Abstract
Seventy-eight poliovirus strains isolated from river water and sewage in Toyama Prefecture, Japan, during 1993 to 1995 were characterized by the PCR-restriction fragment length polymorphism (RFLP) method and by partially sequencing the VP3 and VP1 regions of the viral genome. Of these isolates, 36 were identified as Sabin vaccine strains, and 42 were identified as vaccine variant strains that had less than 1.4% nucleotide divergence from the Sabin strains, including 7 isolates with patterns different from those of Sabin strains as determined by PCR-RFLP analysis. These findings suggest that wild-type poliovirus was not circulating in Toyama Prefecture.
The global polio eradication program has progressed by the initiates of the World Health Organization (4, 5). In Japan, the last outbreak of acute poliomyelitis occurred in 1960, followed by a decline in outbreaks as the result of the introduction of a live oral polio vaccine (OPV) for children in 1961. Since 1963, children have been protected by routine polio vaccination by OPV, and the incidence of clinical polio infection has been drastically reduced. The wild-type strains were detected in one patient with poliomyelitis in 1980 and in two patients with nonacute flaccid paralysis in 1984 and 1993, but since then no wild-type polioviruses have been isolated (11). On such a background, in order to verify polio eradication in Japan it is critical to confirm that the wild-type strains have not been circulating in Japan by characterizing the poliovirus strains excreted by inhabitants to differentiate the possible introduction of wild virus from the agent of vaccine-associated paralytic poliomyelitis. We predicted that the genetic analysis of poliovirus isolated from environmental waters might be a useful way of elucidating this. The viruses isolated from the environmental waters, namely, river water and sewage, correlated well with those excreted by inhabitants (13, 14, 15). Tambini et al. have reported that it is important to study the ecology of polioviruses excreted from human intestines into the environment at the final stage of the polio eradication program (21). We have examined the viruses in river water and sewage to monitor the viral pollution in the environmental waters in Toyama Prefecture from October 1993 to September 1995. In the present study, the genetic characteristics of the poliovirus strains isolated during this examination were compared by sequence analysis of the VP3 and VP1 regions with the wild and Sabin vaccine strains of polioviruses.
Poliovirus isolates from river water and sewage.
Four sampling stations (I, S, O, and G), as shown in Fig. 1, were chosen along the Itachi, Sembo, and Oyabe Rivers and at a sewage disposal plant located alongside the Oyabe River. The river water samples (approximately 700 to 800 ml), squeezed from two cotton pads (50 g each) that were immersed in the stream for 2 days at stations I, S, and O before sampling, were collected twice a month from October 1993 to September 1995. The sewage sample (1 liter) was collected from a sewage settling tank in station G at the same time as the river water samplings. These samples were concentrated by using the filter adsorption and elution method (13). A cellulose nitrate membrane filter (0.45- μm pore size, type TM2; Toyo Roshi, Tokyo, Japan) was used as a virus adsorbent, and the viruses adsorbed on the membrane were eluted into 3% beef extract solution (approximately 10 to 20 ml) by sonic treatment (type K-8814; Kontes). The concentrates thus obtained were inoculated into a total of 30 tube cultures of Vero, RD18S, MA104, and monkey kidney cells. Virus multiplication was checked by cytopathic effect. Isolates were identified by a neutralization test with poliovirus type-specific polyclonal antisera (25 U, rabbit sera). The neutralization test was performed according to standard procedures (24). The antisera were prepared by immunizing rabbits with Sabin type 1, 2, or 3 strains (Denka-Seiken, Tokyo, Japan).
FIG. 1.
Distribution of poliovirus strains isolated from river water and sewage samples. Each dot indicates one poliovirus strain.
