Abstract
Immune checkpoint inhibitors exhibit limited response rates in patients with triple-negative breast cancer (TNBC), suggesting that additional immune escape mechanisms may exist. Here, we performed two-step customized in vivo CRISPR screens targeting disease-related immune genes using different mouse models with multidimensional immune-deficiency characteristics. In vivo screens characterized gene functions in the different tumor microenvironments and recovered canonical immunotherapy targets such as Ido1. In addition, functional screening and transcriptomic analysis identified Lgals2 as a candidate regulator in TNBC involving immune escape. Mechanistic studies demonstrated that tumor cell–intrinsic Lgals2 induced the increased number of tumor-associated macrophages, as well as the M2-like polarization and proliferation of macrophages through the CSF1/CSF1R axis, which resulted in the immunosuppressive nature of the TNBC microenvironment. Blockade of LGALS2 using an inhibitory antibody successfully arrested tumor growth and reversed the immune suppression. Collectively, our results provide a theoretical basis for LGALS2 as a potential immunotherapy target in TNBC.
In vivo CRISPR screens using mouse models with different immune-deficiency features reveal Lgals2 mediates immune evasion in TNBC.
INTRODUCTION
Triple-negative breast cancer (TNBC) is one heterogeneous subtype of breast cancer that lacks expression of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2. Compared with other subtypes of breast cancer, TNBC has a poorer prognosis owing to more malignant biological behavior, higher rate of early relapse, and limited therapeutic options (1, 2). Recently, the advent of cancer immunotherapy using immune checkpoint inhibitors has transformed the treatment paradigm for many malignancies (3). Several phase 3 clinical trials have shown that immune checkpoint inhibitors in combination with adjuvant chemotherapy could have clinical impact on some patients with TNBC (4, 5). However, immune checkpoint inhibitors have undesired side effects and do not achieve sustained clinical response in a large fraction of patients, indicating the importance of discovery of previously unknown genes that promote immune evasion by tumor cells and additional immunotherapeutic strategies (6, 7).
A multitude of studies have been conducted to understand components and interactions between immune cells and tumor cells in the tumor microenvironment (TME) (8, 9). CRISPR screens have markedly enhanced genome editing and made it possible to identify previously unknown genes associated with immunotherapy responses (10–12). Compared with in vitro CRISPR screens, in vivo models could be a more relevant setting to screen for tumor-immune interactions. Furthermore, given the complexity of immune systems, it is better to apply several different immune-selection pressures on tumors in high-throughput in vivo CRISPR screens to identify clinically relevant targets.
In this study, we performed in vivo CRISPR screens targeting disease-related immune genes (DrIM) under different immune-selection pressures in various mouse models of TNBC. We uncovered the dynamics of gene function when engaged in immune activities with different infiltrating immune cells and identified Lgals2 as a key regulator in TNBC involving immune escape. Previous studies revealed that Lgals2, which encodes galectin-2, a member of glycan-binding proteins, is associated with collateral arteriogenesis, myocardial infarction, cell adhesion, and T cell apoptosis (13–16). However, the relationship between Lgals2 and the immunosuppressive phenotype in the TNBC microenvironment remains unclear. Functional and mechanistic studies showed that Lgals2 promotes tumor growth in vivo, not in vitro, by facilitating M2-like polarization and proliferation of macrophages via activation of the Colony-stimulating factor 1 (CSF1)/CSF1 receptor (CSF1R) axis. Our findings suggest that LGALS2 might serve as a potential immunotherapy target in TNBC.
RESULTS
In vivo CRISPR screens targeting disease-related immune genes identify candidate tumor-related immune genes
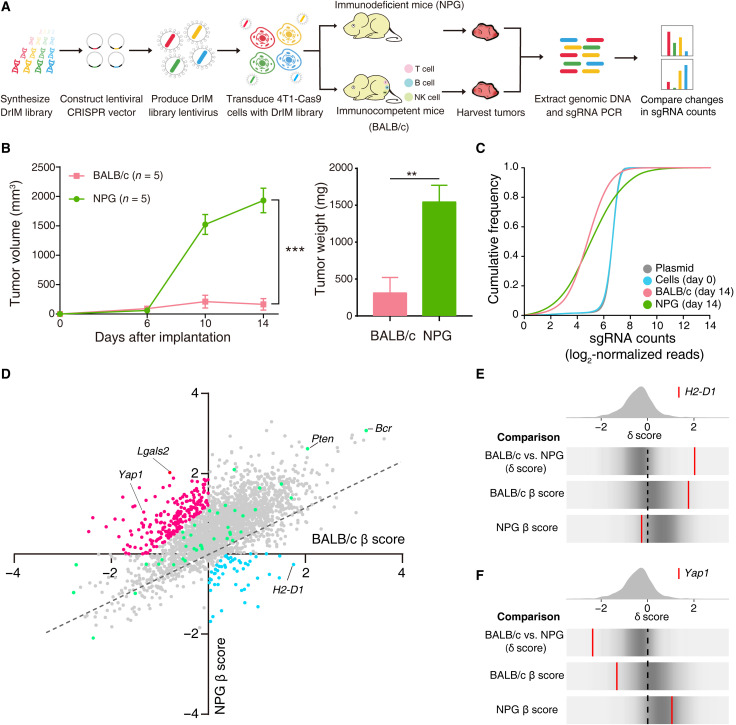
To uncover the immune-related genes in the antitumor or protumor process that might indicate potential therapeutic strategies, we designed and generated a mouse single-guide RNA (sgRNA) library corresponding to all human disease-related immune genes [termed the disease-related immune gene library (DrIM library)]. The DrIM library consisted of 12,000 sgRNAs targeting 2796 genes (4 sgRNAs per gene and 816 nontargeting control sgRNAs) (table S1 and fig. S1A). The mouse TNBC cell line 4T1 with stable expression of Cas9 was generated and transduced with the DrIM library (fig. S1B). After in vitro culture, DrIM-transduced 4T1-Cas9 cells were subcutaneously transplanted into immunocompetent BALB/c mice and immunodeficient nonobese diabetic (NOD)–PrkdcscidII2rgnull (NPG) mice, which lack T cells, B cells, and natural killer (NK) cells, to compare different immune-selection pressures on tumor cells (Fig. 1A). After 14 days, mice were euthanized, and the tumors were harvested for high-throughput sgRNA library sequencing (Fig. 1B and fig. S1C). To decrease bias and increase the accuracy of our screens, two samples were abandoned because of low quality in next-generation sequence. While the library representation of primary plasmid and pretransplanted tumor cells (day 0) followed a log-normal distribution, the sgRNA representation in posttransplanted cells obtained from tumor masses on BALB/c and NPG mice showed a distinct shift (Fig. 1C and fig. S1D). At the individual mouse level, we found that the sgRNA representation between different mice was correlated with each other (fig. S1E).
Fig. 1. In vivo CRISPR screens targeting disease-related immune genes identify candidate tumor-related immune genes.
(A) Schematics of the experimental design. (B) Tumor growth and tumor weight of intramammary fat pad tumors from transplanted DrIM-transduced 4T1-Cas9 cells in BALB/c mice (n = 5) and NPG mice (n = 5). **P < 0.01 and ***P < 0.001. Data are presented as the means ± SEM. (C) Cumulative distribution function plots of DrIM library sgRNAs in the plasmid, cells before transplantation, tumors in BALB/c mice, and tumors in NPG mice. Distributions in each sample type are averaged across individual mice and infection replicates. (D) Scatterplot of the gene essentiality score (β score) in BALB/c versus β score in NPG for all genes after in vivo screening. The dotted line indicates the linear regression trend line. Every dot means a gene in library. The color of the points represents selected genes as candidate in the second round of screen (red, immune escape; blue, immune surveillance; green, control). (E and F) Frequency histograms of the δ score for all sgRNAs. sgRNAs targeting the indicated genes including (E) H2-D1 and (F) Yap1 are shown by the red lines.
By using MAGeCK-MLE (Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout–maximum likelihood estimation) tool, we determined the β scores in BALB/c and NPG separately (table S2). Some of the genes were universally negatively or positively selected in both immunocompetent and severely immunodeficient mice (points of genes fell in the upper right and lower left quadrants) but had different essentialities across conditions (Fig. 1D). In the comparison of screens in BALB/c and NPG mice, we dichotomized the target genes into those that assisted immune escape (δ score < 0) or immune surveillance (δ score > 0). For example, histocompatibility 2, D region locus 1 (H2-D1) (δ score > 0 in our compared selection), also known as major histocompatibility complex class I molecules (MHC-I), could present antigens to CD8+ T cells and engage in immunosurveillance (Fig. 1E). Yap1 (δ score < 0 in the compared selection), which is one of the most important effectors of the Hippo pathway, could influence NK and T cells to facilitate an immunosuppressive TME (Fig. 1F) (17).
In vivo mini-DrIM library screens uncover immune-response features of selected genes under different immune pressures
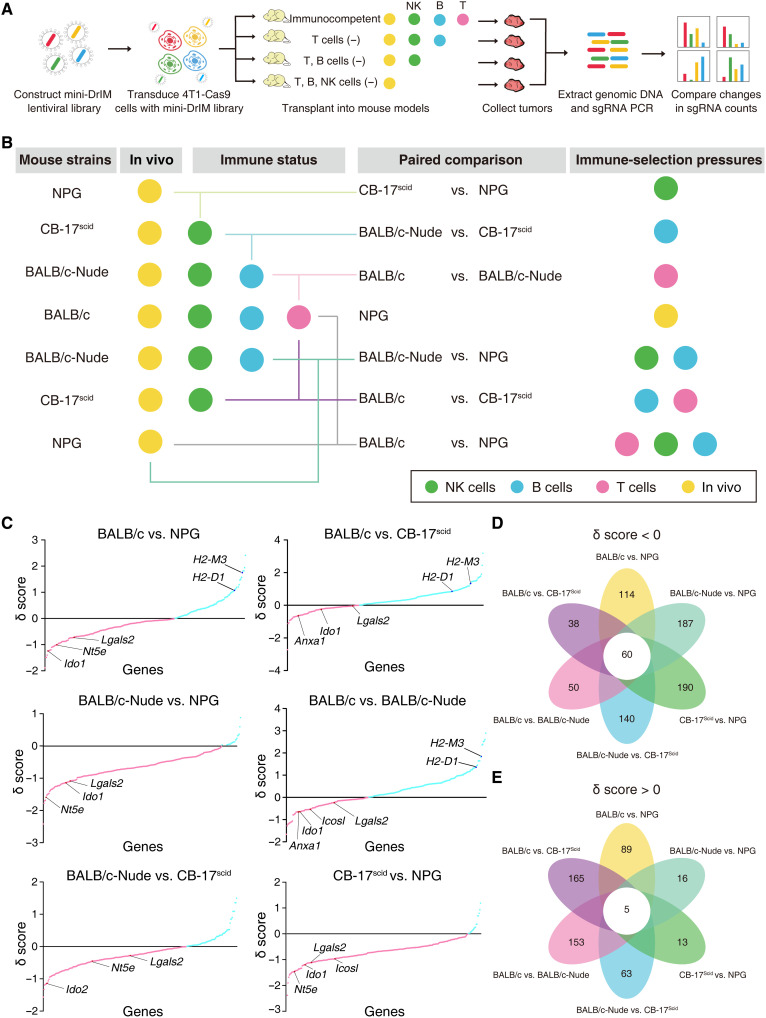
We sought to further assess the essential immune-related genes in the complicated TME, which engaged in antitumor or protumor activities, for potential immunotherapy targets in TNBC. A mini-DrIM library, which included 3546 sgRNAs targeting 227 candidate genes ordered by the absolute value of the δ score from DrIM screening, 46 positive control genes, and nontargeting guides, was generated for the second round of in vivo screening (table S3). To better understand the function of genes in tumor-immunity battles, we designed more sophisticated conditions with different immune-selection pressures: BALB/c mice have a healthy immune system; CAnN.Cg-Foxn1nu/Crl (BALB/c-Nude) mice only lack T cells because of athymia; CB-17/Icr-Prkdcscid/IcrCrl (CB-17scid) mice have normal NK cells, macrophages (Mφ), and granulocytes but lack T cells and B cells; and NPG mice lack T cells, B cells, and NK cells (Fig. 2A). Six kinds of immune-selection pressure were constituted by pairwise comparisons between the four kinds of mice described above with different immune-deficiency features (Fig. 2B).
