Abstract
The soybean is an exotic plant introduced in Paraguay in this century; commercial cropping expanded after the 1970s. Inoculation is practiced in just 15 to 20% of the cropping areas, but root nodulation occurs in most sites where soybeans grow. Little is known about rhizobial diversity in South America, and no study has been performed in Paraguay until this time. Therefore, in this study, the molecular characterization of 78 rhizobial isolates from soybean root nodules, collected under field conditions in 16 sites located in the two main producing states, Alto Paraná and Itapúa, was undertaken. A high level of genetic diversity was detected by an ERIC-REP-PCR analysis, with the majority of the isolates representing unique strains. Most of the 58 isolates characterized by slow growth and alkaline reactions in a medium containing mannitol as a carbon source were clustered with strains representative of the Bradyrhizobium japonicum and Bradyrhizobium elkanii species, and the 16S ribosomal DNA (rDNA) sequences of 5 of those isolates confirmed the species identities. However, slow growers were highly polymorphic in relation to the reference strains, including five carried in commercial inoculants in neighboring countries, thus indicating that the Paraguayan isolates might represent native bradyrhizobia. Twenty isolates highly polymorphic in the ERIC-REP-PCR profiles were characterized by fast growth and acid reactions in vitro, and two of them showed high 16S rDNA identities with Rhizobium genomic species Q. However, two other fast growers showed high 16S rDNA identity with Agrobacterium spp., and both of these strains established efficient symbioses with soybean plants.
The soybean [Glycine max L. (Merrill)] plays an important role in the economy of Paraguay and is one of its chief exports (1). This legume was introduced into the country in the 1920s, but commercial cropping started in the 1970s, with seeds from the United States, Argentina, and Japan, and expanded later, with cultivars from Brazil (1, 9). Inoculation is practiced in just 15 to 20% of the cropping areas, but nodulation occurs in the great majority of the sites in the two main producing areas, the states of Alto Paraná and Itapúa.
Little is known about rhizobial diversity in South America, and no study has been performed in Paraguay until this time. Furthermore, it is important to investigate the genetic diversity and symbiotic effectiveness of the indigenous rhizobial population, in order to better understand the responses to inoculation. Therefore, in this study, the molecular characterization of 78 rhizobial isolates from soybean root nodules, collected under field conditions in 16 sites located in the two main producing states, was undertaken.
Reference strains and soybean rhizobial isolates.
Rhizobial strains used for comparison were Bradyrhizobium japonicum SEMIA 566, SEMIA 5079 (also called CPAC 15; same serogroup as SEMIA 566), and SEMIA 5080 (also called CPAC 7) and Bradyrhizobium elkanii SEMIA 587 and SEMIA 5019 (also called 29w and BR 29); these five strains were or are used in commercial inoculants in the southern region of Brazil (2). B. japonicum strains USDA 6T (also called 3I1b6, ATCC 10324, SEMIA 5052, RCR 3425, or ACCC 15032), USDA 110 (also called 3I1b110, TAL102, RCR3427, 61A89, or SEMIA 5032), and USDA 122 and Sinorhizobium fredii strain USDA 205T (also called LMG 6217 or ATCC 35423) were received from the U. S. Department of Agriculture (USDA, Beltsville, Md.), and S. fredii CCBAU114 was received from the Beijing University of Agriculture (Beijing, People's Republic of China).
Seventy-eight isolates obtained from field-grown soybean nodules of commercial cultivars grown in 16 locations in the states of Alto Paraná and Itapúa were used for genetic characterization. Fifty-eight isolates (isolates 1 to 58) were characterized by alkaline reactions on yeast-mannitol-agar (YMA) medium (15) containing 0.0025% bromothymol blue and showed slow growth (colonies visible only after 5 days of incubation at 28°C). The other 20 isolates (isolates 59 to 78) showed acid reactions on YMA medium and were fast growers (visible growth at 3 days).
ERIC and REP-PCR genomic fingerprintings.
Bacterial DNA (50 ng) was amplified by PCR with enterobacterial repetitive intergenic consensus (ERIC) and repetitive extragenic palindromic (REP) primers according to the methodology of de Bruijn (4), with the cycles slightly modified as described previously (10). Analyses were performed in an MJ Research Inc. PTC-100 thermocycler, and the fragments were visualized after electrophoresis on a 1.5% agarose gel (low EED, type I-A). Clustering analysis of the PCR products was performed using the Bionumerics program (Applied Mathematics, Kortrijk, Belgium), with the UPGMA (unweighted pair-group method, with arithmetic mean) algorithm (11) and the Jaccard (J) coefficient.
