Abstract
Redundancy of multinucleated mature osteoclasts, which results from the excessive fusion of mononucleated preosteoclasts (pOCs), leads to osteolytic diseases such as osteoporosis. Unfortunately, the currently available clinical drugs completely inhibit osteoclasts, thus interfering with normal physiological bone turnover. pOC-specific regulation may be more suitable for maintaining bone homeostasis. Here, circBBS9, a previously unidentified circular RNA, was found to exert regulatory effects via the circBBS9/miR-423-3p/Traf6 axis in pOCs. To overcome the long-standing challenge of spatiotemporal RNA delivery to cells, we constructed biomimetic nanoparticles to achieve the pOC-specific targeted delivery of circBBS9. pOC membranes (POCMs) were extracted to camouflage cationic polymer for RNA interference with circBBS9 (POCM-NPs@siRNA/shRNAcircBBS9). POCM-NPs endowed the nanocarriers with improved stability, accurate pOC targeting, fusogenic uptake, and reactive oxygen species–responsive release. In summary, our findings may provide an alternative strategy for multinucleated cell–related diseases that involves restriction of mononucleated cell multinucleation through a spatiotemporally selective delivery system.
A spatiotemporally selective delivery system restricts osteoclast multinucleation by posttranscriptional regulation.
INTRODUCTION
Hyperactivation of multinucleation is important in numerous pathological processes in mammals, including osteolytic bone disease (1), granuloma tissue destruction in chronic infection (2), HIV spread (3), and the foreign giant cell hyperinflammatory response (4). Osteoclasts (OCs), which are typical multinucleated cells, differentiate from the monocyte/macrophage lineage in response to stimulation by macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor κB ligand (RANKL) (5, 6). Owing to their high transcriptional activity, multinucleated OCs (mOCs) secrete numerous enzymes and acids to exert a main function of bone resorption (7). However, excessive multinucleation of OCs ultimately causes an imbalance in bone reconstruction, leading to osteoporosis, rheumatoid arthritis, and periodontal disease (8, 9). The current first-line treatments for osteolytic diseases, such as bisphosphonate, inhibit OC lineages indiscriminately. This traditional nonselective strategy causes apoptosis of all bone-resorbing cells (10), thus suppressing necessary bone turnover and resulting in atypical femur fractures (11, 12). With a deepening understanding of OC differentiation, the subdivision of OC lineages is becoming clearer. Stepwise of RANKL-induced OC differentiation involves bone marrow macrophages (BMMs), monocytic precursors (MPs), mononucleated preosteoclasts (pOCs), and multinucleated mOCs (13). In contrast to mOCs, pOCs are not multinucleated, and they retain a lower level of bone resorption activity (14) and maintain positive communication with osteoblasts (OBs) to support appropriate osteogenesis (15). Therefore, developing a spatiotemporally selective strategy to restrict the OC lineage in pOCs rather than MPs or mOCs is urgently needed.
OC formation, especially in the pOC stage, exhibits exquisite regulation patterns, including abundant endogenous synthesis of noncoding RNAs (ncRNAs) and plentiful posttranscriptional regulation of bone resorption–specific genes (7, 16). Inhibition of signaling pathways is considered to be the principal regulatory mechanism in OC differentiation (15). MicroRNAs (miRNAs), small RNAs of ~22 nucleotides (nt) in length, interact with “seed” sequences (7 to 8 nt) located within the 3′ untranslated regions (3′UTRs) of target mRNAs to regulate the transcription or translation of the mRNAs. MiRNAs have demonstrated enormous potential in the clinical treatment of several diseases (17, 18). However, introduction of exogenous artificial miRNAs is associated with multiple risks, such as saturation of the RNA processing machinery and off-target effects (19, 20). Circular RNAs (circRNAs), which are covalently closed loops, are back spliced from pre-mRNA and are especially highly expressed during cell development. Because circRNAs are resistant to ribonuclease (RNase) R, they are more stable in cells and exosomes than their corresponding linear transcripts (21, 22). Recently, circRNAs have been reported to counteract miRNA-mediated mRNA inhibition by functioning as natural miRNA “sponges” (23). Because of their unique back-spliced junctions, circRNAs can act as potent endogenous regulators in well-designed RNA interference (RNAi) experiments without affecting their corresponding linear mRNAs (24, 25). However, the precise delivery of RNA-based therapies in vivo has been a long-standing challenge. Although targeted hydroxyapatite delivery modification strategies such as addition of tetracycline and eight repeating sequences of aspartate (D-Asp8) have been studied intensively, approaches for spatiotemporally selective intracellular RNA delivery targeting specific stages of OCs, particularly pOCs, have yet to be developed (26, 27). Moreover, the poor cell-specific targetability and destructive acidic environment in lysosomes prevent intracellular RNAi administration from achieving long-term effects (28). Hence, a spatiotemporally selective RNAi delivery system for pOCs should be established.
Cell membrane coating technology has been widely applied in pursuits ranging from drug delivery and imaging to photoactivatable therapy (29). Biomimetic nanoparticles (NPs) camouflaged with cell membranes inherit the intrinsic functions of cell membranes, which prolongs the circulation of the NPs in the blood and enables them to escape elimination by the immune system (30). Moreover, because they inherit the necessary fusion proteins and adhesion molecules from their source cells, cancer cell membrane–coated NPs can specifically target homologous cells. Thus, homotypic targeted delivery of these NPs is a possible future strategy for disease treatment (31). However, although cell membrane coatings have been used in various fields, targeted delivery with cellular spatiotemporal selectivity during differentiation has not been achieved. pOCs, which are derived from BMMs with 3 days of RANKL treatment, have specific proteins that participate in the circulation, recruitment, cell-to-cell recognition, and fusion of OCs (15). Hence, the membranes of fusogenic cells with specific stage markers are especially suitable for spatiotemporally selective RNAi delivery.
In the present study, we found that circBBS9, a novel and conserved circRNA highly expressed in pOCs, was able to alleviate the mOC multinucleation via accurate RNAi-induced knockdown on day 3 of RANKL treatment. To achieve pOC-targeting delivery of circBBS9, pOC membranes (POCMs) were extracted and used to camouflage a reactive oxygen species (ROS)–responsive charge-switchable cationic polymer and RNA interfering against circBBS9 [small interfering RNA (siRNA)/shRNAcircBBS9]. The cationic polymer 4-(bromomethyl)phenylboronic acid and poly[(2-N,N-diethyl)aminoethyl acrylate] (B-PDEAEA) was previously devised to minimize the side effects of cargoes (32). Then, pOC-specific targeting and fusogenic uptake were confirmed in vitro. The spatiotemporally selective RNA delivery and gene silencing efficiency of POCM-NPs were compared with those of bare NPs and macrophage cell membrane–coated NPs (MM-NPs). Last, spatiotemporally selective delivery to pOCs was verified in mice. A mouse model of osteoporosis revealed notable therapeutic and protective efficacy against osteoporosis without obvious organ damage. Overall, our therapeutic strategy involving posttranscriptional regulation and spatiotemporally selective delivery was able to prevent excessive fusion and function of fusogenic cells.
RESULTS
Characteristics of transcription in different stages of OC multinucleation
Multinucleation involves numerous transcriptional activities. Immunofluorescent (IF) staining was applied to measure the RNA polymerase II and phalloidin content in OCs as well as the C2C12, mouse myoblast cell during the progression of multinucleation. Both OCs and myocytes showed high transcriptional activity after stimulation (fig. S1, A and B). The mechanism of pOC multinucleation during which intermediate forms of mOCs with basic differentiation become multinucleated was explored. The mRNA profiles of pOCs and BMMs were examined by RNA sequencing (RNA-seq) to evaluate RNA transcriptional patterns during multinucleation progression (fig. S1C). Bone resorption–related genes and pOC processes were identified. The gene expression level of dendritic cell–specific transmembrane protein (DC-STAMP), an indispensable protein in OC fusion, was up-regulated by more than 70-fold. The gene expression level of Cathepsin K (CTSK), which promotes extracellular matrix degradation to execute bone resorption of OCs, was also significantly elevated (fig. S1, D and E). Through gene set enrichment analysis (GSEA), genes associated with transcription-related pathways were annotated with Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes pathway terms (fig. S1F). Moreover, as shown in fig. S1 (G and H), in response to RANKL stimulation, pOCs exhibited a highly active state and up-regulation of RNA polymerase genes. ncRNA processing was also enriched according to GSEA, which suggested that ncRNAs were potential targets for regulating the progression of OC multinucleation (fig. S1I).
Stage-specific functions of a new circRNA (circBBS9) in OC multinucleation
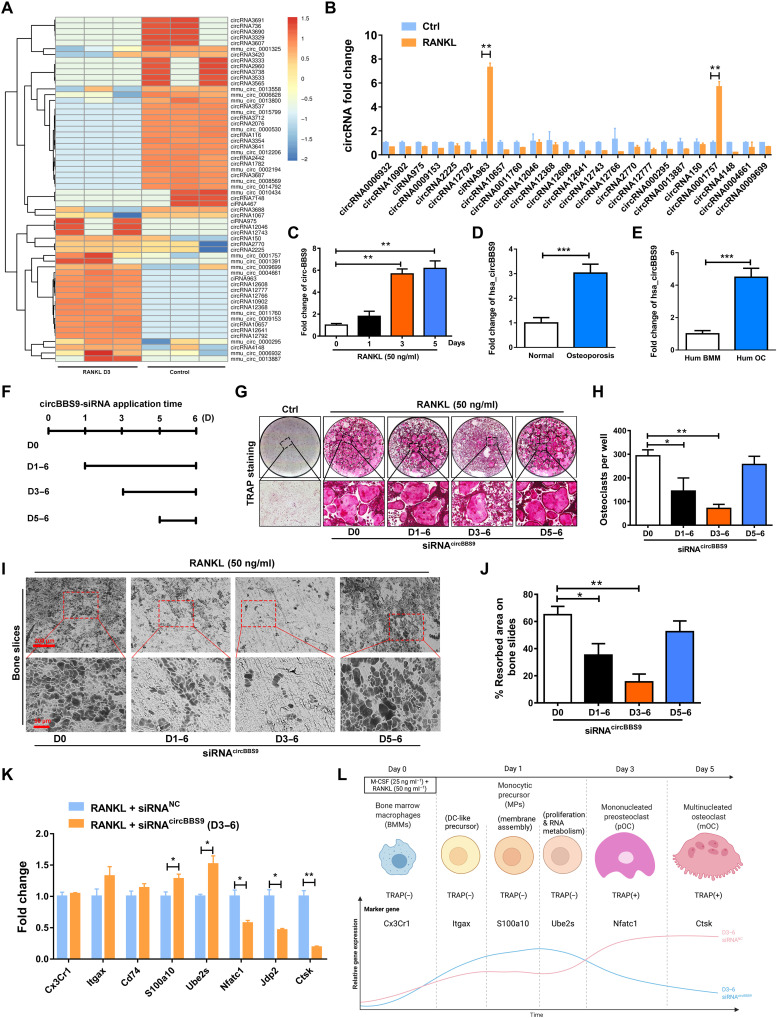
Among the ncRNAs, circRNAs exhibit RNase R resistance, which enables sustainable regulation. Because of their outstanding stability and biological regulatory function, circRNAs have been widely identified as therapeutic targets in various types of cells. Therefore, we attempted to identify circRNAs that act as key developmental regulators during OC multinucleation. Sixty distinct circRNAs with a significant log fold change of >1.5 or <−1.5 (P < 0.05) were identified by RNA-seq in the pOCs compared with the control cells (monocytes), including 40 novel circRNAs and 20 known circRNAs based on CircBase IDs (www.circbase.org/). The heatmap in Fig. 1A shows the 25 up-regulated circRNAs and 35 down-regulated circRNAs. Then, these circRNAs were confirmed by real-time quantitative polymerase chain reaction (RT-qPCR), in which one of the primers spanned the junction site of these circRNAs. Among those circRNAs, mmu_circ_0001757 (designated circBBS9 due to its host gene) stood out (Fig. 1B). Because the ciRNA963 was verified as a wrongly spliced intron-derived circRNA, circBBS9 was chosen to explore its function. Specifically, this circRNA showed a remarkable increase on days 1 to 3 after RANKL stimulation and a stable expression on days 3 to 5, implying that it was strongly promoted during the process of multinucleation (Fig. 1C). We searched CircBase for circBBS9 homologs in human. Unexpectedly, hsa_circ_0134188 (named as hsa_circBBS9) shared 81.69% identity in the alignment fragments of circBBS9. hsa_circBBS9 was high expressed in cancellous bones from the human osteoporosis patients’ spine versus normal people (Fig. 1D). Moreover, the expression of hsa_circ_0134188 was significantly increased in peripheral blood mononuclear cell (PBMC)–derived OCs as detected by qPCR (Fig. 1E).
Fig. 1. CircRNA expression profiling during OC multinucleation and validation of the temporal function of circBBS9 in OCs.
(A) Heatmap analysis of differentially expressed circRNAs from RNA-seq comparing pOCs with BMMs. (B) Up-regulated circRNAs further validated the expression of pOCs by RT-qPCR. (C) Expression of circBBS9 during OC multinucleation. (D) Expression of hsa_circBBS9 in cancellous bones from the human normal and osteoporosis patients’ spine. (E) Expression of hsa_circBBS9 in human BMM and human OCs. (F and G) Representative images and quantification of TRAP-positive mononucleated or multinucleated cells per well (in a 96-well plate) in the presence of M-CSF and RANKL for 5 days after transfection with siRNAcircBBS9 on the indicated days. (H) The number of TRAP-positive cells (>3 nuclei) was quantified. (I and J) Representative images and quantification of the resorbed area in bone slices after BMMs were kept in the presence of M-CSF and RANKL for 5 days after transfection with siRNAcircBBS9 on the indicated days. Scale bars, 200 and 50 μm. (K) Marker gene expression in different stages of OCs was detected 6 days after transfection with siRNAcircBBS9 on day 3 by RT-qPCR. (L) Schematic diagram of the stepwise effects of circBBS9. *P < 0.05, **P < 0.01, and ***P < 0.001. The values and error bars are the means ± SDs. All data were analyzed using Tukey’s multiple comparisons test or Student’s t test after ANOVA.
Then, the properties of circBBS9 were further detected. Sanger sequencing and gel electrophoresis verified the closed-loop structure and transcriptional properties of circBBS9 (fig. S2, A and B). CircBBS9 was resistant to degradation in an RNase R assay (fig. S2C).