Fig. 2. In vivo mini-DrIM library screens uncover immune-response features of selected genes under different immune pressures.
(A) Schematics of the experimental design. (B) Diagram of in vivo screens of the mini-DrIM pool under multiple immune-selection pressures. (C) Scatterplots showing the rank-ordered δ score of all targeted genes in the mini-DrIM library under the indicated immune-selection pressure. X axis shows targeted genes; y axis shows the δ score of each targeted gene. Genes are highlighted in red (δ score < 0) and blue (δ score > 0). (D and E) Flower plots showing the number of genes with (D) δ score < 0 or (E) δ score >0 under each immune-selection pressure and all immune-selection pressures.
Tumors containing mini-DrIM library were developed in all four kinds of mice (n = 10 per group). The library representation and sgRNA distribution of plasmid, pretransplanted tumor cells, and posttransplanted tumor cells were consistent with the first screen (fig. S2, A to C). We found that the dynamics of the sgRNA abundance changed markedly under different selecting conditions. Genes in the mini-DrIM library were ranked by the δ score generated by comparisons between different immune-selection pressure conditions (Fig. 2C and table S4). A total of 60 genes were congruously related with immune evasion in the comparisons of various immune deficiencies (δ score < 0), while 5 genes were identified as potential regulators of immune surveillance in all screening settings (δ score > 0; Fig. 2, D and E, and fig. S2D). The smaller size of the library, the larger numbers of sgRNAs per gene, and multiple immune-selection pressures enabled us to obtain a more comprehensive view of candidate genes. For example, the comparison between immunocompetent BALB/c hosts and T cell–deficient BALB/c-Nude hosts revealed key genes associated with T cell response, and some well-known immunosuppressive genes related to Ido1 and Icosl were on the list of the top-ranked genes relative to immune escape (δ < 0) (18, 19). Cancer-associated Nt5e/Cd73 could accelerate the generation of adenosine and suppress the cytotoxic function of NK cells (20). A similar feature was observed in our screens: Nt5e was negatively selected (δ score < 0) in the comparison between CB-17scid mice (lacking T and B cells) and NPG mice (lacking T, B, and NK cells), which indicated that Nt5e promoted tumor escape from NK cell–mediated killing.
Lgals2 affects tumor growth in vivo, not in vitro
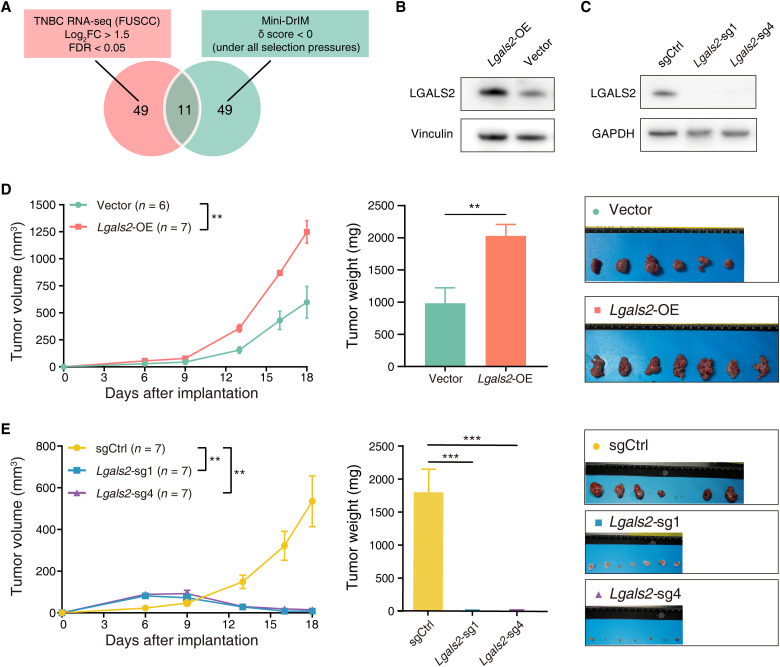
After two rounds of in vivo screens, we further explore putative essential genes mediating immunosuppression in TNBC. We compared mRNA expression levels of genes in the mini-DrIM library between cancerous tissue samples (n = 360) and paired adjacent noncancerous samples (n = 88) using our previously published TNBC database (21). Differentially expressed gene analysis revealed 60 genes with log2(fold change) > 1.5 and false discovery rate (FDR) < 0.05 to be significantly up-regulated in TNBC tumor samples (fig. S3A). A total of 11 genes emerged as candidate hits from the functional screen and transcriptomic analysis (Fig. 3A and table S5). Previously identified immunotherapy targets, such as Ido1 and Cd38, were listed at the top of the 11 genes, which were consistent with their roles in regulating tumor immunity, thereby benchmarking the technical reliability of our in vivo screens (18, 22).
Fig. 3. Lgals2 affects tumor growth in vivo, not in vitro.
(A) Venn diagram of the two criteria to identify the candidate gene hits (significantly enriched in TNBC tumor samples, δ score < 0 under all selection pressures). (B and C) Western blot of Lgals2 protein level in 4T1 cells transduced with (B) either vector or Lgals2–overexpressing (OE) plasmid and (C) either vector control or Lgals2-targeting sgRNAs. (D and E) Tumor growth, tumor weight, and ex vivo images of resected tumors from transplanted 4T1 cells with (D) Lgals2 overexpression or (E) Lgals2 KO in BALB/c mice. ***P < 0.001 and **P < 0.01. Data are presented as the means ± SEM. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
On the basis of the intersection analysis of TNBC cohorts and functional screening readouts, we lastly selected Lgals2, which has few previous reports of its roles in immune escape in breast cancer, for further validation. We used the 4T1 cell line to generate stable overexpression models of Lgals2 (Fig. 3B). To exclude direct effects of Lgals2 on tumor proliferation, we used in vitro proliferation assays to compare cell growth rates of overexpression and control cells. We found that Lgals2 did not have an innate function in tumor proliferation in vitro (P > 0.05; fig. S3B). In contrast, tumors overexpressing Lgals2 grew significantly faster in vivo (P < 0.01; Fig. 3D). Higher expression levels of Lgals2 were correlated with enhanced proliferation of tumor cells in vivo but not in vitro, indicating that immunosuppression mediated by Lgals2 might be dependent on the TME.
Next, we silenced Lgals2 individually in 4T1-Cas9 cell lines with two sgRNAs from the mini-DrIM library. The knockout (KO) efficiency by the CRISPR-Cas9 editing system was confirmed by Western blot (Fig. 3C). Similar to the results in the overexpression group, in vitro proliferation assays suggested no difference between Lgals2-KO cells and control cells (fig. S3B). In vivo experiment revealed that transplanted Lgals2-KO cells showed increased tumor mass in the early stage, whereas tumor volume gradually decreased beginning on day 6 and almost disappeared by harvest on day 18 (P < 0.01; Fig. 3E). The previous literature suggests that Lgals2 is expressed on monocytes and macrophages (15). However, in our immunofluorescence staining of breast cancer tissues, LGALS2 tends to aggregate around CD11b+ cells [including monocytes and macrophages, FITC (fluorescein isothiocyanate)–green] but was not expressed on CD11b+ cells (fig. S3C). These findings prompted us to explore the potential mechanisms by which LGALS2 affects the immune microenvironment in vivo.
Lgals2 is associated with immune cell infiltration in the TME
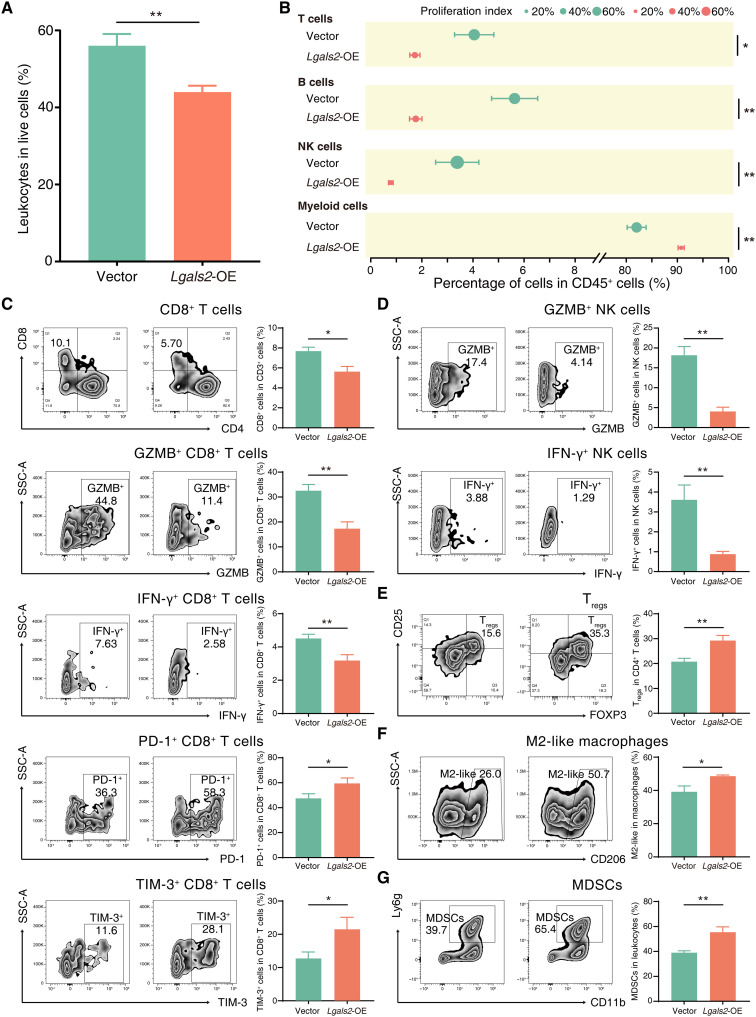
We further used flow cytometry to analyze the infiltrating immune cells in tumors from Lgals2-overexpressing 4T1 cells and control cells transplanted in BALB/c xenografts. Using the pan-leukocyte marker CD45, we found that Lgals2-overexpressing tumors had a lower proportion of leukocytes (CD45+ cells) than vector control tumors (P < 0.01; Fig. 4A). In addition, the composition of different kinds of tumor-infiltrating immune cells (TIICs) was changed, and the percentage of myeloid cells (CD45+CD11b+) in CD45+ cells increased in Lgals2-overexpressing tumors, while T cells (CD45+CD3+), B cells (CD45+CD19+), and NK cells (CD45+CD3−CD49b+) decreased (all P < 0.05; Fig. 4B). The proliferation index was measured by the percentage of Ki67+ proliferating cells among the same kind of cells, and we found a consistent reduction in the proliferation index of T cells, B cells, NK cells, and myeloid cells in Lgals2-overexpressing tumors (all P < 0.05; Fig. 4B). We also observed a significant decrease in the frequency of infiltrating cytotoxic T lymphocytes (CTLs; CD45+CD3+CD8+; P < 0.05), as well as lower levels of cytotoxic factors [interferon-γ (IFN-γ) and granzyme B] and higher expression of exhaustion markers (PD-1 and TIM-3) in single-cell suspensions from Lgals2-overexpressing tumors (Fig. 4C). NK cells also showed a reduction in the frequency and expression of IFN-γ and granzyme B (both P < 0.01; Fig. 4D). Hence, CTLs and NK cells, which are common antitumor TIICs, seemed to be significantly attenuated in numbers and killing ability after Lgals2 overexpression. Furthermore, we detected protumor TIICs in the TME to evaluate the degree of immunosuppression in tumors. Consistent with our predictions, Lgals2-overexpressing tumors contained a higher proportion of regulatory T cells (Tregs; CD45+CD3+CD4+CD25+FOXP3+; P < 0.01), M2-like macrophages (CD45+CD11b+F4/80highCD206+; P < 0.05), and Myeloid-derived suppressor cells (MDSCs) (CD45+CD11b+Ly6g+; P < 0.01) (Fig. 4, E to G).
Fig. 4. Lgals2 is associated with immune cell infiltration in the TME.