16S ribosomal DNA (rDNA) sequence analysis.
Nine isolates representing the main ERIC-REP-PCR clusters were selected for the direct sequencing of PCR fragments obtained by amplification with primers Y1 (16) and Y3 (3′-CTGACCCCACTTCAGCATTGTTCCAT-5′) (J. P. W. Young, unpublished data) at a concentration of 10 pmol · reaction−1. DNA amplification cycles were as described previously (16), except for the annealing temperature, which was increased by 2°C. After amplification, the PCR products were purified using the Concert Rapid PCR Purification System kit (Life Technologies) and were quantified. The PCR fragments (70 ng for Y1 and 40 ng for the other primers) were amplified again using 3.2 pmol of Y1 and Y2 · reaction−1 (16), intermediate primers designed for our strains, and the Big Dye kit (Perkin-Elmer Applied Biosystems), according to the instructions of the manufacturer. The following cycles were used for amplification: 95°C for 2 min, and 30 cycles of denaturing at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 4 min. Samples were then precipitated with 75% isopropanol, left at room temperature for 15 min, and centrifuged at 12,000 rpm for 20 min at 25°C. The supernatant was removed, 250 μl of 75% isopropanol was added, and the tubes were vortexed. Each sample was centrifuged again for 5 min, the supernatant was carefully removed, and the sample was then heated at 90°C for 1 min. Samples were resuspended in 8 μl of buffer (formamide and blue dextran, 5:1 [vol/vol]) and heated for 2 min at 90 to 95°C, followed by a cold shock in ice. For sequencing, 2 μl of each sample was applied to a polyacrylamide gel (Long Ranger Single Packs; FMC Bioproducts), and the sequences were determined in a Perkin-Elmer ABI 377.
GenBank accession numbers and phylogenetic analysis.
The 16S rDNA sequences generated were submitted to the GenBank database to search for significant 16S rRNA alignments. Isolates 1, 40, 42, 49, 52, 62, and 65 were named PRY (Paraguay) strains and received accession numbers AF239842 to AF AF239848; isolates 1, 71, and 73 received accession numbers AF286361 to AF286363, respectively. The partial sequences of the nine isolates were compared to those of the following organisms (with GenBank accession numbers in parentheses): B. japonicum strains USDA 6T (U69638), USDA 110 (Z35330), SEMIA 566 (AF236086), and SEMIA 5080 (AF234889); B. elkanii USDA 76T (U35000), USDA 31 (AF236089), SEMIA 587 (AF234890), and SEMIA 5019 (AF237422); Rhizobium tropici IIB CIAT 899T (U89832); Rhizobium galegae HAMBI 540T (Y12355); Rhizobium genomic species Q strain BDV 5102 (Z94806); S. fredii USDA 205T (M74163); Agrobacterium biovar I strain LMG 11915 (AJ130720); and Agrobacterium radiobacter strain LMG 383 (AJ130719). The sequences were aligned pairwise, and a phylogeny tree was inferred with the UPGMA algorithm using the Bionumerics program.
Genetic diversity of the Paraguayan rhizobial isolates.
All isolates were reinoculated on the soybean cultivar BR-16, and their effectiveness was confirmed. Some of the isolates (e.g., isolates 27, 40, 41, 42, 45, 47, 50, 52, 60, 71, 73, and 77) were as efficient as, or more efficient than, five reference strains used in commercial inoculants in South America: SEMIA 587, SEMIA 5019, SEMIA 5079, SEMIA 5080, and USDA 110 (data not shown).
