Abstract
Amyotrophic lateral sclerosis (ALS) is the third most frequent neurodegenerative disease after Alzheimer’s and Parkinson’s disease. ALS is characterized by the selective and progressive loss of motoneurons in the spinal cord, brainstem and cerebral cortex. Clinical manifestations typically occur in midlife and start with focal muscle weakness, followed by the rapid and progressive wasting of muscles and subsequent paralysis. As with other neurodegenerative diseases, the condition typically begins at an initial point and then spreads along neuroanatomical tracts. This feature of disease progression suggests the spreading of prion-like proteins called prionoids in the affected tissues, which is similar to the spread of prion observed in Creutzfeldt-Jakob disease. Intensive research over the last decade has proposed the ALS-causing gene products Cu/Zn superoxide dismutase 1, TAR DNA-binding protein of 43 kDa, and fused in sarcoma as very plausible prionoids contributing to the spread of the pathology. In this review, we will discuss the molecular and cellular mechanisms leading to the propagation of these prionoids in ALS.
Keywords: amyotrophic lateral sclerosis, prion-like, protein aggregation, physiopathology, therapy
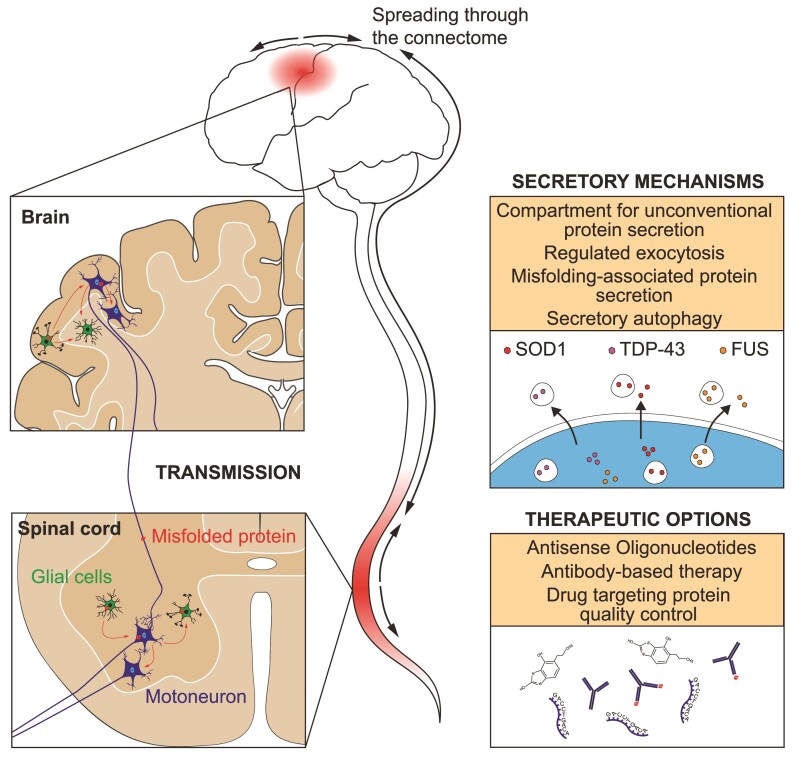
Amyotrophic lateral sclerosis is a fatal neurodegenerative disease characterized by the loss of motoneurons in the brain and spinal cord. Disease onset is anatomically localized and progression occurs by spread to contiguous regions. Raoul et al. discuss the relevance of prion-like propagation as a pathogenic determinant in disease progression.
Graphical Abstract
Graphical Abstract.
