This study applied innovative antibody screening technologies to discover antimalarial antibody variants with enhanced protective potency. Study authors applied precision DNA library generation, high-throughput screening, and analysis of next generation sequencing data to identify mutations that improved malaria protection following in vivo infectious challenge.
Abstract
The monoclonal antibody CIS43 targets the Plasmodium falciparum circumsporozoite protein (PfCSP) and prevents malaria infection in humans for up to 9 mo following a single intravenous administration. To enhance the potency and clinical utility of CIS43, we used iterative site-saturation mutagenesis and DNA shuffling to screen precise gene-variant yeast display libraries for improved PfCSP antigen recognition. We identified several mutations that improved recognition, predominately in framework regions, and combined these to produce a panel of antibody variants. The most improved antibody, CIS43_Var10, had three mutations and showed approximately sixfold enhanced protective potency in vivo compared to CIS43. Co-crystal and cryo-electron microscopy structures of CIS43_Var10 with the peptide epitope or with PfCSP, respectively, revealed functional roles for each of these mutations. The unbiased site-directed mutagenesis and screening pipeline described here represent a powerful approach to enhance protective potency and to enable broader clinical use of antimalarial antibodies.
Introduction
Malaria affects 200 to 400 million people each year, causing nearly 400,000 annual deaths (World Health Organization, 2018). Plasmodium falciparum is the primary parasitic species that causes malaria morbidity and mortality in Africa. Malaria infection is initiated following a mosquito bite in which sporozoites, the infectious form of P. falciparum, are injected into the host and quickly migrate to the liver to infect hepatocytes. A range of treatment and control solutions currently exist, yet global control of malaria remains a persistent challenge and new preventive solutions are urgently necessary to curb continued transmission and disease. Solutions that neutralize initial sporozoite infection have the capacity to fully interrupt the infection cycle, preventing progression to liver stage disease and providing a promising approach for sterilizing immune protection (Julien and Wardemann, 2019).
The P. falciparum circumsporozoite protein (PfCSP) is the most prevalent antigen on the surface of sporozoites and a major target of antibody responses that confer protection against malaria infection, though generally at very high antibody titers (Julien and Wardemann, 2019). PfCSP contains numerous amino acid repeat regions; a dominant “NANP” repeat motif comprises much of the PfCSP central region, a minor “NVDP” repeating sequence occurs four times in the central region, and a unique junctional epitope containing one “NPDP” motif bridges the N-terminal region with the central region. Recently, a number of naturally- or vaccine-elicited human mAbs were revealed to target key regions in PfCSP to provide sterilizing protection against malaria infection following passive transfer in mice (Julien and Wardemann, 2019; Kisalu et al., 2018; Murugan et al., 2020; Tan et al., 2018; Wang et al., 2020). Moreover, in a recent human clinical trial with malaria-naive volunteers, 40 mg/kg of an antimalaria mAb known as “CIS43LS” was intravenously delivered to patients and provided protection against controlled malaria challenge for up to 9 mo in a small number of participants (Gaudinski et al., 2021; Kisalu et al., 2018). CIS43LS targets the PfCSP junctional epitope and was produced by modifying the human CIS43 antibody sequence to include an “LS” amino acid mutation in the Fc region to extend half-life in vivo (Gaudinski et al., 2021).
All together, these prior studies support the utility of CIS43LS and other PfCSP-targeting mAbs for long-acting and effective malaria prophylaxis. However, the production costs of protein drugs such as CIS43LS remain substantially greater than the production cost for small molecules, creating economic barriers for large-scale clinical use of antimalarial antibodies. Improvements in antibody potency should reduce therapeutic dose, thereby providing flexibility in route of administration and potential cost savings and increased global availability of antibody-based antimalarial solutions.
Prior reports by our group (Madan et al., 2021) and others (Adams et al., 2016; Chan et al., 2020; Medina-Cucurella et al., 2018; Van Deventer et al., 2014) have demonstrated that deep mutational scanning using precision screening of site-saturation mutagenesis (SSM) antibody libraries can lead to efficient antibody drug potency improvements, especially when combining multiple mutations for synergistic effects. Importantly, these deep mutational scanning libraries can sample rare variants that are not frequently observed in natural in vivo antibody evolution (Madan et al., 2021; Medina-Cucurella et al., 2018), including variants that require multiple nucleotide substitutions within a single codon, and potentially rare combinations of mutations distributed throughout the entire antibody variable region. This method of iterative precision library generation and in vitro functional screening allows stepwise introduction of potentially beneficial mutations to the parent sequence for isolation of improved variants with fewer modifications than alternative error-prone PCR methods and mouse models (Madan et al., 2021; Medina-Cucurella et al., 2018). Screening with precise SSM antibody libraries maintains high sequence homology with native human antibody genes and can reduce the risk of immune rejection associated with long-term clinical use of highly mutated genes (Cassotta et al., 2019; Jawa et al., 2020; Vaisman-Mentesh et al., 2020). Beyond antibody design and immunogenicity considerations, it has also been shown that most protective anti-PfCSP antibodies recognize nonoverlapping repeat-peptides with high affinity, confounding the native human immune response, and thus far it has been unclear how to optimize the complex protein interactions to discover antimalarial antibodies with sufficient potency for broad clinical use (Julien and Wardemann, 2019; Kisalu et al., 2018; Murugan et al., 2020; Tan et al., 2018; Wang et al., 2020).
To address these challenges in antimalaria antibody development, we report the application of in vitro precision antibody engineering techniques to improve the protective potency of the template antibody CIS43 by enhancing its binding interactions to PfCSP-derived antigens. We found that approximately sixfold improvement of the already potent CIS43 mAb could be achieved with three engineered mutations, and variants with enhanced protection displayed higher affinity to both full-length PfCSP (fl_PfCSP) and the junctional epitope within PfCSP. To probe the mechanism by which identified mutations improved protective function, we determined co-crystal and cryo-electron microscopy (cryo-EM) structures of improved antibodies with junctional peptide epitope and PfCSP, respectively. These data describe a pipeline for efficient antibody engineering with an overall aim of providing substantially improved antibodies that can be used widely for cost-effective prevention of infectious diseases such as malaria.
Results
Antimalarial antibody library screening enhances recognition of CSP antigens
We sought to improve the potency of CIS43 while introducing a minimal number of mutations to the parent sequence. Accordingly, we applied precision antibody library generation and screening strategies for efficient improvement of CIS43 antimalarial activity (Fig. 1). We sequentially generated and screened antibody gene libraries using functional yeast display and next generation sequencing (NGS) to scan the CIS43 heavy and light chain variable regions for potentially beneficial mutations (Fig. 1 a). Comprehensive mutant gene libraries containing all possible single amino acid substitutions in the template sequence were first generated using SSM over the heavy chain variable region gene (VH) and light chain variable region gene (VL) separately (Madan et al., 2021; Wrenbeck et al., 2016). To evaluate the effect of single mutations on antigen-binding performance, the resulting single-mutation DNA libraries, (VH-SSM):VL and VH:(VL-SSM), were cloned into plasmids for yeast surface expression as fragment antigen binding (Fab) libraries while ensuring >100-fold SSM library coverage (Fig. S1 a and Table S1). These plasmid libraries were used to transform genetically engineered yeast to express surface-bound Fab libraries via a galactose-inducible bidirectional promoter followed by FACS based on affinity screening defined by Fab surface expression levels versus Fab recognition of PfCSP-derived antigen probes (Fig. 1 b; Fig. S1, b and c; and Table S2; Madan et al., 2021; Wang et al., 2018). NGS analysis of yeast-transformed single-mutation libraries revealed robust library coverage of single amino acid substitutions prior to screening (Fig. S1 a).
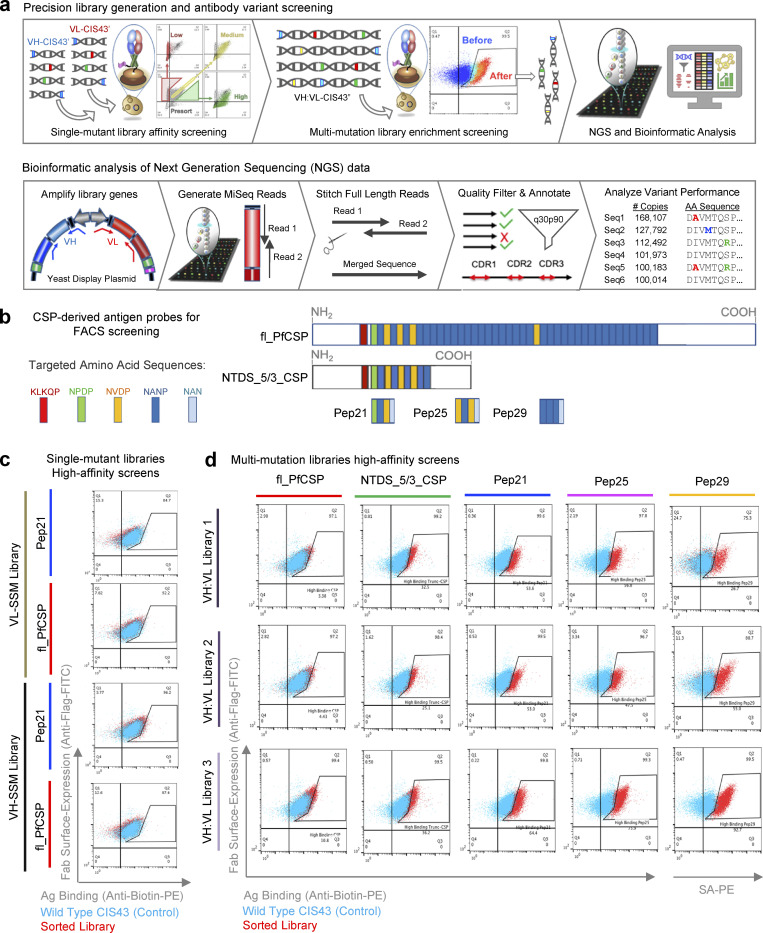
Figure 1.
Precise directed evolution techniques were used to optimize the mAb CIS43 for enhanced binding to PfCSP epitopes. (a) SSM libraries were generated separately for the VH and VL of the antimalarial antibody CIS43 and cloned into yeast Fab display vectors for affinity-based FACS. Plasmid DNA was extracted from high-affinity sorted libraries, and additional combinatorial mutagenesis and screening was performed to select high-affinity multi-mutation variants by FACS. Sorted libraries were analyzed using NGS to quantitatively track each variant across sorting rounds. NGS data were mined to identify improved CIS43 antibody gene variants. After Illumina 2 × 300 bp sequencing, raw fastq sequence files were merged and quality-filtered to obtain high-quality in-frame antibody amino acid sequences. Unique amino acid sequences were compiled into library prevalence values, which were followed to track changes in variant populations across each round of sorting. (b) Five CSP-derived antigen probes were used for yeast display screening. Probes include fl_PfCSP, an N-terminal domain stabilized truncated-CSP format (NTDS_5/3_CSP), and three peptides derived from PfCSP (Pep21, Pep25, and Pep29.). (c) VH-SSM and VL-SSM single-mutation libraries were sorted for high affinity against fl_PfCSP and Pep21 (red) compared to wild-type CIS43 (cyan), which revealed minimal enhancements in library-scale affinity in single-mutation libraries. (d) Flow cytometric analysis of combinatorial multi-mutation Libraries 1, 2, and 3 after three rounds of sorting showed substantially enhanced affinity against fl_PfCSP, NTDS_5/3_CSP, Pep21, Pep25, and Pep29 (red) compared to wild-type CIS43 (cyan).
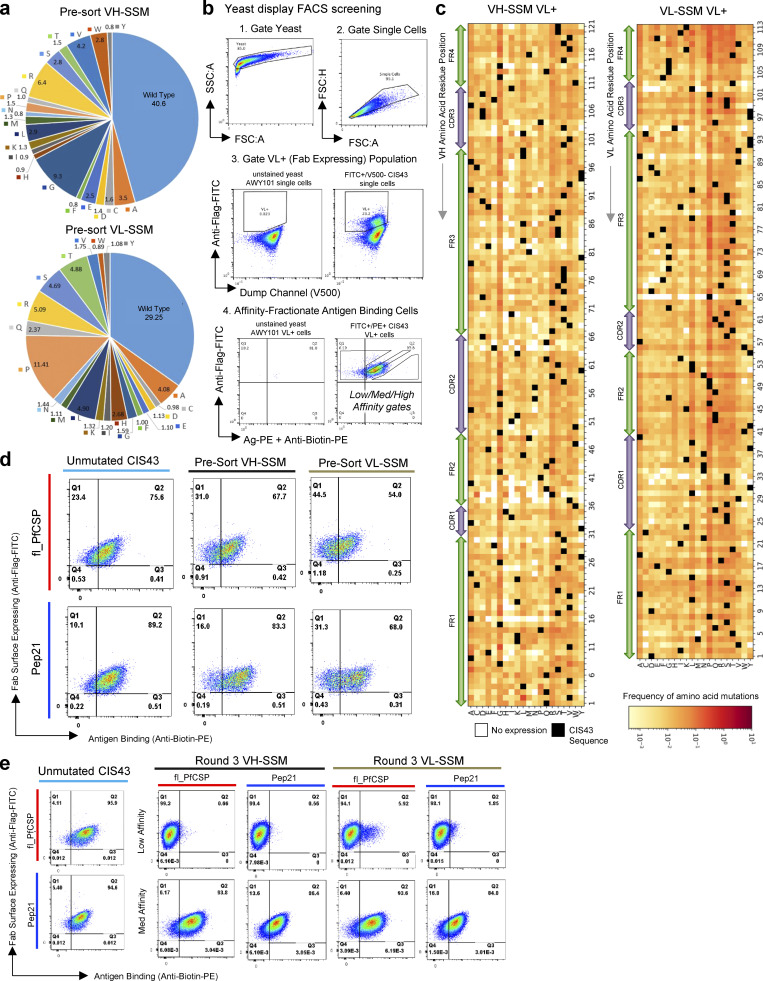
Figure S1.