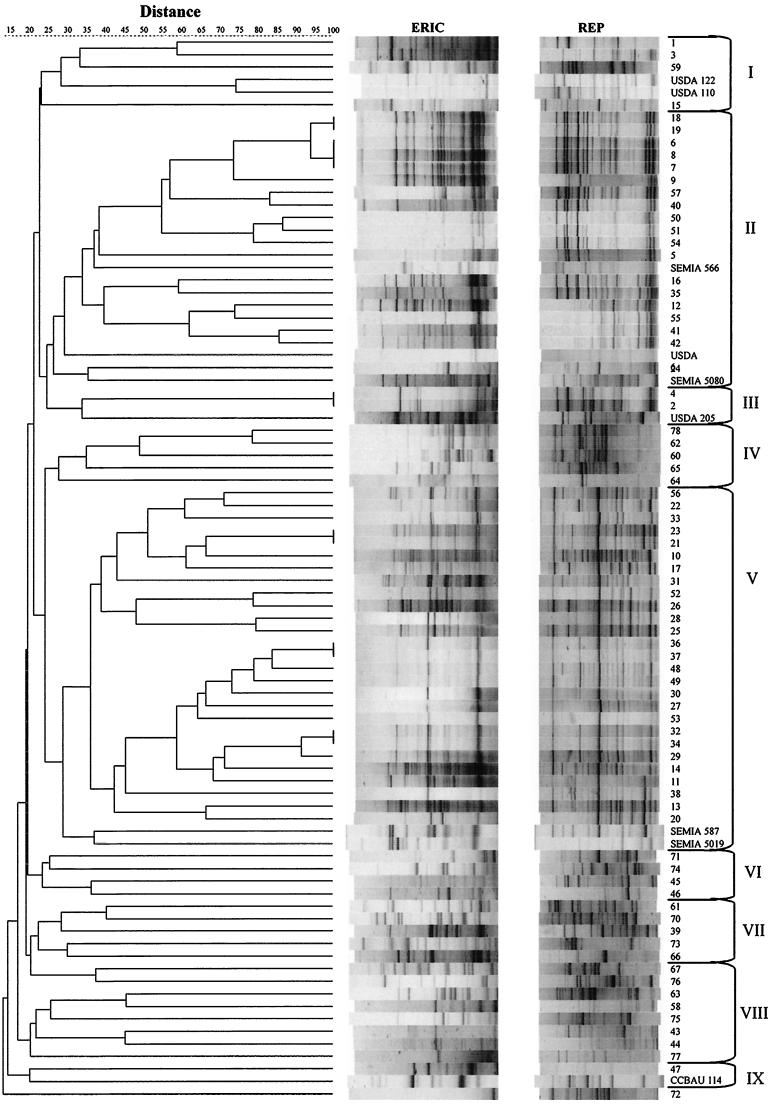
Clustering analysis of the ERIC-REP-PCR products resulted in nine main clusters at a 15% level of similarity, and isolate 72 was linked to those clusters at a level of 14% (Fig. 1). Cluster I included isolates 1, 3, 59, and 15 and B. japonicum strains USDA 122 and USDA 110. Within cluster II were 19 alkalinizing isolates and the B. japonicum reference strains SEMIA 566, USDA 6T, and SEMIA 5080. Cluster III included S. fredii USDA 205T and the alkalinizing isolates 2 and 4. Cluster IV included the acidifying isolates 78, 62, 60, 65, and 64. Cluster V included 27 alkalinizing isolates, as well as the B. elkanii reference strains SEMIA 587 and SEMIA 5019. Clusters VI, VII, and VIII included 17 isolates with high levels of DNA polymorphism, 11 of which were acidifying isolates. Finally, isolate 47 and S. fredii strain CCBAU 114 were joined at a 21% level of similarity in cluster IX.
FIG. 1.
Dendrogram of isolates from field-grown soybean nodules in Paraguay and representative strains of B. japonicum, B. elkanii, and S. fredii, based on cluster analysis of ERIC and REP-PCR products using the UPGMA algorithm and the Jaccard coefficient.
The results obtained with the ERIC-REP-PCR analysis showed a high level of genetic diversity among the Paraguayan soybean rhizobia, since just seven slow growers and none of the fast growers shared similar ERIC-REP-PCR fingerprintings. Therefore, most of the isolates represented, and will be considered as, unique strains. Furthermore, the DNA profiles of the slow growers differed from those of all the reference strains, including those intensively used in soybean commercial inoculants in neighboring countries, indicating that the former strains might represent indigenous bradyrhizobia. However, to confirm the hypothesis of indigenous bradyrhizobia, which are probably symbionts of native legumes that can also nodulate soybean roots, the hypothesis of transference of symbiotic genes from the inoculant strains to indigenous nonsymbiotic bradyrhizobia should first be investigated (12, 13), as should the hypothesis of genetic changes in bacteria due to environmental stress conditions, which frequently occur in tropical regions (5, 10). DNA polymorphism was even higher among the fast growers, indicating a variety of indigenous rhizobial strains able to nodulate soybean roots.