(A) Bar plot comparing the percentage of leukocytes among live cells between vector control and Lgals2-overexpressing 4T1 tumors. **P < 0.01. (B) Forest plot showing different types of infiltrating immune cells in vector controls and Lgals2-overexpressing 4T1 tumors. **P < 0.01; *P < 0.05. Data are presented as means ± SEM. (C to G) The primary tumors of vector control and Lgals2-overexpressing 4T1 tumors of BALB/c mice were harvested for flow cytometry to determine the percentages of (C) CD8+ T cells among CD3+ T cells, granzyme B+ (GZMB+) cells among CD8+ T cells, IFN-γ+ cells among CD8+ T cells, PD-1+ cells among CD8+ T cells, and TIM-3+ cells among CD8+ T cells; (D) GZMB+ cells among NK cells and IFN-γ+ cells among NK cells; (E) FOXP3+CD25+ Tregs among CD4+ T cells; (F) M2-like macrophages among total macrophages; and (G) MDSCs among CD45+ cells. Representative plots of individual tumors are shown on the left, and bar graphs of the summary data for all tumors are shown on the right. **P < 0.01; *P < 0.05; not significant (NS) P ≥ 0.05. Data are presented as the means ± SEM.
To verify the phenotypes of Lgals2 in 4T1 tumors, we constructed an overexpressing model in a second murine TNBC cell line, EMT6 (fig. S4A). Lgals2 overexpression in EMT6 cells was also able to significantly increase tumor growth in vivo but had no effects on proliferation in vitro (fig. S4, B to E). We also measured infiltrating immune cells within tumors by flow cytometry. Similar to that observed in the 4T1 models, overexpression of Lgals2 in EMT6 cells significantly decreased the frequency and cytotoxic function of CTLs and NK cells, while it increased the exhaustion phenotype of CTLs and the population of myeloid cells, especially M2-like macrophages, within the TME (fig. S4, F to M). The effects of Lgals2 in both the 4T1 and EMT6 models suggest that Lgals2 overexpression could enhance immune suppression in TNBC.
Single-cell RNA-seq reveals the Lgals2-induced polarization of macrophages in the TME
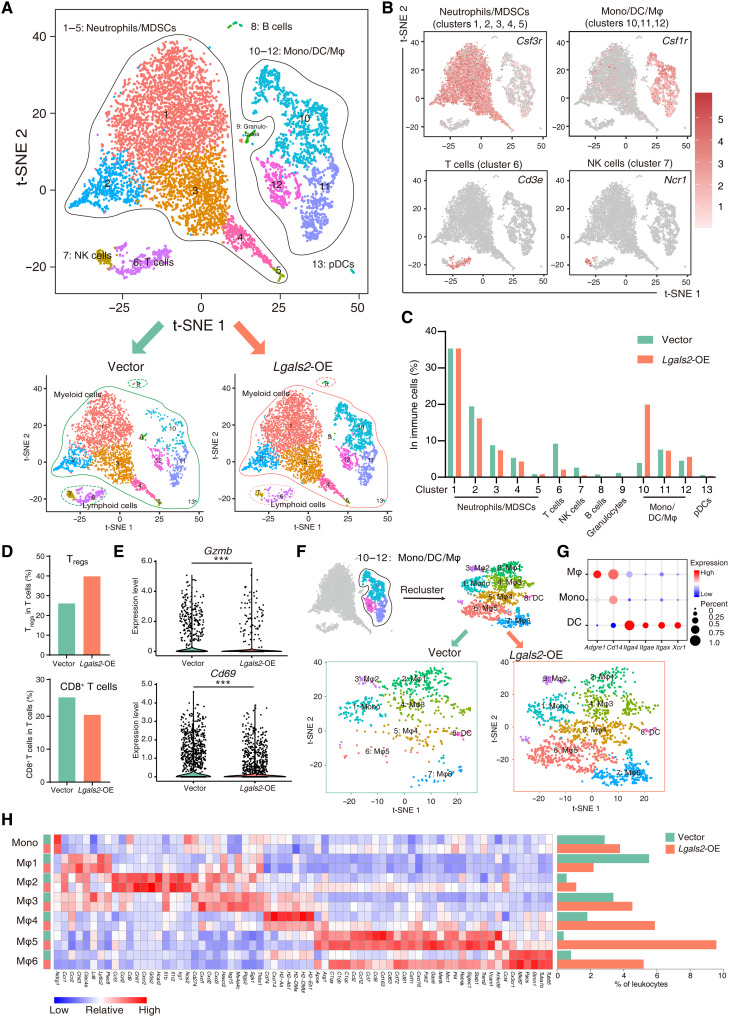
To investigate the unique TME-dependent effects of Lgals2 and in consideration of the abundant cells in the TME, we performed single-cell RNA sequencing (scRNA-seq) to characterize changes in the transcriptome of cells harvested from 4T1 tumor–bearing BALB/c mice. We lastly collected 5548 cells from the Lgals2-overexpressing group and 4802 cells from the vector control group. Cell Ranger and Seurat were applied to classify cells into groups of cell types, and marker genes were used to distinguish immune cells (CD45+) and tumor cells (CD45−KRT18+) (fig. S5A). Tumor cells revealed a significantly higher expression level of Lgals2, which was consistent with our predictions (fig. S5A). To better identify transcriptional clusters consisting of TIICs, we analyzed a dataset of CD45+ cells referring to the known cell type markers and identified 13 clusters, visualized by t-distributed stochastic neighbor embedding (t-SNE): 5 neutrophil/MDSC clusters (#1 to #5), 1 T cell cluster (#6), 1 NK cell cluster (#7), 1 B cell cluster (#8), 1 granulocyte cluster (#9), 3 monocyte/dendritic cell (DC)/macrophage clusters (#10 to #12), and 1 plasmacytoid DC (pDC) cluster (#13) (Fig. 5, A and B). Following overexpression of Lgals2 in tumor cells, the proportion of the lymphoid cell population was decreased, including T cells, B cells, and NK cells (Fig. 5C). The myeloid cell population showed an increase in Lgals2-overexpressing tumors (97.2 versus 87.4%), and the increment was more pronounced in the monocyte/DC/macrophage population (32.8 versus 16.0%), especially in cluster 10 (20.0 versus 3.9%; Fig. 5C).
Fig. 5. Single-cell RNA-seq reveals the Lgals2-induced polarization of macrophages in the TME.
(A) The t-SNE plot of intratumoral immune cells in 4T1 tumors. Cells and clusters are color coded by the major cell type found. (B) t-SNE plot of immune cells displaying marker gene expression. (C) The distribution of immune cell types between control vector and Lgals2-overexpressing tumors. (D) The distribution of Tregs and CD8+ T cells between control vector and Lgals2-overexpressing tumor cells. (E) Violin plot of Gzmb and Cd69 mRNA levels. (F) t-SNE plot of the reclassification of intratumoral monocytes/DCs/macrophages. (G) Expression of marker genes for identifying monocytes, DCs, and macrophages. (H) Heatmap displaying normalized expression of selected genes in each monocyte/DCs/macrophage cluster and histogram displaying the distribution of each cluster between control vector and Lgals2-overexpressing tumor cells.
To accurately define the T cell cluster (#6) identified by scRNA-seq, we computationally separated the T cell cluster (#6) into five clusters according to the expression of classical marker genes: Tregs, T helper 2 (TH2) cells, naïve T cells, CD8+ T cells, and proliferating T cells (fig. S5, B and C). Tregs were increased in Lgals2-overexpressing samples (Fig. 5D). TH2 cells and naïve T cells decreased with the increasing levels of Lgals2 (fig. S5D). CD8+ T cells in Lgals2-overexpressing tumor samples not only decreased in proportion but also expressed decreased levels of Gzmb, Ifng, Prf1, and Gzma, as well as the activation marker Cd69 (Fig. 5, D and E, and fig. S5E).
Unexpectedly, the monocyte/DC/macrophage population showed unexpected complexity and remarkable differences between the scRNA-seq data of Lgals2-overexpressing and control vector samples; hence, we reclustered this population and identified eight distinct clusters (Fig. 5F). One cluster corresponded to monocytes (Adgre−CD14+Itga4−), one cluster to DCs (Adgre−CD14−Itga4+), and six clusters to macrophages (Adgre+CD14+Itga4−) (Fig. 5, F and G). The six macrophage clusters were defined as Mφ1 to Mφ6, and differential genes are presented in Fig. 5H. Chil3, a well-known marker of Tumor-associated macrophages (TAM), was strongly expressed in Mφ1, Mφ2, and Mφ3. Cd274/Pd-l1 and Havcr2/Tim3, both inhibitory immune checkpoints, were enriched in the Mφ3 cell cluster. The Mφ4 cluster expressed a set of markers related to antigen processing—including H2-Aa, H2-Ab1, H2-DMa, H2-DMb, and H2-Eb1—but the expression of these genes decreased when Lgals2 was overexpressed. Mφ5 cluster cells were highly enriched for complement genes (C1qa, C1qb, and C1qc), cytokine genes (Ccl2, Ccl4, Ccl7, Ccl8, and Ccl12), pan-macrophage markers (Cd163 and Mertk), and tissue-resident macrophage markers (Siglec1/Cd169, Cx3cr1, and Trem2). Retnla/Fizz1 and Arg1, which are specific markers of TAM and M2-like macrophages and indicate an immunosuppressive phenotype, were also expressed in Mφ5. Mφ6 was characterized by high expression of Mki67, Stmn1, Pacis, Tuba1b, Tubb5, and Cx3cr1, a series of markers related to proliferation and tissue-resident macrophages (Fig. 5H). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that Mφ4 displayed the pathways associated with antigen processing and presentation, Mφ5 displayed the pathways associated with complement and coagulation cascades and endocytosis, and Mφ6 displayed all the pathways associated with proliferation such as spliceosome, cell cycle, and DNA replication (fig. S5F).
After Lgals2 overexpression, the number and proportion of Mφ1 decreased, whereas the numbers and proportions of Mφ2, Mφ3, Mφ4, Mφ5, and Mφ6 all increased (Fig. 5H). The Mφ4, Mφ5, and Mφ6 clusters all expressed higher levels of the immunosuppressive marker Mrc1/Cd206, which increased significantly after Lgals2 overexpression. The predominant difference between distributions of TIICs in Lgals2 overexpression and vector control samples was observed in the Mφ5 cluster (9.62% of TIICs in Lgals2 overexpression versus 0.41% of TIICs in vector control). In addition, the Mφ6 cluster changed substantially after Lgals2 overexpression: It not only maintained its original characteristics but also expressed all the characteristic genes of the Mφ5 cluster. In summary, the scRNA-seq data indicated remodeling of the macrophage compartments induced by the elevated expression level of Lgals2 within tumor cells.