Single-mutant library synthesis and screening revealed the sequence–function relationship between CIS43 antibody mutants and antimalarial antigen recognition. (a) Distribution of single-mutation VH_NNK and VL_MNN DNA codons in presort SSM libraries, as quantified by NGS. (b) FACS gating strategy for affinity-based sorting of SSM libraries. (c) CIS43 single-mutation heat maps showing library frequencies for mutants expressed in the VL+ Fab-expressing VH-SSM and VL-SSM libraries, prior to sorting for antimalarial antigen recognition. (d) Flow cytometric analysis of CIS43 wild-type surface-displayed Fab and pre-sort single-mutation Fab libraries, stained with Pep21 and fl_PfCSP. (e) Flow cytometric analysis of CIS43 wild-type surface-displayed Fab and Round 3 single-mutation Fab libraries sorted against Pep21 and fl_PfCSP for low-affinity (upper) and medium-affinity (lower) population phenotypes.
We hypothesized that enhanced affinity to CSP antigens would improve antibody potency. Accordingly, Fab-expressing yeast libraries were stained with fluorescently labeled fl_PfCSP and Peptide 21 (Pep21) antigen probes (Fig. 1 b and Table S2) and fractionated en masse into high-, medium-, or low-affinity binding populations against via FACS (Fig. 1 c; Fig. S1 b; and Fig. S1, d and e). Affinity phenotypes were determined at high-throughput for surface-expressed Fab by comparing the relative FACS intensities of Fab expression to antigen binding for yeast library populations (Madan et al., 2021; Wang et al., 2018). Fab surface expression is maintained via conjugation of the Fab heavy chain fragment to the surface of the cell, and appropriate Fab assembly is quantified via a FLAG tag marker on the associated light chain (denoted as variable light positive, or VL+, yeast cells; Wang et al., 2018). High affinity populations were collected by gating cells that produced a higher antigen binding signal relative to Fab surface expression, whereas medium affinity gates collected populations with a signal of Fab expression to antigen recognition similar to the template antibody sequence. Low affinity gates collected cells with high surface Fab expression and lower antigen binding signal (Fig. 1 a inset; Fab expression on the y axis and antigen binding on the x axis).
Bioinformatic analysis of the yeast surface–expressed libraries (measured as the VL+ expressing population; Fig. S1 b) revealed that nearly all 7,488 theoretical single-mutation variants expressed at detectable levels in the Fab-expressing pre-sort libraries (Fig. S1 c). Prior to antigen screening, single-mutation libraries showed generally comparable binding to template CIS43 (Fig. S1 d). After screening, the third round of single-mutation high-affinity sorted libraries showed minimal observable affinity differences in antigen recognition compared to the monoclonal CIS43 control (Fig. 1 c and Fig. S1 e), suggesting that single-mutation variants had a limited capacity to improve antimalarial antibody recognition. In contrast, the low-affinity single-mutation libraries showed clear phenotypic binding differences from monoclonal CIS43 (Fig. S1 e), with a sharp loss of antigen recognition after three rounds of low-affinity sorting. Medium-affinity sorted libraries maintained consistent binding characteristics with template CIS43 control expressed as yeast-surface bound Fab (Fig. S1 e).
Single-mutation variants confer limited improvements in antigen recognition
The phenotypic differences observed by flow cytometry between high, medium, and low affinity sorted libraries suggested that single-mutation libraries had been successfully fractionated according to their antigen binding affinity phenotypes. Libraries were processed and submitted for NGS and bioinformatic analysis of NGS data was used to quantitatively track single amino acid mutations across each sort condition to identify enriched variants of interest (Fig. 2 a; and Fig. S2, a and b). We enumerated the prevalence and enrichment ratio (ER) of each single amino acid mutation to quantify variant performance across sorted populations, binning each single-mutation variant into affinity groups based on ER (Fig. 2; and Fig. S2, a and b). Single-mutation variants with ER >10 in high-affinity sorts and <1 in low-affinity sorts were considered dominant mutants predicted to have improved antigen binding, and with some mutations tested for each screening antigen (Fig. S2, a and b). Affinity-binned groups revealed consistencies in sequences against specific malaria antigens (Fig. 2 a) and most sequences that strongly enriched in high-affinity sorts were depleted in low-affinity sorts and did not strongly enrich in medium affinity sorts (Fig. S2 b). These quantitative bioinformatic data agreed with the qualitative phenotypic observations by flow cytometry, supporting our affinity-based screening approach for fractionating distinct gene variant populations according to antigen recognition phenotypes.
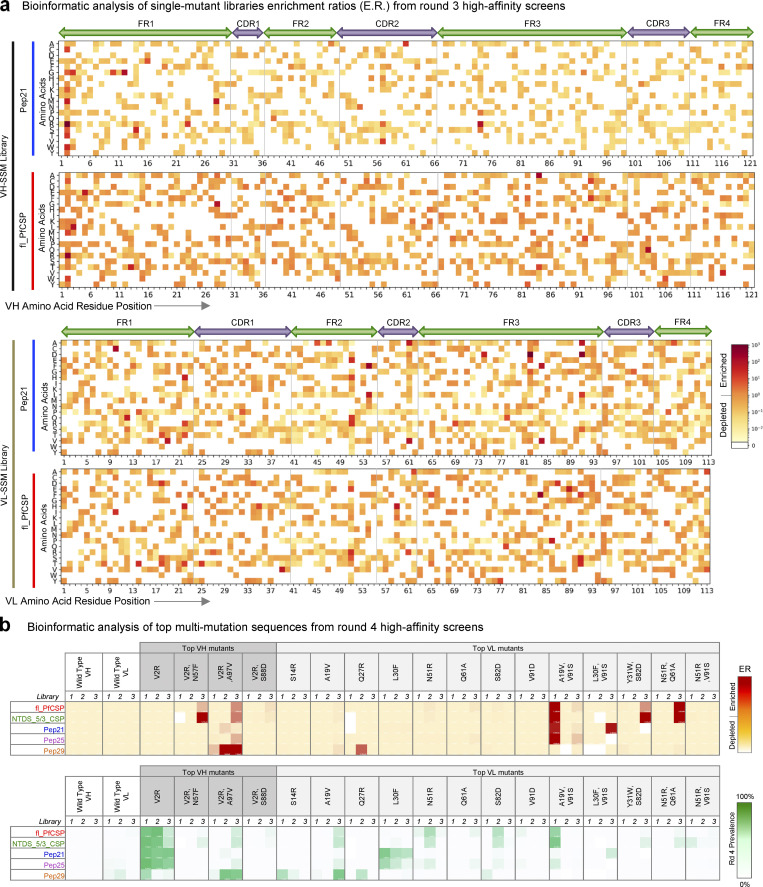
Figure 2.
NGS analysis of FACS-screened libraries revealed single- and multi-mutation variants with enhanced PfCSP antigen recognition. (a) Heat maps revealed the functional impact of single mutations against fl_PfCSP and Pep21 antigens from high-affinity enriched libraries. (b) Bioinformatic analyses of multi-mutation sequences ERs and prevalence values after four rounds of sorting showed that the dominant enriched mutant sequences out-competed unmutated CIS43 template genes for antigen recognition after screening for high-affinity binding against fl_PfCSP, NTDS_5/3_CSP, Pep21, Pep25, and Pep29 antigens.
Figure S2.
Multi-mutation library screening overview. (a) Bioinformatic analyses of single-mutation sequence ER vs. prevalence after three rounds of FACS revealed the relationship between dominant single-mutation sequences (red) versus unmutated CIS43 sequences (cyan) after high-affinity enrichment against fl_PfCSP and Pep21 antigens. (b) Bioinformatic analyses of single-mutation sequence ERs vs. prevalence after three rounds of FACS screening revealed the relationship between dominant single-mutation sequences (red) vs. unmutated CIS43 sequences (cyan) after medium- and low-affinity enrichment against fl_PfCSP and Pep21 antigens. Data are shown for both the low-affinity (upper) and medium-affinity (lower) population phenotypes. (c) Structural mapping of predicted affinity-enhancing mutants based on single-mutation NGS data. (d) ELISA evaluation of expressed and purified single-mutation IgGs.
Bioinformatic analysis of the Round 3 high-affinity libraries identified enriched single mutants predicted for enhanced PfCSP-antigen recognition (Fig. S2, a–c), 26 single-mutation variants, and 4 rationally combined multi-mutation variants (based on highly enriched ER or prevalence metrics) were selected for expression and biophysical characterization as soluble antibodies (Table S3). Notably, multiple substitutions at the second residue in framework region 1 (FR1) of the VH gene showed strong enrichment (Fig. 2 a, top), where a rare valine to arginine substitution (VH_V2R) was highly enriched against Pep21 while the unmutated template CIS43 sequence was simultaneously depleted in this same library (Fig. S2 a, second graph), suggesting that the VH_V2R mutation phenotypically enhanced binding affinity in the libraries characterized via FACS. Top additional VL single mutations of interest included (if different, Kabat numbering [Kabat et al., 1992] is provided in italics): VL_A19V (FR1), VL_N22H (FR1), VL_N51R/N45R (FR2), VL_S82D/S76D (FR3), VL_L84F/L78F (FR3), and VL_V91D/V85D (FR3), each having ERs >10 in the fl_PfCSP and Pep21 high-affinity sorts and <1 in low-affinity sorts (Fig. 2 a, bottom; Fig. S2 b; and Table S3). Notably, we also observed that the unmutated CIS43 template gene was present and/or remained dominant in several of the single-mutation high-affinity screens (Fig. S2 a), suggesting that the single-mutation variants enriched in high affinity screens did not outcompete or substantially enhance antigen affinity over template CIS43.
Structural mapping of FACS-predicted high-affinity single-mutation variants showed most enriched residues were structurally distal to the antigen-binding paratope in the framework regions, and only a selected few mutations were enriched in the complementary determining regions (CDRs; Fig. 2 a and Fig. S2 c, PDB: 6B5M). ELISA analysis of the top FACS-predicted high-affinity single-mutation variants showed comparable antigen recognition for individually expressed IgG single-mutation variants versus parent CIS43 against different CSP-derived antigens (Fig. S2 d), consistent with the limited phenotypic improvements observed in high-affinity FACS populations (Fig. 1 c). These limited improvements to antigen recognition observed via FACS and ELISA suggested that single-mutations would also confer limited improvements in protection, and that multi-mutation combinations would likely be required to enhance antigen recognition.
Multi-mutation variants show improvements in antigen recognition
We next generated and screened multi-mutation gene libraries to identify additional synergistic mutation combinations that improved antigen recognition (Fig. 1, a and d; and Fig. S3). We hypothesized that the enriched variants in high-affinity libraries represented some beneficial progress toward improved antigen recognition and served to eliminate potentially deleterious single mutations, and therefore we used the single-mutation Round 3 high-affinity libraries as a starting template to generate three new multi-mutation libraries (Fig. 1, a and c). Multi-mutation Library 1 was generated by extracting the enriched VH and VL genes from template yeast libraries and subcloning into a shared plasmid backbone to create combinatorial heavy and light chain variants (Fig. S3 a). To increase library diversity, DNA shuffling was also performed on the single-mutation template VH and VL DNA pools, separately, to generate combinations of enriched mutations within the VH and VL genes (Meyer et al., 2014). Shuffled-VH and shuffled-VL genes were then subcloned into a single plasmid backbone to generate a combinatorial shuffled-VH:VL multi-mutation (Library 2). Finally, an additional degree of diversity was included in library design using shuffled-VH:VL genes as template to repeat SSM (“re-SSM”), followed by subcloning the re-SSM genes into a single plasmid backbone, generating a third library consisting of paired VH:VL genes with multiple mutations (Library 3).
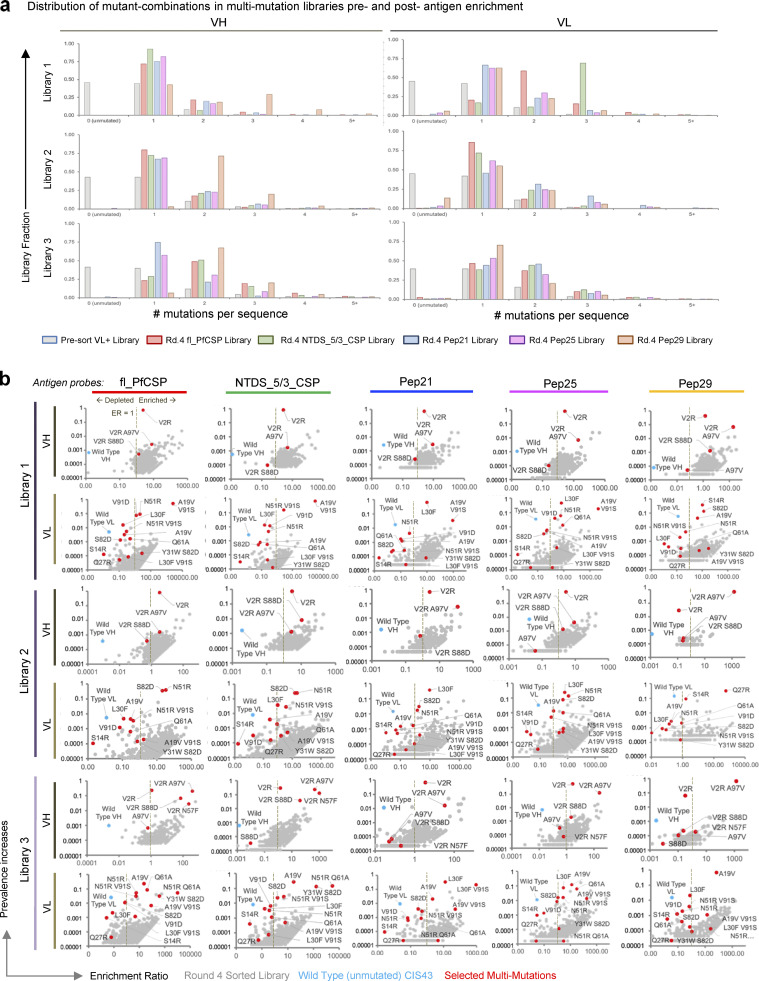
Figure S3.