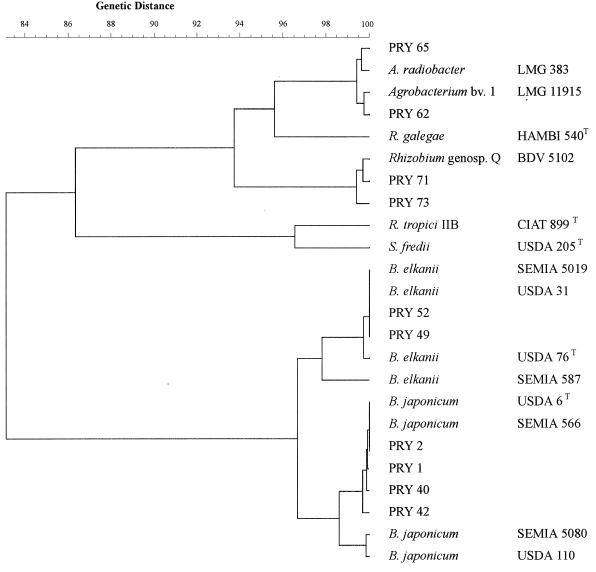
The 16S rDNA sequences obtained with nine Paraguayan strains were compared with those available in the GenBank database, and the phylogenetic tree obtained with the UPGMA algorithm is shown in Fig. 2. The partial 16S rDNA sequences of strain 1 (ERIC-REP-PCR cluster I on Fig. 1) and strains 40 and 42 (cluster II) were 99% similar to that of B. japonicum SEMIA 566 (1,408 of 1,409, 1,407 of 1,410, and 1,394 of 1,399 bp, respectively), which shows complete identity with B. japonicum USDA 6T. The slow-growing strain 2, which had clustered with S. fredii USDA 205 in the ERIC-REP-PCR analysis (Fig. 1), also showed complete 16S rDNA identity with SEMIA 566 (100%; 1,409 of 1,409 bp). Strains 49 and 52 (cluster V, with B. elkanii reference strains) showed 99% (1,411 of 1,412 bp) and 100% (1,411 of 1,411 bp) identities, respectively, with B. elkanii strain SEMIA 5019, which is 99% similar (1,407 of 1,411 bp) to B. elkanii USDA 76T. Therefore, the sequence analyses of the Paraguayan alkalinizing strains confirmed their identities with B. japonicum and B. elkanii species (Fig. 2).
FIG. 2.
Dendrogram built by use of the UPGMA algorithm with the aligned partial 16S rRNA sequences of 9 isolates from field-grown soybean nodules in Paraguay (PRY) and of 14 reference strains belonging to three genera. GenBank accession numbers are given in the text.
Two fast-growing strains, 71 and 73, which grouped into ERIC-REP-PCR clusters VI and VII (Fig. 1), respectively, showed 99% 16S rDNA sequence identity (1,377 of 1,381 and 1,372 of 1,379 bp, respectively) with Rhizobium genomic species Q strain BDV 5102, isolated from a native Australian shrubby legume, Daviesia leptophylla (8). Another strain, PRF 81 (also called SEMIA 4080), isolated from common bean (Phaseolus vulgaris L.) nodules in Brazil and currently commercially recommended in that country (6), has also shown 99% similarity in the partial 16S rDNA sequence with Rhizobium genomic species Q (3).
Two fast-growing strains from cluster IV were also analyzed. Strain 62 showed 99% identity (584 of 585 bp) with Agrobacterium biovar 1 LMG 11915 (AJI30720), and strain 65 showed 99% identity (1,387 of 1,396 bp) with A. radiobacter LMG 383 (AJI30719); both of these Agrobacterium strains were isolated from the root nodules of tropical legumes in Africa (P. de La Judie, A. Willem, G. Nick, T. S. Mohamed, U. Torck, A. Filali-Maltouf, K. Kersters, B. Dreyfus, K. Lindström, and M. Gillis, unpublished data). Other agrobacteria have been isolated from nodules of Acacia spp. in Morocco (7). Agrobacterium spp. are genetically related to Rhizobium genomic species Q (Fig. 2); the genus Agrobacterium also shares several characteristics with, and is genetically closely related to, R. galegae as well as R. tropici, a common bean (P. vulgaris L.) symbiont native to tropical regions of South America (14). Taxonomic classification within those genetically related genera is even more difficult due to the presence of nonsymbiotic rhizobia in soil (7, 12, 13). In this experiment, when the soybean cultivar BR-16 was reinoculated with strains 60 and 62, efficient nodules were formed, in contrast to the agrobacterial strains of Khabaya et al. (7), which were unable to renodulate Acacia spp. Therefore, the results found in this study indicate that native isolates resembling Agrobacterium spp. can be isolated from field-grown nodules and establish an efficient symbiosis with soybean plants, and the ecological importance of those bacteria should be determined. The high level of genetic diversity detected among soybean isolates in Paraguay also strongly encourages other studies of rhizobial diversity in South America.