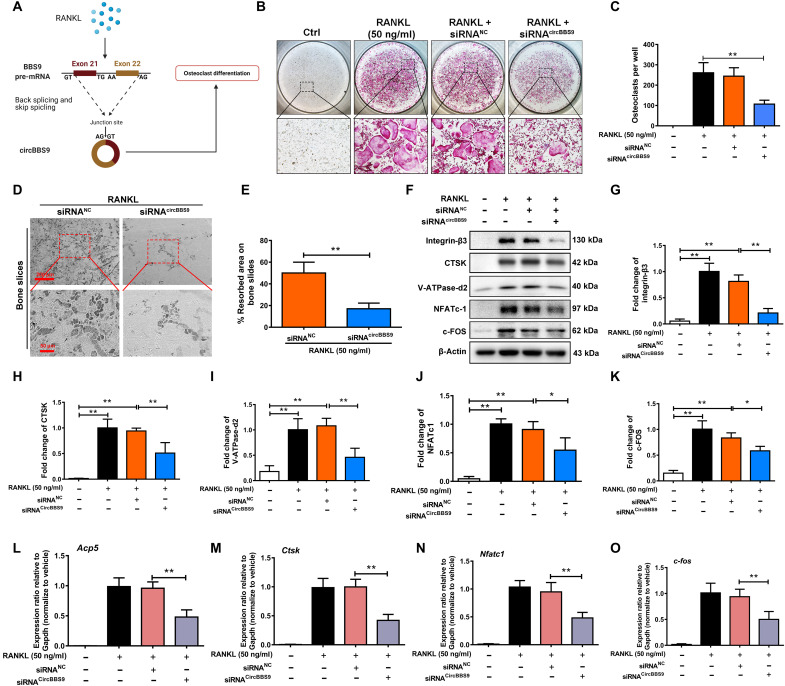
OCs at different stages, from BMMs to mononuclear pOCs to multinucleated mOCs, vary greatly in both their functions and their communication patterns with other cells (33). To explore the exact biological function of circBBS9, we transfected cells with three siRNAs targeting circBBS9. Depletion of circBBS9 was tested by RT-qPCR on day 5. The results indicated that circBBS9 was obviously reduced by transfection with siRNAcircBBS9 without influences on linear BBS9 mRNA (fig. S2, D and E). siRNAcircBBS9 3 was used in further experiments to test the function of circBBS9 in OC multinucleation. tartrate-resistant acid phosphatase (TRAP) buffer staining was performed to assess siRNA transfection on different days after RANKL treatment (Fig. 1, F and G), which demonstrated that siRNA transfection on day 3 inhibited the number of TRAP-positive mOCs and resulted in a certain number of mononuclear pOCs (Fig. 1H). In addition, the pit formation of bone slices caused by OC erosion was significantly attenuated by knockdown on day 3, while knockdown of circBBS9 on other days did not obviously inhibit OC erosion (Fig. 1, I and J). To confirm the specific inhibition of multinucleation, the marker genes of each stage of OC differentiation identified in a previous study were examined (13). When circBBS9 was knocked down on day 3, the expression of genes in the early stage of OC differentiation was maintained, while that of bone resorption–related genes was suppressed (Fig. 1K). These results suggested that the knockdown of circBBS9 on day 3 balanced OC bone resorption function and normal physiological formation (Fig. 1L). Hence, circBBS9 was knocked down on day 3 for further experiments. To verify the effect of circBBS9 (Fig. 2A), we performed TRAP staining and bone slice resorption assays again after transfection of BMMs with negative-control siRNA (siRNANC) or siRNA circBBS9 under RANKL treatment (Fig. 2, B to E). Then, Western blotting (WB) was performed to test the expression of proteins associated with OC maturation and function. The levels of bone resorption–related proteins, such as integrin-β3, CTSK, and V-ATPase-d2 [vacuolar (H+)–adenosine triphosphatase–d2], and key transcription factors, such as nuclear factor of activated T-cells (NFATc1) and c-FOS, were suppressed by siRNAcircBBS9 (Fig. 2, F to K). RT-qPCR assays also proved the decreases in the expression of these genes, implying that circBBS9 knockdown on day 3 effectively alleviated OC multinucleation (Fig. 2, L to O).
Fig. 2. circBBS9 knockdown on day 3 suppressed OC multinucleation.
(A) Assembly process of circBBS9 after RANKL treatment. (B) Representative images and quantification of the TRAP-positive cells per well (in a 96-well plate) in the presence of M-CSF and RANKL for 5 days with transfection of siRNAcircBBS9 or siRNANC on day 3. (C) The number of TRAP-positive cells (>3 nuclei) was quantitatively analyzed. (D and E) Representative images and quantification of the resorbed area in bone slices after BMMs were kept in the presence of M-CSF and RANKL for 5 days after transfection with siRNAcircBBS9 or siRNANC on day 3. Scale bars, 200 and 50 μm. (F) Western blot analysis of the expression of bone resorption–related proteins, including V-ATPase-d2, CTSK, and integrin-β3, and of the transfection factors NFATc1 and c-FOS in OCs with specific antibodies. (G to K) Quantitative analysis of protein expression. The expression of the proteins mentioned above was normalized to that of β-actin. (L to O) The gene expression levels of Nfatc1, c-fos, Acp5, and Ctsk in each group were detected by RT-qPCR and are reported relative to the expression level of Gapdh. The data represent the means ± SD. Significant differences are indicated as follows: *P < 0.05 and **P < 0.01. All data were analyzed using Tukey’s multiple comparisons test or Student’s t test after ANOVA.
Moreover, LVcircBBS9 was established to overexpress circBBS9 (fig. S3A). Treatment with a circBBS9 overexpression lentivirus (LVcircBBS9) but not a negative-control lentivirus (LVNC) significantly increased mOC formation and bone resorption ability (fig. S3, B and C). The expression of Acp5, Ctsk, V-atpase-d2, Traf6, and c-fos were up-regulated by LVcircBBS9 (fig. S3, D to H). In addition, hsa_circ_0134188 knockdown alleviated the expression of these genes in human OCs, demonstrating the potential translatability of the circBBS9 findings (fig. S4, A to E). We established siRNAcircBBS9 in macrophages. siRNAcircBBS9 did not obviously effect M1/M2 polarization or macrophage viability (fig. S5, A to F). The ability of macrophages to clear apoptotic cells was also not distinctly altered after knockdown of circBBS9 (fig. S5G). These data indicated that siRNAcircBBS9 has little impact on macrophages function.
Sponging function of circBBS9 on miR-423-3p to regulate OC multinucleation via the Traf6-mediated axis
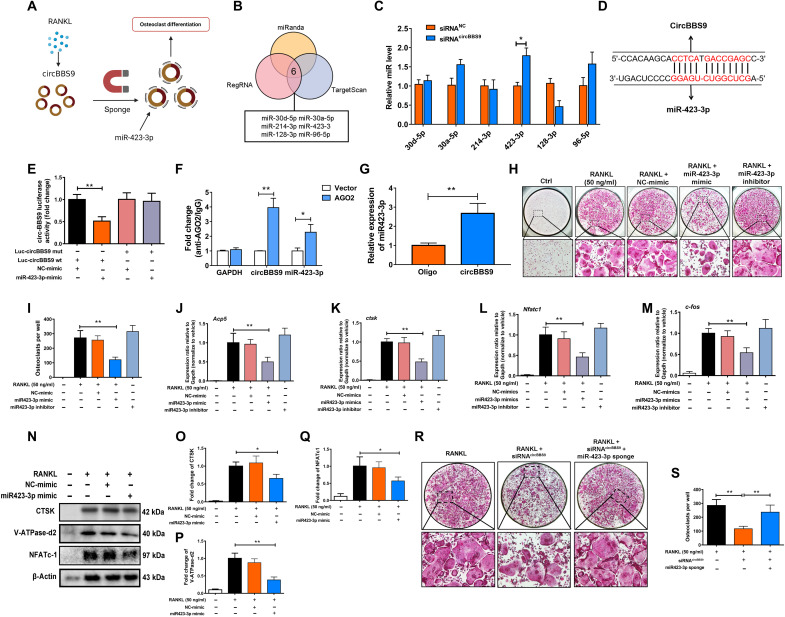
Because the effects of circBBS9 vary in different stages of OC differentiation, we next explored the mechanisms of circBBS9. Given that circRNAs acted as miRNA sponges to regulate gene expression, potential miRNAs associated with circBBS9 targeting were predicted using three databases: TargetScanMouse, miRanda, and regulatory RNA (regRNA) (Fig. 3, A and B). CircBBS9 knockdown increased the expression of miR-423-3p, which was selected for further investigation (Fig. 3C). As detected by luciferase assays, miR-423-3p exhibited reverse complementarity with circBBS9. miR-423-3p mimics significantly decreased the luciferase activity of the wild-type (WT) circBBS9 but not mutant circBBS9, suggesting that miR-423-3p can bind directly to circBBS9 (Fig. 3, D and E). To confirm the regulatory functions of miRNAs and circRNAs, we conducted an anti–Argonaute-2 (AGO2) RNA immunoprecipitation (RIP) assay and a pulldown assay in OCs. In the RIP assay, AGO2 antibodies were used for the training group. CircBBS9 was efficiently immunoprecipitated by the anti-AGO2 antibodies compared with the negative control [immunoglobulin G (IgG)] (Fig. 3F). In the pulldown assay with RT-qPCR detection, compared with a negative probe, a specific biotin-labeled miR-423-3p probe showed distinct enrichment of circBBS9, implying that miR-423-3p and circBBS9 directly bind to each other (Fig. 3G). These results indicate that circBBS9 is the natural endogenous sponge of miR-423-3p.
Fig. 3. miR-423-3p was sponged by circBBS9 and promoted OC multinucleation.
(A) CircBBS9 sponged miR-423-3p to promote OC multinucleation. (B) Six miRNAs were predicted to bind with circBBS9. (C) The expression of the predicted miRNA candidates was tested by RT-qPCR with or without siRNAcircBBS9 treatment. (D) Best predicted binding sites between circBBS9 and miR-423-3p. (E) Luciferase activity of luciferase reporters containing circBBS9 sequences with WT or mutated miR-423-3p binding sites in human embryonic kidney (HEK) 293T cells cotransfected with miR-423-3p mimics or controls. (F) RIP was performed in RAW264.7 cells and was followed by RT-qPCR to detect the ability of AGO2 to immunoprecipitate circBBS9 compared with IgG. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (G) RNA pull-down assays were performed using a circBBS9-specific biotin probe and oligo probe in RAW264.7 cells, followed by RT-qPCR assays to analyze miR-423-3p expression. (H and I) Representative images and quantification of TRAP-positive cells (>3 nuclei) among OCs transfected with miR-423-3p inhibitors, mimics, and controls. (J to M) Gene expression of Nfatc1, c-fos, Acp5, and Ctsk in pOCs after transfection with miR-423-3p inhibitors, mimics, and controls, as analyzed by RT-qPCR. (N to Q) WB was used to analyze the expression of CTSK, V-ATPase-d2, and NFATc1 in pOCs after treatment with miR-423-3p mimics or controls. (R and S) TRAP-positive cells (>3 nuclei) were quantified after treatment with siRNAcircBBS9 alone or combined with miR-423-3p inhibitor. *P < 0.05 and **P < 0.01. All data were analyzed using Tukey’s multiple comparisons test or Student’s t test after ANOVA.
To evaluate the role of miR-423-3p in osteoclastogenesis, mimics or sponges overexpressed or inhibited miR-423-3p in OCs. Compared with the negative-control group, miR-423-3p mimics or inhibitors efficiently hampered or enhanced the formation of OCs, respectively, as shown by TRAP staining (Fig. 3, H and I). RT-qPCR assays of the OC-related genes Acp5, Ctsk, Nfatc1, and c-fos verified the results (Fig. 3, J to M). In addition, the effect of miR-423-3p overexpression on the OC-related proteins CTSK, V-ATPase-d2, and NFATc1 was detected by WB (Fig. 3, N to Q). Rescue experiments were designed to investigate the functional regulation between miR-423-3p and circBBS9 in mOCs. The miR-423-3p sponge reversed siRNAcircBBS9-induced OC-specific gene and protein expression, as revealed by TRAP staining (Fig. 3, R and S).
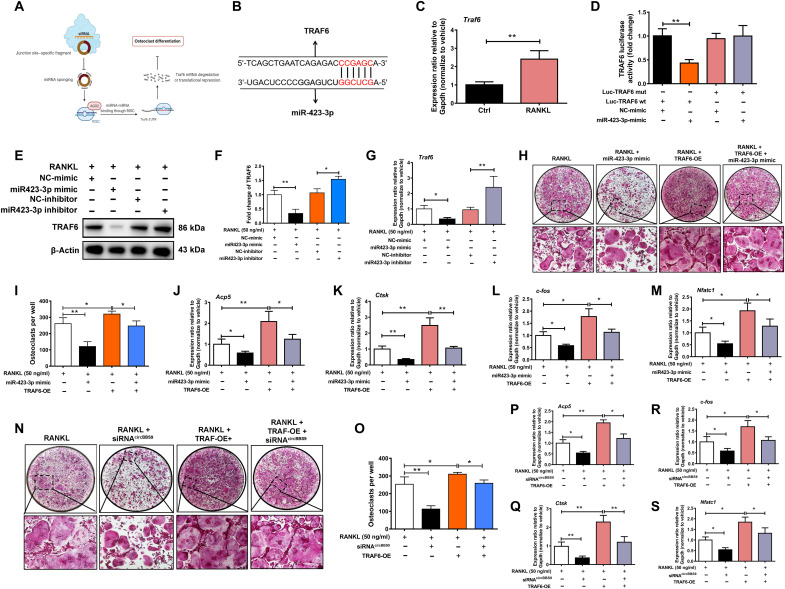
The binding of miRNAs and mRNAs plays a critical role in the circRNA regulation axis (Fig. 4A). According to the binding site prediction of miR-423-3p predicted by TargetScan, the 3′UTR of the Traf6 gene exhibited a strong ability to bind with miR-423-3p (Fig. 4B). Traf6 levels were significantly higher in pOCs than in BMMs (Fig. 4C). A luciferase reporter assay showed that miR-423-3p mimics decreased the activity of the WT luciferase reporter constructed with the predicted binding site compared with the mutant, validating the potential for regulation between Traf6 and miR-423-3p (Fig. 4D). To examine the exact biological function of the miR-423-3p/Traf6 interaction, the protein and gene expression of TRAF6 after miR-423-3p mimic or inhibitor treatment was quantified by WB and RT-qPCR (Fig. 4, E and F). The function of TRAF6 in the circBBS9 regulation axis was also detected. The miR-423-3p mimic decreased the expression of Traf6 mRNA, whereas miR-423-3p inhibitor increased the expression of Traf6 mRNA significantly, implying the regulation between miR-423-3p and Traf6 (Fig. 4G). TRAP staining and RT-qPCR assays were performed to test the rescue of siRNAcircBBS9 and a miR-423-3p mimic with Traf6 overexpression plasmids. These results indicated that the miR-423-3p mimic and siRNAcircBBS9 prevented OC differentiation, while TRAF6 overexpression reversed this effect (Fig. 4, H to S). Collectively, these results imply that the knockdown of circBBS9 in pOCs inhibits OC multinucleation through the circBBS9/miR-423-3p/Traf6 axis, indicating that circBBS9 is an effective posttranscriptional regulator.