Multi-mutation screening enhances understanding of CIS43 sequence-function relationships. (a) Multi-mutation sequence distribution in pre-sort multi-mutation libraries, and after four rounds of screening for high-affinity variants. (b) Bioinformatic analyses of multi-mutation sequence ERs vs. prevalence after four rounds of FACS revealed the relationship between dominant mutant sequences (red) vs. unmutated CIS43 sequences (cyan) after high-affinity enrichment against fl_PfCSP, NTDS_5/3_CSP, Pep21, Pep25, and Pep29 antigens.
Multi-mutation plasmid libraries were used to transform yeast, maintaining at least 1 × 107 library transformants at each step (Table S1). Fab expression was induced in the three separate multi-mutation yeast libraries, and surface-expressed Fabs were screened against five different PfCSP-derived antigens: fl_PfCSP, an N-terminal domain stabilized truncated version of CSP with five major repeats and three minor repeats (NTDS_5/3_CSP), Pep21, Peptide 25 (Pep25), and Peptide 29 (Pep29; Fig. 1, a and b; Fig. 2 b; Fig. S3; and Table S2). We enriched high-affinity variants across four rounds of FACS to generate over 60 sorted multi-mutation library samples (Fig. 1 d). Multi-mutation libraries from the fourth round of high-affinity screening showed significant improvements to antigen binding (Fig. 1 d) and NGS data were mined to identify enriched sequences predicted to enhance antimalarial binding (Fig. 2 b, Fig. S3 b, and Table S3).
A comparison of antigen stains between high-affinity groups with single mutations (Fig. 1 c) and multi-mutations (Fig. 1 d) revealed that multiple mutations dramatically improved binding over single mutants alone. NGS data were mined to quantitatively track multi-mutation variants across sort conditions (Fig. 2 b and Fig. S3). The multi-mutation libraries were produced using single-mutant libraries as starting template, and thus each had a high starting prevalence of the VH_V2R-enriched single mutation (Fig. 2 b, bottom heat map). Over multiple rounds of sorting, the VH_V2R mutation remained highly prevalent in most libraries, and therefore had low enrichment in most sorts (Fig. 2 b). High-affinity sorted multi-mutation libraries showed a higher frequency of single and double mutations on the VH and VL genes in all sorted libraries when compared to the presort population (Fig. S3 a and Table S4), and the unmutated CIS43 template gene was depleted from the sorted population in all multi-mutation sorts showing higher phenotypic affinity performance (Fig. S3 b). Certain multi-mutation variants were enriched across several antigen sort conditions, suggesting that our iterative library screening converged on variants with strongly improved malaria antigen recognition (Fig. 2 b). Bioinformatic evaluation of multi-mutation library NGS data identified several dominant enriched antibody sequences of high interest (Fig. 2 b, Fig. S3 b, and Table S3), and the top 24 multi-mutation variants with ER >10 in multiple library sorts and high prevalence values in Round 4 sorted libraries were selected for expression as soluble antibodies for functional and biophysical characterization.
Multi-mutation variants provided robust protection against malaria challenge in vivo
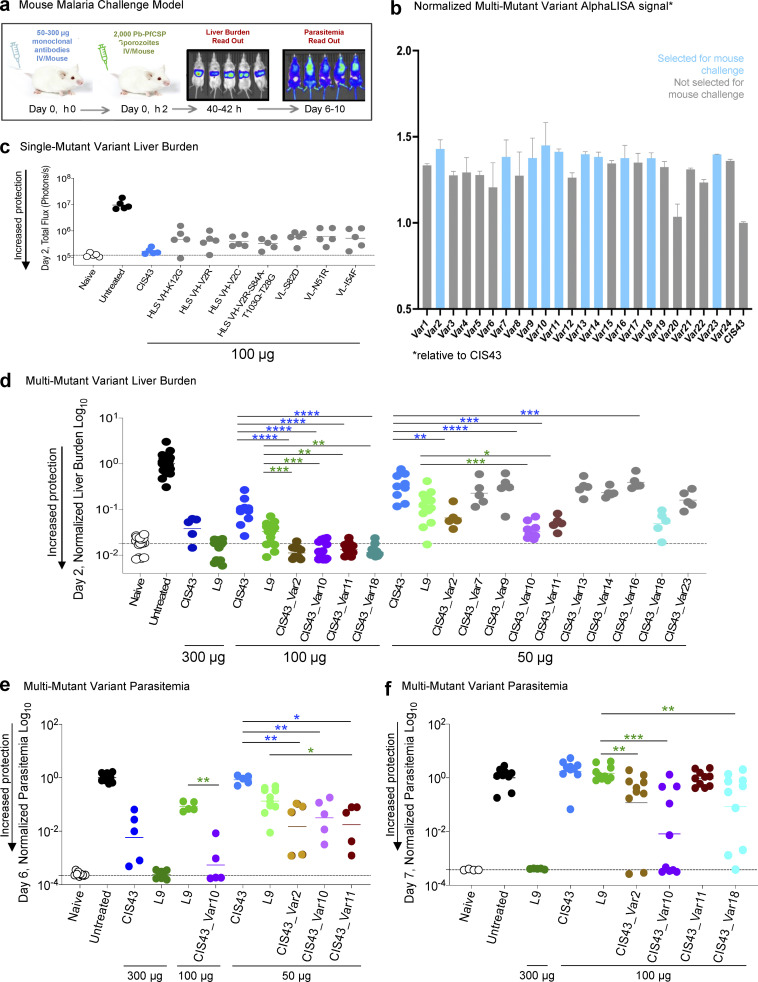
As there are no reliable in vitro functional assays for antimalarial antibody potency, we sought to evaluate the level of protection provided by CIS43 variants with improved affinity using a previously described in vivo murine challenge model to quantify passively administered antibody protection (Wang et al., 2020; Fig. 3 a). To evaluate the capacity of mutant antibody variants to prevent malaria infection, mice were treated with antibodies 2 h prior to controlled intravenous challenge with 2,000 malaria sporozoites that had been genetically modified to encode GFP and luciferase markers. Malaria liver stage infection was quantified at 2 d after challenge by injecting mice with D-luciferin and detecting liver luminescence with IVIS Spectrum. Antibody-treated mice with substantial protection observed at day 2 were maintained and reimaged for total body luminescence 6 d after challenge. The measurements of parasitemia at day 6 provided additional signal amplification for highly sensitive evaluation of the degree of parasitemia in challenged mice, for which differences were often too small to be observed at day 2. As in vivo challenge studies are limited by the number of experimental animal groups evaluated at one time, we prioritized the top variants in our 24 multi-mutation antibody panels based on their affinity to the NPDP19 junctional peptide. Our single-mutation studies and other internal data showed that the affinity against full-length CSP, Pep21, and Pep29 were not reliable correlates of protection for CIS43 variants. However, NPDP19 represented a potential correlate of protection than other available CSP antigens, as revealed by other studies (Kratochvil et al., 2021), and we explored its use further here. Variant affinity against NPDP19 was determined based on the AlphaLISA bead proximity luminescence, and the top ten multi-mutation variants with the highest NPDP19 recognition were selected for challenge evaluation (Fig. 3 b).
Figure 3.
Malaria challenge studies revealed up to 10-fold improved potency for multi-mutation CIS43 variants. (a) A mouse malaria challenge model was used to quantify protective potency of anti-PfCSP mAbs. Anti-PfCSP mAbs were passively transferred into mice 2 h prior to infection with transgenic Pb-PfCSP-GFP/Luc, a rodent malaria parasite (P. berghei, Pb) with the endogenous PbCSP replaced with PfCSP, and which also constitutively expressed luciferase. Luciferin substrate was injected for bioluminescent in vivo imaging by IVIS for an efficient, sensitive, and noninvasive means to quantify liver stage (day 2) and blood stage (day 6) parasite infection. (b) Multi-mutation variant antibodies were screened for functional recognition by apparent AlphaLISA affinity to NPDP19. The top 10 selected antibodies for in vivo protection analyses are shown in light blue. (c) Single-mutation variant liver burden on day 2. Following passive transfer of 100 μg CIS43 (blue) or CIS43 variants (gray), Albino-B6 mice were challenged i.v. with Pb-PfCSP-GFP/LUC SPZ before imaging by IVIS. Total flux indicated the extent of malarial infection in the liver, with naive (background, white) and untreated (maximum burden, black). HLS abbreviation denotes expression as soluble human IgG with a half-life extending “LS” amino acid mutation on the Fc region on the heavy chain. (d) Multi-mutation variant liver burden on day 2. Following passive transfer of 50 μg CIS43, or the indicated multi-mutation variants over several different experiments, the liver burden of each antibody group was normalized by the geometric mean of liver burden values using untreated mice in the same experiment. Statistical differences were assessed using Mann–Whitney test. P values ranges are indicated by stars, with P value <0.05 (*), P value <0.01 (**), P value <0.001 (***), and P value <0.0001 (****). Raw data from this experiment are presented in Fig. S3. (e) Multi-mutation variant parasitemia on day 6. Following passive transfer of CIS43, L9, and the indicated multi-mutation variants at specified doses, the liver burden was assessed at day 2 (captured in Fig. 3 d and Fig. S3, c and d). Mice treated with CIS43_Var2, CIS_Var10, or CIS43_Var11 showed a high degree of protection at day 2 and were followed and reimaged again on day 6 to quantify blood stage infection and/or parasitemia. Statistical differences were assessed using Mann–Whitney test. (f) Multi-mutation variant parasitemia on day 7. Following passive transfer of indicated doses of CIS43, L9, and the indicated multi-mutation variants, the liver burden was assessed at day 2 (captured in Fig. 3 d or Fig. S3 d). Mice were followed and reimaged again at day 7 to assess parasitemia. Statistical differences were assessed using Mann–Whitney test.
These results of these sensitive animal challenge studies showed that single-mutation variants provided some protection against malaria infection (Fig. 3 c), but this protection was not enhanced over the original template CIS43, which was consistent with the limited affinity enhancements observed by flow cytometry and ELISA (Fig. 1 c and Fig. 2 d). In contrast, these protection studies revealed that several multi-mutation variants provided robust protection greater than template CIS43 against malaria infection (Fig. 3 d and Fig. S4) consistent with observed improvements in antigen recognition measured by flow cytometry and AlphaLISA (Fig. 1 d). Multi-mutant variants 2, 10, 11, and 18 were significantly more potent than CIS43 in protecting against liver burden at day 2 after challenge, with CIS43_Var10 showing approximately sixfold improved protective potency versus the parent CIS43 molecule. Variants 2, 10, and 11 also showed statistically significantly lower parasitemia at 6 d after challenge than CIS43 (Fig. 3, e and f).
Figure S4.
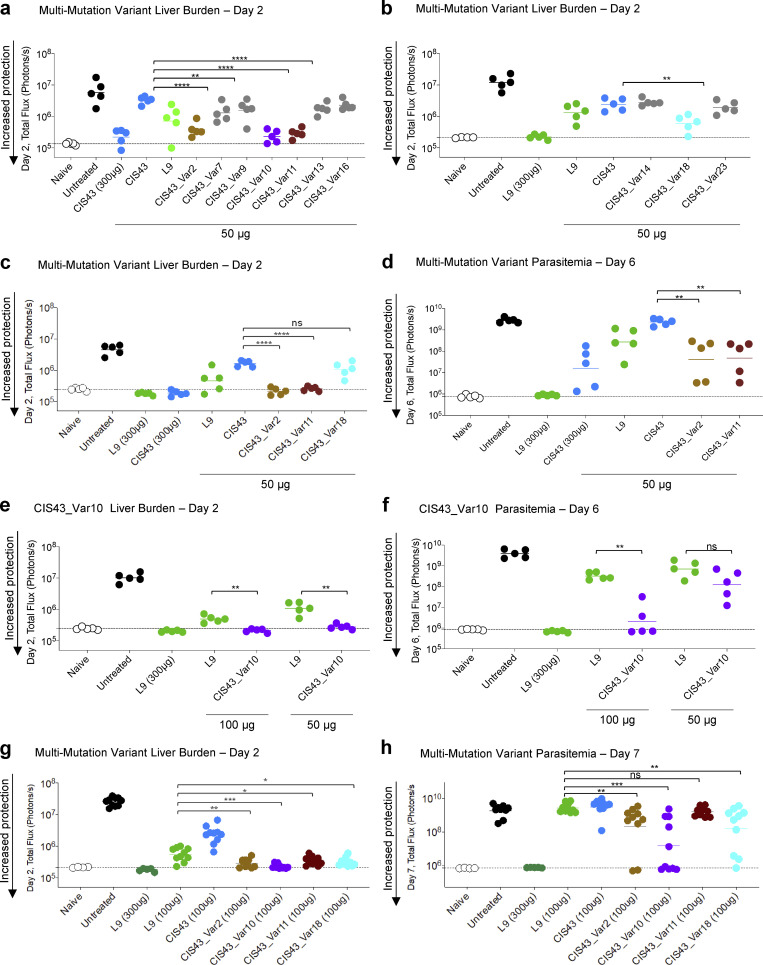
Experimental groups for the analysis of multi-mutation variant protection against malaria challenge. (a and b) Following passive transfer of CIS43, L9 and the indicated multi-mutation variants at the specified doses, Albino-B6 mice were intravenously challenged with 2,000 Pb-PfCSP sporozoites, and the liver burden was assessed at day 2 after challenge. Total flux or bioluminescence expressed as photons/s is a measure of the liver parasite burden and is shown for naive (background, white) and untreated (maximum burden, black) groups. Horizontal lines indicate the geometric mean. Statistical differences were assessed using a two-tailed Mann-Whitney test. P values are indicated by stars, with P value <0.05 (*), P value <0.01 (**), P value <0.001 (***), and P value <0.0001 (****). (c and d) Select multi-mutation variant antibodies were rescreened at the indicated doses and total flux was measured at day 2 for liver burden. Infected mice were euthanized, but protected mice were followed out and reimaged again at day 6; total flux at this timepoint reflects parasitemia or blood stage infection. (e) CIS43_Var10 mediated protection as assessed at day 2, liver burden, and followed to (f) day 6 for parasitemia assessment as compared to L9 at 50 and 100 μg. (g and h) Liver burden, day 2 assessment and followed to (h) day 7 for parasitemia assessment for CIS43_Var2, Var10, Var11, and Var18 at 100 μg compared to CIS43 and L9.