Nucleotide sequence accession numbers.
Isolates 1, 40, 42, 49, 52, 62, and 65 were assigned GenBank accession numbers AF239842 to AF239848, and isolates 1, 71, and 73 were assigned accession numbers AF286361 to AF286363.
Acknowledgments
We thank L. M. O. Chueire for help in several steps of this work; R. J. Campo, D. S. Andrade, G. Andrade Filho, and A. R. J. Eaglesham for discussions and suggestions on the manuscript; and E. M. Souza and L. M. Cruz for designing the primers.
The research described herein was partially financed by FINEP/CNPq/MCT, PRONEX, Group of Excellence in Nitrogen Fixation (41.96.0884.00). L. S. Chen acknowledges an M.Sc. Fellowship from CAPES, and M. Hungria (520396/96-0) acknowledges a research fellowship from CNPq.
Footnotes
Approved by the Head of Research and Development of Embrapa Soja as manuscript 25/2000.
REFERENCES
- 1.Alvarez E R. II Curso Internacional sobre Producción de Soja. Paraguay: MAG, Encarnación; 1989. La soja en el Paraguay, retrospectiva y perspectiva; pp. 551–563. [Google Scholar]
- 2.Boddey L H, Hungria M. Phenotypic grouping of Brazilian Bradyrhizobium strains which nodulate soybean. Biol Fertil Soils. 1997;25:407–415. [Google Scholar]
- 3.Chueire L M O, Bangel E, Ferreira M C, Grange L, Campo R J, Mostasso F L, Andrade D S, Pedrosa F O, Hungria M. Classificação taxonômica, baseada na caracterização molecular, das estirpes de rizóbio recomendadas para as culturas da soja e do feijoeiro. 3. Embrapa Soja, Londrina, Brazil: Embrapa Soja, Boletim de Pesquisa; 2000. [Google Scholar]
- 4.de Bruijn F. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hungria M, Vargas M A T. Environmental factors impacting N2 fixation in legumes grown in the tropics, with emphasis on Brazil. Field Crops Res. 2000;65:151–164. [Google Scholar]
- 6.Hungria, M., D. S. Andrade, L. M. O. Chueire, A. Probanza, F. J. Guttierrez-Mañero, and M. Megías. Isolation and characterization of new efficient and competitive bean (Phaseolus vulgaris L.) rhizobia from Brazil. Soil Biol. Biochem., in press.
- 7.Khabaya B, Neyra M, Normand P, Zerhari Z, Filali-Maltouf A. Genetic diversity and phylogeny of rhizobia that nodulate Acacia spp. in Morocco assessed by analysis of 16S rRNA genes. Appl Environ Microbiol. 1998;64:4912–4917. doi: 10.1128/aem.64.12.4912-4917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafay B, Burdon J J. Molecular diversity of rhizobia occurring on native shrubby legumes in southeastern Australia. Appl Environ Microbiol. 1998;64:3989–3997. doi: 10.1128/aem.64.10.3989-3997.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveri N J, Perucca C E, Morel F. Evolución de cultivares de soja (Glycine max L. Merr.) de origen brasileño. Informe Técnico 35. Misiones, Argentina: Instituto Nacional de Tecnología Agropecuaria—Estación Experimental Agropecuaria INTA; 1981. [Google Scholar]
- 10.Santos M A, Vargas M A T, Hungria M. Characterization of soybean Bradyrhizobium strains adapted to the Brazilian savannas. FEMS Microbiol Ecol. 1999;30:261–272. doi: 10.1111/j.1574-6941.1999.tb00654.x. [DOI] [PubMed] [Google Scholar]
- 11.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman & Co.; 1973. [Google Scholar]
- 12.Sullivan J T, Eardly B D, van Berkum P, Ronson C W. Four unnamed species of nonsymbiotic rhizobia isolated from the rhizosphere of Lotus corniculatus. Appl Environ Microbiol. 1996;62:2817–2825. doi: 10.1128/aem.62.8.2818-2825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan J T, Patrick H N, Lowther W L, Scott D B, Ronson C W. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci USA. 1995;92:8985–8989. doi: 10.1073/pnas.92.19.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Berkum P, Eardly B D. Molecular evolutionary systematics of the Rhizobiaceae. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae—molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–24. [Google Scholar]
- 15.Vincent J M. Manual for the practical study of root nodule bacteria. Oxford, United Kingdom: Blackwell; 1970. [Google Scholar]
- 16.Young J P W, Downer H L, Eardly B D. Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]