Lgals2 facilitates M2-like polarization and proliferation of macrophages through CSF1
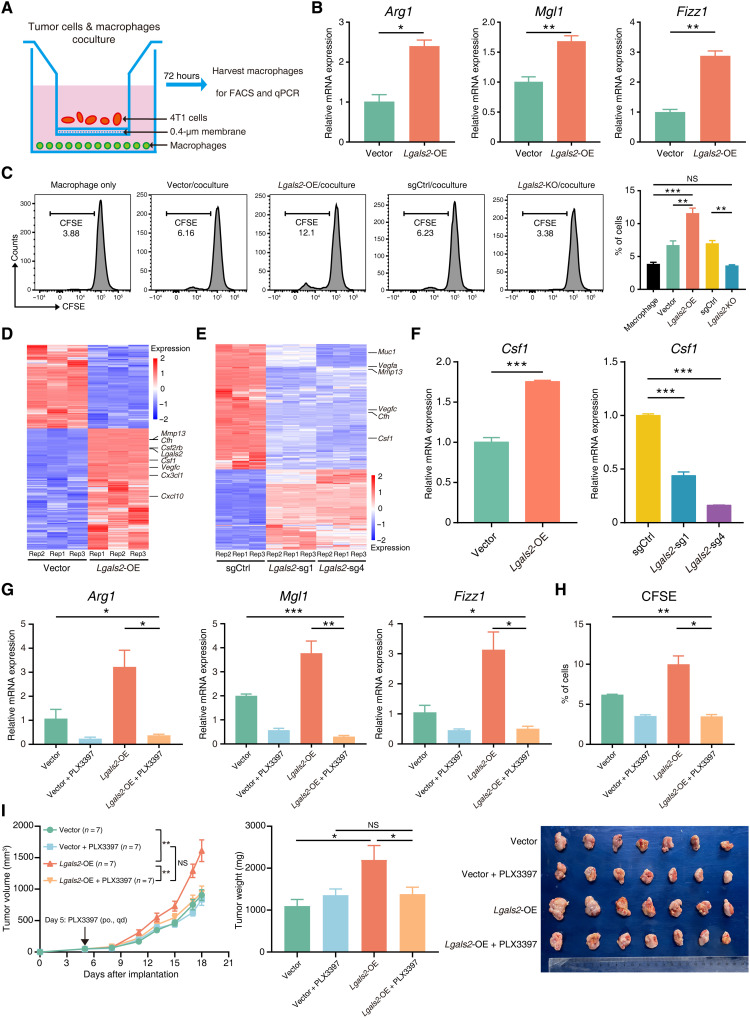
To further validate the association between Lgals2 expression and the macrophage phenotype detected in the flow cytometry analysis and scRNA-seq, we established an in vitro coculture system by culturing 4T1 cells with different levels of Lgals2 expression with mouse peritoneal macrophages (Fig. 6A). Consistent with the in vivo experiments, macrophages cocultured with higher Lgals2 expression of 4T1 cells exhibited up-regulated expression of the M2-like marker Arg1 and other TAM markers, such as Mgl1 and Fizz1, compared with those cocultured with lower Lgals2 expression of 4T1 cells (all P < 0.05; Fig. 6B and fig. S6A). Furthermore, overexpression of Lgals2 in tumor cells stimulated macrophages to exhibit enhanced proliferation ability (Fig. 6C).
Fig. 6. Lgals2 facilitates M2-like polarization and proliferation of macrophages through CSF1.
(A) Schematic showing the 4T1 cells cocultured with mouse macrophages in a Transwell chamber of 0.4-μm pore size. (B) Arg1, Mgl1, and Fizz1 mRNA in mouse macrophages cocultured with Lgals2-overexpressing and vector control 4T1 cells for 72 hours were analyzed by qPCR. (C) CFSE-labeled mouse macrophages were cocultured with indicated 4T1 cells. Macrophage proliferation was quantified using fluorescence-activated cell sorting (FACS) analysis. Representative flow cytometry data are shown on the left, and quantification is shown on the right. (D and E) Heatmap of differentially expressed bulk RNA-seq genes between (D) Lgals2-overexpressing and vector control or (E) Lgals2-KO and vector control 4T1 cells in vitro. (F) qPCR validation of the differentially expressed gene Csf1 in 4T1 cells. (G and H) Mouse macrophages were cocultured with Lgals2-overexpressing and vector control 4T1 cells for 72 hours with/without CSF1R inhibitor PLX3397. (G) Arg1, Mgl1, and Fizz1 mRNA in mouse macrophages were analyzed by qPCR. (H) Macrophage proliferation was quantified using FACS analysis. (I) Tumor growth, tumor weight, and ex vivo images of resected tumors from transplanted 4T1 cells with vector control or Lgals2 overexpression in BALB/c mice following the treatment of PLX3397 (n = 7 each group). The arrow indicates the beginning time of treatment of PLX3397. If not noted otherwise, data are presented as the means ± SEM. ***P < 0.001, **P < 0.01, *P < 0.05, and NS P ≥ 0.05.
To identify genes downstream of Lgals2, we performed transcriptome profiling of in vitro cultured 4T1 cells with different expression of Lgals2, using bulk mRNA-seq with three replicates (Fig. 6, D and E). Using stringent criteria for changes in gene expression, we identified 110 genes up-regulated in Lgals2-overexpressing cells (log2FC > 1, FDR < 0.05) and down-regulated in Lgals2-KO cells (log2FC < −1, FDR < 0.05) (fig. S6B). KEGG pathway analysis revealed that two significantly enriched pathways are cytokine-cytokine receptor interaction and complement coagulation cascades with different expression of Lgals2 (fig. S6C). We validated the expression of five individual genes by quantitative polymerase chain reaction (qPCR), i.e., Csf1, Csf2rb, Vegfc, Cfh, and Muc1, and confirmed that all the results were consistent with the trends indicated by bulk RNA-seq (Fig. 6F and fig. S6D). Among these significant changes in genes, the cytokine Csf1 is a well-known regulator of TAMs in the recruitment to tumor sites and the maintenance of protumor function, and its receptor Csf1r was up-regulated in Mφ5 and Mφ6 cluster cells, as defined by scRNA-seq data accordingly (Fig. 5H) (23).
To determine whether up-regulation of Lgals2 induced tumor cells to release more CSF1 and facilitated M2-like polarization and proliferation of macrophages through the CSF1/CSF1R axis, we applied the CSF1R antagonist pexidartinib (PLX3397) in the coculture assay. Inhibition of the CSF1/CSF1R axis by PLX3397 significantly decreased the expression of Arg1, Mgl1, and Fizz1 in macrophages induced by Lgals2 overexpression in 4T1 cocultures (Fig. 6G). The addition of PLX3397 also abrogated the increased proliferation ability of macrophages when cocultured with Lgals2-overexpressing 4T1 cells as shown in the in vitro coculture assays (Fig. 6H). We further found that depletion of macrophages using PLX3397 in vivo could abrogate the promoting effects that Lgals2-overexpressing 4T1 cells had on tumor growth (Fig. 6I and fig. S6E). In addition, the decreased population and cytotoxic ability of infiltrating CD8+ T cells and NK cells in the Lgals2-overexpressing 4T1 tumors were reversed by PLX3397 (fig. S6, F to K). These findings suggest that the CSF1/CSF1R pathway likely plays a critical role in Lgals2-induced M2-like polarization and proliferation of macrophages.
Blockade of LGALS2 enhances antitumor immune responses and shows the immunotherapeutic potential of targeting LGALS2 in TNBC
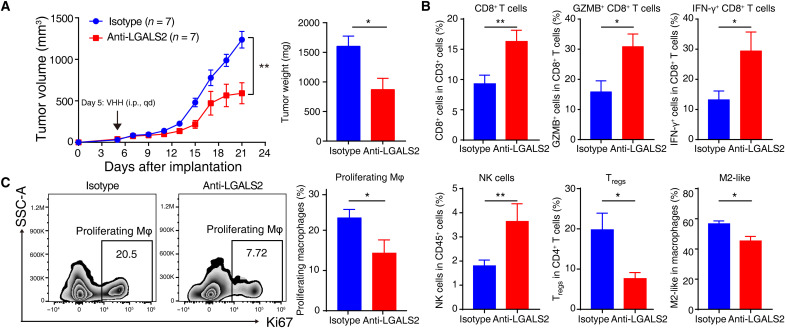
Loss of Lgals2 led to tumor cells being almost eliminated in mouse models; hence, we used a single-domain llama-derived therapeutic antibody to antagonize the LGALS2 protein in mouse models and investigated its potential therapeutic value (16). We observed that growth of tumor was slowed in the mice receiving anti-LGALS2 injection, with significant reductions in tumor volume and weight compared with the values in the control group injected with the isotype (n = 7 per group, P < 0.01; Fig. 7A and fig. S7A). Fewer immunosuppressive cells, including Tregs and M2-like macrophages, infiltrated in the anti-LGALS2 group, while higher percentages of CTLs and NK cells, as well as granzyme B– and IFN-γ–producing CD8+ T cells, were observed (Fig. 7B). After blockade of LGALS2, we observed a robust decrease in the percentage of proliferating macrophages, which might correspond to the Mφ6 subpopulation (with high expression of Mki67) indicated by scRNA-seq (P < 0.01; Fig. 7C). Together, immunosuppression in the TME seemed to be partly reversed by injecting anti-LGALS2 antibody.
Fig. 7. Blockade of LGALS2 enhances antitumor immune responses and shows the immunotherapeutic potential of targeting LGALS2 in TNBC.
(A) Tumor growth and tumor weight of intramammary fat pad tumors from transplanted 4T1 cells in BALB/c mice following isotype (n = 7) or anti-LGALS2 antibody (n = 7) injection. The arrow indicates the beginning time of injection of isotype or anti-LGALS2 antibody. **P < 0.01 and *P < 0.05. Data are presented as means ± SEM. (B) Primary tumors grown from 4T1 cells following different treatments were harvested for flow cytometry to determine the percentages of CD8+ T cells among CD3+ T cells, GZMB+ cells among CD8+ T cells, IFN-γ+ cells among CD8+ T cells, NK cells among CD45+ leukocytes, FOXP3+CD25+ Tregs among CD4+ T cells, and M2-like macrophages among total macrophages. **P < 0.01; *P < 0.05. Data are presented as the means ± SEM. (C) Representative plots and bar graphs of the percentages of proliferating macrophages among total macrophages from the primary tumors grown from 4T1 cells following different treatments. *P < 0.05. Data are presented as the means ± SEM.
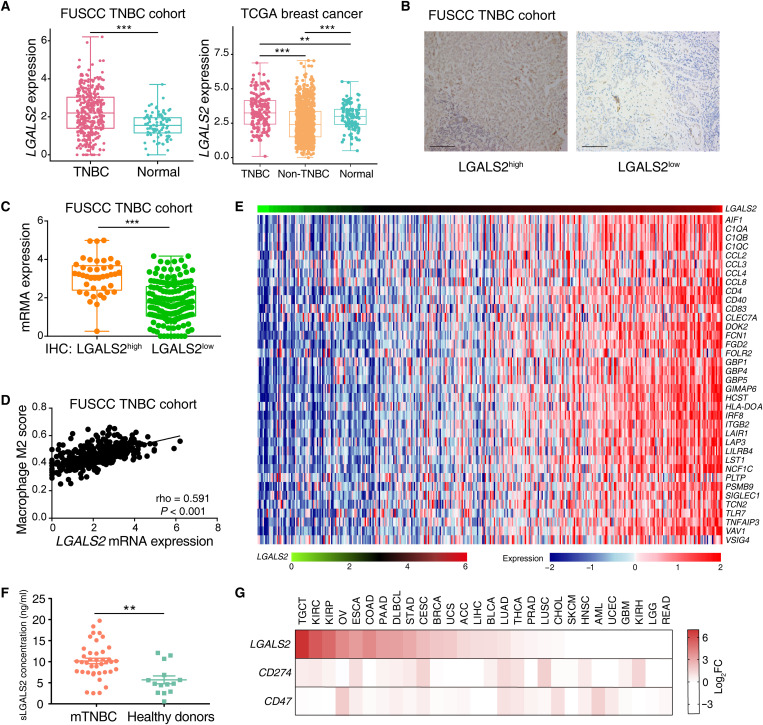
To evaluate the clinical potential of targeting LGALS2, we compared the mRNA expression levels of TNBC samples and normal tissues in our previously published TNBC cohort (21) and found relatively higher human LGALS2 expression in TNBC samples (P < 0.001; Fig. 8A). Transcriptomic data from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) and The Cancer Genome Atlas (TCGA) databases also validated that the expression of LGALS2 was elevated in TNBC samples compared with non-TNBC and normal tissues (all P < 0.01; Fig. 8A and fig. S7B). To estimate LGALS2 protein expression status in human tumor cells, we performed immunohistochemical (IHC) staining for LGALS2 on a tissue microarray (TMA) consisting of 357 TNBC tumor samples from Fudan University Shanghai Cancer Center (FUSCC). Of the 357 specimens, 109 exhibited high expression of LGALS2 protein on tumor cells, and 248 exhibited low expression (Fig. 8B). A total of 193 of 357 specimens from the TMA had corresponding transcriptome profiles, and we found there was a strong correlation between the mRNA expression levels of LGALS2 and the IHC staining results of LGALS2 protein expression levels (P < 0.001; Fig. 8C). Moreover, patients with higher expression of LGALS2 in tumor cells had higher tumor grade and greater tumor size (all P < 0.05; table S6). However, we found that there was no association between LGALS2 expression and relapse-free survival in the TNBC cohort (P = 0.452; fig. S7C).
Fig. 8. LGALS2 is up-regulated in TNBC and associated with M2-like macrophage markers.
