Abstract
Ninety-seven epidemiologically unrelated strains of Listeria monocytogenes were investigated for their sensitivities to quaternary ammonium compounds (benzalkonium chloride and cetrimide). The MICs for seven serogroup 1/2 strains were high. Three came from the environment and four came from food; none were isolated from human or animal samples. All 97 strains carried the mdrL gene, which encodes a multidrug efflux pump, and the orfA gene, a putative transcriptional repressor of mdrL. The absence of plasmids in four of the seven resistant strains and the conservation of resistance after plasmid curing suggested that the resistance genes are not plasmid borne. Moreover, PCR amplification and Southern blot hybridization experiments failed to find genes phylogenetically related to the qacA and smr genes, encoding multidrug efflux systems previously described for the genus Staphylococcus. The high association between nontypeability by phages and the loss of sensitivity to quaternary ammonium compounds are suggestive of an intrinsic resistance due to modifications in the cell wall.
Listeria monocytogenes is the agent of human listeriosis, which is characterized by a variety of severe syndromes, including meningitis, meningoencephalitis, and sepsis, which mostly affects old and immunosuppressed individuals and pregnant women (20). This bacterium is frequently present in soil and surface water samples, and it has been found in a wide range of dairy products, meats, and seafood (3, 18). It is generally believed that the consumption of contaminated food is the principal route of infection, especially since the increase in industrial food production (21).
Despite the application of rigorous procedures of cleaning and disinfection of the processing environment in the food industry, processed food has been contaminated by L. monocytogenes even when the raw ingredients were free of the pathogen (4, 23). L. monocytogenes can attach to various kinds of surfaces, and it has been found in biofilms in meat and dairy processing environments (9). Various types of dairy and other food plant sanitizers are widely used. Quaternary ammonium compounds (QACs) are employed both as disinfectants for manual processing lines and surfaces in the food industry and as antiseptics in human medicine. It is possible that some strains of L. monocytogenes may have acquired resistance to these disinfectants.
No mechanism of resistance to QACs has been described for L. monocytogenes, but one such mechanism is well known for the genus Staphylococcus. It is a multidrug efflux system encoded by the qacA and smr genes, found on both conjugative and nonconjugative plasmids (17). Moreover, the recent identification of a new locus in L. monocytogenes involved in cellobiose-dependent repression of hly expression led to the discovery of a gene named mdrL. This gene codes for a putative protein homologous (21 to 24% identity) to a member of the multidrug resistance efflux pump family of Bacillus subtilis. Another gene, named orfA, may produce a repressor of mdrL (8).
The aims of our study were the following: (i) to establish the levels of sensitivity to QACs of L. monocytogenes isolates from various ecosystems, (ii) to evaluate the distribution of the orfA and mdrL genes in the different listerial populations, and (iii) to examine whether L. monocytogenes strains contain plasmid genes closely related to qacA and smr as a possible cause of low sensitivity to QACs.
Sensitivity of L. monocytogenes strains to QACs.
Ninety-seven epidemiologically unrelated strains were selected to represent various L. monocytogenes ecosystems: the environment (n = 19), food products (n = 41), and human (n = 19) and animal (n = 18) pathological samples. All isolates were biochemically characterized by conventional identification methods (1). Antisera 1/2 and 4 were used for serogrouping according to the instructions of the manufacturer (Difco, Detroit, Mich.). MICs were determined by a dilution method on Mueller-Hinton agar medium (bioMérieux). Aliquots of 0.3 μl of bacterial inoculum adjusted to a turbidity of 0.5 McFarland unit were spotted onto agar containing the disinfectants to be tested (5 × 104 bacteria per spot). The following disinfectants were tested: benzalkonium chloride (1 to 20 mg/liter), cetrimide (2 to 40 mg/liter), chlorhexidine digluconate (0.5 to 10 mg/liter), acriflavine (5 to 500 mg/liter), and ethidium bromide (5 to 125 mg/liter). Agar plates were incubated at 37°C for 18 h. For benzalkonium chloride and cetrimide, dilutions were at 1-mg/liter steps. Staphylococcus aureus A-83 (harboring the qacA gene), A-82 (harboring the smr gene), and A-84 (sensitive to QACs) were included as positive and negative controls (Centre National de Référence des Staphylocoques, Lyon, France).