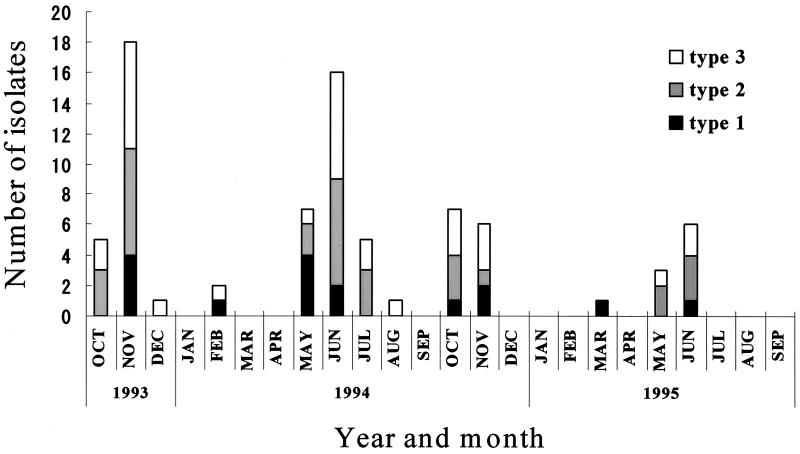
A total of 78 poliovirus strains were isolated from the samples from stations I, S, O, and G. Of these isolates, 6 (3 of type 2 and 3 of type 3) were isolated from the river water and 72 (16 of type 1, 28 of type 2, and 28 of type 3) were from the sewage samples (Fig. 1). OPV vaccination for children has been routinely given in May and October every year in Toyama Prefecture. Most isolates were found during the period following vaccination (Fig. 2). It has been reported that most excretion periods for poliovirus after OPV immunization of children are from 1 to 2 months (1, 23). In our study, the isolates had circulated in the environmental water for up to 3 months after OPV immunization. Though the excretion periods of type 2 and 3 strains were considered to be longer than that of type 1 (19), the differences in excretion periods among these three types of isolates were not observed.
FIG. 2.
Poliovirus isolates from river water and sewage samples.
Reference polioviruses.
The isolates were compared with the following strains: Sabin (types 1, 2, and 3), Mahoney (type 1), Lansing (type 2), Saukett (type 3), and polio vaccine viruses (F115, type 1; F209, type 2; F313, type 3) used in Japan during 1993 to 1995. Sabin and wild-type strains were obtained from the National Institute of Infectious Diseases, Tokyo, Japan. Vaccine viruses of Japan were prepared in the Japan Poliomyelitis Research Institute, Tokyo, Japan.
PCR-restriction fragment length polymorphism (PCR-RFLP) analysis.
RNA of poliovirus was extracted by treatment with RNAzol B (Tel-Test, Inc., Friendswood, Tex.) and phenol-chloroform. The cDNA of the viral RNA was synthesized with reverse transcriptase (Perkin-Elmer, Foster City, Calif.) at 37°C for 90 min and then amplified by PCR. The PCR was performed on a DNA thermal cycler (PJ2000; Perkin-Elmer) with 35 cycles of denaturation (94°C, 30 s), annealing (45°C, 1 min), and elongation (72°C, 1 min). The downstream primer (UC1) had the sequence 5′-GAATTCCATGTCAAATCTAGA-3′, and the upstream primer (UG1) had the sequence 5′-TTTGTGTCAGCGTGTAATGA-3′ (2). The amplified fragments (474 to 480 bp) were then separately digested with the restriction enzyme DdeI (5 U), HpaII (14 U), or HaeIII (10 U) (all from Pharmacia Biotech) at 37°C for 2 h. Digested products were electrophoresed on a 3.5% agarose gel, stained with ethidium bromide, and visualized by UV illumination.
According to the results of PCR-RFLP analysis for all isolates, 71 had restriction patterns similar to those of the homotypic Sabin strains and 7 had different patterns. Among type 1 isolates, strains G4-2, G4-12, G26-11, and G28-3 had restriction patterns different from that of Sabin 1 with HaeIII (Fig. 3A). Among those of type 2, strains O41-1 and G27-14 had patterns different from those of Sabin 2 with DdeI and HaeIII, respectively (Fig. 3B). The type 3 isolate, strain G19-4, had a pattern different from that of Sabin 3 with DdeI, and the patterns obtained with three restriction enzymes were similar to those of the Saukett strain (Fig. 3C). RFLP patterns different from those of Sabin strains were confirmed by subsequent sequence analysis of those strains (Table 1). The RFLP patterns of vaccine strains used in Japan were similar to those of Sabin strains.