Fig. 4. The circBBS9/miR-423-3p/Traf6 axis regulated OC multinucleation.
(A) miR-423-3p inhibited Traf6 mRNA to repress OC multinucleation. RISC, RNA-induced silencing complex. (B) Predicted binding sites between circBBS9 and miR-423-3p. (C) RT-qPCR showed the expression of Traf6 mRNA in pOCs and BMMs. (D) Luciferase activity of luciferase reporters containing Traf6 sequences with WT or mutated miR-423-3p binding sites in HEK293T cells cotransfected with miR-423-3p mimics or controls. (E to G) The protein expression and mRNA expression of TRAF6 in pOCs after transfection with miR-423-3p inhibitors, mimics, and controls were assessed by WB and RT-qPCR, respectively. (H and I) Representative images and quantification of TRAP-positive cells (>3 nuclei) after treatment with miR-423-3p mimics or a TRAF6 overexpression plasmid alone or in combination. (J to M) Gene expression of Acp5, Ctsk, Nfatc1, and c-fos in pOCs after transfection with miR-423-3p mimics and a TRAF6 overexpression plasmid. (N and O) Representative images and quantification of TRAP-positive cells (>3 nuclei) after treatment with siRNAcircBBS9 or a TRAF6 overexpression plasmid alone or in combination. (P to S) Gene expression of Acp5, Ctsk, Nfatc1, and c-fos in pOCs after transfection with siRNAcircBBS9 and TRAF6 overexpression plasmids. *P < 0.05 and **P < 0.01. The values and error bars are the means ± SDs. All data were analyzed using Tukey's multiple comparisons test or Student's t test after ANOVA.
Characterization of spatiotemporally selective delivery systems
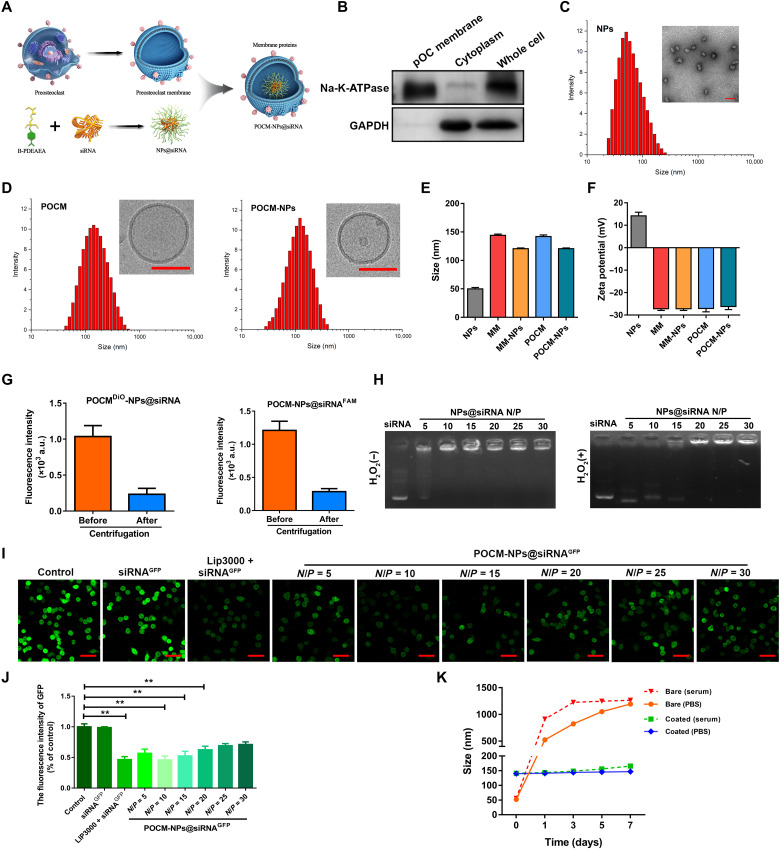
We previously confirmed that the therapeutic effects of circBBS9 relied on the specific stage of OC differentiation, emphasizing that the spatiotemporally selective release of nucleic acids was a prerequisite. As the ROS levels in pOCs were increased (fig. S6A), we adjusted our previously devised ROS-triggered cationic polymer B-PDEAEA and applied it to encapsulate nucleic acids (NPs) to achieve delivery in vitro (fig. S6B). RAW264.7 cells transfected with luciferase plasmids were encapsulated with different N/P ratios and then cultured in 0 or 10% fetal bovine serum (FBS)–containing medium. The NPs with an N/P ratio of 10 exhibited the highest transfection efficiency in 0% FBS medium (fig. S6C). Furthermore, the ROS-dependent transfection efficacy of NPs was evaluated according to luciferase expression levels after different H2O2 treatments (fig. S6D).
pOCs exhibit exquisite patterns of migration, recognition, and fusion that are attributed to the interactions of membrane proteins after RANKL-induced differentiation. Thus, we extracted the cell membranes of BMMs (MMs) and pOCs (POCMs) to package the B-PDEAEA/RNA core (NPs) for further investigation (Fig. 5A). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Na-K-ATPase protein levels in the cytoplasm, POCMs, and whole cell were detected by WB analysis, respectively, which verified the successful isolation of POCMs (Fig. 5B). NPs were synthesized with an average diameter of 50.0 nm and zeta potential of 14.1 mV. The POCM coating increased their size, with the final POCM-NP formulation exhibiting an average diameter of approximately 141.9 nm, while its zeta potential (−26.2 mV) was close to that of pOC vesicles (−27.1 mV). The size and zeta potential of MM and MM-NPs was similar with POCM and POCM-NPs, respectively, indicating successful surface coating with cell membrane (Fig. 5, C to F, and fig. S7A). Furthermore, after POCM-NPs@siRNAFAM (containing FAM-labeled siRNA) or POCMDiO-NPs@siRNA (containing 3,3’-Dioctadecyloxacarbocyanine perchlorate (DiO)-labeled cell membrane) were centrifuged, the fluorescence intensity in the supernatant decreased markedly, further indicating the high efficiency of the membrane coating (Fig. 5G). The characteristics of ROS release in OCs were established. H2O2 (5 mM) significantly promoted nucleic acid escape from NPs@siRNA and NPs@shRNA, according to DNA gel electrophoresis (Fig. 5H and fig. S6E). The transfection efficiency of NPs with POCM encapsulation at different N/P ratios was examined in green fluorescent protein (GFP)–transfected RAW264.7 cells. The optimal concentration of Lipofectamine 3000 was explored in fig. S6 (F and G). Compared with bare siRNAGFP, Lipofectamine 3000 alone, and the NPs created with the other N/P ratios, the POCM-NPs@siRNAGFP with 10 and 15% N/P ratios exhibited relatively high silencing efficiency (Fig. 5, I and J). To balance cell cytotoxicity and gene transfection efficiency (34), an optimized N/P ratio of 10 was used in subsequent experiments. The long-term stability of POCM-NPs was assessed in phosphate-buffered saline (PBS) and serum-containing medium for 1 week. While the POCM-NPs maintained their initial size during this period, the bare NPs swelled considerably, highlighting the advantage of using a camouflaged cell membrane to stabilize NPs in a physiological environment (Fig. 5K).
Fig. 5. The preparation and characterization of POCM-NPs.
(A) Schematic of RNA loading into B-PDEAEA (NPs) with camouflage by POCM coating. (B) WB for membrane protein Na-K-ATPase and plasma protein GAPDH in isolated POCMs, pOC cytoplasm, and pOC cell lysate. (C) Size and transmission electron microscopy micrograph of NPs. Scale bar, 100 nm. (D) Size and cryo–electron microscopy micrograph of POCM and POCM-NPs. Scale bars, 100 nm. (E) Mean diameters of NPs, MM, MM-NPs, POCM, and POCM-NPs after construction. (F) Zeta potentials of NPs, MM, MM-NPs, POCM, and POCM-NPs. (G) Fluorescence intensity in the supernatant of POCM-NP@siRNAFAM (containing FAM-labeled siRNA) or POCMDiO-NP@siRNA (containing DiO-labeled cell membrane) after centrifugation. a.u., arbitrary units. (H) Gel retardation assay of polyplexes at designated N/P ratios after 1 hour of incubation with or without H2O2 at 37°C. (I) Visualization of the transfection efficiency in GFP-transduced RAW264.7 cells after coculture with control, siRNAGFP, lipo3000-siRNAGFP, or POCM-NPs@siRNAGFP at the indicated N/P ratio for 48 hours. Scale bars, 50 μm. GFP, green. (J) Quantification of transfection efficiency of control, siRNAGFP, lipo3000-siRNAGFP, or POCM-NPs@siRNAGFP at the indicated N/P ratio. (K) Stability of POCM-NPs@siRNA and NPs@siRNA over time in PBS- or serum-containing medium. **P < 0.01. The values and error bars are the means ± SDs. All data were analyzed using Tukey’s multiple comparisons test or Student’s t test after ANOVA. .
Self-selective binding by the POCM camouflage
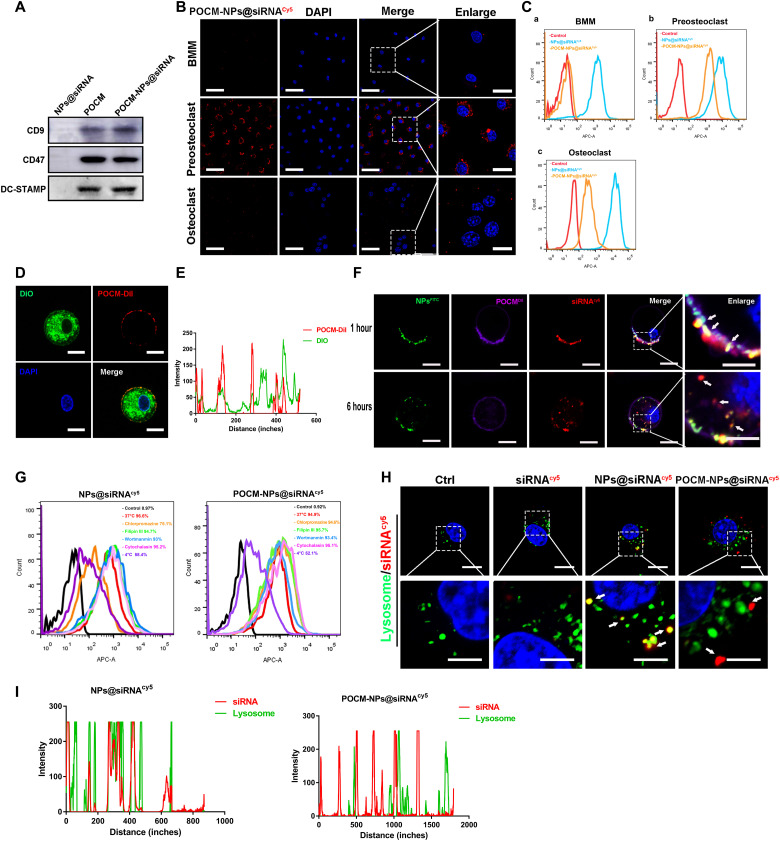
Previous studies have reported that fusion recognition proteins are expressed in pOCs upon RANKL stimulation, which enables the homotypic fusion of OCs (35, 36). To confirm the components of the extracted membrane, the proteomic profile of the RANKL-induced POCMs was established and compared with that of the MMs. The proteins were also classified by GO enrichment (fig. S7, B and C). For functional reasons, proteins participating in migration and targeting, such as CXCR4, CDC42, and RAC2, were clearly highly expressed in pOCs as were fusion-related proteins such as CD44 and OSCAR (fig. S7, D and E). The crucial OC proteins DC-STAMP, CD47, and CD9 were identified by WB in the POCMs and POCM-NPs (Fig. 6A). Together, the above results suggested that a pOC-derived membrane could be efficiently produced, establishing POCM-NPs as a biomimetic platform to deliver NPs to pOCs.
Fig. 6. Self-recognized binding and fusion-mediated intracellular release process of POCM-NPs@siRNA.
(A) Western blot images of POCM proteins in POCMs and POCM-NPs@siRNA, where NPs@siRNA were applied as a control. (B) Representative fluorescence visualization showing self-recognized binding of POCM-NPs@siRNAcy5 in BMMs, pOCs, and mOCs. Scale bars, 50 μm. Enlarged scale bars, 20 μm. siRNA, red. DAPI, 4′,6-diamidino-2-phenylindole. (C) Cy5-positive BMM, pOC, and OC measured by flow cytometry after treatment of POCM-NPs@siRNAcy5. (D and E) Fluorescence images and intensity of DiO-labeled pOCs and DiI-labeled POCMs, indicating the interaction between pOCs and POCMs. Scale bars, 10 μm. POCs, green; POCM, red; nuclei, blue. (F) Fluorescence visualizations showing the cellular uptake and subcellular distribution after incubation of FITC-labeled B-PDEAEA, DiI-labeled POCM, and Cy5-labeled siRNA incubation with pOCs for 1 and 6 hours. Scale bars, 10 μm. Enlarged scale bars, 5 μm. NPs, green; POCM, purple; siRNA, red; nuclei, blue. (G) Cy5-positive cells measured by flow cytometry after treatment with POCM-NPs@shRNAcy5 for 2 hours. pOCs were cooled at 4°C or separately pretreated with endocytosis-related inhibitors for 0.5 hours at 37°C. (H and I) Fluorescent visualization of siRNAcy5 and lysosome localization in pOCs after incubation with POCM-NPs@siRNAcy5 for 2 hours and intensity profiles across the cell. Scale bars, 10 μm. Enlarged scale bars, 5 μm. siRNA, red; nuclei, blue; lysosome, green.
The cell-selective targeting of POCM-NPs was observed by using NPs@siRNAcy5, MM-NPs@siRNAcy5, or POCM-NPs@siRNAcy5 (siRNA labeled with Cy5). First, the pOC-selective binding of POCM-MPs to OC lineage cells was verified. OC cell lines were incubated with NPs@siRNAcy5, MM-NPs@siRNAcy5, and POCM-NPs@siRNAcy5 for 6 hours, and bare siRNAcy5 was applied as control. Notably, in contrast to the inefficient delivery of MM-NPs@siRNAcy5 and NPs@siRNAcy5, POCM-NPs exhibited OC-selective binding. The results of flow cytometric analysis and fluorescence microscopy suggested that the entrance efficiency of mOCs was lower than that of pOCs (Fig. 6, B and C, and fig. S8, A and B). Second, we assessed whether the uptake of POCM-NPs was specific to the bone microenvironment. Notably, there were no obvious red puncta in OBs, bone mesenchymal stem cells (MSCs), or human umbilical vein endothelial cells (HUVECs), after 6 hours of incubation with POCM-NPs@siRNAcy5 (fig. S8C). The efficiency of bare siRNAcy5, NPs@siRNAcy5, and POCM-NPs@siRNAcy5 in this cell uptake was measured by flow cytometric analysis (fig. S8D). NPs@siRNAcy5 showed a high transfection efficiency in all type of cells, whereas POCM@siRNAcy5 could just enter into pOC. These findings suggested that compared to MM-NPs, POCM-camouflaged NPs exhibited a spatiotemporally selective delivery ability in terms of the bone microenvironment and the stage of OC multinucleation.