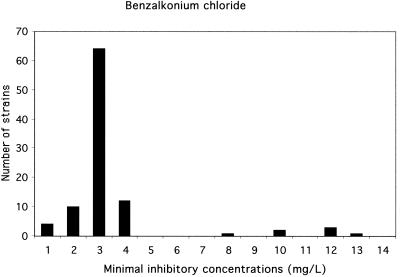
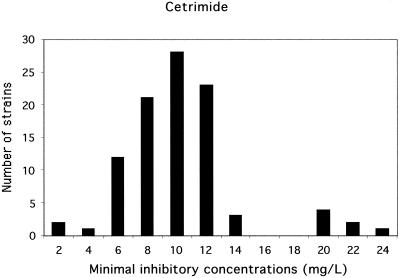
Two distinct populations were identified (Fig. 1 and 2). Ninety strains were scored as susceptible: the MICs of benzalkonium chloride were under 4 mg/liter and the MICs of cetrimide were under 14 mg/liter. Seven strains were scored as less susceptible: the MICs of benzalkonium chloride were over 7 mg/liter and the MICs of cetrimide were over 18 mg/liter. For these seven strains, the MICs of QACs and of chlorhexidine were also high. No significant association was found between the high MICs of QACs and the MICs of ethidium bromide or acriflavine. Thus, the MICs of QACs in vitro were high for 7% of our L. monocytogenes strains. This poor sensitivity may explain the persistence of some L. monocytogenes strains on manual processing lines and surfaces in food industry plants despite strict application of cleaning and disinfecting procedures (15). Consequently, the use of two different sanitizers employed alternately for the cleaning of food plants and the food industry environment may be beneficial.
FIG. 1.
Distribution of the 97 L. monocytogenes strains according to the MICs of benzalkonium chloride. Two populations were observed. One group, scored as susceptible, consisted of 90 strains for which the MICs were ≤4 mg/liter; the second group, scored as less susceptible, consisted of 7 strains for which the MICs were ≥8 mg/liter. These seven strains were also less susceptible to cetrimide.
FIG. 2.
Distribution of the 97 L. monocytogenes strains according to the MICs of cetrimide. Two populations were observed. One group scored as susceptible, consisted of 90 strains for which the MICs were ≤14 mg/liter. The second group, scored as less susceptible, consisted of 7 strains for which the MICs were ≥20 mg/liter. These seven strains were also less susceptible to benzalkonium chloride.
The seven resistant strains belong to serovars 1/2a and 1/2c. None of the 37 serogroup 4 strains were resistant to QACs (Table 1). Three of these seven resistant strains of L. monocytogenes were from environmental samples from food industry sites and 4 were from food products (Table 1). None came from animals, humans, or other environment samples. Therefore, the existence of strains poorly sensitive to QACs in food samples does not appear to be a major cause of human contamination, unless the resistance of the environmental and food isolates is lost upon infection of a human.
TABLE 1.
Origins and serogroups of the 97 epidemiologically unrelated strains of L. monocytogenes studied
| Serogroup | No. of strains from:
|
Total | |||
|---|---|---|---|---|---|
| Environment | Animals | Food products | Humans | ||
| 1/2 | 16a | 9 | 27b | 8 | 60 |
| 4 | 3 | 9 | 14 | 11 | 37 |
| All | 19 | 18 | 41 | 19 | 97 |
Including three strains resistant to QACs.
Including four strains resistant to QACs.
Identification of the mdrL and orfA genes by PCR.