(A) Box plot showing LGALS2 mRNA expression levels across TNBC, non-TNBC, and normal samples in the FUSCC and TCGA cohorts. Whiskers indicate the minimum and maximum values. ***P < 0.001; **P < 0.01. (B) Representative images of IHC staining of LGALS2 in TNBC tumor samples. Scale bars, 200 μm. (C) Box plot comparing LGALS2 mRNA expression level between high and low IHC scores of LGALS2 for TNBC in which mRNA expression and IHC were both available in the FUSCC cohort (n = 193). Whiskers indicate the minimum and maximum values. ***P < 0.001. (D) The association between LGALS2 mRNA expression levels and M2-like macrophage scores in TNBC patient samples from FUSCC (n = 360). The M2-like macrophage scores were computed as described by Xiao et al. (60). (E) Heatmap of the association between LGALS2 mRNA levels and a list of TAM-related genes in the FUSCC TNBC cohort (n = 360). FDR < 0.05 and Spearman rho > 0.3 were used as the criteria to select the most significant TAM markers for generating the heatmap. (F) Comparison of serum LGALS2 concentration between patients with metastatic TNBC and healthy donors. **P < 0.01. Whiskers indicate the means ± SEM. (G) Heatmap of LGALS2 tumor–to–matched normal mRNA expression ratios (log2FC) compared to known immune checkpoints in many different types of tumors.
Consistent with the animal studies, we observed a strong correlation between human LGALS2 levels and M2-like macrophage scores (rho = 0.591, P < 0.001; Fig. 8D). The expression of LGALS2 and CSF1 also showed a significant positive correlation (rho = 0.424, P < 0.001; fig. S7D). We ranked TNBC samples from FUSCC according to the mRNA levels of LGALS2 and revealed a strong correlation between LGALS2 and the recently published breast TAM signature of aggressive breast cancer subtypes (24), which included protumor TAM markers (CCL8 and SIGLEC1), chemokine-related genes (CCL2, CCL4, and CCL8), and complement-related genes (C1QA, C1QB, and C1QC) (Fig. 8E).
Previous studies reported that mammalian LGALS2 protein could be secreted extracellularly (25); hence, we tested the concentration of serum LGALS2 protein (sLGALS2) in tumor-bearing mice and humans by enzyme-linked immunosorbent assay (ELISA). The concentrations of sLgals2 were shown to be obviously decreased in Lgals2-KO 4T1 cell–induced tumor-bearing mice (both P < 0.05; fig. S7E). In murine models, Lgals2 expression levels in tumor cells seemed to be associated with the concentration of sLGALS2. When compared with 14 healthy donors, 40 patients with metastatic breast cancer had significantly increased concentrations of sLGALS2 (P < 0.01; Fig. 8F).
In addition, pan-cancer RNA-seq data from the TCGA, Therapeutically Applicable Research to Generate Effective Treatment Program (TARGET), and Genotype Tissue Expression Project (GTEX) databases revealed that relatively high expression of LGALS2 seemed to be a generalizable phenomenon in other tumors compared with normal samples, such as testicular germ cell tumors, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, and ovarian serous cystadenocarcinoma (Fig. 8G and fig. S7F), demonstrating that the therapeutic potential for LGALS2 blockade could be investigated in other cancer types.
DISCUSSION
To capture potential immunotherapy targets in complicated interactions between the immune system and tumor cells, we developed a customized DrIM library and a more precise mini-DrIM library for in vivo CRISPR screening. Immunocompetent mice and mice with various degrees of immunodeficiency were selected as screening models to generate different immune-selection pressures. Our customized in vivo CRISPR screen based on disparities of mouse immune systems aimed to observe the function shifts of immune-related genes across various TMEs and identify the interconversion of genes assisting between immune surveillance and immune escape. We successfully identified Lgals2 as a potential immunotherapy target for TNBC. Compared with the lack of discrepancy in vitro, the tumor-promoting effect of Lgals2 overexpression and the tumor-eliminating effect of Lgals2-KO in vivo most likely depended on the Lgals2-induced remodeling of TME. The usage of anti-LGALS2 antibody may provide a new strategy for immunotherapy of TNBC.
T cells are the main force in the antitumor response and turning points in immunotherapy. The effective blockade of immune checkpoints in clinical treatment, including CTLA4 and PD-1, is also related to T cells (26). Hence, previous CRISPR-Cas9 screens were conducted on cancer cells or T cells to identify potential immunotherapy targets or genes sensitizing tumors to immunotherapy (10, 27–29). Cancer cells are essential components of the TME, and tumor antigens are indispensable for immune systems to generate effective antitumor responses (30). Distinct from injecting mice with drugs to stimulate the immune system provisionally, our screens provided a new approach in seeking new immunotherapeutic targets based on unrest of inherent immune systems. In addition, accompanied by advances in immunotherapy, additional immune checkpoints were found and even extended to innate immune checkpoints such as CD47 and TIGIT (31, 32). Recently, B cells were also reported to affect immunotherapy responses. Our screens under different immune pressures characterized the function of genes among the complicated TME with the infiltration of various immune cells. Genes assisting immune surveillance or the immune escape of tumor cells were identified individually under pairwise comparisons between four kinds of mouse models with different TMEs. An exciting aspect of our screens is that well-known genes related to T cell exhaustion, such as IDO1 and ICOSL, emerged as assisting immune escape under the comparison between wild-type immunocompetent hosts and nude T cell–deficient mice, and NT5E emerged as assisting immune escape associated with NK cells in CB-17scid hosts relative to NPG hosts. However, in addition to cancer cell–intrinsic TME regulators, it is possible that many cell-extrinsic factors, including tumor-derived cytokines and chemokines, could also have effects on the TMEs. The in vivo screenings applied in additional cell types and more mouse models are likely to identify additional targets that can regulate the TME. Furthermore, a recent research pointed out that CRISPR vector components were immunogenic and may cause rejection in several mouse cancer models, which should be considered when performing in vivo CRISPR screening (33). Thus, pooled in vivo CRISPR screens free of the liability of iatrogenic neoantigen expression to tumor models should be performed in the subsequent CRISPR screening to reveal additional immunotherapy targets.
In two rounds of screens, Lgals2 was consistently correlated with immune escape and ranked top in the list of immune escape under all selection pressure. Lgals2 is a member of the galectin family and has a distinct expression in different tissues (34–36). It plays different roles in inflammation, immune response, and arteriogenesis, but its function in cancer has been only scarcely studied (13–16). Individual report showed that serum galectin-2 was increased in patients with colon and breast cancer, and elevated circulating galectin-2 levels were associated with increased mortality in patients with colorectal cancer (37). However, galectin-2 was found to play a suppressive role in colorectal tumor growth in another study (36). Moreover, previous researches revealed that the prognostic role of galectin-2 was tumor type dependent, with higher expression predicting worse prognosis in urothelial tumors while better outcomes in non–small cell lung cancer (38, 39). Although other members of the galectin family, such as galectin-1, galectin-3, and galectin-9, have been well investigated in cancers and clinical immunotherapy (40), the biological effect and function of galectin-2 in TNBC remain unclear. Given that Lgals2 was associated with tumor cell escape beyond surveillance by T cells, B cells, and NK cells, we hypothesized that the immunosuppression mechanism mediated by Lgals2 may be related to another kind of important immune cells, TAMs. Expansion of TAMs plays a prominent role in the evasion of tumors from established immune surveillance (41). In this study, we confirmed that Lgals2 could significantly influence the percentages of TAMs among TIICs in mouse models. Our results from the in vitro coculture experiment demonstrated that Lgals2 facilitates M2-like polarization and proliferation of macrophages. Moreover, analysis of our clinical TNBC dataset also supported a positive association between LGALS2 expression, M2-like macrophage activation, and TAM infiltration.
Transcriptional profiling revealed that Lgals2 depletion or overexpression led to the significant change of a variety of factors, including CSF1. Previous studies have confirmed that multiple tumors could secret CSF1 into the TME (42, 43). CSF1 controls the recruitment, proliferation, differentiation, survival, and function of monocytes and macrophages. CSF1R signaling in TAMs may promote their acquisition of an immunosuppressive and protumorigenic M2-like phenotype (41, 44). Our data revealed that CSF1/CSF1R blockade could reverse the M2-like polarization and proliferation of macrophages induced by Lgals2 overexpression in TNBC. Thus, CSF1 secretion by tumors may play an important role for the increased abundance of M2-like macrophages in Lgals2-overexpressing tumors.
Although our study focused on the macrophage population because of its huge change from the scRNA-seq analysis of TIIC populations, we could not exclude the possibility that other types of immune cells may also be involved in the immune escape in Lgals2-overexpressing tumors. For example, DCs, which decreased in the Lgals2-overexpressing tumors, could enhance antitumor immune response by presenting antigen and priming T cells (45). In contrast, tumor-associated neutrophils could inhibit an antitumor immune response (46). Thus, it is still needed to be further explored whether Lgals2 could directly influence the activity of other immune cells in the TME, which is ongoing in our laboratory.
Our study also has important implications for clinical translations. We showed that LGALS2 is a potential target, and its depletion could restrain cancer development with enhanced antitumor immunity. A single-domain llama-derived antibody has been developed to inhibit the LGALS2 protein in the cardiovascular field (16). Our study demonstrated that the tumor growth and the immunosuppression phenotype in the TME could be inhibited by applying this therapeutic antibody, suggesting its potential value for clinical applications. However, further effort is needed to test its side effect and optimize its targeting and pharmacodynamic efficacy. More efficient and selective LGALS2 inhibitors are also needed to be developed for preclinical and clinical evaluation of the feasibility of targeting LGALS2 in TNBC and other cancers.
In summary, our screens provided a systematic and efficient approach to uncover potential targets of immunotherapy targets. The validation and characterization of a candidate target, Lgals2, proved that perturbation of this gene in cancer cells could obtain remarkable antitumor activity in vivo. However, the deeper mechanisms are still unknown, and future work is needed. In addition to TNBC, further studies regarding the role of Lgals2 involving multiple cancers would be worthwhile. Our findings have direct implication for revisiting the interaction of tumor cells and the immune system, which may serve as a blueprint to develop new drugs and therapeutic strategies.
MATERIALS AND METHODS
Cell lines and culture conditions
We used murine breast cancer cell lines 4T1 (gift from Y. Kang of Princeton University) and EMT6 (from Nanjing Cobioer), and human embryonic kidney (HEK) 293T cells (from the Shanghai Cell Bank Type Culture Collection Committee) in this study. All cell lines were grown in complete growth medium and cultured in a humidified incubator at 37°C and 5% CO2 according to standard protocols. Each cell line identity was verified by short tandem repeat profiling. Only cells within 6 months of thawing were used for the current study. For mouse peritoneal macrophage isolation, BALB/c mice were injected intraperitoneally with 3 ml of 3% thioglycollate. Four days after injection, the peritoneal cavity was washed with 5 ml of Dulbecco’s modified Eagle’s medium (DMEM) with 1% fetal bovine serum. Cells from the peritoneal exudates were collected and seeded on dishes. Adherent cells were incubated with DMEM supplemented with 10% fetal bovine serum.
DrIM and mini-DrIM library design
To construct the DrIM library, we used the Human-Mouse: Disease Connection search tool to select all 2796 genes, which corresponded to genes related to human diseases and immune system, from the Mouse Genome Informatics database (www.informatics.jax.org/). We chose four sgRNAs for each gene. The final DrIM library contained a total of 12,000 sgRNAs, including 11,184 sgRNAs targeting 2796 immune-related mouse genes and 816 nontargeting control sgRNAs (table S1).
According to the screening results from the DrIM library, 273 candidate genes were selected for the second-round screening. Each candidate gene was matched with 10 sgRNAs, with the addition of 816 nontargeting control sgRNAs, resulting in a total of 3546 sgRNAs in the mini-DrIM library (table S3).
Array oligo synthesis and pooled library cloning
DNA oligonucleotide library synthesis was performed on a microarray as recommended by the manufacturer. Full-length oligonucleotides were amplified by PCR using Q5 High-Fidelity DNA Polymerase (NEB) and cloned by Gibson ligation reaction (NEB) assembly into the lentiGuide-Puro vector to generate the DrIM and mini-DrIM libraries. To ensure no loss of representation, an estimated library coverage of >300× was achieved by electroporation. The library was subsequently purified and sequenced on a HiSeq 2500 (Illumina) to monitor the change in the abundance of each sgRNA between the initial and final cell populations.