Based on the promising functional data reported for CIS43_Var10 in these animal studies, we next sought to compare its efficacy to other reported protective mAbs. A recently isolated CSP-targeting human mAb, L9 (Wang et al., 2020), is two- to threefold more potent than CIS43 and was considered as the “best-in-class” benchmark for protective capacity of antimalarial antibodies. In our in vivo functional challenge study, CIS43_Var10 was approximately twofold more potent than the antibody L9 (Wang et al., 2020) when comparing parasitemia levels at 6 d after challenge (Fig. 3, d and f and Fig. S4). These P. falciparum challenge studies demonstrated that our antibody engineering pipeline could improve CIS43 mAb potency by approximately sixfold with only three introduced mutations (VH_V2R, VL_L30F/L27cF, and VL_V91S/V85S), and confirmed that several other multi-mutation combinations could substantially improve antibody performance with very few mutations introduced to the CIS43 template antibody (Fig. 4).
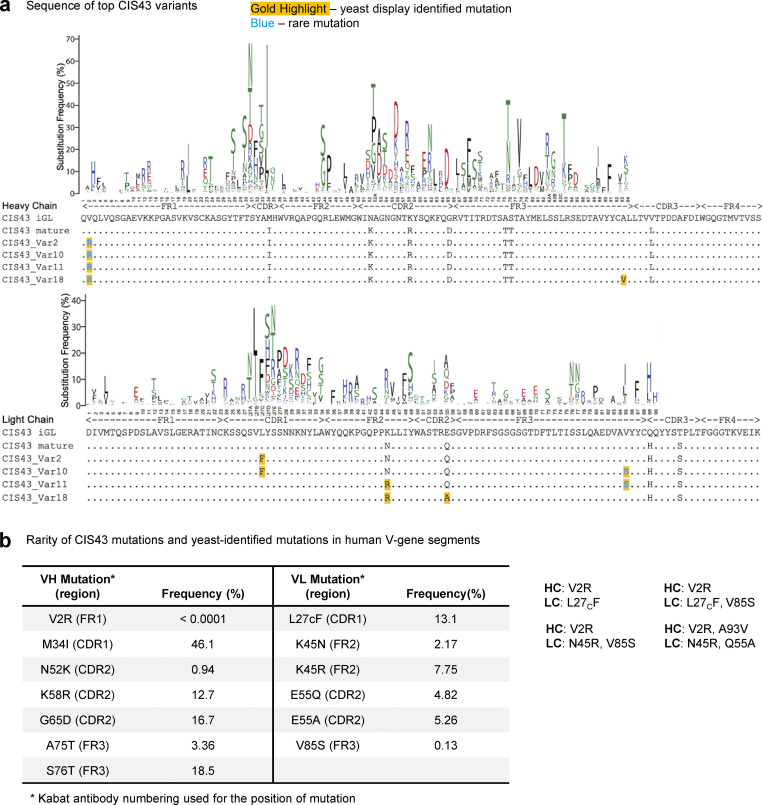
Figure 4.
Location and natural frequency of mutations improving CIS43. (a) Sequences of improved CIS43 variant antibodies are shown along with the heavy (top) and light (bottom) antibody V-gene mutational profiles. We define a rare mutation as a frequency <0.5% in human immune repertoires. (b) Frequencies of amino acid mutations in the context of their respective germline V-gene, reported from cAb-Rep server (Guo et al., 2019).
When comparing the sequences of the top four CIS43 variants, we observed high-sequence homology and redundancy in dominant mutations (Fig. 4 a). We reported the frequencies of amino acid substitutions in the context of their respective germline V-gene using the cAb-Rep server (Guo et al., 2019) and defined rare mutations as having a frequency of <0.5% in human repertoires. Template CIS43 varies from the inferred paired germline sequence by 11 amino acid substitutions, with substitutions ranging in observed frequency from 0.94 up to 18.5%. The top four CIS43 variants (variants 2, 10, 11, and 18) comprised of two, three, or four substitutions made by unique combinations of six different substitutions to the template CIS43 sequence (VH_V2R, VH_A97V/A93V, VL_L30F/L27cF, VL_N51R/N45R, VL_Q61A/Q55A, VL_V91S/V85S) identified by yeast display (Fig. 4 a, gold highlights) two of which (VH_V2R and VL_V91S/V85S; Fig. 4 a, blue font) represent rare mutations infrequently observed in natural human repertoires (Guo et al., 2019). The VH_V2R and VL_V91S/V85S rare mutations have natural frequencies of <0.0001 and 0.13%, respectively, requiring at least two nucleotide substitutions in a single codon to mutate the germline residue. These adjacent DNA point mutations are not frequently produced by antibody gene editing machinery, making them less accessible mutations via natural antibody development but easily accessible for functional screening via SSM.
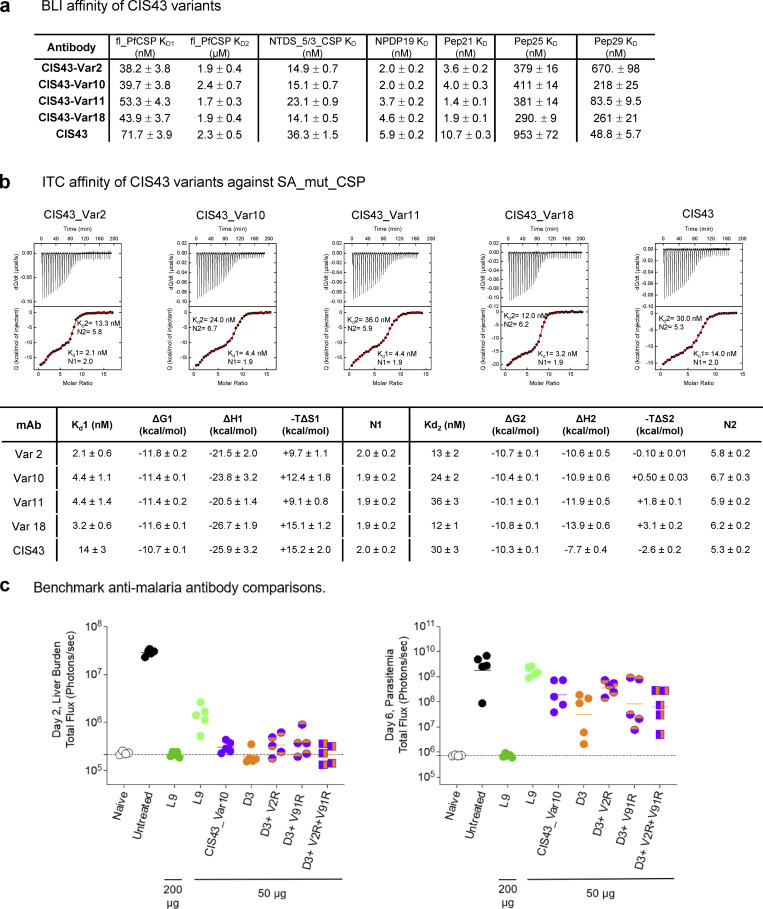
Another recent study on CIS43-derived antibody variants used an engineered mouse model reported an antimalarial antibody iGL-CIS43.D3 (D3) that surpassed the protective potency of L9 (Kratochvil et al., 2021). iGL-CIS43.D3 differs from the template CIS43 mAb by nine amino acid substitutions, whereas the top CIS43 variant CIS43_Var10 varies by three amino acids from the template mAb. We tested additional CIS43 variants by grafting the unique CIS43_Var10 mutations onto the D3 template and compared this panel of antibodies head-to-head against L9 (Fig. S5 c). Despite sharing one VL_L30F mutation, the additional mutations in CIS43_Var10 did not synergize with D3 mutations to provide additional protective benefits. CIS43_Var10, D3, and combined variations of the two were each approximately twofold more potent than antibody L9, yet there were no statistically significant differences detected between CIS43_Var10, D3, or any of the three combination variants, even at the highly sensitive day 6 parasitemia measurement timepoint (Fig. S5 c). These results defined the mutations in CIS43_Var10 and D3 as independent improvement pathways for enhanced potency of the CIS43 template antibody.
Figure S5.
Screening of antibody multi-mutation variants by AlphaLISA, BLI, and ITC. (a) BLI affinity of improved CIS43 variants; Var2, Var10, Var11, and Var18 were determined against fl_PfCSP, NTDS_5/3_CSP, NPDP19, Pep21, Pep25, and Pep29. (b) Isothermal titration calorimetry of CSP_SA_mut with various improved CIS43 variants. The affinities and stoichiometry are shown for both KD1 and KD2, respectively. (c) Comparison of the protection capacity of L9, CIS43_Var10, D3, and combination mutant variants.
Structural basis of improved antibody-based protection
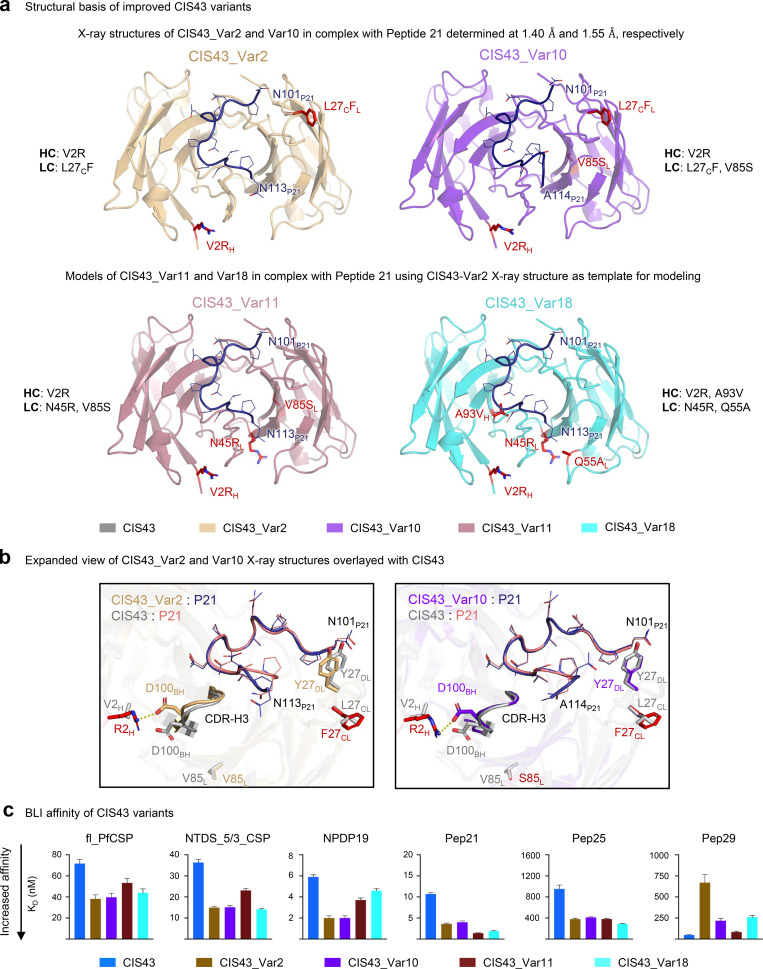
After identifying four substantially improved CIS43 antibodies (variants 2, 10, 11, and 18), we next sought to determine the functional and structural mechanisms that enabled improved efficacy (Fig. 5). Variants 2, 10, 11, and 18 contained combinations of six unique substitutions: VH_V2R, VH_A97V/A93V, VL_L30F/L27cF, VL_N51R/N45R, VL_Q61A/Q55A, VL_V91S/V85S (Table S3, template numbering/Kabat numbering). We co-crystallized CIS43_Var2 and CIS43_Var10 with Pep21 containing the PfCSP major antigenic NPDP junctional epitope and determined the structures of CIS43_Var2 and CIS43_Var10 in complex with Pep21 at 1.40 and 1.55 Å resolution, respectively (Fig. 5 a and Table S5). We analyzed these data, along with modelled structures of CIS43_Var11 and CIS43_Var18, to compare structural features of antigen recognition (Fig. 5 a).
Figure 5.
Structural and functional analysis of the mechanisms of improved antimalaria protection. (a) X-ray structures of CIS43_Var2 and CIS43_Var10 in complex with the junctional epitope (Pep21) determined at 1.40 and 1.55 Å resolution. CIS43_Var11 and CIS43_Var18 structures were modeled using CIS43_Var2 as template. The location of the mutations is shown in red. The Pep21 numbering corresponds to PfCSP sequence. (b) Overlay of CIS43_Var2 with CIS43 showed interactions of Arg2 with Asp100B, resulting in significant movement of CDR-H3 (left); CIS43_Var10 overlayed with CIS43 showed similar interactions between Arg2 and Asp100B, without substantial changes to the CDR-H3 (right). (c) BLI affinity of improved CIS43 variant Fabs was measured against fl_PfCSP, NTDS_5/3_CSP, NPDP19, Pep21, Pep25, and Pep29 antigens. The affinity of multi-mutation variants was improved over CIS43 for all antigens except Pep 29.