FIG. 3.
RFLP patterns of the reference strains and isolates. Reference strains: Sabin 1, 2, and 3, Mahoney, Lansing, and Saukett. Isolates: G17-21, G4-2, G3-1, O41-1, G27-14, G4-18, and G19-4. UC, uncut; MW, DNA molecular weight marker (MspI digest of pBR322). Strains G17-21, G3-1, and G4-18 had restriction patterns similar to those of Sabin strains. G4-2, O41-1, G27-14, and G19-4 had patterns different from those of Sabin strains.
TABLE 1.
Differences in nucleotide sequences between the genomes of Sabin strains and isolates
| Sabin strain | Region and positiona | Change from Sabin to isolate
|
Isolated strainb | |
|---|---|---|---|---|
| Nucleotide | Amino acidc | |||
| Type 1 | VP3 | |||
| 2451 | C→T | T→I (229) | G28-9 | |
| 2456 | A→G | I→V (231) | G4-2 | |
| 2466 | A→T | K→I (234) | G4-16, G10-4, G15-12 | |
| VP1 | ||||
| 2545 | G→A | G28-3 | ||
| 2602 | T→C | G16-6 | ||
| 2608 | A→G | G28-9 | ||
| 2728 | G→A | G18-5 | ||
| 2737 | C→T | G3-11 | ||
| 2743d | C→T | G4-2, G4-12, G26-11, G28-3 | ||
| 2749 | A→G | I→M (90) | G28-3 | |
| 2774 | A→G | K→E (99) | G3-11 | |
| 2776 | G→T | K→N (99) | G28-9 | |
| 2795 | A→G | T→A (106) | G4-2, G4-12, G41-6, G10-4, G15-12, G18-5, G28-3, G28-9, G42-7 | |
| 2815 | T→C | G28-9 | ||
| 2839 | G→A | G28-9 | ||
| 2854 | C→T | G4-16, G10-4, G15-12 | ||
| Type 2 | VP3 (2468) | A→T | E→V (234) | G42-21 |
| VP1 | ||||
| 2520 | T→C | G15-8 | ||
| 2537 | T→C | V→A (19) | G18-2 | |
| 2546 | C→T | T→I (22) | O18-1 | |
| 2547 | T→C | G27-14 | ||
| 2548 | T→C | S→P (23) | G40-4 | |
| 2550 | C→T | G39-8 | ||
| 2566 | G→A | D→N (29) | G18-2 | |
| 2568 | C→T | G18-2 | ||
| 2576d | C→T | P→L (32) | O41-1 | |
| 2597 | A→G | K→R (39) | G4-9 | |
| 2626 | A→G | T→A (49) | G41-2 | |
| 2634d | T→C | G27-14 | ||
| 2687 | G→A | R→K (69) | G25-6, G25-7 | |
| 2790 | A→G | G18-10 | ||
| 2796 | T→C | G2-13 | ||
| 2820 | A→G | G40-4 | ||
| Type 3 | VP1 | |||
| 2479 | T→C | G4-19 | ||
| 2535 | A→G | K→R (20) | G2-19, G2-24, G27-9 | |
| 2560d | T→C | G19-4 | ||
| 2592 | T→C | V→A (39) | S25-1 | |
| 2636 | G→A | A→T (54) | G5-1, G17-6, G19-14 | |
| 2637 | C→T | A→V (54) | S9-2, G3-12, G3-14, G4-18, G4-19, G27-3 | |
| 2674 | A→G | G4-18 | ||
| 2707 | A→G | G3-12 | ||
| 3713 | C→T | G5-1 | ||
| 2776 | G→A | S9-2 | ||
| 2785 | T→C | G3-8, G3-16 | ||
| 2790 | T→C | M→T (105) | G4-18, G5-1, G27-3 | |
| 2815 | A→G | G5-1 | ||
Nucleotide and amino acid positions numbered by Nomoto et al., Pollard et al., Stanway, et al., and Toyoda et al. (17, 18, 20, 22) were used.
G, sewage sample; O, sample from Oyabe River; S, sample from Sembo River.