Virus production and transduction
To generate cells stably expressing Cas9, we introduced lentiCas9-Blast (Addgene), pMD2.G (Addgene), and psPAX2 (Addgene) constructs into HEK293T cells for lentivirus packaging. Forty-eight hours after virus transfection, 4T1 cells were selected with blasticidin (5 μg/ml) for 7 days to obtain stably integrated cells. Subsequently, we used the DrIM or mini-DrIM library, with pMD2.G and psPAX2 constructs for virus packaging. Cells were infected at a low multiplicity of infection (MOI = 0.3) to ensure that each cell received approximately one viral copy with high probability. For virus pool infection, a total of 5 × 107 cells were plated in 12-well dishes with 3 × 106 cells per well and centrifuged in the presence of viral supernatant and polybrene for 2 hours at 2000 rpm. Forty-eight hours after infection, the stably integrated cells were selected with puromycin (5 μg/ml) for 7 days. After 7 days, 3 × 107 cells were spun down and frozen for genomic DNA extraction. At the same time, cells were washed twice in sterile phosphate-buffered saline (PBS) and resuspended at 1.5 × 107 cells/ml in PBS for transplantation.
For genetic KO of Lgals2, sgRNAs targeting individual genes were cloned into the lentiGuide-Puro vectors. Single sgRNA virus was generated by the transfection of HEK293T cells using the procedure described for the library virus. After harvest, the viruses were introduced into Cas9 cells. Forty-eight hours after infection, the stably integrated cells were selected with puromycin (5 μg/ml) for 7 days. The KO efficiency was confirmed by Western blot.
To express Lgals2 in the cells of interest, we generated a lentivirus expression construct for the genes of interest by cloning the corresponding DNA fragments into a pCDH vector. We transfected HEK293T cells with pCDH, pMD2.G, and psPAX2 constructs to generate the lentivirus. The supernatant medium containing the virus was collected and used to infect 4T1 and EMT6 cells, and stably integrated cells were selected with puromycin (5 μg/ml) for 7 days.
Xenograft models
All in vivo experiments used 6-week-old female mice maintained under pathogen-free conditions. NPG mice were obtained from Beijing Vitalstar Biotechnology, and BALB/c, BALB/c-Nude, and CB-17scid mice were obtained from Beijing Vital River Laboratory Animal Technology. All animal experiments were performed according to protocols approved by the Research Ethical Committee of FUSCC.
In vivo mouse studies
For the first-round CRISPR screening, DrIM-transduced 4T1-Cas9 cells were injected subcutaneously into the mammary fat pad region of BALB/c or NPG mice at 3 × 106 cells per mouse. For the second-round CRISPR screening, mini-DrIM–transduced 4T1-Cas9 cells were injected subcutaneously into the mammary fat pad region of BALB/c, BALB/c-nude, CB-17scid, or NPG mice at 3 × 106 cells per mouse. For the single-gene validation and drug treatment experiment, gene-edited 4T1 or EMT6 cells or control cells were injected subcutaneously into the mammary fat pad region of BALB/c mice at 1 × 106 cells per mouse. In the treatment experiment, after the tumor volume reached 100 mm3, mice were randomly assigned to the control group and treatment group. For the macrophage depletion experiment, mice were treated with daily oral doses CSF1R inhibitor PLX3397 (40 mg/kg; Selleck). For the nanobody treatment experiment, anti-LGALS2 VHH (2C10, QVQ) or anti-HIV VHH (1F10, QVQ) as isotype was injected intraperitoneally every day as previously described (16). The treatment consisted of a charging dose directly after randomization, followed by a daily maintenance dose. The investigator was blinded to the treatment groups during the experiment and when assessing the outcome. Mice were monitored daily and euthanized by CO2 asphyxiation and cervical dislocation before any sign of distress. After mice were euthanized, primary tumors were dissected for further analysis.
Genomic DNA extraction and sgRNA library readout
We performed next-generation sequencing to determine the abundance of sgRNAs in the primary tumors. We also sequenced the library input plasmid and the pretransplantation library-transduced baseline cells. A QIAamp DNA Mini Kit (Qiagen) was applied to extract genomic DNA from cells and xenografts according to the manufacturer’s protocol. A two-step PCR amplification procedure was performed on genomic DNA using NEBNext High-Fidelity 2× PCR Master Mix (NEB). For the first PCR, the amount of input genomic DNA for each sample was calculated to achieve 300× library coverage, which resulted in 23.76 μg of DNA per sample for DrIM library and 7.02 μg of DNA per sample for mini-DrIM library. A region containing the sgRNA cassette was amplified using primers specific to the sgRNA expression vector: first PCR primer forward, AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG; first PCR primer reverse, CTTTAGTTTGTATGTCTGTTGCTATTATGTCTACTATTCTTTCC.
The first PCR products for each sample were pooled and used for amplification with barcoded second-step PCR primers. The second PCR was performed in a 20-μl reaction volume using 0.125 ng of the product from the first PCR. The primers used for the second PCR include an 8–base pair barcode for the multiplexing of different biological samples: second PCR primer forward, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT (index) TCTTGTGGAAAGGACGAAACACCG; second PCR primer reverse, CAAGCAGAAGACGGCATACGAGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTACTATTCTTTCCCCTGCACTGT. The amplicons resulting from the second PCR were purified with beads (Beckman), quantified, mixed, and sequenced using a HiSeq 2500 system (Illumina).
CRISPR screening analysis
The raw FASTQ files were demultiplexed using Geneious 8.0 (Biomatters Inc.). The number of reads of each unique sgRNA for a certain sample was normalized by the total reads for each sample and then log transformed. We used the MAGeCK algorithm to quantify the enrichment of candidate genes (47). The MAGeCK algorithm using an MLE approach (“MAGeCK-MLE”) can model complex experimental designs. In MAGeCK-MLE, the read count of an sgRNA was modeled as a negative binomial random variable, and the effects of different conditions were represented as β scores. The β scores reflect the extent of selection in each condition: βgr > 0 (or < 0) means that g is positively (or negatively) selected in condition r, and the default control condition is cells before transplantation with library transduced. The values of β, together with the information of whether an sgRNA is efficient, could be estimated by maximizing the joint log-likelihood of observing all sgRNA read counts of g on all different samples and were optimized using an expectation-maximization algorithm. Large absolute β scores mean that genes have enormous effects during screens, and β scores close to zero mean that minor effects happen. The δ score was defined as the difference in the β scores of a single gene between two conditions.
Human tumor bulk RNA-seq data
Cohorts of patients with breast cancer from FUSCC, METABRIC, and TCGA have been previously described (21, 48, 49). Expression and clinical data of METABRIC and TCGA cohorts were downloaded from the website (www.cbioportal.org/). All samples were from previously untreated primary breast cancers. TNBC sample selection of METABRIC was based on the expression data because METABRIC data lack estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2.
Expression data of pan-cancer cohorts collected by TCGA, TARGET, and GTEX were downloaded as log2(normalized counts + 1) values from UCSC Xena (https://xenabrowser.net/). Tumor types with fewer than nine individual patients for either tumor or healthy tissues were excluded. Abbreviations and full names of tumor types and the number of samples analyzed are listed in table S7.
In vitro cell proliferation assays
Short-term viability assays were performed by plating cells in 100 μl of their corresponding growth medium using optimal seeding densities per well in 96-well plates. Optimal seeding densities were established for each cell line to reach 75 to 80% confluence at the end of the assay. Cell viability was assessed with the Cell Counting Kit-8 (Dojindo) at the indicated time points. The absorbance was measured at 450 nm.
Western blot
For immunoblots, total protein was extracted with T-PER Tissue Protein Extraction Reagent (Thermo Fisher) with 2% SDS and 1× protease inhibitor (Biosharp). BCA Protein Assay Kit (Solarbio) was used to determine the protein concentration. Protein samples were normalized for protein content, resolved by SDS–polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes (Millipore), and incubated overnight at 4°C with the appropriate primary antibodies. Signals were detected using peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch), and images were captured using Amersham Imager 600 (Cytiva).
Flow cytometry analysis
For flow cytometry analysis of in vivo experiments, mouse tumors and spleens were quickly excised and then mechanically dissociated with scissors in sterile PBS. Splenocytes were filtered in 70-μm cell strainers. Tumor fragments were digested in serum-free RPMI + deoxyribonuclease I (20 μg/ml; Roche) + collagenase D (1 mg/ml; Roche) + collagenase I (1 mg/ml; Sigma-Aldrich) for 30 to 60 min at 37°C with rotation to promote dissociation. Single-cell suspensions were passed through a 70-μm cell strainer. Red blood cells in both tumor and spleen samples were then lysed with red blood cell lysis buffer (eBioscience) for 5 min at room temperature. The lysis reactions were quenched by the addition of 20 ml of PBS, and samples were centrifuged at 300g for 5 min at 4°C. Cells were then washed in D-PBS and stained with BD Horizon Fixable Viability Stain 700 (BD Biosciences) at a 1:3000 dilution in D-PBS for 15 min at 4°C. A monoclonal antibody to CD16/32 (BioLegend) was used to block cells for 15 min at 4°C before staining with antibody panels. To determine cytokine production, cells were cultured in 0.5 ml of complete RPMI medium and stimulated at 37°C with 1 μl of Cell Activation Cocktail (with brefeldin A) (BioLegend) in 24-well plates for 4.5 to 6 hours. After stimulation, cells were stained with fluorescently labeled antibodies to the surface proteins at a 1:100 dilution in Cell Staining Buffer (BioLegend) for 30 min at 4°C. For intracellular protein analyses, cells were then fixed and permeabilized by using Fixation Buffer (BioLegend) and Intracellular Staining Permeabilization Wash Buffer (BioLegend) according to the manufacturers’ instructions. Transcription Factor Buffer Set (BD Biosciences) was applied if intranuclear transcription factors were planned to be detected. Permeabilized cells were then incubated with fluorescently labeled antibodies to the intracellular proteins for 30 min at 4°C. A CytoFLEX S Flow Cytometer (Beckman Coulter) was used for flow cytometry data acquisition, and data were analyzed with FlowJo software (version 10.5.3, TreeStar).
Immunofluorescence
We performed immunofluorescence on formalin-fixed and paraffin-embedded (FFPE) sections (4 μm thick) of 4T1 murine tumors from BALB/c mice. The FFPE slides were deparaffinized in xylene and then rehydrated successively in 100, 90, and 70% alcohol. The antigen was retrieved by EDTA buffer (pH 9.0) at 95°C for 10 min. Before staining, slides were blocked with 5% goat serum in PBS. After blocking, primary antibodies were incubated overnight at 4°C, and secondary antibodies were incubated for 1 hour at room temperature in 0.1% bovine serum albumin (BSA) in PBS solution. After the slides were washed three times with PBS for 5 min, slides were incubated with 4′,6-diamidino-2-phenylindole (DAPI) solution for another 10 min at room temperature. Images were collected using a confocal microscope (Leica). Images of three nonoverlapping optical fields covering the surface of the tumor sections were captured.
Single-cell RNA-seq
Single-cell suspensions were generated using the Mouse Tumor Dissociation Kit (Miltenyi Biotec) according to the manufacturer’s protocol. A Dead Cell Removal Kit (Miltenyi Biotec) was applied to eliminate dead cells from the single-cell suspensions. The cell suspension was loaded into Chromium microfluidic chips to establish single-cell gel beads in emulsion (GEMs) for the directed retrieval of approximately 5000 cells and barcoded with the Chromium Controller (10x Genomics). RNA from the barcoded cells was subsequently reverse transcribed, and sequencing libraries were constructed with reagents from a Chromium Single Cell 3’ Reagent Kit v3 (10x Genomics) according to the manufacturer’s instructions. Sequencing was performed with the Illumina sequencing platform (NovaSeq) in Novogene.
Raw reads were demultiplexed and mapped to the reference genome (mm10, GRCm38) by the 10x Genomics Cell Ranger pipeline (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger). For each gene and each cell barcode filtered by Cell Ranger, unique molecule identifiers (UMIs) were counted to construct digital expression matrices. The resulting analysis files for each sample were aggregated using the cellranger aggr pipeline, which performed a between-sample normalization step and merged two samples into one.