While two of the six mutations providing the basis for the top four improved variant function occurred in CDRs: VL_L30F/L27cF (CDR-L1) VL_Q61A/Q55A (CDR-L2), none of the six mutations made direct contact with Pep21 (Fig. 5 b). In addition, two of the six mutations (VH_V2R and VL_V91S/V85S) were rare mutations infrequently observed in natural antibody sequences (Fig. 4). A comparison of CIS43_Var2 and CIS43_Var10 structures over parent CIS43 showed few structural differences; in CIS43_Var2, the arginine in VH_V2R interacted with Asp100B, which appeared to alter the position of the CDR-H3 by 2.2 Å (as measured at the β-carbons of Asp 100B), with a resulting change in the orientation of the C-terminal region of Pep21 to accommodate this movement (Fig. 5 b, left). Notably, in CIS43_Var10, VH_V2R induced a smaller alteration in the CDR-H3 with Pep21 assuming its canonical position (Fig. 5 b, right).
Improved performance for the most protective antibody variants appeared to be associated with increased binding affinity against the junctional NPDP epitope. Overall, we observed little modification in stoichiometry for the second binding event against the minor repeat NANPNVDP contained in Pep21 and Pep25 (Fig. 5 c and Fig. S5). Affinity assays quantified the degree of improved binding for top variants, with bio-layer interferometry (BLI) showing tighter affinity to fl_PfCSP, NTDS_5/3, Peptide NPDP19, Pep21, and Pep25, but not to the major repeat NANP sequence contained in Pep29 for the most improved variants (Fig. 5 c and Fig. S5 a). Binding analysis of CIS43 and engineered antibody variants to CSP by isothermal titration calorimetry (ITC) revealed improved affinities of the top four variants with respect to KD,1 (the high affinity site associated with the junctional epitope) compared to CIS43 (Fig. S5 b). Interestingly, CIS43_Var18, which contained both key mutations that occurred in the CDRs (VL_L30F/L27cF in CDR-L1, and VL_Q61A/Q55A in CDR-L2), showed substantially improved affinity to Pep21, but only marginally improved affinity to NPDP19 (Fig. 5 c), indicating differential improvement in affinity to highly similar peptides (Table S2). Furthermore, we observed CIS43_Var2 (containing VH_V2R and VL_L30F/L27cF) and CIS43_Var10 (containing VH_V2R, VL_L30F/L27cF, and VL_V91S/V85S) to be closely matched to one another in affinity for Pep21 and Pep25. However, the more functionally improved variant, CIS43_Var10, had stronger affinity to Pep29 than CIS43_Var2, suggesting that synergistic contributions from the VL_V91S/V85S substitution were necessary for potent antimalaria function related to recognition of the major NANP repeat.
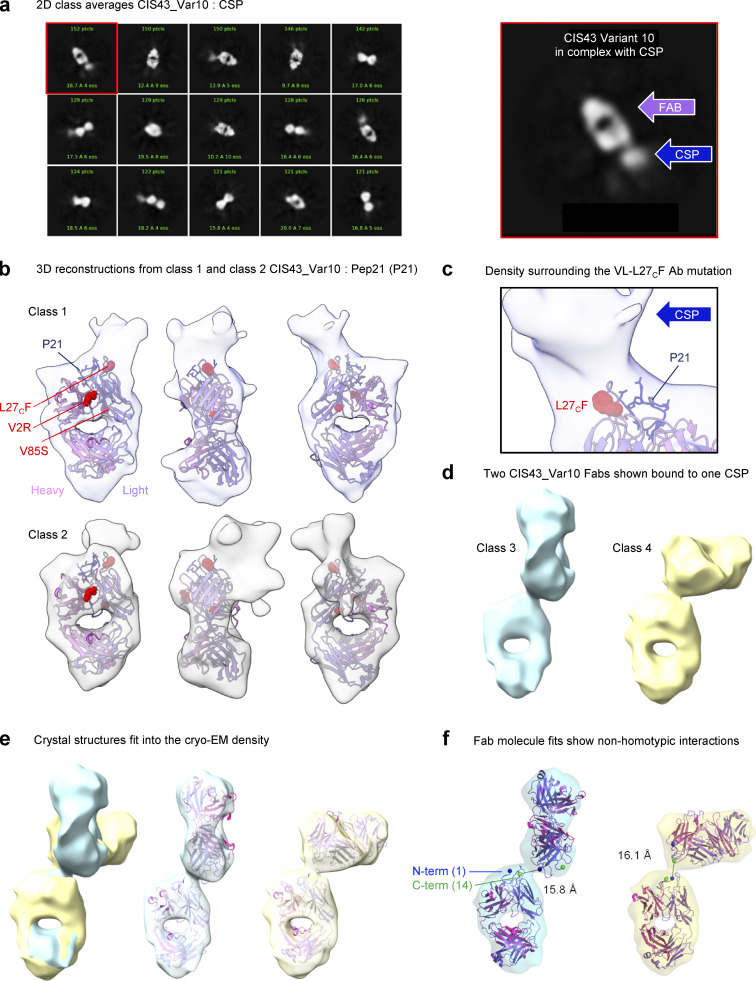
While the crystal structure revealed the VH_V2R mutation to orient the position of the CDR-H3, the importance of the VL_L30F/L27cF was not immediately clear as it faced away from the bound peptide. To further investigate possible interactions, we analyzed CIS43_Var10 Fab in complex with fl_PfCSP by cryo-EM (Fig. 6). Two dimensional (2D)–class averages showed clear Fab signal, with additional signal shown extending from the tip of the Fv region corresponding to bound, but disordered fl_PfCSP (Fig. 6 a). 3D reconstructions for multiple classes were determined at 6.1–10.9 Å resolution. While these cryo-EM reconstructions with fl_PfCSP showed much lower resolution than the Pep21-co-crystal structures, these new data revealed important insights regarding full-length antigen recognition that could not be determined from crystallography data with peptides alone (Fig. 6 b and Table S6). Two of the classes observed by cryo-EM were used for rigid-body fitting with the Pep21-bound crystal structure of CIS43_Var10 and showed the VL_L30F/L27cF facing away from the peptide, but directly towards reconstruction density attributed to fl_PfCSP (Fig. 6, b and c). Multiple classes were also obtained for two Fabs linked by their co-binding of the same fl_PfCSP in close proximity (Fig. 6 d). The two primary classes of linked Fabs displayed unique angles of attachment (Fig. 6 e) and, although the chirality may not be certain at this resolution, all combinations showed similar results. Fitting the crystal structure to the linked classes indicated that the Fabs were too distant to interact with one another, notably the distance between the peptide termini in both cases was ∼16 Å (Fig. 6 f). Overall, these structures reveal that epitope recognition by the highly protective CIS43_Var10 antibody encompasses additional interactions beyond Pep21. These cryo-EM data support malaria intervention strategies that seek to improve affinity to the junctional epitope of CIS43 specifically in the context of full-length CSP, and highlight the junctional epitope presented by fl_PfCSP as an important target for antibody-based sterilizing immune protection.
Figure 6.
Cryo-EM structures of CIS43_Var10 in complex with CSP. (a) 2D class averages are shown for CIS43_Var10 in complex with CSP. The Fab shows additional signal from the CSP protruding from the Fv region. Right: A panel is highlighted showing the two components of the complex. (b) 3D reconstructions from two classes are shown fit with the crystal structures of CIS43_Var10 bound to Pep21. The ∼6–7 Å resolution maps both show density related to the CSP beyond the bound peptide and encompassing the VL-L27CF mutation on the antibody. (c) A closer view of the density surrounding the VL-L27CF mutation. The map indicates that regions of CSP beyond Pep21 extend in the direction of the mutation suggesting additional interactions. (d) Two classes showed two Fabs linked through binding a single CSP, indicating that two Fabs can bind close to one another in the reconstructions of 10–11 Å resolution. (e) Classes 3 and 4 were aligned by class 1; surfaces demonstrate distinct angles between the two fabs in each class. Fits were also assessed for various combinations in chirality with similar results. The crystal structure was fit into the cryo-EM density to assess any potential contacts. (f) Atomic models of the crystal structure fit to the density of the 10–11 Å maps indicate no Fab–Fab contacts, notably the N and C-termini of the CSP peptide in the crystal structure (blue and green spheres, respectively) are located ∼16 Å from each other in both cases.
Discussion
We applied precision antibody engineering efforts to greatly enhance the potency of the CIS43 antimalarial antibody. CIS43_Var10 provided approximately sixfold improved protective potency against malaria challenge compared to CIS43, and these substantial functional improvements were achieved with minimal mutations to the CIS43 template antibody. Our study demonstrates the power of precision library generation and yeast display screening to improve antibody-based protection against malaria infection, and we define several critical molecular features of PfCSP as vaccine and antibody targets.
The complex features of anti-PfCSP antibody responses have made it historically challenging to discover antibodies with optimized molecular interactions sufficient for clinical use. Most protective anti-PfCSP antibodies recognize nonoverlapping repeat peptides with high affinity and low SHM (Julien and Wardemann, 2019; Kisalu et al., 2018; Murugan et al., 2020; Tan et al., 2018; Wang et al., 2020); prior studies have suggested little correlation between SHM and immune protection for PfCSP-specific antibodies (Julien and Wardemann, 2019; Murugan et al., 2019). Structural diversity and sequence degeneracy associated with the long-range, extended spiral conformation of PfCSP’s repeat regions have also been shown to induce homotypic Fab–Fab interactions in potent natural antibodies (Imkeller et al., 2018; Oyen et al., 2020; Oyen et al., 2018; Pholcharee et al., 2020). Considering the molecular challenges for antimalarial immune recognition described by these studies, our antibody improvement campaigns demonstrated that enhanced binding affinity to the junctional epitope and fl_PfCSP together can improve antimalarial protective potency while introducing fewer mutations to the parent antibody sequence than some alternative techniques, even for naturally potent antibodies like CIS43 (Kratochvil et al., 2021). Our screening and mutant characterization efforts defined key PfCSP epitopes as potential antigens for vaccine and antibody targets. In particular, the junctional epitope of PfCSP site is a promising target for additional potency improvements, with additional important contacts on full-length CSP revealed by cryo-EM.
We found that the introduction of precision mutations for functional screening, paired with combinatorial shuffling, can lead to strong potency improvements by eliciting rare combinations of mutations that target critically important sites. Other groups have reported techniques to identify mutant antibodies with improved function (Kratochvil et al., 2021; Wang et al., 2021a), and we show here that SSM and subsequent DNA shuffling can provide with generally similar protective potency to the best antimalarial antibodies currently reported with only three mutations to the template gene.
The most improved variant, CIS43_Var10, comprised just three mutations (VH_V2R, VL_L30F/L27cF, and VL_V91S/V85S) and provided approximately sixfold improved protective potency against malaria challenge compared to the parent mAb CIS43. Notably, while one of these mutations occurred in a CDR (VL_L30F/L27cF, CDR-L1), the crystal structure revealed that none of the three mutations made direct contact with Pep21. Cryo-EM analysis of VL_L30F/L27cF facing towards density attributed to the PfCSP suggests that this mutation interacts beneficially with a region not observed in the peptide crystal structure; the density extended from the Fv region of the Fab and is indicative of fl_PfCSP maintaining some structure in the areas close to or directly interacting with the Fab but becoming structurally disordered as it extends outward. Furthermore, the observation of multiple classes with two Fabs co-binding closely to the same fl_PfCSP strongly suggests that the two arms of an IgG could display bivalent binding that would provide higher avidity.
Two CIS43_Var10 mutations (VH_V2R and VL_V91S/V85S) are both rare mutations that require multiple nucleotide substitutions within a single codon and are infrequently observed in natural antibody sequences (Fig. 4; Guo et al., 2019). Natural antibody development samples a heavily biased sequence space due to SHM targeting preferences, which impedes the natural development of effective antibodies against various diseases (Julien and Wardemann, 2019; Murugan et al., 2020; Richardson and Moore, 2019; Schramm and Douek, 2018). Prior studies showed minimal correlation between SHM and immune protection for PfCSP-specific antibodies, and our findings support a hypothesis that natural antimalarial antibody development may be limited by incomplete sampling of rare mutations in vivo (Schramm and Douek, 2018; Shen et al., 2020; Sheng et al., 2017; Fig. 4). Rare mutations can play an important role in protective antibody development for infections with repeated antigen exposure like malaria and HIV-1 (Julien and Wardemann, 2019; Murugan et al., 2020; Saunders et al., 2019; Shen et al., 2020) where chronic or repeated infection can sometimes enable the development of improbable mutations which achieve remarkable protective activity (Richardson and Moore, 2019; Saunders et al., 2019).
Our data highlight the advantages of in vitro antibody library screens using SSM library generation techniques to easily sample rare mutations throughout the entire variable region. While rare mutations may be beneficial for antigen recognition and improved protection, a rare mutation can potentially enhance antibody immunogenicity. The risk of forming antidrug immune responses increases with each mutation to the natural template protein, especially if a modification creates or modifies a CD4+ T-cell epitope (Cassotta et al., 2019; Jawa et al., 2020; Vaisman-Mentesh et al., 2020). Further immunogenicity studies are needed to explore the immunogenic potential and immune tolerance outcomes for rare amino acid substitutions in human antibody drugs, including the rare substitutions identified in this report.
The comparatively small library size for SSM of the antibody variable regions (∼104) permits facile screening with adequate coverage in yeast display, and any mutations that are deleterious to binding (up to 50% of possible single mutations; Madan et al., 2021) can be eliminated prior to combinatorial DNA shuffling. Thus, detrimental mutations are removed in early rounds of screening while promising mutations are enriched for subsequent recombination. By beginning recombination of mutations using a smaller, high-quality single-mutation pool, the DNA shuffling and combinatorial mutagenesis steps have greater efficiency than alternative techniques via an enhanced probability of identifying synergistic mutations. In contrast, traditional and less precise library generation techniques like error-prone PCR have limited control over the amount of mutational variation in each sequence per round, increasing the likelihood of incorporating deleterious or nonbeneficial mutations into a library (Madan et al., 2021). Error-prone PCR has difficulty sampling important rare mutations in isolation, some of which may even require three adjacent nucleotide substitutions, but which are easily sampled via SSM (Madan et al., 2021).