Fusion-mediated and intracellular uptake of POCM-NPs
The intracellular transfer of the POCM-NPs revealed the mechanism behind their spatiotemporally selective delivery ability. The POCMDiI-NPs [POCM labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI)] were incubated with pOCs (labeled with DiO) for 1 hour. As shown in Fig. 6 (D and E), POCMs (the red signal) accumulated mainly in the outer cellular layer. To further investigate the intracellular transfer of siRNA, POCM-NPs@siRNAcy5 (siRNA labeled with Cy5) were established. The POCM and B-PDEAEA were labeled with DiI (the purple signal) and fluorescein isothiocyanate (FITC) (the green signal), respectively. After incubation of the pOCs with the nanoformulations for 1 hour, most fluorescence still overlapped, and yellow puncta were observed by confocal microscopy. After incubation with pOCs for 6 hours, the numbers of separate red and purple puncta increased, indicating that the NPs efficiently dissociated under oxidative stress conditions (Fig. 6F). Considering the colocalization of the POCM-NPs and their original membrane, we then verified whether endocytosis participated in the cellular uptake of POCM-NPs. Endocytosis inhibitors including chlorpromazine, filipin III, wortmannin, and cytochalasin D did not affect the cellular uptake analyzed by fluorescence-activated cell sorting (FACS) (Fig. 6G) or luciferase expression (fig. S9A) of POCM-NPs. These results indicated that the cellular internalization of the POCM-NPs did not occur via endocytosis, micropinocytosis, or phagocytosis but instead occurred through a membrane fusion mechanism. When cultured with lysosomal marker (LysoTracker Green)–stained pOCs, the POCM-NPs@siRNAcy5 and NPs@siRNAcy5 showed enhanced red fluorescence intensity, and the former exhibited a lower overlap with green puncta than bare siRNAcy5 and NPs@siRNAcy5 after 2 hours of cellular uptake (Fig. 6, H and I). In the meanwhile, lysosome and siRNA showed few colocalization in whole stage (1, 3, and 6 hours), furtherly indicating a lysosome bypass manner of POCM (fig. S8, E and F). In addition, shRNAcy5 was confirmed to a lysosome bypass manner via intracellular tracking (fig. S9B). These results demonstrate a delicate hierarchy of POCM-NPs@siRNA/shRNA (short hairpin–mediated RNA) delivery after the pOC-specific binding with fusogenic uptake and lysosomal degradation bypass. The findings provide insights for the development of a universal platform to efficiently deliver and release RNA therapeutics.
Temporal specificity of gene silencing by POCM-NPs
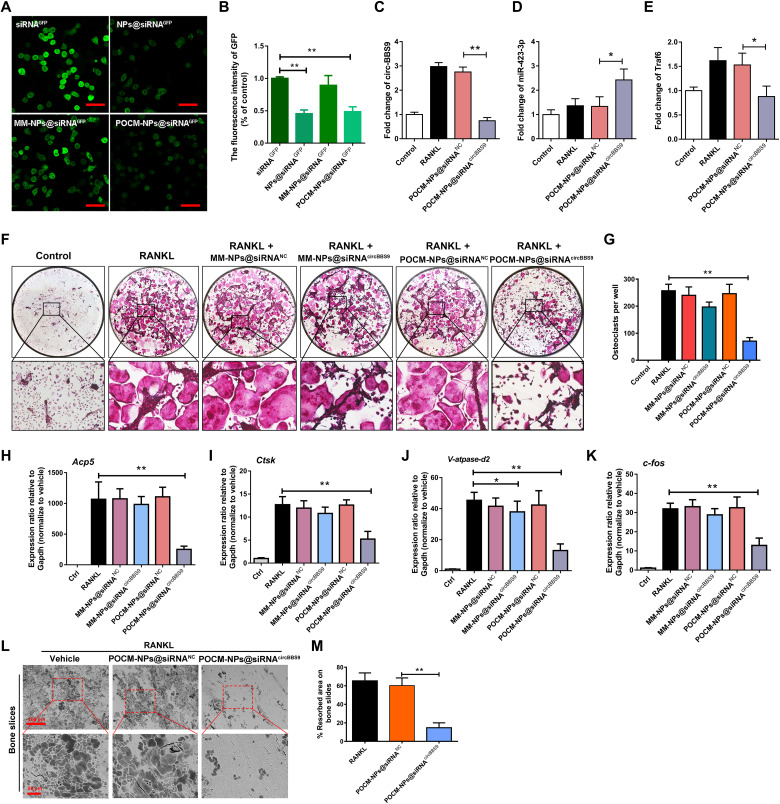
It has been proven that a spatiotemporally selective delivery system targeting pOCs can potentially provide a platform for conducting circBBS9 stage-specific knockdown to restrain OC multinucleation. pOCs transfected with GFP were used to quantify the knockdown efficiency of POCM-NPs@siRNA. Then, pOCs were incubated with bare siRNA against GFP (siRNAGFP), NPs with siRNAGFP (NPs@siRNAGFP), MM-NPs with siRNAGFP (MM-NPs@siRNAGFP), and POCM-NPs with siRNAGFP (POCM-NPs@siRNAGFP) for 48 hours. The fluorescence intensity of the cells was assessed by confocal fluorescence microscopy (Fig. 7, A and B). Notably, the results showed that the pOC-derived spatiotemporally selective delivery system exhibited higher gene silencing efficiency than NPs and MM-NPs.
Fig. 7. POCM-NPs@siRNA enhanced gene silencing efficiency.
(A to B) Visualization of transfection efficiency in GFP-transduced RAW264.7 cells with RANKL treated for 3 days after coculture with siRNAGFP, NPs@siRNAGFP, MM-NPs@siRNAGFP, or POCM-NPs@siRNAGFP. Scale bars, 50 μm. GFP, green. (C to E) Expression of circBBS9, miR-423-3p, and Traf6 as detected by RT-qPCR. (F and G) Representative images and quantification of TRAP-positive mononucleated or multinucleated cells per well (in a 96-well plate) in the presence or absence of M-CSF and RANKL for 5 days with initial incubation with MM-NPs@siRNANC, MM-NPs@siRNAcircBBS9, POCM-NPs@siRNANC, or POCM-NPs@siRNAcircBBS9. (H to K) Gene expression levels of V-atpase-d2, c-fos, Acp5, and Ctsk in each group were assessed by RT-qPCR and reported relative to the expression level of Gapdh. (L and M) Representative images and quantification of the resorbed area in bone slices after OC treatment in each group. Scale bars, 200 and 50 μm. *P < 0.05 and **P < 0.01. The values and error bars are the means ± SDs.All data were analyzed using Tukey’s multiple comparisons test or Student’s t test after ANOVA. .
Consistent with GFP inhibition, the delivery of POCM-NPs@siRNAcircBBS9 on day 3 resulted in significantly greater regulation of the circBBS9/miR-423-3p/Traf6 axis than delivery of POCM-NPs@siRNAcircBBS9 or POCM-NPs@siRNANC (Fig. 7, C to E). TRAP staining revealed that after the silencing of circBBS9, the number and size of mOCs were decreased; consequently, the bone resorption ability was reduced. However, an appreciable quantity of mononuclear pOCs remained (Fig. 7, F and G). As shown in Fig. 7 (H to K), RANKL-induced up-regulation of mOC-specific marker genes was suppressed by treatment with POCM-NPs@siRNAcircBBS9. MM-NPs@siRNAcircBBS9 were applied as controls and failed to inhibit OC multinucleation. Bovine bone slice resorption assays also showed that the POCM-NPs@siRNAcircBBS9 hampered the bone resorption ability of OC (Fig. 7, L and M). Moreover, shRNAcircBBS9 was applied for genome-integrated knockdown, which resulted in suppression of mOC formation via the circBSS9/miR-423-3p/Traf6 axis similar to that elicited by siRNAcircBBS9 (fig. S10, A to I). These data suggested that the POCM-NPs enabled siRNA/shRNAcircBBS9 to achieve spatiotemporally selective knockdown that maintained the balance between mOC inhibition and pOC protection.
Homotypic targeting of POCM-NPs@siRNAcircBBS9
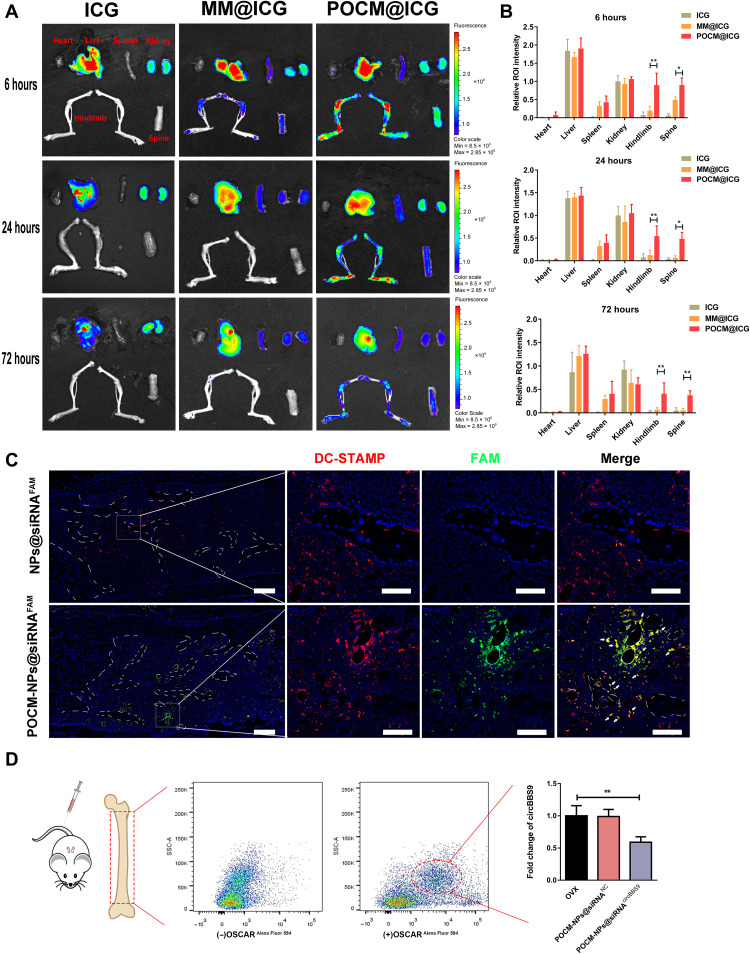
After verifying that POCM-NPs@siRNA/shRNAcircBBS9 exhibited efficient and selective delivery in vitro, we further explored their pOC homing ability in vivo. To evaluate the biodistribution of POCM-NPs, we injected free indocyanine green (ICG) solution, ICG-labeled MM@ICG, and POCM@ICG into 18 mice (three groups with 6 mice per group). All of the mice were euthanized at 6, 24, or 72 hours after injection. Their feet, spines, livers, kidneys, spleens, and hearts were collected to test the fluorescence intensity (Fig. 8, A and B). POCM@ICG significantly accumulated in the hindlimb and spine at 6 hours, and the accumulation was still evident at 72 hours, exhibiting a pronounced bone homing ability. In contrast, MM@ICG exhibited the lowest penetration due to lack of pOCs homotypic targeting. The high-degree accumulation at all time points in the liver and kidney suggested that these NPs were liver-dependent elimination of POCM-NPs (Fig. 8A). The quantitative analysis of the fluorescence signals further confirmed the ability for POCM to be recruited at the bone compared to MM (Fig. 8B). Next, FAM-labeled siRNA was used to explore the pOC-specific binding of POCM, POCM-NPs@siRNAFAM and NPs@siRNAFAM were injected into OVX mice, and the femurs of the mice were extracted for IF analysis. An anti–DC-STAMP antibody was applied to identify the pOCs in the bone marrow and on the bone surface (37). As shown in Fig. 8C, the green puncta (FAM) overlapped with the red puncta (DC-STAMP) both on the bone surface and in the bone marrow, implying the pOC targeting of POCM-NPs. The pOC-specific knockdown in vivo was further explored. Three-month-old OVX mice were intravenously injected twice with POCM-NPs@siRNAcircBBS9 or POCM-NPs@siRNANC 2 weeks after ovariectomy. Then, the mice were euthanized 5 days later, and their bone marrow cells were extracted. OSCAR+ cells deemed pOCs in bone marrow were sorted by FACS. The expression of circBBS9 was distinctly decreased in pOCs treated with POCM-NPs@siRNAcircBBS9 but not in pOCs treated with POCM-NPs@siRNANC (Fig. 8D). Collectively, the results of radiography, histological examination, and FACS suggested that POCM-NPs@siRNAcircBBS9 were able to efficiently deliver siRNA into pOCs in vivo.
Fig. 8. Homotypic targeting of POCM-NPs@siRNAcircBBS9.
(A to B) Representative images of the biodistribution of different NPs in the heart, liver, spleen, kidneys, hindlimb, and spine 6, 24, and 72 hours after intravenous administration of ICG solution, MM@ICG, or POCM@ICG. ROI, region of interest. (C) Fluorescence images of bone sections stained with DC-STAMP antibody after treatment with POCM-NPs@siRNAFAM or NPs@siRNAFAM. Scale bars, 200 μm. Enlarged scale bars, 50 μm. FAM, green; nuclei, blue; DC-STAMP, red. (D) pOCs in bone marrow were selected by OSCAR antibody. The expression of circBBS9 in pOCs was measured by RT-qPCR in OSCAR+ cells. *P < 0.05 and **P < 0.01. The values and error bars are the means ± SEMs. All data were analyzed using unpaired Student’s t tests.