The Seurat package was applied to the combined dataset (50). A gene with expression in more than three cells was considered to be expressed, and each cell was required to have at least 200 expressed genes. Raw UMI counts were normalized, and most variable genes were detected by the FindVariableFeatures function. Principal components analysis (PCA) was performed using variable genes. For clustering, we used the function FindClusters, which implements shared nearest neighbor based on PCA using the first 20 principal components with a resolution of 0.6. A t-SNE dimensional reduction analysis was performed to obtain a two-dimensional representation of the cell states. The FindAllMarkers function was applied to identify marker genes, which were used with the Wilcoxon rank sum test to determine significant genes. These marker genes were used to assign cluster identity to individual cell types based on the CellMarker database (51) and existing literature (52–54).
To examine T cells, clusters expressing Cd3e were extracted from aggregated samples. Cells expressing Csf1r were also extracted for reanalysis. Variable gene selection analysis, PCA, clustering, t-SNE dimensional analysis, and marker selection analysis of T cell cluster and monocyte/DC/macrophage population were performed as described above.
qPCR and bulk RNA-seq
Total RNA was extracted using the RNeasy Mini Kit (Qiagen) following the protocol in the kit. Complementary DNA (cDNA) for qPCR was generated using HiScript III RT SuperMix for qPCR (Vazyme). Afterward, cDNA was subjected to qPCR using ChamQ SYBR qPCR Master Mix (Vazyme). Specific primers are listed in table S8. Samples were processed using the Applied Bioscience 7900HT Fast Real-Time PCR System, and relative mRNA expression was normalized to Actb and was determined via the ΔΔCt method.
Triplicate total RNA samples from each group were subjected to bulk RNA-seq library preparation. The samples with RNA integrity number ≥ 7 were subjected to subsequent analysis. RNA libraries were constructed using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) according to the manufacturer’s protocol. The libraries were sequenced on the Illumina sequencing platform (HiSeq X Ten) in Shanghai OE Biotech.
Bulk RNA-seq processing and analysis
Raw data were processed using Trimmomatic (55), and the reads containing ploy-N and the low-quality reads were removed to obtain the clean reads. Then, the clean reads were mapped to a mouse reference genome (mm10, GRCm38). The read counts of each gene were obtained by htseq-count (56), and the fragments per kilobase of exon model per million mapped fragments (FPKM) values were calculated using cufflinks (57). Differentially expressed genes were identified using DESeq2 (58).
Coculture assay
The 24-well Transwell chambers with 0.4-μm-pore polycarbonate membrane were applied for the coculture assay. Mouse peritoneal macrophages (5 × 105) were seeded in the lower chamber 24 hours before coculture, and 1000 tumor cells were added in the upper chamber. After 72 hours, macrophages were collected for subsequent analyses including qPCR and flow cytometry. To detect the macrophage proliferation ability, the macrophages were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) following the kit’s instructions (CFSE Cell Division Tracker Kit, BioLegend). CFSE-labeled macrophages were cocultured with the indicated cells. Macrophage proliferation was quantified using flow cytometry analysis.
Serum samples and TNBC TMAs
Serum samples were collected from patients with metastatic TNBC and healthy donors at FUSCC. All samples were immediately stored in a −80°C refrigerator and thawed on ice before analysis. FFPE tissue samples from 357 female patients with histologically confirmed unilateral stage I to III primary TNBC who underwent mastectomy at FUSCC were examined and used to generate TMAs. Written informed consent was received from the participants before their inclusion in the study. This study was approved by the Research Ethical Committee of FUSCC.
IHC analysis
The TMA sections were generated by the Department of Pathology at the FUSCC. IHC staining of TMAs was performed as previously described (59). Briefly, the TMA sections were placed at 70°C for 1 hour, deparaffinized in xylene, and then rehydrated successively in 100, 90, and 70% alcohol. The antigen was retrieved by citric acid buffer (pH 6.0) at 95°C for 20 min. The inactivation of endogenous peroxidase and the blockage of nonspecific sites were achieved using a two-step protocol (GTVision III). The TMAs were incubated overnight at 4°C with the primary antibody rabbit polyclonal anti-LGALS2 (LSBio). After the sections were washed twice with PBS for 5 min, the antigen-binding sites were visualized using the GTVision III detection system/Mo&Rb according to the manufacturer’s protocol. TMAs representing duplicate samples from each case were stained and scored semiquantitatively. The staining was graded on the basis of the staining intensity (0, negative; 1, weak; 2, strong) and the percentage of stained cells (0, 0 to <5%; 1, 5 to <25%; 2, 25 to <75%; 3, 75 to 100%). A score ranging from 0 to 6 was calculated by multiplying the staining extent score by the intensity score. Tumors with a score greater than or equal to 3 were considered to exhibit high LGALS2 expression, whereas those with a score less than 3 were classified as showing low LGALS2 expression. The score used for all the subsequent analyses was the average of the available scores, and the scores were reviewed in parallel by two experienced breast disease pathologists who were blinded to all clinical data.
Enzyme-linked immunosorbent assay
To measure the concentration of LGALS2 in the peripheral blood, serum samples from patients with metastatic breast cancer, healthy donors, and BALB/c mice transplanted with 4T1 cells with vector control or sgLgals2 were used to perform ELISA assays with LGALS2 antibody (Signalway Antibody) according to the manufacturer’s protocol. The experiments were conducted three times.
Statistic summary
All statistical analyses were performed with R software (www.R-project.org, version 3.5.2) or GraphPad Prism software (version 7.0b), unless noted otherwise in the method details. The experimenters were blinded to the group assignments and outcome assessments. All cell-based in vitro experiments were independently repeated three times in triplicate. For the in vivo experiments, n represents the number of animals used for each condition; the animals were randomly assigned to either the experimental or control group using the random number table method. Student’s t test, Mann-Whitney test, and Kruskal-Wallis test were applied to compare continuous variables and ordered categorical variables where appropriate. Survival curves were constructed using the Kaplan-Meier method and compared with the log-rank test. Correlation analysis was conducted with Spearman’s correlation. All results are shown as the means ± SEM, unless otherwise indicated. Two-sided P values less than 0.05 were considered statistically significant. FDR correction was used in multiple tests to decrease false-positive rates.
Acknowledgments
We are thankful to the TCGA Research Network, the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), the Therapeutically Applicable Research to Generate Effective Treatment Program (TARGET), and the Genotype Tissue Expression Project (GTEX) for providing the data analyzed in this study. We thank H. Wang and Y. Zhu for helpful suggestions and technical assistance.
Funding: This work was supported by grants from the National Natural Science Foundation of China (81672601, 81872137, and 82103369), the Ministry of Science and Technology of China (2018YFE020160), and the National Science and Technology Major Project (2020ZX09201-013). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions: Z.-M.S., X.H., and G.-H.D. participated in the conception and design of the study. P.J., Y.G., M.-L.J., and X.H. developed the methodology. P.J., Y.G., M.-L.J., H.-L.W., L.-W.G., W.-J.C., Y.L., and X.-Y.M. performed the experiments. P.J., Y.G., M.-L.J., H.-L.W., and Y.-C.P. performed bioinformatic and statistical analysis of the data. P.J., Y.G., M.-L.J., Y.-Z.J., G.-H.D., X.H., and Z.-M.S. wrote, reviewed, and/or revised the manuscript. G.-H.D., X.H., and Z.-M.S. supervised the study. All authors approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The results shown here are based, in part, on data generated by the FUSCC, TCGA, METABRIC, TARGET, and GTEX cohorts and are available in a public repository from the https://xenabrowser.net/ and www.cbioportal.org/ websites. Our sequencing datasets are deposited in the NCBI Sequence Read Archive (SRA) database under the accession codes PRJNA742668, PRJNA743653, and PRJNA743660. All data can be viewed in the SRA website (www.ncbi.nlm.nih.gov/sra) by pasting the accession number (PRJNA742668, PRJNA743653, and PRJNA743660) into the text search box or through the following hyperlinks: https://trace.ncbi.nlm.nih.gov/Traces/study/?acc=, PRJNA742668; https://trace.ncbi.nlm.nih.gov/Traces/study/?acc=, PRJNA743653; and https://trace.ncbi.nlm.nih.gov/Traces/study/?acc=, PRJNA743660. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S8
REFERENCES AND NOTES
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Denkert C., Liedtke C., Tutt A., von Minckwitz G., Molecular alterations in triple-negative breast cancer—The road to new treatment strategies. Lancet 389, 2430–2442 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Wilky B. A., Immune checkpoint inhibitors: The linchpins of modern immunotherapy. Immunol. Rev. 290, 6–23 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Schmid P., Rugo H. S., Adams S., Schneeweiss A., Barrios C. H., Iwata H., Dieras V., Henschel V., Molinero L., Chui S. Y., Maiya V., Husain A., Winer E. P., Loi S., Emens L. A.; IMpassion130 Investigators , Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 21, 44–59 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Schmid P., Cortes J., Pusztai L., McArthur H., Kummel S., Bergh J., Denkert C., Park Y. H., Hui R., Harbeck N., Takahashi M., Foukakis T., Fasching P. A., Cardoso F., Untch M., Jia L., Karantza V., Zhao J., Aktan G., Dent R., O’Shaughnessy J.; KEYNOTE-522 Investigators , Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 382, 810–821 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Schoenfeld A. J., Hellmann M. D., Acquired resistance to immune checkpoint inhibitors. Cancer Cell 37, 443–455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy L. B., Salama A. K. S., A review of cancer immunotherapy toxicity. CA Cancer J. Clin. 70, 86–104 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Kon E., Benhar I., Immune checkpoint inhibitor combinations: Current efforts and important aspects for success. Drug Resist. Updat. 45, 13–29 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Azizi E., Carr A. J., Plitas G., Cornish A. E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., Choi K., Fromme R. M., Dao P., McKenney P. T., Wasti R. C., Kadaveru K., Mazutis L., Rudensky A. Y., Pe’er D., Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308.e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manguso R. T., Pope H. W., Zimmer M. D., Brown F. D., Yates K. B., Miller B. C., Collins N. B., Bi K., LaFleur M. W., Juneja V. R., Weiss S. A., Lo J., Fisher D. E., Miao D., Van Allen E., Root D. E., Sharpe A. H., Doench J. G., Haining W. N., In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelson T., Heckl D., Ebert B. L., Root D. E., Doench J. G., Zhang F., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber J., Braun C. J., Saur D., Rad R., In vivo functional screening for systems-level integrative cancer genomics. Nat. Rev. Cancer 20, 573–593 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Ozaki K., Inoue K., Sato H., Iida A., Ohnishi Y., Sekine A., Sato H., Odashiro K., Nobuyoshi M., Hori M., Nakamura Y., Tanaka T., Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-alpha secretion in vitro. Nature 429, 72–75 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Sturm A., Lensch M., Andre S., Kaltner H., Wiedenmann B., Rosewicz S., Dignass A. U., Gabius H. J., Human galectin-2: Novel inducer of T cell apoptosis with distinct profile of caspase activation. J. Immunol. 