Numbers in parentheses are positions.
Nucleotide positions showed RFLP patterns that were different from those of Sabin strains.
Sequence analysis.
An appropriate DNA-sized band of PCR product was extracted from a 1.5% SeaKem GTG agarose gel (FMC Bioproducts, Rockland, Maine) and purified by passage through SUPREC-01 (TaKaRa Shuzo Co., Ltd., Shiga, Japan). The purified PCR product was sequenced by using a dRhodamine Terminator cycle sequencing reaction with a DNA sequencing kit (PE Applied Biosystems) and a genetic analyzer (ABI Prism 310; Perkin-Elmer). The sequence data (type 1, positions 2422 to 2860; type 2, 2424 to 2862; type 3, 2419 to 2851) on the VP3 and VP1 regions were aligned by using the software Gene Works version 2.3.1 (IntelliGenetics Inc.), and amino acid sequences were deduced.
Of 78 isolates, the nucleotide sequences of 36 isolates (4 of type 1, 17 of type 2, and 15 of type 3) were identical to those of homotypic Sabin strains. The other 42 isolates (12 of type 1, 14 of type 2, and 16 of type 3) were Sabin variants that had less than 1.4% nucleotide divergence from homotypic Sabin in the VP3 and VP1 region (Table 1). On the other hand, the nucleotide sequences of vaccine viruses used in Japan were identical to those of homotypic Sabin strains. Therefore, it is suggested that the mutations in the variant isolates occurred after administration of the vaccine. The mutation of the poliovirus genome during replication in human intestine or in vitro has been reported previously (7, 8, 9, 12, 16). Thirty-two of these variants had one to three amino acid substitutions.
Among the 12 type 1 variants, mutations of one to six nucleotides per strain were found. Strain G28-3 had four nucleotide mutations, 3 (at positions 2545, 2749, and 2795) for which there was reversion toward the Mahoney strain. Nine variants had mutation A→G at nucleotide position 2795, which was a reversion toward the Mahoney strain. According to Bouchard et al. (3), the mutation in this position has been related to attenuation. It should be noted that this back mutation has occurred in many isolates. Moreover, the mutation induced the amino acid substitution Thr106→Ala in VP1, which is located near the neutralization antigenic site (N-Ag 1). Other mutations close to N-Ag 1 were found at amino acid position 90 (Ile→Met) in strain G28-3 and at position 99 (Lys→Glu) in G3-11 and in G28-9 (Lys→Asn). In order to clarify the effect of amino acid substitution close to N-Ag 1, the neutralization titers of type-specific polyclonal antibody against type 1 isolates were assayed (Table 2). Sabin variants were easily neutralized at approximately the titer for the Sabin type 1 strain by type 1 antibody but not by type 2 and 3 antibodies. This suggests that OPV immunization might give protective immunity against infection with vaccine variants. Among 14 type 2 variants, mutations of one to three nucleotides per strain were found at random positions. No mutation near N-Ag 1 was found. Among 16 type 3 variants, mutations of one to four nucleotides per strain were found. A mutation close to N-Ag 1 was found at amino acid position 105 (Met→Thr) in strains G4-18, G5-1, and G27-3. These three variants were neutralized at approximately the same titer as the Sabin type 3 strain by type 3 antibody (data not shown).
TABLE 2.
Neutralizing-antibody titers of Sabin type 1 polyclonal antiserum against poliovirus type 1 strains
| Straina | Antibody titer |
|---|---|
| Sabin type 1 | 2,560 |
| Mahoney | 2,560 |
| G3-11, G16-1, G26-11, G28-9, G35-10, G42-7 | 1,780 |
| G4-2, G4-12, G4-16, G16-2, G17-21, G18-5, G28-3 | 2,560 |
| G16-6 | 5,120 |
Strains beginning with G are poliovirus type 1 isolates from sewage samples.
Concluding remarks.
The present study shows that poliovirus isolates were all derived from the Sabin vaccine strains, suggesting that Toyama Prefecture, Japan, is a wild-poliovirus-free area.