Therapeutic effect of POCM-NPs@siRNAcircBBS9 on osteoporosis
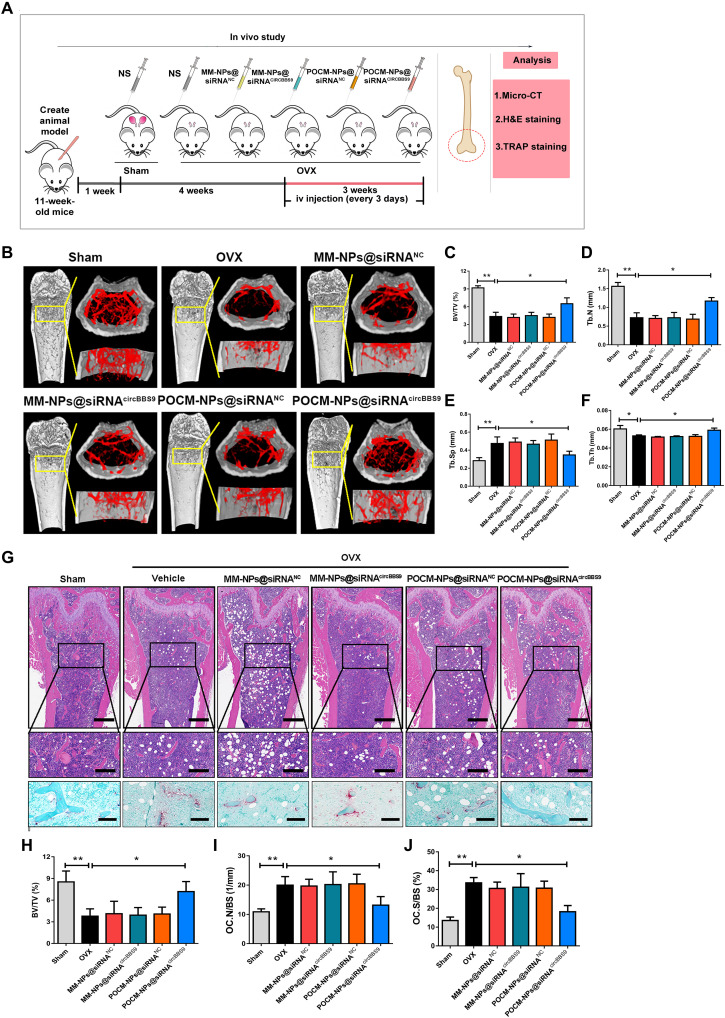
To further investigate the in vivo protective and therapeutic effects, mice with OVX-induced osteoporosis or sham-operated mice were treated with NPs beginning 1 week or 4 weeks after surgery (38, 39). The OVX mice were intravenously injected with saline, MM-NPs@siRNANC, MM-NPs@siRNAcircBBS9, POCM-NPs@siRNANC, or POCM-NPs@siRNAcircBBS9 every 3 days for a total of six injections to evaluate the therapeutic effects of these formulations. All mice were euthanized 8 weeks after surgery (Fig. 9A). Micro–computed tomography (micro-CT) showed that, compared to the OVX group, the group intravenously injected with POCM-NPs@siRNAcircBBS9 exhibited higher bone volume/total volume (BV/TV), trabecular thickness (Tb.Th), and trabecular number (Tb.N) values but a lower relative trabecular separation (Tb.Sp) value (Fig. 9, B to F). Micro-CT suggested that MM-NPs@siRNAcircBBS9, MM-NPs@siRNANC, and POCM-NPs@siRNANC could not inhibit the decrease in bone mass effectively, whereas POCM-NPs@siRNAcircBBS9 stopped the loss of bone mass in both regimens. Hematoxylin and eosin (H&E) and TRAP staining showed that the OVX procedure increased the number of OCs in vivo, while POCM-NPs@siRNAcircBBS9 but not MM-NPs@siRNANC, MM-NPs@siRNAcircBBS9, and POCM-NPs@siRNANC reversed this trend. In addition, the values of several bone parameters, such as the percentage of the bone surface covered with OCs (Oc.S/BS) and OC number (N.Oc/BS), were also markedly diminished under treatment with POCM-NPs@siRNAcircBBS9 compared to the other treatment (Fig. 9, G to J). The treatments started at 1 week after OVX also showed the similar effect of 5 weeks after OVX, suggesting the protective ability of POCM-NPs@siRNAcircBBS9 (fig. S11).
Fig. 9. Antiosteoporosis effect of POCM-NPs@siRNAcircBBS9.
(A) Schematic illustration of the process and group division of animal experiments. H&E, hematoxylin and eosin; micro-CT, micro–computed tomography. (B) Micro-CT evaluation of the femur and tibia bone masses in each group (n = 6 samples for each group). (C to F) Quantitative measurements of bone microstructure-related parameters, such as BV/TV, Tb.Sp, Tb.N, and Tb.Th, in the sham, OVX, MM-NPs@siRNANC, MM-NPs@siRNAcircBBS9, POCM-NPs@siRNANC, and POCM-NPs@siRNAcircBBS9 groups. (G to J) H&E and TRAP staining images and quantitative statistics of BV/TV, OC.N/BS, and OC.S/BS values in each group. Scale bars, 400 μm. Enlarged scale bars, 200 μm. *P < 0.05 and **P < 0.01. The values and error bars are the means ± SEMs. All data were analyzed using unpaired Student’s t tests.
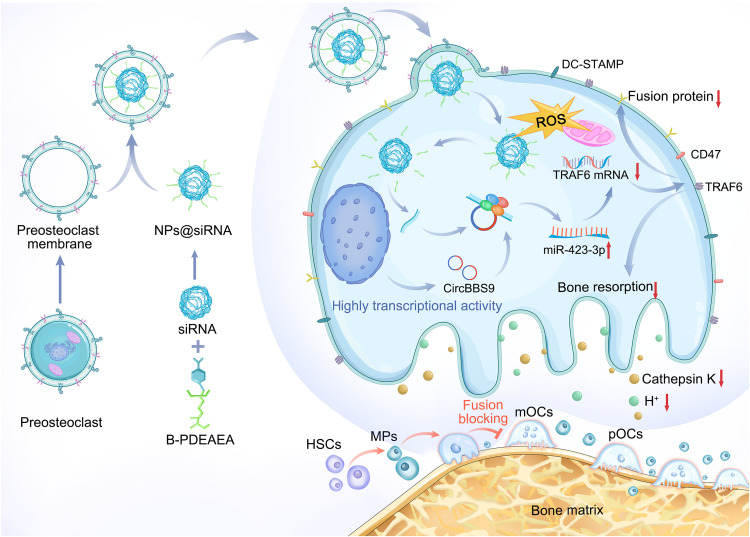
Collectively, the results of these experiments demonstrate that the POCM-NPs@siRNA/shRNAcircBBS9 spatiotemporally selective delivery system achieves stable bone targeting and antiosteoporosis effects, as shown in the schematic diagram (Fig. 10).
Fig. 10. Schematic illustration of the mechanism of circBBS9 and its associated delivery systems.
siRNAcircBBS9 was encapsulated with B-PDEAEA to form NPs. The POCM was extracted to camouflage the NPs, and then POCM-NPs@siRNAcircBBS9 were recruited to pOCs in the bone microenvironment. Upon recognition of the POCMs and fusion into pOCs, the intermediate forms in the OC lineage, the NPs entered the cytosol and released the siRNA in a ROS-triggered manner. The siRNAcircBBS9 down-regulated the expression of circBBS9 via the RISC, leading to inhibition of OC multinucleation and bone resorption via the circBBS9/miR-423-3p/Traf6 axis. HSCs, hematopoietic stem cells.
DISCUSSION
Cell fusion–mediated multinucleation plays a vital role in reproduction and development. Under normal physiological conditions, skeletal muscle cells exist in a fused multinucleated syncytium to control movement. However, excessive multinucleation often leads to an unstable internal environment. Disease-related multinucleation, including inflammation in response to foreign material, bacterial infection, and the cancer, progression is a process of excessive cell fusion to perform a pathological function. With increasing research using high-resolution sequencing and imaging techniques, the development of OCs has been clarified both in vivo and in vitro. Recent studies have revealed that OCs are recycled by fusion and fission to maintain their dynamic bone resorption activity in vivo (40). Single-cell RNA-seq has indicated that the stages of RANKL-induced OC differentiation in vitro can be divided into MPs, the MP stage, the DC-like precursor stages, membrane raft assembly, the initiation of RNA metabolism, the terminal differentiation of pOCs, and the formation of mOCs according to the expression of specific gene markers (13). In addition, while mononuclear OCs have a certain bone resorption function, they can also maintain normal endocrine functions (33). These new perspectives on OCs suggest that multinucleation inhibitors with pOC selectivity are potential agents for treatment of osteolytic diseases. This key concept can even be extended to other multinucleation-related diseases.
Our data show that, during multinucleation, cells have high transcriptional activity that lays the foundation for complex biological processes such as fusion and function in mature multinucleated cells. Therefore, posttranscriptional regulation at the mononucleated stage to restrain terminal multinucleation may be an ideal way to treat multinucleation-related diseases. An increasing number of circRNA functions have been revealed in the development of various cell types, and circRNAs have been shown to be ideal targets to regulate these processes (22). As determined by experiments using natural miRNA sponges, circRNAs are embedded in regulatory networks of ncRNAs and mRNAs in osteoporosis (24, 41). In this study, we identified a novel OC-promoting circRNA called circBBS9 in OVX mice and its conserved circRNA, hsa_circ_0134188, in patients with osteoporosis. To achieve the best bone resorption–inhibiting effect and preserve certain pOCs, we limited the timing of siRNA transfection to day 3. Because of the susceptibility of siRNAs to nuclease degradation (42), the abundance of siRNAcircBBS9 after transfection on day 0 may have been insufficient to achieve a therapeutic concentration when circBBS9 was expressed in pOCs. Moreover, the miR-423-3p/Traf6 axis–mediated inhibitory effect relies on TRAF6, the major regulator of RANKL signaling, which dominates the key stage of mOC differentiation (43). Knockdown of circBBS9 at day 5 failed to inhibit bone resorption. This result indicates that circBBS9 acts as a multinucleation inhibitor rather than a bone resorption suppressor. Although siRNAcircBBS9 is considered a precise regulator in pOCs, an appropriate tool to enable practical delivery is urgently needed.
Various engineering strategies in the field of in vivo exogenous RNA delivery have been developed to overcome challenges such as efficient distribution to targeted tissue, cellular uptake by specific cells, and timely release in the cytosol or nucleus. Cell membrane–coated NP delivery systems have attracted rapidly increasing attention. Membrane components, such as membrane receptor proteins, adhesion proteins, and fusion partners, perform complex cell functions (44), which has led to the development of a series of cell membrane–coated NPs in different fields (29). The major strengths of the spatiotemporally selective delivery system in this work can be split into two parts. Spatial selectivity is defined by the persistent circulation and substantial distribution of pOCs. Temporal selectivity is defined by the selective binding to pOCs and the release of cargoes during the process of OC multinucleation. The specific protein CD47 is widely expressed in monocytes and can bind to signal regulatory protein (SIRP) to enable cells to escape phagocytosis by macrophages (45). This property is inherited by pOCs, allowing the long-term circulation of POCM-NPs (46). Another prerequisite for effective pOCs targeting is adequate POCM-NPs perfusion of bone. Bone tissue is highly vascularized, and there are numerous fenestrations in the vessel walls that increase POCM-NP permeability in this region (47). Moreover, chemokines, such as CXCL12, are highly secreted by specific stromal cells enriched in perivascular regions in the bone marrow cavity and recruit NPs with CXCR4 to achieve bone homing (48). These distinct features of the bone microenvironment are assumed to contribute to the delivery of POCM-NPs.
In our study, POCM-NPs were preferentially taken up by pOCs instead of mOCs, which was fortuitous. This homogeneous mode was different from the normal pattern by which pOCs join mOCs (49, 50). Studies have found that large differentiated mOCs (>10 nuclei) express higher levels of bone-resorbing proteins such as integrins αv and β3, cathepsin K, matrix metalloproteinase–9, and RANK than small mOCs (<5 nuclei) but lower levels of the fusion-related molecule SIRP (51). Rather than inhibiting macrophage-mediated phagocytosis, SIRP promotes OC fusion and plays an indispensable role. As a receptor, SIRP is connected to CD47 and MAC-1 and mediates adhesion and fusion between cells. In view of the reduced expression of SIRP in multinucleated OCs, we speculate that SIRP may be an efficient molecule; that is, it may be partly responsible for the efficient fusion of POCMs after recognition (52, 53).
Here, negatively charged POCMs were fusogenically internalized by pOCs, allowing both siRNA and shRNA to bypass endocytosis and lysosomal degeneration for successful delivery. We suppose that the intracellular uptake of POCM-NPs is similar to that of liposomes, which fuse with the membrane and then release their contents into the cytosol (54). These results reduce our concerns about lysosomal inhibition of RNA treatment efficacy (55). Both shRNA and siRNA exhibited significant enrichment in vitro, implying that POCM-NPs were able to reach the cytosol and nucleus. These unique targeting properties provide a spatiotemporally selective delivery system for the regulation of mOC formation and function.
In conclusion, on the basis of the different properties of each spatiotemporally specific cell type, we have established a proof of concept for customization of membrane shells and elaboration of the posttranscriptional regulation of multinucleated cell lines. This concept will undoubtedly yield precise therapeutics.
MATERIALS AND METHODS
Cell culture
All animal and human experiments used were approved by the Institutional Animal Ethics Committee of Sir Run Run Shaw Hospital of Zhejiang University (SRRSH20200321 and 20210817-33). Human and animal subjects were followed with Institutional Review Board and Institutional Animal Care and Use Committee guidelines. Fresh BMMs from 6- to 8-week-old C57BL/6J mice were isolated as reported (56). Briefly, the tibia- and femur-derived bone marrow was flushed and then cultured in α-minimum essential medium (α-MEM) supplemented with 10% FBS (Gibco, USA) and 1% penicillin-streptomycin (complete α-MEM). To obtain pure BMMs, nonadherent cells were collected and cultured in complete α-MEM plus M-CSF (25 ng/ml) (R&D Systems, USA). After 3 days of culture, the attached cells were used for experiments. Then, BMMs were stimulated with M-CSF and RANKL (50 ng/ml) (R&D Systems, USA) for 3 days to obtain pOCs and 5 days to obtain mOCs. C2C12 cells, RAW 264.7 cells, 3T3 cells, and MSCs were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences, Shanghai Institute of Cell Biology, Chinese Academy of Sciences. HUVECs and Jurkat (human T lymphocytes) cells were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd. GFP-expressing RAW264.7 (RAW/GFP) cells were purchased from Shanghai Jisidenuo Biological Technology Co., Ltd. C2C12 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) containing 10% FBS and supplemented with 1% penicillin-streptomycin and were differentiated by switching the medium to DMEM with 2% horse serum (Solarbio Life Science, Tongzhou District, Beijing, China) with antibiotics. RAW264.7 cells were cultured in RPMI 1640 (Gibco, USA) supplemented with 10% FBS and 1% penicillin-streptomycin. MSCs and RAW/GFP cells were cultured in DMEM (Gibco, USA) supplemented with 10% FBS and 1% penicillin-streptomycin. 3T3 cells were cultured in complete α-MEM. HUVECs were cultured in endothelial cell medium (ScienCell, Carlsbad, CA, USA) supplemented with 5% FBS (ScienCell, Carlsbad, CA, USA), 1% ECGS (ScienCell, Carlsbad, CA, USA), and antibiotics [penicillin (100 U/ml), ScienCell and streptomycin (100 mg/ml), Carlsbad, CA, USA]. Jurkat cells were maintained in DMEM (Gibco, USA) supplemented with 10% FBS and 1% penicillin-streptomycin. All cells were expanded and maintained in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Monocyte isolation, macrophage differentiation, and OC generation
The relevant experiments were performed as reported (57). Briefly, human whole blood was mixed with an equal volume of PBS, and PBMCs were isolated with a kit for human mononuclear cell isolation (catalog no. P8680; Solarbio Life Science, Tongzhou District, Beijing, China). The samples were centrifuged for 20 min at 800g and room temperature with the acceleration set to 3 and the deceleration set to 2. The intermediate buffy coat layers containing PBMCs were washed three times with PBS by centrifuging for 5 min at 250g and room temperature with the acceleration set to 9 and the deceleration set to 5. Thereafter, the PBMCs were counted and differentiated into macrophages by adding macrophage differentiation medium. The cells were differentiated for up to 7 days with one medium change. For OC differentiation, cells were seeded at a density of 2.5 × 106 cells per well in six-well cell culture plates and differentiated toward OCs for up to 14 days. The medium was changed twice per week. The cells were cultured in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
RNA polymerase II and actin-ring staining
At the end of the culture period, the cells were fixed with 4% paraformaldehyde (PFA) for 20 min. Next, the cells were washed in PBS three times for 2 min each. The cells were incubated with 5% BSA in PBS containing 0.1% Triton X-100 at 37°C for 30 min and then incubated overnight at 4°C with a primary antibody against RNA polymerase II (1:100, Abcam). The cells were washed three times with PBS and incubated with Alexa Fluor 488 donkey anti-rabbit secondary antibodies (1:1000) for 1 hour at 37°C. The cells were rinsed three times with PBS and incubated with DAPI Fluoromount-G for 10 min (Yeasen, Shanghai). For staining of F-actin, rhodamine-coupled phalloidin was used (Yeasen, Shanghai). All images were captured on a confocal fluorescence microscope (Nikon, A1 PLUS, Tokyo, Japan).