DNA shuffling of heavy and light chains for beneficial single mutants also provides an ability to generate new combinations of mutations that may not be readily produced during B cell–based antibody discovery methods, due to difficulties sampling rare mutations in vivo. By screening both the variable heavy and light single-mutation landscapes separately by SSM, pools of enriched single mutations can be randomly combined across the paired heavy and light chain variable regions, and within each individual variable region, using DNA shuffling. The process of SSM, screening, and DNA shuffling generates a library of beneficial multi-mutation variants with an enhanced probability of synergistic effects. The improved multi-mutant antibodies produced using this approach contained fewer mutations to the template human mAb compared to alternative methods, which may minimize drug rejection risks (Cassotta et al., 2019; Vaisman-Mentesh et al., 2020). Limiting mutational divergence from human genes is a critical design parameter for antibody drugs (and generally for protein therapeutics), especially for drugs with a long circulation half-life administered to immunocompetent individuals, which is a major use case for highly potent antimalarial antibodies like we describe here. Additional follow-up rounds of precision mutational screening for CIS43_Var10 and other clones could yield further potency benefits, beyond the improvements achieved in this study.
Future antibody engineering efforts may lead to further enhanced potency, both for CIS43 and other antimalarial antibodies such as L9 (Wang et al., 2020), particularly in combination with advanced methods for antibody discovery and screening (Dosenovic et al., 2015; Kratochvil et al., 2021; Tian et al., 2016; Wang et al., 2021b). Improved potency of antimalarial mAbs should enable lower dosage requirements and subcutaneous injections (versus current higher dose intravenous injection), with major associated reductions in production, delivery, storage, and drug administration costs. In addition, the use of Fc mutations to enhance half-life could lead to protection over a period of 6–12 mo using only a single subcutaneous injection (Gaudinski et al., 2021), which could achieve seasonal and perennial control in infants and children. Transient antibody expression technologies, including mRNA-based gene delivery (Kose et al., 2019; Polack et al., 2020) and vectored immunoprophylaxis (Balazs et al., 2014; Zhan et al., 2021), could provide additional delivery strategies, and the improved potency obtained here with CIS43_Var10 may enable clinical antibody protection from multiple delivery methods. We expect that precision antibody engineering strategies will be increasingly used to enhance antibody drug potency against infectious diseases, and that improved molecules and new antibody drug delivery platforms will make broadly accessible antibody-based prophylaxis an impactful strategy to curb malaria transmission in the coming decades.
Materials and methods
SSM library generation and cloning into yeast display
Single-mutation plasmid libraries were constructed using a previously reported one-pot SSM protocol (Wrenbeck et al., 2016). CIS43 VH and VL gene libraries containing a single amino acid substitution were separately generated using mutagenic primers that contained degenerate single codons to express all 20 amino acids at each residue of the VH and VL. A process of single-strand nicking, exonuclease digestion, and degenerate oligonucleotide-primed PCR was used to generate each library (VH and VL) containing the comprehensive set of single-residue substitutions across the CIS43 variable regions (Medina-Cucurella et al., 2019). The separate (VH-SSM):VL and VH:(VL-SSM) libraries were cloned into a yeast display vector containing a FLAG-marker to quantify Fab surface expression, as well as a leucine zipper and protein disulfide isomerase expression for enhanced expression of diverse antibody libraries (Wang et al., 2018). The plasmid DNA libraries were used to transform yeast populations, and library sizes of at least 4 × 105 were maintained in all cloning steps, as described previously (Madan et al., 2021).
FACS of single-mutation libraries
Transformed yeast libraries were cultured in SGDCAA medium to induce Fab surface expression, (20 g/liter galactose, 6.7 g/liter yeast nitrogen base, 5 g/liter casamino acids, 5.4 g/liter Na2HPO4, 8.6 g/liter NaH2PO4.H2O; SGCAA from TEKnova) supplemented with 2 g/lliter dextrose (SGDCAA) for 36 h at 20°C and 225 rpm. Antibody Fabs were fused to a FLAG tag linked at the light chain constant region to quantify protein surface display. Yeast libraries were washed and stained with an anti-FLAG FITC monoclonal to quantify Fab expression (F4049, Clone M2; Sigma-Aldrich). fl_PfCSP and Pep21 (NPDPNANPNVDPNAN) antigen probes were generated by fluorescently labeling the biotinylated constructs with an anti-biotin-PE label (12-9895-82, clone BK-1/39; Thermo Fisher Scientific). To screen single-mutation Fab libraries for antigen-recognition, anti-FLAG-FITC–labeled libraries were co-stained with fluorescently conjugated antigen probes. In the first round of FACS, 3e7 yeast cells were stained and sorted using three gates to sort and collect low, medium, and high-affinity yeast library groups by the ratio of Fab surface expression versus Ag binding for the cell population, as described previously (Fahad et al., 2021; Madan et al., 2021). For all sorting experiments, control libraries of FITC+ yeast were also collected to track the identity and initial prevalence of library variants with successful Fab expression. Collected cells were grown in low-pH SDCAA (20 g/liter dextrose, 6.7 g/liter yeast nitrogen base, 5 g/liter casamino acids, 10.4 g/liter trisodium citrate, and 7.4 g/liter citric acid monohydrate, pH 4.5) for 24–48 h in a 30°C incubator shaking at 225 rpm. After sorted yeast collection and culture, each sort was repeated (using low, medium, or high affinity, to match the initial Round 1 sort) for an additional two rounds.
Multi-mutation library design and screening
Plasmid libraries were isolated from sorted yeast cells using previously described DNA extraction methods (Medina-Cucurella et al., 2019). VH and VL genes were amplified from library plasmids using Kapa Hifi HotStart ReadyMix (Kapa Biosystems; Roche; Wang et al., 2018). Three VH:VL paired combinatorial multi-mutation libraries were designed using template DNA from the enriched single-mutation high-affinity screens. Library 1 was generated by combinatorial pairing of enriched single-mutations VH and VL genes using restriction enzyme digest and subcloning. Library 2 was generated by performing DNA shuffling with enriched single-mutation VH and VL gene libraries, separately (Meyer et al., 2014). Template DNA was fragmented with DNAseI, followed by homologous reassembly and reamplification of the shuffled genetic material_ENREF_7. The separate shuffled-VH and shuffled-VL genes were combined in a single shuffled-VH:VL library via restriction enzyme digest and subcloning. Library 3 was generated with an additional round of SSM performed on the separate shuffled-VH and shuffled-VL genes. The re-SSM-shuffled genes were then combined into a single VH:VL library via restriction enzyme digest and subcloning.
Yeast libraries were next generated using multi-mutation plasmids, as described above for the single-mutation library screens, and library sizes exceeding 1 × 107 were maintained in all cloning steps. Expression was induced and stained as above using an anti-FLAG FITC mAb (F4049, Clone M2; Sigma-Aldrich) and a fluorescently labeled antigen probe. A truncated version of PfCSP (NTDS_5/3_CSP) displaying five of the major repeat peptides (NANP) with three of the minor repeat peptides (NVDP) was also used for screening. NTDS_5/3_CSP was based on the 3D7 clone and contains four N-terminal domain stabilizing (NTDS) mutations that prevent dimer formation by mutating an N-terminal free cysteine in the original sequence from “QEYQC” to “QEYQS” and increase expression by mutating the charged residues (bold) in the sequence: “KKNSRSLGE” to “SSNSASLGE” immediately upstream of the “RSLGE” PEXEL II NTD sequence (underlined; Wang et al., 2020). Biotinylated Pep25 (NVDPNANPNVDPNAN) and Avi-tagged Pep29 (NANPNANPNANPNAN) antigen probes were also generated by fluorescently labeling the constructs with an anti-biotin-PE label (12-9895-82, clone BK-1/39; Thermo Fischer Scientific) and streptavidin-PE (SA-PE; Thermo Fischer Scientific), respectively. Multi-mutation libraries were enriched for high-affinity binding as described above for single-mutation library screening; 3 × 107 yeast cells were stained and the 0.1% highest affinity variants in the library, as determined by the highest ratio of antigen binding to Fab expression (Fahad et al., 2021; Madan et al., 2021; Wang et al., 2018). Control libraries of FITC+ yeast cells were also collected to track the identity and initial prevalence of multi-mutation library variants with successful Fab surface expression. Multi-mutation libraries were enriched for high-affinity antigen binders for an additional three rounds using FACS. Collected cells were cultured in low-pH SDCAA (20 g/liter dextrose, 6.7 g/liter yeast nitrogen base, 5 g/liter casamino acids, 10.4 g/liter trisodium citrate, and 7.4 g/liter citric acid monohydrate, pH 4.5) for 24–48 h at 30°C and 225 rpm.
NGS and bioinformatic analysis of sorted antibody libraries
VH and VL genes were extracted and amplified from sorted yeast libraries and submitted for NGS on a 2 × 300 bp Illumina MiSeq platform. First, plasmids were isolated from cultured yeast cells as described previously (Medina-Cucurella et al., 2019). VH and VL genes were amplified from library plasmids using Kapa Hifi HotStart ReadyMix (Kapa Biosystems; Roche) as described previously (DeKosky et al., 2016; Madan et al., 2021; Wang et al., 2018). Amplified libraries were prepared for NGS with an addition round of PCR to incorporate barcodes and adapters for Illumina sequencing.
Raw Illumina fastq sequence reads were processed as described previously (DeKosky et al., 2016; Lee et al., 2016; Madan et al., 2021). NGS reads were quality-filtered for a score of 30 over 90% of the raw reads using Fastxtoolkit (v0.0.14; http://hannonlab.cshl.edu/fastx_toolkit/). Filtered reads were then processed using IgBlast software to reference the IMGT database and determine complete variable region gene alignments (Brochet et al., 2008; Ye et al., 2013). Once variable region alignments were obtained, mutant CIS43 sequences were aligned to the template CIS43 antibody sequence using Usearch software. Mutations were defined by determining the percent identity match to the template CIS43 gene and denoting the substituted amino acid residue according to the template CIS43 gene sequence, with Base 1 corresponding to the start of the variable framework region. Kabat numbering identifiers are also listed for key mutations and referred to in structural data (Figs. 4 and 5 and Table S3). The number of reads for unique sequences was enumerated in each library, and these data were used to determine the prevalence, or frequency, of each variant in the sorted libraries:
We defined a variant’s ER as the change in sequence prevalence from the initial Fab expressing (VL-FITC+) sorted control library to the experimentally screened library:
Each single-mutation variant was binned into high, medium, and low affinity screened groups by comparing prevalence and ER values across different screening conditions, as we reported previously (Fahad et al., 2021; Madan et al., 2021). Multi-mutation variants were analyzed by their ER in high-affinity sorted library screens, using the multi-mutation VL-FITC+ prevalence in the denominator.
Antibody expression
Antibody variable heavy chain and light chain sequences were codon optimized, synthesized, and cloned into a VRC8400 (CMV/R expression vector)-based IgG1 vector as previously described (Kong et al., 2019). The variants were expressed by transient transfection in Expi293 cells (Thermo Fisher Scientific) using Turbo293 transfection reagent (SPEED BioSystems) according to the manufacturer’s recommendation. About 50-μg plasmid encoding heavy-chain and 50-μg plasmid encoding light-chain variant genes were mixed with the transfection reagents, added to 100 ml of cells at 2.5 × 106/ml, and incubated in a shaker incubator at 120 rpm, 37°C, 9% CO2. At 5 d after transfection, cell culture supernatant was harvested and purified with a Protein A (GE Healthcare) column. The antibody was eluted using IgG Elution Buffer (Thermo Fisher Scientific) and were brought to neutral pH with 1 M Tris-HCl, pH 8.0. Eluted antibodies were dialyzed against PBS overnight and were confirmed by SDS-PAGE before use.
ELISA characterization of CIS43 variants
For fl_PfCSP, ELISA proceeded using Immulon 4HBX flat bottom microtiter plates (Thermo Scientific Nunc) coated with 100 μl of 1 μg/ml of PfCSP in bicarbonate buffer overnight at 4°C. Coated plates were blocked with 200 μg of PBS + 10% FBS for 2 h at room temperature (RT), followed by incubation for 2 h at 37°C with 100 μl of single-mutant IgG or control mAbs at varying concentrations (0.000002–20.0 μg/ml, 10-fold serial dilutions). Plates were washed six times with PBS-Tween-20 between each step. To detect antigen bound antibody, plates were incubated with 100 μl/well of 0.1 μg/ml HRP-labeled goat antihuman IgG (Bethyl Laboratories Inc). After a final wash, samples were incubated for about 15 min with 1-Step Ultra TMB-ELISA Substrate (Thermo Fisher Scientific Inc.) before adding 100 μl/well of stopping solution (2N sulfuric acid, 100 ml/well) and reading the optical density at 450 nm. Similarly, for Peptide ELISA, PierceTM streptavidin-coated high-capacity plates (Thermo Fisher Scientific Inc.) were coated with 100 μl per well of 0.05 μg/ml biotinylated 15-mer peptides (GenScript Biotech Corporation) diluted in wash buffer (25 mM Tris, 150 mM NaCl, pH 7.2 Tris-buffered saline, 0.1% BSA, and 0.05% Tween-20). Coated plates were incubated for 2 h at 37 C with 100 μl of single-mutant IgG or control mAbs at varying concentrations (0.000002–20.0 μg/ml, 10-fold serial dilutions); and detection of antigen bound antibody was performed using HRP-conjugated goat antihuman IgG as detailed above.
AlphaLISA characterization of CIS43 variants
AlphaLISA (PerkinElmer) is a bead-based proximity assay in which singlet oxygen molecules, generated by high-energy irradiation of Donor beads, transfer to Acceptor beads, which are within a distance of ∼200 nm. It is a sensitive high-throughput screening assay that does not require washing steps. A cascading series of chemical reactions results in a chemiluminescent signal. Purified antibodies were diluted to 100 nM in AlphaLISA buffer (PBS + 0.05% Tween-20 + 0.5 mg/ml BSA). Subsequently, 5 μl of the IgGs were transferred to an OptiPlate-384 assay plate (white opaque; PerkinElmer), mixed with 10 μl (10 nM final conc.) of biotinylated peptide probe and 10 μl (10 μg/ml final conc.) of anti-human IgG (Fc specific; PerkinElmer) acceptor beads. After an hour of incubation at RT, nonshaking, 25 μl (40 μg/ml final conc.) of streptavidin donor beads (PerkinElmer) were added. The plate was then incubated for 30 min at RT in the dark before the AlphaLISA signal was detected using a SpectraMax i3x multi-mode microplate reader (Molecular Devices).