It has been reported that RFLP analysis was useful for intratypic differentiation between wild and vaccine strains (2, 10, 25). Seven of 78 isolates were differentiated as non-Sabin-like strains by RFLP analysis. On the other hand, sequencing analysis showed that 42 of those isolates were Sabin variants that had less than 1.4% nucleotide divergence from Sabin strains, including the 7 isolates differentiable by RFLP patterns. Results of RFLP and sequencing analyses revealed little difference in identifying identical and mutated isolates between the two methods. Variants were found in types 1, 2, and 3 after OPV immunization. Though the RFLP analysis is useful for intratypic differentiation among isolates, it should be applied more carefully to vaccine variants appearing immediately after OPV immunization of children.
Though the sewage system served an average of 50% of the population in Toyama Prefecture during 1993 to 1995, sanitary conditions were supposed to be relatively good because of improved domestic drainage disposal and lifestyle, including the use of paper diapers in baby care. Of 78 isolates, 6 were isolated from river water samples and the rest were from sewage samples. Therefore, it is suggested that the most polioviruses from humans had accumulated in sewage. Because of the difficulty of giving the general population access to the sewage system, the risk of infection with vaccine variants in the environment may be considered low. However, it will be necessary to assay the possible neurovirulence of vaccine variants from the environment. Since the year 2000, an inactivated polio vaccine immunization program in the United States has started to reduce the risk of infection by revertants excreted from humans (6). Likewise, in terms of infection from environmental isolates, the introduction of an inactivated polio vaccine would contribute to the final stage of the polio eradication program.
Acknowledgments
We are grateful to T. Yoneyama and A. Hagiwara (National Institute of Infectious Diseases) for distribution of the polioviruses.
REFERENCES
- 1.Alexander J P, Jr, Gary H E, Jr, Pallansch M A. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J Infect Dis. 1997;175(Suppl. 1):S176–S182. doi: 10.1093/infdis/175.supplement_1.s176. [DOI] [PubMed] [Google Scholar]
- 2.Balanant J, Guillot S, Candrea A, Delpeyroux F, Crainic R. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology. 1991;184:645–654. doi: 10.1016/0042-6822(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 3.Bouchard M J, Lam D-H, Racaniello V R. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J Virol. 1995;69:4972–4978. doi: 10.1128/jvi.69.8.4972-4978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Progress toward global eradication of poliomyelitis, 1996. Morb Mortal Wkly Rep. 1997;46:579–584. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Progress toward global eradication of poliomyelitis, 1997. Morb Mortal Wkly Rep. 1998;47:414–419. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Recommendations of the Advisory Committee on Immunization Practices: revised recommendations for routine poliomyelitis vaccination. Morb Mortal Wkly Rep. 1999;48:590. [PubMed] [Google Scholar]
- 7.Christodoulou C, Colbere-Garapin F, Macadam A, Taffs L F, Marsden S, Minor P, Horaud F. Mapping of mutations associated with neurovirulence in monkeys infected with Sabin 1 poliovirus revertants selected at high temperature. J Virol. 1990;64:4922–4929. doi: 10.1128/jvi.64.10.4922-4929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chumakov K M, Norwood L P, Parker M L, Dragunsky E M, Ran Y, Levenbook I S. RNA sequence variants in live poliovirus vaccine and their relation to neurovirulence. J Virol. 1992;66:966–970. doi: 10.1128/jvi.66.2.966-970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chumakov K M, Dragunsky E M, Norwood L P, Douthitt M P, Ran Y, Taffs R E, Ridge J, Levenbook I S. Consistent selection of mutations in the 5′-untranslated region of oral poliovirus vaccine upon passaging in vitro. J Med Virol. 1994;42:79–85. doi: 10.1002/jmv.1890420115. [DOI] [PubMed] [Google Scholar]