RNA-seq and bioinformatic analysis
Total RNA of BMMs and pOCs (1 × 106 per well) was extracted in accordance with the TRIzol reagent (Invitrogen, Shanghai, China) instructions. Following library preparation according to the manufacturer’s protocol, paired-end sequencing was performed on an Illumina HiSeq 4000 (LC Bio, China) in accordance with the vendor’s recommended protocol. The numbers of reads mapped to each gene and circRNA are presented as the fragments per kilobase of transcript per million fragments mapped values. The differentially expressed circRNAs were selected with thresholds of a log2 (fold change) >1 or log2 (fold change) <−1 and statistical significance (P < 0.05). The RNA-seq data included in this study are in data file S1. GSEA was performed to identify the significant pathways and gene sets between control and RANKL-treated pOCs according to the normalized gene expression using the GSEA v4.1.0 tool (www.broad.mit.edu/gsea/). The Kolmogorov-Smirnov test was applied to analyze the differences between the two groups.
TRAP staining
Cells were stained for TRAP using an acid phosphatase leukocyte diagnostic kit (Cosmo Bio, CA, USA) after fixation with 4% PFA as recommended by the manufacturer. Multinucleated OCs (>3 nuclei) and mononucleated TRAP-positive cells were detected in duplicate wells of 48-well plates. Micrographs of staining were observed at the original magnification ×100.
Bone slice resorption assay
Freeze-dried beef bone slices (Φ 0.5 mm by 6 mm) washed with 0.9% NaCl solution three times were used to detect bone resorption capacity. BMMs at a density of 2 × 105 cells per well were cultured in six-well collagen-coated plates (Corning Inc., Corning, NY, USA) and stimulated with M-CSF (25 ng/ml) in complete α-MEM. Then, RANKL (50 ng/ml) was applied to each well to stimulate osteoclastogenesis at 0, 1, 3, and 5 days. For bare siRNA/shRNA, POCM-NPs, and MM-NPs, the cells were stimulated with the POCM-NP@siRNA/shRNAcircBBS9/NC or MM-NP@siRNA/shRNAcircBBS9/NC solution (5.0 μg per well) in the presence of siRNA or shRNA (5.0 μg per well) for 4 hours. The medium was then replaced, and the cells were incubated for another 48 hours. After dissociation of the cells using cell dissociation solution (Sigma-Aldrich, Australia), the cells on the surfaces of the bone tablets were scraped off with a brush. A scanning electron microscope (TM-1000, Hitachi, Tokyo, Japan) was used to observe bone slices sputter-coated with Au-Pd to obtain images as previously described (58).
Real-time quantitative polymerase chain reaction
According to the manufacturer’s protocol, an RNA kit was used (Qiagen, Valencia, CA, USA) to isolate total RNA from the cells. Reverse transcription reagents (Promega, Madison, WI, USA) were used to reverse transcribe RNA into complementary DNA. The RT-qPCR system (catalog no. A6002, Promega, WI, USA) was applied for qPCR according to the manufacturer’s instructions. The relative gene expression was quantified by a Rotor-Gene Q (Qiagen, Hilden, Germany) RT-qPCR system. For the RNase R assay, total RNA was incubated with or without RNase R Geneseed (3 U/mg) (Guangzhou, China) and detected by RT-qPCR. Divergent primers were designed to amplify circBBS9. The amplification products were detected by agarose gel electrophoresis and sequencing. For circRNA and mRNA, Gapdh was used as an internal control, and U6 was used as an internal control of miRNA. siRNAcircBBS9-treated and nontreated BMMs were stimulated with lipopolysaccharide and interleukin-4 and then assayed mRNA by RT-qPCR. RT PCR primers were purchased from Sangon Biotech (Shanghai, China). All primers are listed in table S1.
Isolation of total RNA from human surgical trabecular bone
Total RNA was extracted from the surgical trabecular bone as previously described (59). Briefly, the spine cancellous bones of osteoporosis and normal patients were sawed in pieces of 0.5 cm by 0.5 cm by 0.1 cm. The cancellous fragments were pestled in nitrogen, and the pulverized bone was used for RNA extraction with TRIzol. The total RNA was isolated using the RNA kit (Qiagen, Valencia, CA, USA). The nucleic acid purity and RNA quantity were measured by the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA), and samples meeting quality control were used for RT-qPCR experiments. Informed consent for using these specimens was obtained before surgery.
Knockdown or overexpression of circRNAs, miRNAs, or mRNAs
The siRNAs, shRNAs, overexpression lentiviruses targeting of circBBS9, miRNA mimics, and miRNA inhibitors or sponges were all purchased from GenePharma (Shanghai GenePharma Co., Ltd., China). BMMs stimulated with RANKL were cultured in six-well plates (2 × 105 cells per well) for 0, 1, 3, and 5 days. The cells were treated with siRNAcircBBS9 for 4 hours on the indicated days. For the other knockdown or overexpression experiments, cells were transfected at day 0. Then, the medium was replaced with the corresponding medium containing 10% FBS, and the cells were incubated until 6 days after the initial stimulation with RANKL. For transfection, Lipofectamine 3000 (Invitrogen, CA, USA) was applied according to the manufacturer’s instructions. The efficiency of knockdown or overexpression was detected by WB and RT-qPCR on day 6 after RANKL treatment and by TRAP staining and bone slice resorption assays as mentioned above. All nucleotide sequences are listed in table S2.
Western blot analysis
SDS–polyacrylamide gel electrophoresis and WB were carried out accordingly to detect expression of protein in cell or cell membrane lysate as standard protocol. Antibodies are listed in table S3.
Gene knockdown cell activity experiment
A phagocytosis experiment was performed as reported (60). Briefly, macrophages with or without knockdown were plated on 24-well plates (2 × 105 cells per well) and cultured for 24 hours. BCECF-2’,7’-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein (AM)–stained apoptotic Jurkat cells were added to macrophages at a ratio of 5 to 10:1 and incubated for 2 hours in DMEM containing 2% FBS. The cells were then washed with PBS and fixed in 4% PFA. F-Actin in macrophages was stained with rhodamine-coupled phalloidin (Yeasen, Shanghai) for 1 hour at room temperature. The cells were rinsed three times with PBS and incubated with DAPI Fluoromount-G for 10 min (Yeasen, Shanghai). Images were captured with a confocal fluorescence microscope.
Luciferase reporter assay
pRL Renilla Luciferase Control Reporter Vectors (E2241, Promega, USA), GV272 luciferase reporter vectors with WT or mutated binding sites of miR-423-3p on the TRAF6 3′UTR or circBBS9, and an internal control were cotransfected into human embryonic kidney 293 cells. Control cells were transfected with miR-423-3p mimics or a negative control. After culture for 48 hours, a Dual-Luciferase Reporter Assay Kit (E1910, Promega) was used to perform a luciferase reporter assay on the cell lysates. The relative firefly luciferase activity was evaluated by quantifying the bioluminescence with a Varioskan LUX microplate reader (Thermo Fisher Scientific) and normalizing it to that of Renilla luciferase.
RNA immunoprecipitation
A RIP assay was carried out by using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA). Lysis buffer with an RNase inhibitor and protease inhibitor cocktail was used to lyse cells. The magnetic beads were preincubated with an Ago2 antibody or mouse IgG antibody for 1 hour at room temperature, and the OC lysates were immunoprecipitated with the magnetic beads overnight at 4°C. TRIzol was used to extract RNA for RT-qPCR.
Biotinylated RNA pull-down assay
A biotinylated DNA probe synthesized by GeneChem (Shanghai, China) for circBBS9 (sequence: GAGTACAGCCACCTCCAGAGATGAG) and a negative-control sequence (sequence: CTCATCTCTGGAGGTGGCTGTACTC) were dissolved in deoxyribonuclease-/RNase-free water to 100 μM. The OC lysate was incubated with the probe at 37°C for 4 hours. Then, streptavidin-coated magnetic beads (Dynabeads MyOne Streptavidin C1, 65,001, Invitrogen) were added, and the mixture was incubated for an additional 30 min. The pull-down complex was eluted from the C1 beads with wash buffer, from which RNAs were purified and subjected to RT-qPCR analysis.
ROS quantitative assay
The probe H2DCFDA was applied to examine intracellular ROS levels. Cells were treated with RANKL (50 ng/ml) for 3 days. Then, they were tested with a fluorescent probe (1:1000) and incubated in an incubator for 30 min after being washed and centrifuged carefully. Images were obtained on a confocal fluorescence microscope (Nikon, A1, Japan).
Preparation and characterization of NPs
PDEAEA was synthesized as previously reported (32). Briefly, AIBN (0.048 g, 0.29 mmol or 0.024 g, 0.14 mmol) and DEAEA (5 g, 29 mmol) were placed in a 25-ml flask and deoxygenated at room temperature with nitrogen and then polymerized at 65°C for 24 hours. The viscous liquid was dissolved in CH2Cl2 and precipitated in cold n-hexane. The resulting PDEAEA was purified by reprecipitation three times and dried. PDEAEA (20 K, 0.3 g, 1.8-mmol tertiary amines) was reacted with 4-(bromomethyl)phenylboronic acid (0.56 g, 2.6 mmol) in 20 ml of N,N′-dimethylformamide at room temperature for 24 hours. The resulting solutions were dialyzed overnight against deionized water using a dialysis bag with a molecular weight cutoff of 3500 Da and then lyophilized to afford the product B-PDEAEA (0.5 g, yield 92%). Hepes buffer solution (10 mm, pH 7.4) was used to dilute plasmid DNA, and diethyl pyrocarbonate–treated water was used to dilute siRNA to a concentration of 40 μg/ml. B-PDEAEA was dissolved in Hepes buffer solution (10 mm, pH 7.4) at various concentrations according to the preset N/P ratios (N/P = 5, 10, 15, 20, 25, and 30). Then, equal volumes of shRNA and siRNA solution were added to the B-PDEAEA solution. The mixture was immediately vortexed for 10 s for shRNA or for 1 min for siRNA and then statically incubated for 30 min for shRNA or for 20 min for siRNA to obtain NPs. The sizes and zeta potentials of the polyplexes were measured by dynamic light scattering (DLS) (Malvern, UK). The encapsulation efficiency of polyplexes with various N/P ratios and ROS responsiveness was evaluated by gel retardation assay. The polyplexes were electrophoresed on a 1% agarose gel at 100 V for 30 min. Gel Red (Biotium) was used in the agarose gel for shRNA or siRNA detection. The morphology of polyplexes stained with water-soluble phosphotungstic acid was visualized using a transmission electron microscope (JEM-1400plus).
For ROS responsiveness assessment, siRNA polyplexes with different N/P ratios were treated with 5 mM H2O2 at 37°C for 1 hour and then subjected to electrophoresis. shRNA polyplexes at an N/P ratio of 10 were incubated with the indicated H2O2 concentrations (0, 0.1, 0.25, 0.5, 1, 2.5, and 5 mM) for 1 hour at 37°C. shRNA and bare shRNA incubated with 5 mM H2O2 were used as controls. The solutions were then subjected to electrophoresis as described above.
Isolation of MM and POCM
Macrophages and pOCs were suspended at a density of 5.0 × 106 cells/ml in ice-cold TM buffer [50 mM Tris HCl and 10 mM Magnesium (pH 7.5)] solution and subsequently extruded through a mini-extruder 40 times to disrupt the cells. Sucrose (1 M) was subsequently mixed with the cell homogenate to a final concentration of 0.25 M sucrose, and the mixture was centrifuged at 4°C and 2000g for approximately 10 min. The resulting supernatant was collected via further centrifugation at 3000g for an additional 35 min. The cell membranes were collected and rinsed with ice-cold TM buffer in 0.25 M sucrose for purification. Macrophage and POCM vesicles were obtained by extruding the purified cell membranes through the same set of porous membranes. A bicinchoninic acid (BCA) protein assay was used to analyze the total protein content in the purified MMs and POCMs. The membrane material was stored at −80°C for future study. The protease-digested membrane protein samples from macrophages and pOCs were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) to obtain a raw file of the original MS results. The result of LC-MS/MS is listed in data files S2.