173, 3825–3837 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Yildirim C., Vogel D. Y., Hollander M. R., Baggen J. M., Fontijn R. D., Nieuwenhuis S., Haverkamp A., de Vries M. R., Quax P. H., Garcia-Vallejo J. J., van der Laan A. M., Dijkstra C. D., van der Pouw Kraan T. C., van Royen N., Horrevoets A. J., Galectin-2 induces a proinflammatory, anti-arteriogenic phenotype in monocytes and macrophages. PLOS ONE 10, e0124347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollander M. R., Jansen M. F., Hopman L., Dolk E., van de Ven P. M., Knaapen P., Horrevoets A. J., Lutgens E., van Royen N., Stimulation of collateral vessel growth by inhibition of galectin 2 in mice using a single-domain llama-derived antibody. J. Am. Heart Assoc. 8, e012806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata M., Ham K., Hoque M. O., A time for YAP1: Tumorigenesis, immunosuppression and targeted therapy. Int. J. Cancer 143, 2133–2144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith C., Chang M. Y., Parker K. H., Beury D. W., DuHadaway J. B., Flick H. E., Boulden J., Sutanto-Ward E., Soler A. P., Laury-Kleintop L. D., Mandik-Nayak L., Metz R., Ostrand-Rosenberg S., Prendergast G. C., Muller A. J., IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Discov. 2, 722–735 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solinas C., Gu-Trantien C., Willard-Gallo K., The rationale behind targeting the ICOS-ICOS ligand costimulatory pathway in cancer immunotherapy. ESMO Open 5, e000544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chambers A. M., Lupo K. B., Matosevic S., Tumor microenvironment-induced immunometabolic reprogramming of natural killer cells. Front. Immunol. 9, 2517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y. Z., Ma D., Suo C., Shi J., Xue M., Hu X., Xiao Y., Yu K. D., Liu Y. R., Yu Y., Zheng Y., Li X., Zhang C., Hu P., Zhang J., Hua Q., Zhang J., Hou W., Ren L., Bao D., Li B., Yang J., Yao L., Zuo W. J., Zhao S., Gong Y., Ren Y. X., Zhao Y. X., Yang Y. S., Niu Z., Cao Z. G., Stover D. G., Verschraegen C., Kaklamani V., Daemen A., Benson J. R., Takabe K., Bai F., Li D. Q., Wang P., Shi L., Huang W., Shao Z. M., Genomic and transcriptomic landscape of triple-negative breast cancers: Subtypes and treatment strategies. Cancer cell 35, 428–440 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Chen L., Diao L., Yang Y., Yi X., Rodriguez B. L., Li Y., Villalobos P. A., Cascone T., Liu X., Tan L., Lorenzi P. L., Huang A., Zhao Q., Peng D., Fradette J. J., Peng D. H., Ungewiss C., Roybal J., Tong P., Oba J., Skoulidis F., Peng W., Carter B. W., Gay C. M., Fan Y., Class C. A., Zhu J., Rodriguez-Canales J., Kawakami M., Byers L. A., Woodman S. E., Papadimitrakopoulou V. A., Dmitrovsky E., Wang J., Ullrich S. E., Wistuba I. I., Heymach J. V., Qin F. X., Gibbons D. L., CD38-mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov. 8, 1156–1175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batool A., Wang Y. Q., Hao X. X., Chen S. R., Liu Y. X., A miR-125b/CSF1-CX3CL1/tumor-associated macrophage recruitment axis controls testicular germ cell tumor growth. Cell Death Dis. 9, 962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassetta L., Fragkogianni S., Sims A. H., Swierczak A., Forrester L. M., Zhang H., Soong D. Y. H., Cotechini T., Anur P., Lin E. Y., Fidanza A., Lopez-Yrigoyen M., Millar M. R., Urman A., Ai Z., Spellman P. T., Hwang E. S., Dixon J. M., Wiechmann L., Coussens L. M., Smith H. O., Pollard J. W., Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 35, 588–602.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes R., Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim. Biophys. Acta 1473, 172–185 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Hoos A., Development of immuno-oncology drugs - From CTLA4 to PD1 to the next generations. Nat. Rev. Drug Discov. 15, 235–247 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Parnas O., Jovanovic M., Eisenhaure T. M., Herbst R. H., Dixit A., Ye C. J., Przybylski D., Platt R. J., Tirosh I., Sanjana N. E., Shalem O., Satija R., Raychowdhury R., Mertins P., Carr S. A., Zhang F., Hacohen N., Regev A., A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel S. J., Sanjana N. E., Kishton R. J., Eidizadeh A., Vodnala S. K., Cam M., Gartner J. J., Jia L., Steinberg S. M., Yamamoto T. N., Merchant A. S., Mehta G. U., Chichura A., Shalem O., Tran E., Eil R., Sukumar M., Guijarro E. P., Day C. P., Robbins P., Feldman S., Merlino G., Zhang F., Restifo N. P., Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong M. B., Wang G., Chow R. D., Ye L., Zhu L., Dai X., Park J. J., Kim H. R., Errami Y., Guzman C. D., Zhou X., Chen K. Y., Renauer P. A., Du Y., Shen J., Lam S. Z., Zhou J. J., Lannin D. R., Herbst R. S., Chen S., Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178, 1189–1204.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanmamed M. F., Chen L., A paradigm shift in cancer immunotherapy: From enhancement to normalization. Cell 175, 313–326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng M., Jiang W., Kim B. Y. S., Zhang C. C., Fu Y. X., Weissman I. L., Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat. Rev. Cancer 19, 568–586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q., Bi J., Zheng X., Chen Y., Wang H., Wu W., Wang Z., Wu Q., Peng H., Wei H., Sun R., Tian Z., Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 19, 723–732 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Dubrot J., Lane-Reticker S. K., Kessler E. A., Ayer A., Mishra G., Wolfe C. H., Zimmer M. D., Du P. P., Mahapatra A., Ockerman K. M., Davis T. G. R., Kohnle I. C., Pope H. W., Allen P. M., Olander K. E., Iracheta-Vellve A., Doench J. G., Haining W. N., Yates K. B., Manguso R. T., In vivo screens using a selective CRISPR antigen removal lentiviral vector system reveal immune dependencies in renal cell carcinoma. Immunity 54, 571–585.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Nio-Kobayashi J., Tissue- and cell-specific localization of galectins, β-galactose-binding animal lectins, and their potential functions in health and disease. Anat. Sci. Int. 92, 25–36 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Thijssen V. L., Hulsmans S., Griffioen A. W., The galectin profile of the endothelium: Altered expression and localization in activated and tumor endothelial cells. Am. J. Pathol. 172, 545–553 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Zhao L., Lau Y. S., Zhang C., Han R., Genome-wide CRISPR screen identifies LGALS2 as an oxidative stress-responsive gene with an inhibitory function on colon tumor growth. Oncogene 40, 177–188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrow H., Guo X., Wandall H. H., Pedersen J. W., Fu B., Zhao Q., Chen C., Rhodes J. M., Yu L. G., Serum galectin-2, -4, and -8 are greatly increased in colon and breast cancer patients and promote cancer cell adhesion to blood vascular endothelium. Clin. Cancer Res. 17, 7035–7046 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Langbein S., Brade J., Badawi J. K., Hatzinger M., Kaltner H., Lensch M., Specht K., Andre S., Brinck U., Alken P., Gabius H. J., Gene-expression signature of adhesion/growth-regulatory tissue lectins (galectins) in transitional cell cancer and its prognostic relevance. Histopathology 51, 681–690 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Uso M., Jantus-Lewintre E., Calabuig-Farinas S., Blasco A., Garcia Del Olmo E., Guijarro R., Martorell M., Camps C., Sirera R., Analysis of the prognostic role of an immune checkpoint score in resected non-small cell lung cancer patients. Onco. Targets. Ther. 6, e1260214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou F. C., Chen H. Y., Kuo C. C., Sytwu H. K., Role of galectins in tumors and in clinical immunotherapy. Int. J. Mol. Sci. 19, 430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X., Liu R., Su X., Pan Y., Han X., Shao C., Shi Y., Harnessing tumor-associated macrophages as aids for cancer immunotherapy. Mol. Cancer 18, 177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin W., Xu D., Austin C. D., Caplazi P., Senger K., Sun Y., Jeet S., Young J., Delarosa D., Suto E., Huang Z., Zhang J., Yan D., Corzo C., Barck K., Rajan S., Looney C., Gandham V., Lesch J., Liang W. C., Mai E., Ngu H., Ratti N., Chen Y., Misner D., Lin T., Danilenko D., Katavolos P., Doudemont E., Uppal H., Eastham J., Mak J., de Almeida P. E., Bao K., Hadadianpour A., Keir M., Carano R. A. D., Diehl L., Xu M., Wu Y., Weimer R. M., DeVoss J., Lee W. P., Balazs M., Walsh K., Alatsis K. R., Martin F., Zarrin A. A., Function of CSF1 and IL34 in macrophage homeostasis, inflammation, and cancer. Front. Immunol. 10, 2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi Y. W., Kim Y. H., Oh S. Y., Suh K. W., Kim Y. S., Lee G. Y., Yoon J. E., Park S. S., Lee Y. K., Park Y. J., Kim H. S., Park S. H., Kim J. H., Park T. J., Senescent tumor cells build a cytokine shield in colorectal cancer. Adv. Sci. 8, 2002497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y., Knolhoff B. L., Meyer M. A., Nywening T. M., West B. L., Luo J., Wang-Gillam A., Goedegebuure S. P., Linehan D. C., DeNardo D. G., CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 74, 5057–5069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West R. B., Rubin B. P., Miller M. A., Subramanian S., Kaygusuz G., Montgomery K., Zhu S., Marinelli R. J., De Luca A., Downs-Kelly E., Goldblum J. R., Corless C. L., Brown P. O., Gilks C. B., Nielsen T. O., Huntsman D., van de Rijn M., A landscape effect in tenosynovial giant-cell tumor from activation of CSF1 expression by a translocation in a minority of tumor cells. Proc. Natl. Acad. Sci. U.S.A. 103, 690–695 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim I. S., Gao Y., Welte T., Wang H., Liu J., Janghorban M., Sheng K., Niu Y., Goldstein A., Zhao N., Bado I., Lo H. C., Toneff M. J., Nguyen T., Bu W., Jiang W., Arnold J., Gu F., He J., Jebakumar D., Walker K., Li Y., Mo Q., Westbrook T. F., Zong C., Rao A., Sreekumar A., Rosen J. M., Zhang X. H., Immuno-subtyping of breast cancer reveals distinct myeloid cell profiles and immunotherapy resistance mechanisms. Nat. Cell Biol. 21, 1113–1126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W., Xu H., Xiao T., Cong L., Love M. I., Zhang F., Irizarry R. A., Liu J. S., Brown M., Liu X. S., MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Curtis C., Shah S. P., Chin S. F., Turashvili G., Rueda O. M., Dunning M. J., Speed D., Lynch A. G., Samarajiwa S., Yuan Y., Graf S., Ha G., Haffari G., Bashashati A., Russell R., McKinney S.; METABRIC Group, Langerod A., Green A., Provenzano E., Wishart G., Pinder S., Watson P., Markowetz F., Murphy L., Ellis I., Purushotham A., Børresen-Dale A. L., Brenton J. D., Tavaré S., Caldas C., Aparicio S., The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Network , Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., Luo T., Xu L., Liao G., Yan M., Ping Y., Li F., Shi A., Bai J., Zhao T., Li X., Xiao Y., CellMarker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47, D721–D728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gubin M. M., Esaulova E., Ward J. P., Malkova O. N., Runci D., Wong P., Noguchi T., Arthur C. D., Meng W., Alspach E., Medrano R. F. V., Fronick C., Fehlings M., Newell E. W., Fulton R. S., Sheehan K. C. F., Oh S. T., Schreiber R. D., Artyomov M. N., High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell 175, 1014–1030.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hollern D. P., Xu N., Thennavan A., Glodowski C., Garcia-Recio S., Mott K. R., He X., Garay J. P., Carey-Ewend K., Marron D., Ford J., Liu S., Vick S. C., Martin M., Parker J. S., Vincent B. G., Serody J. S., Perou C. M., B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell 179, 1191–1206.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H. D., Krishnan I., Maroni G., Meyerovitz C. V., Kerwin C. M., Choi S., Richards W. G., De Rienzo A., Tenen D. G., Bueno R., Levantini E., Pittet M. J., Klein A. M., Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng Y.-Z., Xue M.-Z., Shen H.-J., Li X.-G., Ma D., Gong Y.-R., Liu Y., Qiao F., Xie H.-Y., Lian B., Sun W.-L., Zhao H.-Y., Yao L., Zuo W.-J., Li D.-Q., Wang P., Hu X., Shao Z.-M., PHF5A epigenetically inhibits apoptosis to promote breast cancer progression. Cancer Res. 78, 3190–3206 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Xiao Y., Ma D., Zhao S., Suo C., Shi J., Xue M. Z., Ruan M., Wang H., Zhao J., Li Q., Wang P., Shi L., Yang W. T., Huang W., Hu X., Yu K. D., Huang S., Bertucci F., Jiang Y. Z., Shao Z. M.; AME Breast Cancer Collaborative Group , Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin. Cancer Res. 25, 5002–5014 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S8