- 10.Furione M, Guillot S, Otelea D, Balanant J, Candrea A, Crainic R. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology. 1993;196:199–208. doi: 10.1006/viro.1993.1468. [DOI] [PubMed] [Google Scholar]
- 11.Infectious Agents Surveillance Center. Poliomyelitis, Japan, 1962–1995. Infect Agents Surveillance Rep. 1997;18:1–2. [Google Scholar]
- 12.Lu Z, Rezapkin G V, Douthitt M P, Ran Y, Asher D M, Levenbook I S, Chumakov K M. Limited genetic changes in the Sabin 1 strain of poliovirus occurring in the central nervous system of monkeys. J Gen Virol. 1996;77:273–280. doi: 10.1099/0022-1317-77-2-273. [DOI] [PubMed] [Google Scholar]
- 13.Matsuura K, Hasegawa S, Nakayama T, Morita O, Uetake H. Viral pollution of the rivers in Toyama City. Microbiol Immunol. 1984;28:575–588. doi: 10.1111/j.1348-0421.1984.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 14.Matsuura K, Ishikura M, Nakayama T, Hasegawa S, Morita O, Uetake H. Ecological studies on reovirus pollution of rivers in Toyama Prefecture. Microbiol Immunol. 1988;32:1221–1234. doi: 10.1111/j.1348-0421.1988.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura K, Ishikura M, Nakayama T, Hasegawa S, Morita O, Katori K, Uetake H. Ecological studies on reovirus pollution of rivers in Toyama Prefecture. II. Molecular epidemiological study of reoviruses isolated from river water. Microbiol Immunol. 1993;37:305–310. doi: 10.1111/j.1348-0421.1993.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 16.Mulders M N, Reimerink J H J, Stenvik M, Alaeddinoglu I, van der Avoort H G A M, Hovi T, Koopmans M P G. A Sabin vaccine-derived field isolate of poliovirus type 1 displaying aberrant phenotypic and genetic features, including a deletion in antigenic site 1. J Gen Virol. 1999;80:907–916. doi: 10.1099/0022-1317-80-4-907. [DOI] [PubMed] [Google Scholar]
- 17.Nomoto A, Omata T, Toyoda H, Kuge S, Horie H, Kataoka Y, Genba Y, Nakano Y, Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci USA. 1982;79:5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard S R, Dunn G, Cammack N, Minor P D, Almond J W. Nucleotide sequence of a neurovirulent variants of the type 2 oral poliovirus vaccine. J Virol. 1989;63:4949–4951. doi: 10.1128/jvi.63.11.4949-4951.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pöyry T, Stenvik M, Hovi T. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl Environ Microbiol. 1988;54:371–374. doi: 10.1128/aem.54.2.371-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanway G, Hughes P J, Mountford R C, Reeve P, Minor P D, Schild G C, Almond J W. Comparison of the complete nucleotide sequences of the genomes of the neurovirulent poliovirus P3/Leon/37 and its attenuated Sabin vaccine derivative P3/Leon 12a1b. Proc Natl Acad Sci USA. 1984;81:1539–1543. doi: 10.1073/pnas.81.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tambini G, Andrus J K, Marques E, Boshell J, Pallansch M, de Quadros C A, Kew O. Direct detection of wild poliovirus circulation by stool surveys of healthy children and analysis of community wastewater. J Infect Dis. 1993;168:1510–1514. doi: 10.1093/infdis/168.6.1510. [DOI] [PubMed] [Google Scholar]
- 22.Toyoda H, Kohara M, Kataoka Y, Suganuma T, Omata T, Imura N, Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984;174:561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- 23.Vaccine Administration Subcommittee, Japan Live Poliovaccine Research Commission. Evaluation of Sabin live poliovirus vaccine in Japan. II. Clinical, virologic and immunologic effects of vaccine in children. Jpn J Med Sci Biol. 1966;19:277–291. doi: 10.7883/yoken1952.19.277. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization. Manual for the virological investigation of poliomyelitis, C-1211. Geneva, Switzerland: Expanded program on immunization and division of communicable diseases. World Health Organization; 1992. [Google Scholar]
- 25.Yoshida H, Li J, Yoneyama T, Yoshii K, Shimizu H, Thanh N T H, Toda K, Long N T, Tu P V, Miyamura T, Hagiwara A. Two major strains of type 1 wild poliovirus circulating in Indochina. J Infect Dis. 1997;175:1233–1237. doi: 10.1086/593677. [DOI] [PubMed] [Google Scholar]