Evaluation of protective antibody performance in mouse models of malaria challenge
Female 6- to 8-wk-old B6(Cg)-Tyrc-2J/J albino and Balb/c mice were obtained from The Jackson Laboratory. All animals were maintained and cared for in accordance with the American Association for Accreditation of Laboratory Animal Care Standards. All mouse procedures were performed according to protocols approved by the Institutional Animal Care and Use Ethics Committees of the National Institute of Allergy and Infectious Diseases (Animal Study Protocol VRC-20-0855).
To generate sporozoites, transgenic Plasmodium berghei (strain ANKA 676m1C11, MRA-868) expressing fl_PfCSP and a GFP/luciferase fusion protein (Pb-PfCSP-GFP/Luc-SPZ or Pb-PfCSP-SPZ) were obtained from salivary glands of infected mosquitoes, as previously described (Flores-Garcia et al., 2019). Briefly, Anopheles stephensi (Nijmegan) mosquitoes were obtained and reared from a colony maintained at the Laboratory of Malaria and Vector Research (National Institute of Allergy and Infectious Diseases, National Institutes of Health). Female mosquitoes were allowed to feed on 6- to 8-wk-old female Balb/c mice infected with blood-stage Pb-PfCSP- GFP/LUC parasites. After infection, mosquitoes were maintained in an incubator at 19–20°C and supplied with a sterile cotton pad soaked in 10% sucrose, changed every 48 h. 18 to 21 d following mosquito infections, salivary glands were dissected and ground in 400 μl of L-15 medium (Millipore-Sigma), and viable sporozoites (SPZs) were counted in a Neubauer chamber.
To assess the protective efficacy of anti-PfCSP mAbs in vivo, varying concentrations ranging 50–300 μg of PfCSP mAbs were diluted in sterile filtered 1× PBS (pH 7.4; total volume 200 μl/mouse) and administered into the tail veins of female 6- to 8-wk-old B6(Cg)-Tyrc-2J/J albino mice (n = 5 or n = 10 mice per group, The Jackson Laboratory). After 2 h, mice were intravenously challenged in the tail vein with 2,000 freshly harvested Pb-PfCSPGFP/Luc-SPZ in Leibovitz’s L-15 medium (Thermo Fisher Scientific).
To assess malaria infection, mice were injected i.p. with 150 μl D-luciferin (30 mg/ml; PerkinElmer), anesthetized with isoflurane and imaged with an IVIS Spectrum in vivo imaging system (PerkinElmer) 10 min after luciferin injection. Parasite liver load was assessed 40–42 h after challenge, whereas parasitemia was measured 6 d following challenge. Parasite load was quantified by analyzing a region of interest in the upper abdominal region for liver stage, or whole animal for parasitemia; bioluminescence or the total flux (photons/second; p/s) was measured using the manufacturer’s software (Living Image 4.5; PerkinElmer).
CIS43 variants were compared to an equivalent dose of CIS43 or L9 as benchmark controls, using the two-tailed Mann-Whitney test. Statistical differences were estimated on the parasite load (liver burden or parasitemia) raw bioluminescence measurements (Fig. S3) or on the log10(normalized liver burden or parasitemia) values. To enable comparison of parasite infection between different studies, the liver burden (or parasitemia) of each group was normalized based on the geometric mean of the liver burden/parasitemia values from the untreated mice in the same experiment. Data were plotted and graphed using GraphPad Prism, unless otherwise stated. P values <0.05 were considered significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) as indicated in the figures.
Affinity measurements by BLI
Antibody Fab binding affinity to various ligands was measured using biolayer interferometry on an Octet Red384 instrument (fortéBio) with streptavidin capture biosensors (fortéBio) in solid black tilt-well 96-well plates (Greiner Bio-One). Assays were performed with agitation at 30°C. Immobilization of biotinylated fl_PfCSP, NTDS_5/3_CSP, NPDP19, Pep21, Pep25, and Pep29 was performed for 60 s, followed by a 60-s baseline in buffer (PBS + 1% BSA). Association with Fab (serially diluted from 1,000 to 62.5 nM) was done for 60 s, followed by a dissociation step in buffer for 180 s. In all Octet measurements, parallel correction to subtract systematic baseline drift was carried out by subtracting the measurements recorded for a loaded sensor incubated in PBS. Data analysis was carried out using Octet software, version 9.0. Experimental data were fitted globally with a 1:1 Langmuir model of binding for all the antigens except fl_PfCSP which was fitted with a 2:1 Langmuir model of binding.
ITC
For ITC experiments, an N-terminal domain stabilized version of fl_PfCSP (based on 3D7 clone of the NF54 strain (PlasmoDB ID: PF3D7_0304600.1) with increased expression was used. Four amino acid mutations (as indicated in Table S2) were introduced in the N-terminal domain of recombinant fl_PfCSP (Kisalu et al., 2018) to generate NTDS_fl_CSP (PfCSP_SAmut_C5S; Wang et al., 2020). These changes remove an enzymatic processing site and help prevent dimerization upon solubilization through the conversion of a cysteine to a serine resulting in increased yields and consistent analyses.
ITC was carried out using a MicroCal VP-ITC microcalorimeter from Malvern Panalytical. Prior to the experiments, both PfCSP and different antibodies were exhaustively dialyzed against PBS, pH 7.4. The antibody solution, prepared at a concentration of ∼25 µM (expressed per antigen binding site), was injected in 7-µl aliquots into the calorimetric cell containing PfCSP at a concentration of ∼0.3 µM. All titrations were performed at 25°C. The exact concentrations of the reactants in each experiment were determined from the protein absorbance at 280 nm. The heat evolved upon each injection of antibody was obtained by integration of the calorimetric signal, dQ/dt. The heat associated with binding to PfCSP was obtained by subtracting the heat of dilution from the heat of reaction. The separate heats were plotted against the molar ratio, and values for the enthalpy change, ΔH, the association constant, Ka (the dissociation constant, Kd = 1/Ka), and the stoichiometry, N, were obtained by nonlinear regression of the data to a binding polynomial accounting for the binding to two sets of sites with different binding affinities (Freire et al., 2009). Gibbs energy, ΔG, was calculated from the relation ΔG = −RTlnKa, where R is the universal gas constant (1.987 cal/(K × mol)) and T is the absolute temperature in kelvin. The entropy contribution to Gibbs energy, −TΔS, was calculated from the known relation ΔG = ΔH − TΔS. The results were expressed per mole of antigen binding sites and the stoichiometry, N, denotes the number of antigen binding sites per mole of PfCSP.
Crystallization and structural analysis
Antibody Fab and Pep21 (PfCSP residues 101–115) complexes were prepared by mixing 1:2 M ratio to a concentration of 15 mg/ml. Crystallization conditions were screened in Hampton Research screening kits, Wizard screening kits, Precipitant Synergy screening kits using a mosquito robot. Crystals initially observed from the wells were manually reproduced. The CIS43_Var2: P21 complex crystal grew in 0.2 M ammonium sulfate and 28% wt/vol polyethylene glycol 4,000; the CIS43_Var10: P21 complex crystal grew in 15% isopropanol, 0.2 M ammonium citrate pH 3.5 and 25% wt/vol polyethylene glycol 3,350; crystals were cryoprotected in 20% glycerol and flash-frozen in liquid nitrogen. Data were collected at a temperature of 100 K and a wavelength of 1.00 Å at the SER-CAT beamline ID-22 (Advanced Photon Source; Argonne National Laboratory). Diffraction data were processed with the HKL2000 suite_ENREF_13. Structure solution was obtained by molecular replacement with Phaser using CIS43 Fab structures (PDB ID: 6B5M) as a search model. Model building was carried out with Coot (Emsley and Cowtan, 2004). Refinement was carried out with Phenix (Liebschner et al., 2019). Ramachandran statistical analysis indicated that the final structures contained no disallowed residues or no more than 0.22% disallowed residues. Data collection and refinement statistics are shown in Table S5.
Cryo-EM structures of CIS43_Var10 in complex with fl_PfCSP
CIS43_Var10 was mixed at a 1:1 M stoichiometry with fl_PfCSP and the complex was deposited on a C-flat grid (Protochips). The grid was vitrified in ethane with an FEI Vitrobot Mark IV set to a wait time of 30 s, blot time of 3 s, and blot force of 1. Automated data collection on a Titan Krios electron microscope was performed with Leginon (Suloway et al., 2005) with a Gatan K3 direct detection device. Exposures were collected in movie mode for a 2 s with the total dose of 51.15 e–/Å2 fractionated over 50 raw frames. Images were preprocessed using Appion (Lander et al., 2009; Voss et al., 2009) during collection; individual frames were aligned and dose-weighted using MotionCor2 (Zheng et al., 2017) and CTFFind4 (Rohou and Grigorieff, 2015; Zhang, 2016) was used to estimate the CTF. CryoSPARC 2.15 (Punjani et al., 2017) was used for 2D classifications, ab initio 3D reconstruction, homogeneous refinement, and nonuniform 3D refinement. Initial 3D reconstruction and final refinements were performed using C1 symmetry. Atomic coordinates were rigid-body fit in Chimera and distances were assessed in Pymol. Overlapping analysis for the linked-Fab classes was assessed with the maps and their Z-flipped counterparts to confirm no combination showed a similar angle. We note that there were several minor classes with additional Fab–Fab angles. Similar methods to collect an 8:1 stoichiometry of the Fab:fl_PfCSP complex resulted in primarily empty grid holes on two grids under similar conditions. The few particles that were in the holes appeared as single Fabs or aggregates, from which we were not able to obtain sufficient structural definition. Data collection and refinement statistics are shown in Table S6.
Online supplemental material
Fig. S1 shows sequence features of antibody recognition. Fig. S2 shows the multi-mutation library screening overview. Fig. S3 provides data indicating significant multi-mutation sequence-function relationships. Fig. S4 shows experimental animal groups for analysis of multi-mutation variant protection against malaria challenge. Fig. S5 shows data characterizing multi-mutation variants by AlphaLISA, BLI, and ITC. Table S1 provides experimental data collected from library synthesis. Table S2 lists amino acid sequences of antigens used to engineer enhanced antibody protection. Table S3 lists antibody variants expressed as soluble IgG. Table S4 shows multi-mutation library mutant distribution data corresponding to Fig. S3 a. Table S5 shows x-ray crystallography data collection and refinement statistics for Fig. 5. Table S6 shows cryo-EM data collection and reconstruction statistics for Fig. 6.
Supplementary Material
provides cloning and transformation results and calculation of theoretical library coverage for CIS43 gene variant libraries.
lists sequences of PfCSP-dervied antigen probes used to engineer and assess enhanced antibody protection.
lists CIS43 multi-mutation variants mined from the data in Fig. 2 and expressed as soluble human IgG1 antibodies in HEK293 cells for detailed functional evaluation.
shows multi-mutation library mutant distribution data corresponding to Fig. S3 a.
lists x-ray crystallography data collection and refinement statistics for Fig. 5.
lists cryo-EM data collection and reconstruction statistics for Fig. 6.
Acknowledgments
We thank Jennifer Hackett for help with sequencing and Lawrence Wang for providing PfCSP antigen.
This work was supported by a University of Kansas Self Graduate Fellowship (B.B. Banach), the Departments of Chemical Engineering and Pharmaceutical Chemistry, the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy at the New York Structural Biology Center, the Simons Foundation (SF349247), the New York State Foundation for Science, Technology and Innovation, the National Institutes of Health National Institute of General Medical Sciences (GM103310), the National Cancer Institute (HHSN261200800001E to A. Schön), National Institutes of Health grants DP5OD023118, P20GM103418, R01AI141452, and R21AI143407, and the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Author contributions: Designed the experiments: B.B. Banach, B. Bonilla, P. Tripathi, T.D. Nguyen, M. Pancera, A.H. Idris, R.A. Seder, P.D. Kwong, and B.J. DeKosky. Performed the experiments: B.B. Banach, P. Tripathi, L. Da Silva Pereira, J. Gorman, M. Dillon, P.K. Kiyuka, T.D. Nguyen, B. Bonilla, B. Flynn, J.R. Francica, N.K. Hurlburt, N.K. Kisalu, T. Liu, L. Ou, R. Rawi, A. Schön, C.-H. Shen, I.-T. Teng, and B. Zhang. Analysed the data: B.B. Banach, P. Tripathi, J. Gorman, A.S. Fahad, T.D. Nguyen, J.R. Francica, R. Rawi, A. Schön, C.-H. Shen, A.H. Idris, R.A. Seder, P.D. Kwong, and B.J. DeKosky. Wrote the manuscript: B.B. Banach, P.D. Kwong, and B.J. DeKosky. Reviewing & editing: all authors.
Data availability
Structural data have been deposited in the Protein Data Bank under accession codes 7SG5 and 7SG6.