Recently, a multidrug resistance efflux pump, MdrL, was identified in L. monocytogenes. Its amino acid sequence presents 21 to 24% identity with the Bmr and Blt efflux pumps of B. subtilis (8). Insertion mutagenesis of the reference strain L028 demonstrated that this efflux pump is responsible for substrate extrusion from L. monocytogenes. MICs of macrolides and heavy metals for the mutant strain were lower than those for the wild type (J. C. Perez-Diaz, M. T. Mata, M. C. Negri, and F. Baquero, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 671, 1999). This mechanism is a possible cause of the reduced sensitivity to QACs observed in our resistant strains. PCR amplification with primers lltb1 and lltb2 (Table 2) was used to identify the mdrL gene in our population of listeriae. The PCR mixture consisted of a buffer of 10 mM Tris-HCl–50 mM KCl–2.5 mM MgCl2 (pH 8.3) (Perkin-Elmer), a 100 μM concentration of each of the four deoxyribonucleoside triphosphates (Boehringer, Mannheim, Germany), 20 pmol of each of the two primers, 25 ng of DNA, and 0.5 U of AmpliTaq DNA polymerase (Perkin-Elmer) in a total volume of 25 μl. The reaction procedure consisted of an initial denaturation step at 94°C for 120 s followed by 30 cycles of denaturation at 94°C for 60 s, primer annealing at 50°C for 60 s, and extension at 72°C for 90 s (10 min for the last extension). Amplicons of the expected size (1,136 bp) were obtained with all 97 strains, suggesting that the mdrL gene is present in all isolates. It is thus unlikely that the QAC resistance of some L. monocytogenes strains is due to the acquisition of mdrL gene, which appears to be ubiquitous.
TABLE 2.
Nucleotide sequences of primers used for PCR amplification of the genes orfA, mdrL, qacA, and smr
| Gene | Primera | Sequence (5′ to 3′) | Amplicon size (kb) |
|---|---|---|---|
| orfA | orf1 | AAATGATTGCTCGTGAAGCT | 0.467 |
| orf2 | CGCACACCATTTTAATTCTG | ||
| mdrL | lltb1 | AAATGGATAACAGCGGCAG | 1.136 |
| lltb2 | TGTAAGGTAAAATGTGCTGG | ||
| qacA | qac3 | ACTACTGATATGATGACATCA | 1.512 |
| qac4 | AGTTATATCAAGTGATTTGGG | ||
| smr | smr1 | ATAGCCATAAGTACTGAAGTT | 0.291 |
| smr2 | ACCGAAAATGTTTAACGAAAC |
The modulation of the efflux rate by regulation of multidrug efflux genes involving specific trans-acting repressor proteins may be the basis for the high MICs of QACs (6). No specific regulator has been described for the MdrL pump. Nevertheless, a second putative protein, OrfA, which was identified at the same time as MdrL, may be a transcriptional repressor of mdrL gene expression (8). PCR amplification with primers orf1 and orf2 (Table 2) was used under the conditions described above to identify the orfA gene in our L. monocytogenes population. A fragment of the expected size (467 bp) was obtained for all 97 strains, suggesting that orfA is also ubiquitous.
Detection of plasmid DNA and plasmid curing.
To assess whether QAC resistance is plasmid associated, we studied the plasmid contents of all strains and cured them by heat treatment. Strains were cultured on heart brain broth (Oxoïd, Dardilly, France) at 30°C. Plasmids were extracted by the alkaline method as described by Birnboim and Doly (2). Escherichia coli V517 was used as a standard plasmid-containing strain (plasmid DNA of 55, 7.4, 5.7, 4, 3.1, 2.8, and 2.2 kb) and was included in parallel with each extraction. Plasmid DNA was detected in only three of the seven QAC-resistant strains. These three QAC-resistant strains were cured of plasmids by heat treatment as previously described (11). L. monocytogenes 88-1710 and 89-367 containing cadmium resistance plasmids were included to confirm that the method used allowed plasmid curing by the observation of the loss of cadmium resistance. No differences were found between the MICs of QACs before and after plasmid curing. This, and the observation that four resistant strains were plasmid free, suggests that the genes responsible for the QAC resistance of our seven strains can be easily transferred among L. monocytogenes strains. Therefore, the spread of this resistance may be limited in the environment and in food products.