Introduction
A common feature of neurodegenerative diseases is a scenario of events that leads to specific neuronal dysfunctions in a defined neuroanatomical region of the brain. The progressive failure of proteostasis in the CNS leads to the accumulation, aggregation and propagation of a subset of pathogenic proteins.1,2 These diseases commonly progress from focal sites and spread to neighbouring regions, as well as to the functionally connected, more distant areas of the CNS. The different stages of disease progression have been well described for Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis (ALS).3,4 In infectious prion diseases, the progression of neuronal degeneration in the CNS correlates with the spreading of a prion protein (PrP), and similar mechanisms of toxic protein propagation have been proposed for other neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease and ALS.1,5 The molecular basis of this proteotoxic spreading is the capacity of the deleterious scrapie PrP to pass its peculiar conformation onto other PrP species.6 Accumulating evidence indicates that α-synuclein, tau and Aβ peptides in Parkinson’s disease and Alzheimer’s disease have similar PrP properties and can seed and propagate within the CNS.7,8 For instance, prion-like proteins are found in insoluble deposits, referred to as amyloid fibrillary structures, such as α-synuclein in Lewy bodies in Parkinson’s disease and amyloid plaques in Alzheimer’s disease.9,10 The term prion-like proteins or prionoids is used because the associated neurodegenerative diseases are not likely to be infectious, as prion diseases are.7–9 In ALS, several proteins, including Cu/Zn superoxide dismutase 1 (SOD1), TAR DNA-binding protein of 43 kDa (TDP-43), and fused in sarcoma (FUS), harbour some features of prionoids.10–12 In this review, we will discuss recent findings on the spread of prion-like proteins in ALS and the current debates that highlight that there may also be a differential vulnerability of functionally interconnected neurons to neurotoxic prionoids, illustrating the complexity of pathogenic mechanisms.
SOD1, TDP-43 and FUS proteinopathies in ALS
ALS is characterized by the selective loss of both upper and lower motoneurons that irremediably leads to progressive paralysis. This evolves rapidly into generalized wasting that irrevocably causes death by respiratory failure. The median survival time from the clinical onset is about 3 years. Approximately 90% of ALS cases are sporadic with largely unknown aetiology, but an increasing number of genes have been linked to the 10% of cases that show a familial inheritance of the disease.13,14 Some of these genes are also mutated in frontotemporal dementia (FTD), suggesting that these two diseases have overlapping physiopathological mechanisms.15 Both sporadic (sALS) and familial (fALS) forms of the disease are histologically characterized by the presence of multiple types of protein deposits in the CNS, which defines ALS as a proteinopathy. Currently, mutations in genes coding for C9ORF72, SOD1, TDP-43 and FUS are the most frequently found mutations in fALS.13,14 Whereas many genes (more than 30) are linked to fALS, only relatively few proteins (TDP-43, SOD1, FUS, C9ORF72 dipeptides, optineurin, ubiquitin and p62) are present in diverse types of inclusions in the spinal cords and brains of patients with ALS.14,16 Together, TDP-43 and FUS intracellular protein inclusions can be found in more than 90% of post-mortem tissues from patients with ALS and in more than 50% of tissues from patients with FTD, reflecting the pathological overlap that exists between these two disorders.17 SOD1 inclusions are predominantly found in patients with pure ALS with SOD1 mutations. Although mutant forms of TDP-43, FUS and SOD1 can be associated with distinct clinical manifestations,18 they share common pathogenic features that lead to ALS. The presence of these proteins in intracellular protein inclusions and aggregates in ALS might be explained by their low magnitude of solubility, which leads to an increased susceptibility to becoming insoluble when a slight cellular proteostasis disturbance occurs.19,20 Furthermore, TDP-43 proteinopathies are observed in other neurodegenerative diseases, such as Alzheimer’s disease and Parkinson’s disease.21–23 Structurally, although the inclusions are less well characterized in ALS than those found in Alzheimer’s disease or Parkinson’s disease, they are classified as non-amyloid or amyloid-like, as they do not fulfil the criteria of the highly ordered amyloid fibrils of proteins typically found in other neurodegenerative diseases, such as PrP, Aβ peptide, tau and α-synuclein.9