References
- Adams, R.M., Mora T., Walczak A.M., and Kinney J.B.. 2016. Measuring the sequence-affinity landscape of antibodies with massively parallel titration curves. Elife. 5:e23156. 10.7554/eLife.23156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs, A.B., Ouyang Y., Hong C.M., Chen J., Nguyen S.M., Rao D.S., An D.S., and Baltimore D.. 2014. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med. 20:296–300. 10.1038/nm.3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet, X., Lefranc M.P., and Giudicelli V.. 2008. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36:W503–W508. 10.1093/nar/gkn316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassotta, A., Mikol V., Bertrand T., Pouzieux S., Le Parc J., Ferrari P., Dumas J., Auer M., Deisenhammer F., Gastaldi M., et al. 2019. A single T cell epitope drives the neutralizing anti-drug antibody response to natalizumab in multiple sclerosis patients. Nat. Med. 25:1402–1407. 10.1038/s41591-019-0568-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., and Procko E.. 2020. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 369:1261–1265. 10.1126/science.abc0870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky, B.J., Lungu O.I., Park D., Johnson E.L., Charab W., Chrysostomou C., Kuroda D., Ellington A.D., Ippolito G.C., Gray J.J., and Georgiou G.. 2016. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. USA. 113:E2636–E2645. 10.1073/pnas.1525510113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic, P., von Boehmer L., Escolano A., Jardine J., Freund N.T., Gitlin A.D., McGuire A.T., Kulp D.W., Oliveira T., Scharf L., et al. 2015. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell. 161:1505–1515. 10.1016/j.cell.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., and Cowtan K.. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fahad, A.S., Timm M.R., Madan B., Burgomaster K.E., Dowd K.A., Normandin E., Gutierrez-Gonzalez M.F., Pennington J.M., De Souza M.O., Henry A.R., et al. 2021. Functional profiling of antibody immune repertoires in convalescent zika virus disease patients. Front. Immunol. 12:615102. 10.3389/fimmu.2021.615102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Garcia, Y., Herrera S.M., Jhun H., Perez-Ramos D.W., King C.R., Locke E., Raghunandan R., and Zavala F.. 2019. Optimization of an in vivo model to study immunity to Plasmodium falciparum pre-erythrocytic stages. Malar. J. 18:426. 10.1186/s12936-019-3055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire, E., Schon A., and Velazquez-Campoy A.. 2009. Isothermal titration calorimetry: General formalism using binding polynomials. Methods Enzymol. 455:127–155. 10.1016/S0076-6879(08)04205-5 [DOI] [PubMed] [Google Scholar]
- Gaudinski, M.R., Berkowitz N.M., Idris A.H., Coates E.E., Holman L.A., Mendoza F., Gordon I.J., Plummer S.H., Trofymenko O., Hu Z., et al. 2021. A monoclonal antibody for malaria prevention. N. Engl. J. Med. 385:803–814. 10.1056/NEJMoa2034031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Chen K., Kwong P.D., Shapiro L., and Sheng Z.. 2019. cAb-rep: A database of curated antibody repertoires for exploring antibody diversity and predicting antibody prevalence. Front. Immunol. 10:2365. 10.3389/fimmu.2019.02365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkeller, K., Scally S.W., Bosch A., Martí G.P., Costa G., Triller G., Murugan R., Renna V., Jumaa H., Kremsner P.G., et al. 2018. Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope. Science. 360:1358–1362. 10.1126/science.aar5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawa, V., Terry F., Gokemeijer J., Mitra-Kaushik S., Roberts B.J., Tourdot S., and De Groot A.S.. 2020. T-cell dependent immunogenicity of protein therapeutics pre-clinical assessment and mitigation–updated consensus and review 2020. Front. Immunol. 11:1301. 10.3389/fimmu.2020.01301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien, J.-P., and Wardemann H.. 2019. Antibodies against Plasmodium falciparum malaria at the molecular level. Nat. Rev. Immunol. 19:761–775. 10.1038/s41577-019-0209-5 [DOI] [PubMed] [Google Scholar]
- Kabat, E.A., Te Wu T., Perry H.M., Foeller C., and Gottesman K.S.. 1992. Sequences of Proteins of Immunological Interest. Diane Publishing Company. [Google Scholar]
- Kisalu, N.K., Idris A.H., Weidle C., Flores-Garcia Y., Flynn B.J., Sack B.K., Murphy S., Schön A., Freire E., Francica J.R., et al. 2018. A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite. Nat. Med. 24:408–416. 10.1038/nm.4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, R., Duan H., Sheng Z., Xu K., Acharya P., Chen X., Cheng C., Dingens A.S., Gorman J., Sastry M., et al. 2019. Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell. 178:567–584.e19. 10.1016/j.cell.2019.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose, N., Fox J.M., Sapparapu G., Bombardi R., Tennekoon R.N., de Silva A.D., Elbashir S.M., Theisen M.A., Humphris-Narayanan E., Ciaramella G., et al. 2019. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 4:eaaw6647. 10.1126/sciimmunol.aaw6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochvil, S., Shen C.H., Lin Y.C., Xu K., Nair U., Da Silva Pereira L., Tripathi P., Arnold J., Chuang G.Y., Melzi E., et al. 2021. Vaccination in a humanized mouse model elicits highly protective PfCSP-targeting anti-malarial antibodies. Immunity. 54:2859–2876.e7. 10.1016/j.immuni.2021.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, G.C., Stagg S.M., Voss N.R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P.W., et al. 2009. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166:95–102. 10.1016/j.jsb.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Boutz D.R., Chromikova V., Joyce M.G., Vollmers C., Leung K., Horton A.P., DeKosky B.J., Lee C.H., Lavinder J.J., et al. 2016. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med. 22:1456–1464. 10.1038/nm.4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner, D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in phenix. Acta Crystallogr. D Struct. Bio. 75:861–877. 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan, B., Zhang B., Xu K., Chao C.W., O’Dell S., Wolfe J.R., Chuang G.-Y., Fahad A.S., Geng H., Kong R., et al. 2021. Mutational fitness landscapes reveal genetic and structural improvement pathways for a vaccine-elicited HIV-1 broadly neutralizing antibody. Proc. Natl. Acad. Sci. USA. 118:e2011653118. 10.1073/pnas.2011653118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Cucurella, A.V., Zhu Y., Bowen S.J., Bergeron L.M., and Whitehead T.A.. 2018. Pro region engineering of nerve growth factor by deep mutational scanning enables a yeast platform for conformational epitope mapping of anti-NGF monoclonal antibodies. Biotechnol. Bioeng. 115:1925–1937. 10.1002/bit.26706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Cucurella, A.V., Steiner P.J., Faber M.S., Beltran J., Borelli A.N., Kirby M.B., Cutler S.R., and Whitehead T.A.. 2019. User-defined single pot mutagenesis using unamplified oligo pools. Protein Eng. Des. Sel. 32:41–45. 10.1093/protein/gzz013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, A.J., Ellefson J.W., and Ellington A.D.. 2014. Library generation by gene shuffling. Curr. Protoc. Mol. Biol. 105:Unit 15.12. 10.1002/0471142727.mb1512s105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan, R., Scally S.W., Costa G., Mustafa G., Thai E., Decker T., Bosch A., Prieto K., Levashina E.A., Julien J.-P., and Wardemann H.. 2019. Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs. bioRxiv:798769. [DOI] [PubMed] [Google Scholar]
- Murugan, R., Scally S.W., Costa G., Mustafa G., Thai E., Decker T., Bosch A., Prieto K., Levashina E.A., Julien J.P., and Wardemann H.. 2020. Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs. Nat. Med. 26:1135–1145. 10.1038/s41591-020-0881-9 [DOI] [PubMed] [Google Scholar]
- Oyen, D., Torres J.L., Cottrell C.A., Richter King C., Wilson I.A., and Ward A.B.. 2018. Cryo-EM structure of P. falciparum circumsporozoite protein with a vaccine-elicited antibody is stabilized by somatically mutated inter-Fab contacts. Sci. Adv. 4:eaau8529. 10.1126/sciadv.aau8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyen, D., Torres J.L., Aoto P.C., Flores-Garcia Y., Binter Š., Pholcharee T., Carroll S., Reponen S., Wash R., Liang Q., et al. 2020. Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein. PLoS Pathog. 16:e1008373. 10.1371/journal.ppat.1008373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pholcharee, T., Oyen D., Torres J.L., Flores-Garcia Y., Martin G.M., González-Páez G.E., Emerling D., Volkmuth W., Locke E., King C.R., et al. 2020. Diverse antibody responses to conserved structural motifs in plasmodium falciparum circumsporozoite protein. J. Mol. Biol. 432:1048–1063. 10.1016/j.jmb.2019.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack, F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. 2020. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 383:2603–2615. 10.1056/nejmoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani, A., Rubinstein J.L., Fleet D.J., and Brubaker M.A.. 2017. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 14:290–296. 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- Richardson, S.I., and Moore P.L.. 2019. The antibody response in HIV-1-infected donors. Curr. Opinion HIV AIDS. 14:233–239. 10.1097/COH.0000000000000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou, A., and Grigorieff N.. 2015. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192:216–221. 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, K.O., Wiehe K., Tian M., Acharya P., Bradley T., Alam S.M., Go E.P., Scearce R., Sutherland L., Henderson R., et al. 2019. Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science. 366:eaay7199. 10.1126/science.aay7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm, C.A., and Douek D.C.. 2018. Beyond hot spots: Biases in antibody somatic hypermutation and implications for vaccine design. Front. Immunol. 9:1876. 10.3389/fimmu.2018.01876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C.-H., DeKosky B.J., Guo Y., Xu K., Gu Y., Kilam D., Ko S.H., Kong R., Liu K., Louder M.K., et al. 2020. VRC34-Antibody lineage development reveals how a required rare mutation shapes the maturation of a broad HIV-neutralizing lineage. Cell Host Microbe. 27:531–543.e6. 10.1016/j.chom.2020.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, Z., Schramm C.A., Kong R., NISC Comparative Sequencing Program, Mullikin J.C., Mascola J.R., Kwong P.D., Shapiro L., Benjamin B., Bouffard G., Brooks S., et al. 2017. Gene-specific substitution profiles describe the types and frequencies of amino acid changes during antibody somatic hypermutation. Front. Immunol. 8:537. 10.3389/fimmu.2017.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway, C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., and Carragher B.. 2005. Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151:41–60. 10.1016/j.jsb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Tan, J., Sack B.K., Oyen D., Zenklusen I., Piccoli L., Barbieri S., Foglierini M., Fregni C.S., Marcandalli J., Jongo S., et al. 2018. A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein. Nat. Med. 24:401–407. 10.1038/nm.4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M., Cheng C., Chen X., Duan H., Cheng H.-L., Dao M., Sheng Z., Kimble M., Wang L., Lin S., et al. 2016. Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell. 166:1471–1484.e18. 10.1016/j.cell.2016.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman-Mentesh, A., Gutierrez-Gonzalez M., DeKosky B.J., and Wine Y.. 2020. The molecular mechanisms that underlie the immune biology of anti-drug antibody formation following treatment with monoclonal antibodies. Front. Immunol. 11:1951. 10.3389/fimmu.2020.01951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deventer, J.A., and Wittrup K.D.. 2014. Yeast surface display for antibody isolation: Library construction, library screening, and affinity maturation. In Monoclonal Antibodies. Ossipow V., and Fischer N., editors. Humana Press, Totowa, NJ. 151–181. [DOI] [PubMed] [Google Scholar]
- Voss, N.R., Yoshioka C.K., Radermacher M., Potter C.S., and Carragher B.. 2009. DoG picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166:205–213. 10.1016/j.jsb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., DeKosky B.J., Timm M.R., Lee J., Normandin E., Misasi J., Kong R., McDaniel J.R., Delidakis G., Leigh K.E., et al. 2018. Functional interrogation and mining of natively paired human VH:VL antibody repertoires. Nat. Biotechnol. 36:152–155. 10.1038/nbt.4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.T., Pereira L.S., Flores-Garcia Y., O’Connor J., Flynn B.J., Schön A., Hurlburt N.K., Dillon M., Yang A.S.P., Fabra-García A., et al. 2020. A potent anti-malarial human monoclonal antibody targets circumsporozoite protein minor repeats and neutralizes sporozoites in the liver. Immunity. 53:733–744.e8. 10.1016/j.immuni.2020.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.T., Pereira L.S., Kiyuka P.K., Schon A., Kisalu N.K., Vistein R., Dillon M., Bonilla B.G., Molina-Cruz A., Barillas-Mury C., et al. 2021a. Protective effects of combining monoclonal antibodies and vaccines against the Plasmodium falciparum circumsporozoite protein. PLoS Pathog. 17:e1010133. 10.1371/journal.ppat.1010133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Ray R., Kratochvil S., Melzi E., Lin Y.C., Giguere S., Xu L., Warner J., Cheon D., Liguori A., et al. 2021b. Multiplexed CRISPR/CAS9-mediated engineering of pre-clinical mouse models bearing native human B cell receptors. EMBO J. 40:e105926. 10.15252/embj.2020105926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2018. World Malaria Report 2018. World Health Organization, Geneva. [Google Scholar]
- Wrenbeck, E.E., Klesmith J.R., Stapleton J.A., Adeniran A., Tyo K.E., and Whitehead T.A.. 2016. Plasmid-based one-pot saturation mutagenesis. Nat. Methods. 13:928–930. 10.1038/nmeth.4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J., Ma N., Madden T.L., and Ostell J.M.. 2013. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41:W34–W40. 10.1093/nar/gkt382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, W., Muhuri M., Tai P.W.L., and Gao G.. 2021. Vectored immunotherapeutics for infectious diseases: Can rAAVs be the game changers for fighting transmissible pathogens? Front. Immunol. 12:673699. 10.3389/fimmu.2021.673699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. 2016. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193:1–12. 10.1016/j.jsb.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., and Agard D.A.. 2017. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 14:331–332. 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides cloning and transformation results and calculation of theoretical library coverage for CIS43 gene variant libraries.
lists sequences of PfCSP-dervied antigen probes used to engineer and assess enhanced antibody protection.
lists CIS43 multi-mutation variants mined from the data in Fig. 2 and expressed as soluble human IgG1 antibodies in HEK293 cells for detailed functional evaluation.
shows multi-mutation library mutant distribution data corresponding to Fig. S3 a.
lists x-ray crystallography data collection and refinement statistics for Fig. 5.
lists cryo-EM data collection and reconstruction statistics for Fig. 6.
Data Availability Statement
Structural data have been deposited in the Protein Data Bank under accession codes 7SG5 and 7SG6.