Preparation and characterization of MM-NPs and POCM-NPs
After isolating the MMs and POCMs, the membranes were suspended in Hepes buffer solution (1 to 2 mg/ml) and then added to an equal volume of NPs. Then, the samples were sonicated for 5 min before being extruded through 400- and 200-nm polycarbonate porous membranes with an Avanti mini extruder (Xi’an Ruixi Biological Technology Co., Ltd., China) at least 10 times to obtain MM-coated NPs (MM-NPs) and POCM-coated NPs (POCM-NPs). MM and POCM vesicles were prepared by extruding purified cell membranes through the same set of porous membranes. The expression of the specific surface markers on the POCMs and POCM-NPs was determined by WB. The sizes and zeta potentials of the membrane-coated NPs were measured by DLS. Morphological examination of the membrane-coated NPs was performed using a Talos F200C 200-kV transmission electron microscope (Thermo Fisher Scientific, USA). The coating efficiency of POCMs on NPs was determined by monitoring the fluorescence intensity of POCM-NPs@siRNAFAM or POCMDiO-NPs@siRNAFAM (100 μg/ml) before and after centrifugation (10,000g, 10 min). For a stability study, the samples were stored at room temperature in PBS or the corresponding medium, and size was measured periodically by DLS. A similar method was applied to prepare NPs@siRNAcy5, NPs@siRNAFAM, NPs@siRNAcircbbs9, ICG-loaded MMs, and POCMs by replacing the NPs.
DNA transfection efficiency
For luciferase gene transfection, pGL4.13 (Promega) was used as the plasmid DNA expressing luciferase. Cells were cultured on a 96-well plate at a density of 15,000 cells per well in 200 μl of corresponding medium and incubated overnight to reach 80% confluence. The medium was replaced with fresh medium (containing 0 or 10% FBS). Polyplex solutions (25 μl) and POCM-NPs (50 μl) were added at a dose of 1 μg of siRNA or shRNA per well, and the cells were incubated for 4 hours. Then, the transfection medium was replaced with 200 μl of fresh medium containing 10% FBS. The cells were cultured for an additional 44 hours. The expression level of the luciferase plasmid was determined according to the standard protocol illustrated in the manufacturer’s manual (Promega). The protein content of the cell lysis solution was determined with a BCA assay kit. The luciferase activity was normalized with respect to the protein concentration (relative luciferase luminescence units per milligram of protein).
GFP knockdown assay
For GFP silencing research, RAW/GFP cells were used as a model. Bare siRNAGFP, siRNAGFP with a different volume of Lipofectamine 3000, POCM-coated siRNAGFP complexes with different N/P ratios, MM-NPs@siRNAGFP, and POCM-NPs@siRNAGFP were delivered to cells as described above. GFP silencing was analyzed by using confocal microscopy and ImageJ software to calculate the reduction in green fluorescence of GFP.
Cell Counting Kit-8 (CCK-8) assay
The cytotoxicity of siRNAcircBBS9 was evaluated by CCK-8 assay using BMMs. Generally, the cells were cultured at a density of 1 × 104 cells per well in a 96-well plate in 200 μl of the corresponding medium. After 1 day of incubation, the cells were transfected with siRNAcircBBS9 and cultured for 48 hours. When that incubation was complete, the medium was replaced with 110 μl of fresh medium containing 10 μl of CCK-8 solution. After an additional 4 hours of incubation, the absorbance of each well at 450 nm was measured with a microplate reader (Thermo Fisher Scientific, USA).
Cellular uptake assay
The cellular uptake of NPs were examined by confocal microscopy and flow cytometry for 3T3 cells, MSCs, HUVECs, macrophages, pOCs, and mOCs. shRNA was labeled with Cy5 (Mirus Bio) according to the manufacturer’s instructions, and fluorescein-tagged negative-control siRNA (siRNAcy5) (Shanghai GenePharma Co., Ltd., China) was mixed with B-PDEAEA at 10 N/P ratios before use, respectively. MM-NPs and POCM-NPs were prepared as described above. Cells were seeded onto glass-bottom petri dishes at 50,000 cells per dish in 1.5 ml of cell culture medium before use. Then, the medium was replaced with fresh medium. Solutions (50 μl) of polyplexes, MM-NPs, and POCM-NPs were added at a dose of 1 μg of siRNA or shRNA per dish. After a timed incubation, the nuclei were stained with two drops of Hoechst 33342 (Molecular Probes, Carlsbad, CA) per milliliter of medium for 15 min. Then, cells were analyzed using flow cytometry to obtain the Cy5-positive cell percentage (10,000 cells were counted per treatment). Images were obtained on a confocal fluorescence microscope (Nikon, A1, Japan). Fluorescence measurements were analyzed with ImageJ software as previously reported.
Fusogenic uptake of POCMs and pOCs
The POCMs were labeled with DiI for 15 min. DiI-POCMs were isolated by centrifugation at 10,000g for 5 min, washed twice with PBS, and resuspended in PBS at 1 mg/ml. pOCs were cultured as described above in glass-bottom petri dishes at a density of 50,000 cells per dish before use. pOCs were labeled with DiO for 15 min. After being washed twice with PBS, the cells were incubated with complete medium. Then, DiI-POCM solution was added to the medium, and the mixture was incubated for 1 hour. Images were acquired on a confocal fluorescence microscope (Nikon, A1, Japan).
Effects of endocytosis inhibitors on cellular uptake
pOCs were seeded in six-well plates at a density of 200,000 cells per well in 1.5 ml of medium and cultured overnight. The medium was then replaced with 1.5 ml of fresh medium. Cytochalasin D (an actin polymerization inhibitor), wortmannin (a phosphatidylinositol 3-kinase-mediated micropinocytosis inhibitor), filipin III (a caveolae-mediated endocytosis inhibitor), and chlorpromazine (a clathrin-mediated endocytosis inhibitor) were separately added to the medium at concentrations of 5, 5, 7.5, and 50 μM, respectively. After a 30-min incubation, polyplex and POCM-NP solutions were added at a dose of 30 ng of Cy5-labeled siRNA (siRNAcy5) per well and cultured with the cells for 2 hours. The medium was removed, and the cells were washed with PBS, detached with trypsin, isolated, rinsed three times with PBS, resuspended in PBS, and immediately analyzed using flow cytometry to obtain the Cy5-positive cell percentage (10,000 cells were counted per treatment). The effects of inhibitors on luciferase gene transfection were assessed as described below.
Cells were incubated in 96-well plates at a density of 15,000 cells per well in 0.2 ml of medium overnight until they reached 70 to 80% cell confluence. The medium was then replaced with fresh medium. The cells were pretreated with cytochalasin D, wortmannin, filipin III, or chlorpromazine for 30 min at the concentrations indicated above; polyplex solutions were added at a dose of 0.5 μg DNA per well; and the cells were cultured following the steps described above.
Subcellular distribution analysis
FITC-B-PDEAEA was synthesized as previously described (32). siRNAcy5 was complexed with FITC-B-PDEAEA at an N/P ratio of 10 at room temperature for 20 min before use, and POCM-NPs were prepared as described above. Then, POCM-NPs were labeled with DiI at a 1/1000 dilution for 15 min to obtain POCMDiI-NPs. Solutions of POCM-NPs labeled with different fluorescent dyes were added at a dose of 1 μg of siRNA per dish. After timed incubation, the medium was replaced with fresh medium, and then the nuclei were stained with Hoechst 33342 for 15 min. Cells were washed and observed by confocal fluorescence microscopy.
To label the lysosomes, the cells were further incubated with LysoTracker Green (Molecular Probes, Carlsbad, CA) at a concentration of 200 nM for 15 min after incubation of POCM-NPs@siRNA for 1, 2, 3,or 6 hours and POCM-NPs@shRNA for 1 and 6 hours, respectively, and then the nuclei were stained with two drops of Hoechst 33342 per milliliter of medium for 15 min. The cells were rinsed three times with PBS before observation by confocal fluorescence microscopy. The peak of fluorescence of lysosomes and siRNA was analyzed by ImageJ.
Inhibition of circBBS9, miR-423-3p, Traf6, and OC-related gene expression by POCM-NPs and MM-NPs
RT-qPCR was used to analyze the expression of genes related to circBBS9. BMMs stimulated with RANKL were cultured in six-well plates (2 × 105 cells per well) for 3 days. The cells were treated with the different polyplexes for 4 hours. Then, the medium was replaced with the corresponding medium containing 10% FBS, and the cells were incubated for another 48 hours. The expression of circBBS9, miR-423-3P, Traf6, and OC-related genes was detected by RT-qPCR. Bone slice resorption assays and TRAP staining were also performed to examine the function of mOCs.
In vivo bone targeting experiment
The specific bone binding of POCMs was confirmed using an IVIS Spectrum In Vivo Imaging System. ICG-loaded MMs or POCMs were prepared as described above. Mice were intravenously injected with ICG, ICG-MM, and ICG-POCM solutions (10 μl/g) into the tail vein. The biodistribution analysis of ICG was performed with a 710-nm excitation wavelength and a 785-nm filter at 6 hours, 1 day, and 3 days after injection. Furthermore, the heart, liver, spleen, kidneys, spine, and hindlimb were harvested and analyzed in vitro for the distribution of ICG-MMs and ICG-POCMs. Living Image software was used to quantify the fluorescence intensity.
To further prove the specific bone binding of POCMs, MM-NPs@siRNAFAM and POCM-NPs@siRNAFAM were injected into the tail veins of mice 2 weeks after OVX. Bone tissue sections were prepared to evaluate the distributions of MMs and POCMs in vivo. Briefly, femur samples were obtained 1 day after injection. All femurs were postfixed in 4% PFA for 1 day, decalcified in 14% EDTA for 12 hours at 37°C, and embedded in paraffin for sectioning. They were then incubated with DC-STAMP primary antibodies overnight at 4°C. After primary antibody incubation, the sections were washed with PBS with Tween 20 (PBST) for 4 × 5 min and then incubated with Alexa Fluor 594 goat anti-rabbit secondary antibodies for 1 hour at room temperature. The sections were rinsed three times with PBST and incubated with DAPI Fluoromount-G for 10 min. The images were captured with a confocal fluorescence microscope.
To determine the knockdown of circBBS9 in vivo, the tibia- and femur-derived bone marrow from mice 2 weeks after OVX was flushed after 5 days of twice injection of POCM-NPs@siRNANC or POCM-NPs@siRNAcircBBS9 and blocked with 5% BSA in PBS for 30 min. Then, the cells were stained with OSCAR (MA5-33374, Invitrogen) for 30 min at 4°C, washed with PBS three times, and incubated with Alexa Fluor 594 goat anti-rat secondary antibodies (ab150160, Abcam) for 1 hour. FACS was performed using a FACSAria III sorter and FACSDiva software (BD Biosciences). pOCs gated as OSCAR+ were sorted for RT-qPCR.
OVX-induced osteoporosis mouse model
As shown in Fig. 9A and fig. S11A, C57BL/6J mice (80 in total) were randomly divided into 10 groups: (i) the sham group, (ii) the OVX treatment group, (iii) the OVX plus MM-NP@siRNANC group, (iv) the OVX plus MM-NP@siRNAcircBBS9 group, (v) the OVX plus POCM-NP@siRNANC group, (vi) the OVX plus POCM-NP@siRNAcircBBS9 group, and (vii to x) the same drug of (iii) to (vi) with different administration times (n = 6 per group).Then, the mice in groups (ii) to (x) underwent a bilateral ovariectomy under chloral hydrate anesthesia to induce postmenopausal osteoporosis, while the ovaries of sham group underwent exteriorized but not resected for mice. After 5 weeks of recovery from surgery, the mice of (iii) to (vi) group were given MM-NPs@siRNANC, MM-NPs@siRNAcircBBS9, POCM-NPs@siRNANC, or POCM-NPs@siRNAcircBBS9 at siRNA (0.90 mg/kg) per mouse via tail vein injection. The same administration of NPs above was applied to mice of (vii) to (x) group that had 1 week of recovery from surgery. These mice were administered the same treatment on the basis of their groups every 3 days for 3 weeks, while the mice from the sham and OVX groups received tail injections of saline as a control. Eight weeks after surgery, all mice were humanely euthanized to assess the bone protection effect of POCM-NPs@siRNAcircBBS9.
Micro-CT scanning and analysis
Mice were euthanized to obtain multiple bone tissues, including femur and tibia tissues. After the bone tissues were fixed in 10% neutral buffered formalin for 24 hours, the left femur and left tibia tissues were selected for micro-CT (SkyScan 1275, Bruker, USA) scanning with the following parameters: a 50-kV source voltage, a 450-μA source current, an aluminum 0.5-mm filter, a 9-μm pixel size, and a 0.4° rotation step. Furthermore, the acquired images were reconstructed using NRecon software (Bruker micro-CT, Kontich, Belgium) with the following parameters: a ring artifact correction of 8, a smoothing setting of 2, and a beam hardening correction of 30%. The trabeculae were further analyzed by selecting a refined volume of 1 mm in height in the range of 0.5 mm distal to the vertebral body and the growth plate of the femur or tibia. After setting a constant threshold of 80 to 255, the BV/TV, Tb.N, and Tb.Sp were further analyzed with the CTAn program (Bruker micro-CT, Kontich, Belgium).
Evaluation of bone histomorphometry
The above bone tissues were used for histological assessment and metabolism evaluations after micro-CT analysis. Then, the bone tissues were placed in 14% EDTA (Sigma-Aldrich, Germany) for decalcification at 37°C for 7 days and subsequently embedded in paraffin for sectioning. Furthermore, the obtained sections were separately subjected to TRAP activity staining and H&E staining to assess OC formation and distribution in vivo, as well as trabecular bone distribution in vivo.
BIOQUANT OSTEO software (Bioquant Image Analysis Corporation, Nashville, TN, USA) was then used to quantitatively assess bone tissue morphology from the obtained slice images.
Statistical analysis
All experiments were performed with at least 3 replicates (in the figure legends). All quantitative data are presented as the means ± SD. Furthermore, Tukey’s multiple comparisons test or Student’s t test after ANOVA was applied to assess the statistical significance of our experimental results. A result was considered statistically significant when the P values satisfied one the following conditions: *P < 0.05, **P < 0.01, or ***P < 0.001.
Acknowledgments
Funding: The study was supported by the National Nature Science Fund of China (grant nos. 82072414, 82102313, and 52003234), the Natural Science Fund of Zhejiang Province (grant nos. LQ21H060009, LY21H060002, and LQ21E030007), the Innovative Talent Support Program Project of Zhejiang Provincial Health Commission (grant no. 2021433298), the Public Projects of Zhejiang Province (LGF19H060013), the Zhejiang Provincial Program for the Cultivation of High-Level Innovative Health Talents, and Hangzhou Qianjiang Distinguished Expert and Zhejiang Provincial Program for the Cultivation of High-Level Innovative Health Talents.
Author contributions: Methodology: Q.W., H.W., H.T., Y.W., X. Liu, and R.T. Experiment: Q.W., H.W., H.Y., W.Y., Z.D., P.C., Z.L., and C.J. Supervision: X. Lin and S.F. Writing—original draft: X. Lin, Q.W., H.W., and Y.W. Writing—draft revision and proofreading: Q.W., X. Lin, and X. Liu.
