerence Katoba, VMD, Hôpital Général Sendwe, Lubumbashi, DRC; Lamande Kitambala, BS, Centre Hospitalier de Kingasani, Kinshasa, DRC; and Jamie Lewis, BS, Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, United States Centers for Disease Control and Prevention, Atlanta, GA 30333.
Funding for this work was provided by the Bill and Melinda Gates Foundation through the SURVAC Project (Grant number 51214), the World Health Organization, the U.S. Centers for Diseases Control and Prevention, the CDC Foundation and the Ministry of Health of the Democratic Republic of the Congo. The authors have no other funding or conflicts of interest to disclose.
REFERENCES
- 1.Tate JE, Burton AH, Boschi-Pinto C, et al. ; WHO-coordinated Global Rotavirus Surveillance Network. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:136–141. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Estimated rotavirus deaths for children under 5 years age: 2008, 453000. 2012. Available at: http://www.who.int/immunization_monitoring/burden/rotavirus_estimates/en/index.html. Accessed November 15, 2012.
- 3.CIA. Africa: Congo, Democratic Republic of the. 2012. Available at: https://www.cia.gov/library/publications/the-world-factbook/geos/cg.html. Accessed September 20, 2012.
- 4.The Laboratory Working Group for SURVAC. Subregional training on diagnostics for bacterial meningitis and rotavirus gastroenritis. Global Immunization News. 2011:9–11. [Google Scholar]
- 5.Prasad BV, Burns JW, Marietta E, et al. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature. 1990;343:476–479. [DOI] [PubMed] [Google Scholar]
- 6.Matthijnssens J, Ciarlet M, Rahman M, et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol. 2008;153:1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshino Y, Sereno MM, Midthun K, et al. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc Natl Acad Sci USA. 1985;82:8701–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthijnssens J, Bilcke J, Ciarlet M, et al. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol. 2009;4:1303–1316. [DOI] [PubMed] [Google Scholar]
- 9.Rahman M, Matthijnssens J, Yang X, et al. Evolutionary history and global spread of the emerging g12 human rotaviruses. J Virol. 2007;81: 2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabue JP, Peenze I, de Beer M, et al. Characterization of human rotavirus recovered from children with acute diarrhea in Kinshasa, Democratic Republic Of Congo. J Infect Dis. 2010;202(suppl):S193–S197. [DOI] [PubMed] [Google Scholar]
- 11.WHO. African Rotavirus Surveillance Network, Draft Standard Operating Procedure. WHO Regional office for Africa; 2006. [Google Scholar]
- 12.Gentsch JR, Glass RI, Woods P, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das BK, Gentsch JR, Cicirello HG, et al. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO. Global Rotavirus Information and surveillance Bulletin. Rotavirus bulletin. 2012;5:1–11. [Google Scholar]
- 15.Mwenda JM, Ntoto KM, Abebe A, et al. Burden and epidemiology of rotavirus diarrhea in selected African countries: preliminary results from the African Rotavirus Surveillance Network. J Infect Dis. 2010;202(suppl): S5–S11. [DOI] [PubMed] [Google Scholar]
- 16.Fischer TK, Valentiner-Branth P, Steinsland H, et al. Protective immunity after natural rotavirus infection: a community cohort study of newborn children in Guinea-Bissau, west Africa. J Infect Dis. 2002;186:593–597. [DOI] [PubMed] [Google Scholar]
- 17.Cunliffe NA, Ngwira BM, Dove W, et al. Epidemiology of rotavirus infection in children in Blantyre, Malawi, 1997–2007. J Infect Dis. 2010;202(suppl): S168–S174. [DOI] [PubMed] [Google Scholar]
- 18.Moïsi JC, Nokes DJ, Gatakaa H, et al. Sensitivity of hospital-based surveillance for severe disease: a geographic information system analysis of access to care in Kilifi district, Kenya. Bull World Health Organ. 2011;89:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkwood CD. Genetic and antigenic diversity of human rotaviruses: potential impact on vaccination programs. J Infect Dis. 2010;202(suppl): S43–S48. [DOI] [PubMed] [Google Scholar]