Detection of the qacA and smr genes.
Multidrug resistance efflux pumps, QacA and Smr, have been found in the genus Staphylococcus. They confer resistance to a number of classes of antimicrobial organic cations, including QACs (13, 16). Around 13% of Staphylococcus strains isolated from the food industry show resistance to QACs, a finding similar to the 7% observed in our population of listeriae (7). To assess whether any genes related to the qacA and smr genes are implicated in the QAC resistance of our strains, PCR and Southern blot hybridization were used. PCR was performed as described above with primers qac3 and qac4, flanking the qacA gene, and with primers smr1 and smr2, flanking the smr gene (Table 2). DNA from six of the seven resistant strains gave a 2.7-kb fragment with the qac3 and qac4 primers, although a 1.4-kb fragment was expected. These PCR products were sequenced using the ThermoSequenase dye terminator cycle sequencing premix kit (Amersham Life Sciences, Cleveland, Ohio) and the Abi Prism 377 DNA sequencer (Perkin-Elmer). The 500 bases at the 5′ and 3′ ends of the amplified 2.7-kb fragment were sequenced. The sequences were dissimilar to any sequences in the qacA gene and also to any other known bacterial nucleotide sequence. PCRs with primers smr1 and smr2 flanking the smr gene did not amplify any fragment. Southern blot hybridization was performed as previously described (5). qac and smr probes were produced with DNA-amplified products of S. aureus A-83 DNA and S. aureus A-82 labeled with alkaline phosphatase-conjugated antibody of the AlkPhos direct labeling kit (Amersham Life Sciences). No hybridization was observed in Southern blotting experiments with the DNA of the seven strains. Therefore, the QAC resistance of L. monocytogenes does not appear to be due to a multidrug efflux system encoded by genes phylogenetically related to qac or smr genes.
QAC resistance and phage typing.
Phage typing was done as previously described using the international set of phages and experimental phages (1). Of the 97 strains studied, 15 were nontypeable by the entire set of phages used. Five of the seven QAC-resistant strains were nontypeable. Therefore, QAC resistance was significantly associated with nontypeability (P = 0.0008, Student's t test). This is suggestive of structural changes in the walls of the resistant strains. Such a phage-resistant phenotype in experimentally mutated strains of L. monocytogenes has already been obtained and was the result of the lack of N-acetylglucosamine in the teichoic acid of the cell wall (22). Therefore, an explanation for the poor sensitivity of some L. monocytogenes strains may be an intrinsic resistance arising from modifications of the thickness and the degree of cross-linking of peptidoglycan in the cell wall (14). This type of phenomenon has already been described for the genus Bacillus and for mucoid strains of S. aureus (10, 14).
In conclusion, high MICs of QACs were observed for some L. monocytogenes strains isolated from the environment and from food products. This resistance is not plasmid associated and is not due to the qacA and smr genes or to closely related genes. Overexpression of the MdrL protein encoded by the mdrL gene, ubiquitous in L. monocytogenes, or an intrinsic resistance may explain the high MICs of QACs displayed in vitro by 7% of the strains studied.
Acknowledgments
We thank M. E. Reverdy (Centre National de Référence des Staphylocoques) for kindly providing S. aureus strains A-82, A-83, and A-84. We also thank A. Fenneteau for technical assistance in phage typing.