Competing interests: Xianfeng Lin, Qingqing Wang, Haoli Wang are inventors on an invention disclosure of the patents (CN202210471073.3, CN202210470915.3, CN202210471542.1 and CN202210471542.1; filed May 1, 2022) related to this work. Xin Liu is the inventor on an invention disclosure of a patent (CN202210471542.1; filed May 1, 2022) related to this work. The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 to S3
Figs. S1 to S11
Other Supplementary Material for this manuscript includes the following:
Data S1 and S2
REFERENCES AND NOTES
- 1.Maurizi A., Rucci N., The osteoclast in bone metastasis: Player and target. Cancers (Basel) 10, 218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrtwich L., Nanda I., Evangelou K., Nikolova T., Horn V., Sagar, Erny D., Stefanowski J., Rogell L., Klein C., Gharun K., Follo M., Seidl M., Kremer B., Münke N., Senges J., Fliegauf M., Aschman T., Pfeifer D., Sarrazin S., Sieweke M. H., Wagner D., Dierks C., Haaf T., Ness T., Zaiss M. M., Voll R. E., Deshmukh S. D., Prinz M., Goldmann T., Hölscher C., Hauser A. E., Lopez-Contreras A. J., Grün D., Gorgoulis V., Diefenbach A., Henneke P., Triantafyllopoulou A., DNA damage signaling instructs polyploid macrophage fate in granulomas. Cell 167, 1264–1280.e18 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Bracq L., Xie M., Benichou S., Bouchet J., Mechanisms for cell-to-cell transmission of HIV-1. Front. Immunol. 9, 260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson J. M., Rodriguez A., Chang D. T., Foreign body reaction to biomaterials. Semin. Immunol. 20, 86–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers T. J., The birth of the osteoclast. Ann. N. Y. Acad. Sci. 1192, 19–26 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Boyle W. J., Simonet W. S., Lacey D. L., Osteoclast differentiation and activation. Nature 423, 337–342 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Boissy P., Saltel F., Bouniol C., Jurdic P., Machuca-Gayet I., Transcriptional activity of nuclei in multinucleated osteoclasts and its modulation by calcitonin. Endocrinology 143, 1913–1921 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Roodman G. D., Windle J. J., Paget disease of bone. J. Clin. Invest. 115, 200–208 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baum R., Gravallese E. M., Bone as a target organ in rheumatic disease: Impact on osteoclasts and osteoblasts. Clin Rev Allergy Immunol 51, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers M. J., Crockett J. C., Coxon F. P., Mönkkönen J., Biochemical and molecular mechanisms of action of bisphosphonates. Bone 49, 34–41 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Armamento-Villareal R., Napoli N., Panwar V., Novack D., Suppressed bone turnover during alendronate therapy for high-turnover osteoporosis. N. Engl. J. Med. 355, 2048–2050 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Black D. M., Geiger E. J., Eastell R., Vittinghoff E., Li B. H., Ryan D. S., Dell R. M., Adams A. L., Atypical femur fracture risk versus fragility fracture prevention with bisphosphonates. N. Engl. J. Med. 383, 743–753 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukasaki M., Huynh N. C.-N., Okamoto K., Muro R., Terashima A., Kurikawa Y., Komatsu N., Pluemsakunthai W., Nitta T., Abe T., Kiyonari H., Okamura T., Sakai M., Matsukawa T., Matsumoto M., Kobayashi Y., Penninger J. M., Takayanagi H., Stepwise cell fate decision pathways during osteoclastogenesis at single-cell resolution. Nat. Metab. 2, 1382–1390 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Dou C., Ding N., Luo F., Hou T., Cao Z., Bai Y., Liu C., Xu J., Dong S., Graphene-based MicroRNA transfection blocks preosteoclast fusion to increase bone formation and vascularization. Adv. Sci. (Weinh) 5, 1700578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kodama J., Kaito T., Osteoclast multinucleation: Review of current literature. Int. J. Mol. Sci. 21, 5685 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dou C., Cao Z., Yang B., Ding N., Hou T., Luo F., Kang F., Li J., Yang X., Jiang H., Xiang J., Quan H., Xu J., Dong S., Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci. Rep. 6, 21499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beg M. S., Brenner A. J., Sachdev J., Borad M., Kang Y.-K., Stoudemire J., Smith S., Bader A. G., Kim S., Hong D. S., Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 35, 180–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reid G., Pel M. E., Kirschner M. B., Cheng Y. Y., Mugridge N., Weiss J., Williams M., Wright C., Edelman J. J. B., Vallely M. P., McCaughan B. C., Klebe S., Brahmbhatt H., MacDiarmid J. A., van Zandwijk N., Restoring expression of miR-16: A novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. 24, 3128–3135 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Khan A. A., Betel D., Miller M. L., Sander C., Leslie C. S., Marks D. S., Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 27, 549–555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm D., Streetz K. L., Jopling C. L., Storm T. A., Pandey K., Davis C. R., Marion P., Salazar F., Kay M. A., Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Li X., Yang L., Chen L.-L., The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71, 428–442 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Enuka Y., Lauriola M., Feldman M. E., Sas-Chen A., Ulitsky I., Yarden Y., Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang R., Xing L., Zheng X., Sun Y., Wang X., Chen J., The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol. Cancer 18, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Miao F., Yin B. H., Zhang X., Xue D. D., Ma C., CircRNA_009934 induces osteoclast bone resorption via silencing miR-5107. Eur. Rev. Med. Pharmacol. Sci. 24, 7580–7588 (2020). [DOI] [PubMed] [Google Scholar]
- 25.He A. T., Liu J., Li F., Yang B. B., Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 6, 185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X., Wang Q., Gu C., Li M., Chen K., Chen P., Tang Z., Liu X., Pan H., Liu Z., Tang R., Fan S., Smart nanosacrificial layer on the bone surface prevents osteoporosis through acid-base neutralization regulated biocascade effects. J. Am. Chem. Soc. 142, 17543–17556 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Liu J., Dang L., Li D., Liang C., He X., Wu H., Qian A., Yang Z., Au D. W. T., Chiang M. W. L., Zhang B.-T., Han Q., Yue K. K. M., Zhang H., Lv C., Pan X., Xu J., Bian Z., Shang P., Tan W., Liang Z., Guo B., Lu A., Zhang G., A delivery system specifically approaching bone resorption surfaces to facilitate therapeutic modulation of microRNAs in osteoclasts. Biomaterials 52, 148–160 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Yin H., Kanasty R. L., Eltoukhy A. A., Vegas A. J., Dorkin J. R., Anderson D. G., Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Fang R. H., Kroll A. V., Gao W., Zhang L., Cell membrane coating nanotechnology. Adv. Mater. 30, e1706759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao L., Bu L.-L., Xu J.-H., Cai B., Yu G.-T., Yu X., He Z., Huang Q., Li A., Guo S.-S., Zhang W.-F., Liu W., Sun Z.-J., Wang H., Wang T.-H., Zhao X.-Z., Red blood cell membrane as a biomimetic nanocoating for prolonged circulation time and reduced accelerated blood clearance. Small 11, 6225–6236 (2015). [DOI] [PubMed] [Google Scholar]
- 31.He Z., Zhang Y., Feng N., Cell membrane-coated nanosized active targeted drug delivery systems homing to tumor cells: A review. Mater. Sci. Eng. C Mater. Biol. Appl. 106, 110298 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Liu X., Xiang J., Zhu D., Jiang L., Zhou Z., Tang J., Liu X., Huang Y., Shen Y., Fusogenic reactive oxygen species triggered charge-reversal vector for effective gene delivery. Adv. Mater. 28, 1743–1752 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Takito J., Nakamura M., Heterogeneity and actin cytoskeleton in osteoclast and macrophage multinucleation. Int. J. Mol. Sci. 21, 6629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang J., Liu X., Zhou Z., Zhu D., Zhou Q., Piao Y., Jiang L., Tang J., Liu X., Shen Y., Reactive oxygen species (ROS)-responsive charge-switchable nanocarriers for gene therapy of metastatic cancer. ACS Appl. Mater. Interfaces 10, 43352–43362 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Helming L., Gordon S., The molecular basis of macrophage fusion. Immunobiology 212, 785–793 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Møller A. M. J., Delaissé J.-M., Søe K., Osteoclast fusion: Time-lapse reveals involvement of CD47 and Syncytin-1 at different stages of nuclearity. J. Cell. Physiol. 232, 1396–1403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu Y.-H., Mensah K. A., Schwarz E. M., Ju Y., Takahata M., Feng C., McMahon L. A., Hicks D. G., Panepento B., Keng P. C., Ritchlin C. T., Regulation of human osteoclast development by dendritic cell-specific transmembrane protein (DC-STAMP). J. Bone Miner. Res. 27, 79–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song L., Bi Y.-N., Zhang P.-Y., Yuan X.-M., Liu Y., Zhang Y., Huang J.-Y., Zhou K., Optimization of the time window of interest in ovariectomized imprinting control region mice for antiosteoporosis research. Biomed. Res. Int. 2017, 8417814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grassi F., Tyagi A. M., Calvert J. W., Gambari L., Walker L. D., Yu M., Robinson J., Li J.-Y., Lisignoli G., Vaccaro C., Adams J., Pacifici R., Hydrogen sulfide is a novel regulator of bone formation implicated in the bone loss induced by estrogen deficiency. J. Bone Miner. Res. 31, 949–963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonald M. M., Khoo W. H., Ng P. Y., Xiao Y., Zamerli J., Thatcher P., Kyaw W., Pathmanandavel K., Grootveld A. K., Moran I., Butt D., Nguyen A., Corr A., Warren S., Biro M., Butterfield N. C., Guilfoyle S. E., Komla-Ebri D., Dack M. R. G., Dewhurst H. F., Logan J. G., Li Y., Mohanty S. T., Byrne N., Terry R. L., Simic M. K., Chai R., Quinn J. M. W., Youlten S. E., Pettitt J. A., Abi-Hanna D., Jain R., Weninger W., Lundberg M., Sun S., Ebetino F. H., Timpson P., Lee W. M., Baldock P. A., Rogers M. J., Brink R., Williams G. R., Bassett J. H. D., Kemp J. P., Pavlos N. J., Croucher P. I., Phan T. G., Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption. Cell 184, 1330–1347.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z., Li C., Huang P., Hu F., Jiang M., Xu X., Li B., Deng L., Ye T., Guo L., CircHmbox1 targeting miRNA-1247-5p is involved in the regulation of bone metabolism by TNF-α in postmenopausal osteoporosis. Front. Cell Dev. Biol. 8, 594785 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu B., Weng Y., Xia X.-H., Liang X.-J., Huang Y., Clinical advances of siRNA therapeutics. J. Gene Med. 21, e3097 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Gohda J., Akiyama T., Koga T., Takayanagi H., Tanaka S., Inoue J.-i., RANK-mediated amplification of TRAF6 signaling leads to NFATc1 induction during osteoclastogenesis. EMBO J. 24, 790–799 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim W. A., June C. H., The principles of engineering immune cells to treat cancer. Cell 168, 724–740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rebres R. A., Kajihara K., Brown E. J., Novel CD47-dependent intercellular adhesion modulates cell migration. J. Cell. Physiol. 205, 182–193 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Chen H.-Y., Deng J., Wang Y., Wu C.-Q., Li X., Dai H.-W., Hybrid cell membrane-coated nanoparticles: A multifunctional biomimetic platform for cancer diagnosis and therapy. Acta Biomater. 112, 1–13 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Nagasawa T., Microenvironmental niches in the bone marrow required for B-cell development. Nat. Rev. Immunol. 6, 107–116 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Gronthos S., Zannettino A. C. W., The role of the chemokine CXCL12 in osteoclastogenesis. Trends Endocrinol. Metab. 18, 108–113 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Pánczél Á., Nagy S. P., Farkas J., Jakus Z., Győri D. S., Mócsai A., Fluorescence-based real-time analysis of osteoclast development. Front. Cell Dev. Biol. 9, 657935 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacome-Galarza C. E., Percin G. I., Muller J. T., Mass E., Lazarov T., Eitler J., Rauner M., Yadav V. K., Crozet L., Bohm M., Loyher P. L., Karsenty G., Waskow C., Geissmann F., Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature 568, 541–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trebec D. P., Chandra D., Gramoun A., Li K., Heersche J. N. M., Manolson M. F., Increased expression of activating factors in large osteoclasts could explain their excessive activity in osteolytic diseases. J. Cell. Biochem. 101, 205–220 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Podolnikova N. P., Hlavackova M., Wu Y., Yakubenko V. P., Faust J., Balabiyev A., Wang X., Ugarova T. P., Interaction between the integrin Mac-1 and signal regulatory protein α (SIRPα) mediates fusion in heterologous cells. J. Biol. Chem. 294, 7833–7849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han X., Sterling H., Chen Y., Saginario C., Brown E. J., Frazier W. A., Lindberg F. P., Vignery A., CD47, a ligand for the macrophage fusion receptor, participates in macrophage multinucleation. J. Biol. Chem. 275, 37984–37992 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Kunisawa J., Masuda T., Katayama K., Yoshikawa T., Tsutsumi Y., Akashi M., Mayumi T., Nakagawa S., Fusogenic liposome delivers encapsulated nanoparticles for cytosolic controlled gene release. J. Control. Release 105, 344–353 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Varkouhi A. K., Scholte M., Storm G., Haisma H. J., Endosomal escape pathways for delivery of biologicals. J. Control. Release 151, 220–228 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Chen K., Qiu P., Yuan Y., Zheng L., He J., Wang C., Guo Q., Kenny J., Liu Q., Zhao J., Chen J., Tickner J., Fan S., Lin X., Xu J., Pseurotin A inhibits osteoclastogenesis and prevents ovariectomized-induced bone loss by suppressing reactive oxygen species. Theranostics 9, 1634–1650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Husch J. F. A., Stessuk T., den Breejen C., van den Boom M., Leeuwenburgh S. C. G., van den Beucken J. J. J. P., A practical procedure for the in vitro generation of human osteoclasts and their characterization. Tissue Eng. Part C Methods 27, 421–432 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Stapleton M., Sawamoto K., Alméciga-Díaz C. J., Mackenzie W. G., Mason R. W., Orii T., Tomatsu S., Development of bone targeting drugs. Int. J. Mol. Sci. 18, 1345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsiklauri L., Werner J., Kampschulte M., Frommer K. W., Berninger L., Irrgang M., Glenske K., Hose D., El Khassawna T., Pons-Kühnemann J., Rehart S., Wenisch S., Müller-Ladner U., Neumann E., Visfatin alters the cytokine and matrix-degrading enzyme profile during osteogenic and adipogenic MSC differentiation. Osteoarthr. Cartil. 26, 1225–1235 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Cai W., Dai X., Chen J., Zhao J., Xu M., Zhang L., Yang B., Zhang W., Rocha M., Nakao T., Kofler J., Shi Y., Stetler R. A., Hu X., Chen J., STAT6/Arg1 promotes microglia/macrophage efferocytosis and inflammation resolution in stroke mice. JCI Insight 4, e131355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3
Figs. S1 to S11
Data S1 and S2