REFERENCES
- 1.Audurier A, Martin C. Phage typing of Listeria monocytogenes. Int J Food Microbiol. 1989;8:251–257. doi: 10.1016/0168-1605(89)90022-6. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genigeorgis C A, Dutulescu D, Fernandez J. Prevalence of Listeria spp. in poultry meat at the supermarket and slaughterhouse level. J Food Prot. 1989;52:618–624. doi: 10.4315/0362-028X-52.9.618. [DOI] [PubMed] [Google Scholar]
- 5.Gousset N, Rosenau A, Sizaret P-Y, Quentin R. Nucleotide sequences of genes coding for fimbrial proteins in a cryptic genospecies of Haemophilus spp. isolated from neonatal and genital tract infections. Infect Immun. 1999;67:8–15. doi: 10.1128/iai.67.1.8-15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grkovic S, Brown M H, Roberts N J, Paulsen I T, Skurray R A. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem. 1998;273:18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 7.Heir E, Sundheim G, Holck A L. Resistance to quaternary ammonium compounds in Staphylococcus spp. isolated from the food industry and nucleotide sequence of the resistance plasmid pST827. J Appl Bacteriol. 1995;79:149–156. doi: 10.1111/j.1365-2672.1995.tb00928.x. [DOI] [PubMed] [Google Scholar]
- 8.Huillet E, Larin S, Pardon P, Berche P. Identification of a new locus in Listeria monocytogenes involved in cellobiose-dependent repression of hly expression. FEMS Microbiol Lett. 1999;174:265–272. doi: 10.1111/j.1574-6968.1999.tb13578.x. [DOI] [PubMed] [Google Scholar]
- 9.Jeong D K, Frank J F. Growth of Listeria monocytogenes at 10°C in biofilms with microorganisms isolated from meat and dairy processing environments. J Food Prot. 1994;57:576–586. doi: 10.4315/0362-028X-57.7.576. [DOI] [PubMed] [Google Scholar]
- 10.Kolawole D O. Resistance mechanisms of mucoid-grown Staphylococcus aureus to the antimicrobial action of some disinfectants and antiseptics. FEMS Microbiol Lett. 1984;25:205–209. [Google Scholar]
- 11.Lebrun M, Loulergue J, Chaslus-Dancla E, Audurier A. Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl Environ Microbiol. 1992;58:3183–3186. doi: 10.1128/aem.58.9.3183-3186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Littlejohn T G, Diberardino D, Messerotti L J, Spiers S J, Skurray R A. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene. 1991;101:59–66. doi: 10.1016/0378-1119(91)90224-y. [DOI] [PubMed] [Google Scholar]
- 13.Littlejohn T G, Paulsen I T, Gillepsie M T, Tennent J M, Midgley M, Jones I G, Purewal A S, Skurray R A. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol Lett. 1992;74:259–265. doi: 10.1016/0378-1097(92)90439-u. [DOI] [PubMed] [Google Scholar]
- 14.McDonnell G, Russell A D. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999;12:147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mustapha A, Liewen M B. Destruction of Listeria monocytogenes by sodium hypochlorite and quaternary ammonium sanitizers. J Food Prot. 1989;52:306–311. doi: 10.4315/0362-028X-52.5.306. [DOI] [PubMed] [Google Scholar]
- 16.Paulsen I T, Brown M H, Littlejohn T G, Mitchell B A, Skurray R A. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci USA. 1996;93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux system. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocourt J, Cossart P. Listeria monocytogenes. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology. Washington, D.C.: American Society for Microbiology; 1997. pp. 337–352. [Google Scholar]
- 19.Rouch D A, Cram D S, DiBerardino D, Littlejohn T G, Skurray R A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 20.Schuchat A, Swaminathan B, Broome C V. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tappero J W, Schuchat A, Deaver K A, Mascola L, Wenger J D. Reduction in the incidence of human listeriosis in the United States. Effectiveness of prevention efforts. JAMA. 1995;273:1118–1122. doi: 10.1001/jama.1995.03520380054035. [DOI] [PubMed] [Google Scholar]
- 22.Tran H L, Fiedler F, Hodgson D A, Kathariou S. Transposon-induced mutations in two loci of Listeria monocytogenes serotype 1/2a result in phage resistance and lack of N-acetylglucosamine in the teichoic acid of the cell wall. Appl Environ Microbiol. 1999;65:4793–4798. doi: 10.1128/aem.65.11.4793-4798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker R L, Jensen L H, Kinde H, Alexander A V, Owen L S. Environment survey for Listeria species in frozen milk plants in California. J Food Prot. 1991;54:178–182. doi: 10.4315/0362-028X-54.3.178. [DOI] [PubMed] [Google Scholar]