SOD1 is a cytosolic metalloenzyme of 154 amino acids (aas) involved in cell detoxification by catalyzing the conversion of the superoxide radical into oxygen and hydrogen peroxide.24SOD1 was the first gene found to be linked to fALS, and mutations in this gene are found in ∼20% of cases.25 There are several lines of evidence indicating that the primary pathogenic mechanism of the disease is a toxic gain of function of SOD1 mutants. Although Sod1 null mice showed age-associated muscle denervation and increased susceptibility to oxidative stress, the loss of SOD1 function did not lead to typical ALS.26,27 In addition, ablation or overexpression of wildtype SOD1 does not affect SOD1 mutant-mediated motoneuron disease in mice.28 The concentration and enzymatic activity of SOD1 mutants observed in patients with fALS does not correlate with the age of onset or disease severity.29,30 The toxic gain of function seen with SOD1 mutants is associated with a higher propensity of these mutants to form aggregates when cellular quality control and protein catabolism decline.28 Several studies have shown an increase in misfolded wildtype SOD1, not necessarily in an aggregated state, in the spinal cord tissues of sALS and fALS patients.31–35 Misfolded SOD1 is present in upper and lower motoneurons and glia cells in sALS, supporting the fact that a global decline in protein homeostasis is a general feature in ALS. However, the contribution of misfolded wildtype SOD1 to pathology in sALS is still a matter of debate.32,36
TDP-43 is an RNA-binding protein (RBP), member of the heterogeneous nuclear ribonucleoprotein family, and is involved in several functions related to the regulation of RNAs, such as pre-mRNA splicing, mRNA stability and transport and translation.23,37 TDP-43 is a nuclear protein but may actively shuttle into the cytoplasm when associated with RNAs export complex.37 Several mutations in the TARDBP gene are linked with dominant fALS-FTD (4% of cases) and with <1% of fFTD cases.38 These TDP-43 mutants mislocalize in the cytoplasm, where they form aggregates.39 Both loss of function, due to TDP-43 nuclear depletion, and toxic gain of function, owing to proteostasis failure, contribute to the disease.40,41 In both sporadic ALS and FTD, wildtype TDP-43 is largely found in cytoplasmic inclusions that are mainly composed of post-translational modified TDP-43, including truncated, phosphorylated and ubiquitinated forms.42–44
FUS, like TDP-43, is an RBP involved in DNA repair, RNA splicing and RNA regulation, and is mainly localized in the nucleus.35 Mutations in FUS account for about 4–5% of fALS cases and occur very rarely in fFTD.16,45 Cytoplasmic aggregates of FUS are observed in the CNS of patients harbouring FUS mutations,46,47 but recently it has been shown that mislocalization and not necessarily aggregates of wildtype FUS might be present in sALS.48
Basic cellular events of prion-like protein propagation
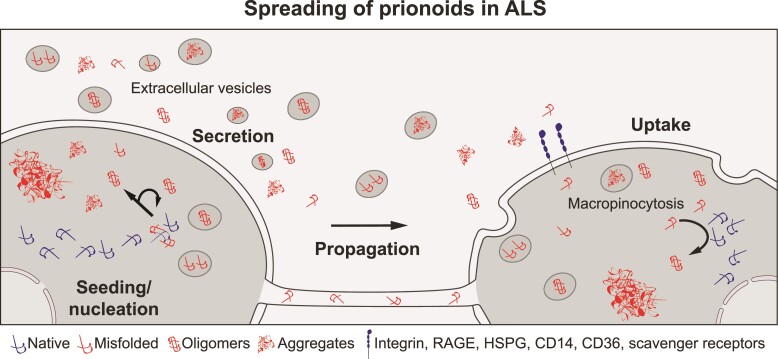
The cellular pathways implicated in the propagation of prion-like proteins can be schematically divided into three steps: the release of prion-like proteins from cells, uptake by neighbouring cells and seeding and nucleation in recipient cells (Fig. 1). Although it is well documented that the spread of prion-like proteins in different neurodegenerative diseases involves interconnected neurons, it is, however, not always clear if the anatomical pattern of cellular alterations reflects the real spread of a toxic protein or if it is also due to a differential vulnerability of a neuronal population.2 The contribution of glial cells as an intermediary between populations of neurons is also to be considered in the dissemination of prion-like proteins.50–52 The mechanism of propagation of prion-like proteins in different neurodegenerative diseases may implicate different processes: proteotoxicity takes place extracellularly for Aβ and PrP and intracellularly for tau, α-synuclein and ALS-linked prion-like proteins (SOD1, TDP-43 and FUS).53 Eukaryotic cells have developed machineries based on chaperon-mediated quality control, proteasomal and autophagosomal protein degradation systems that prevent intracellular aberrant protein accumulation.54,55 However, the primary molecular events leading to deployment of misfolded harmful proteins in a subset of cells are still poorly understood, especially for sporadic neurodegenerative diseases. The progressive deterioration of proteostasis associated with ageing56 and poorly characterized environmental cellular stressors might favour intracellular seeding and nucleation of prion-like proteins, especially for those whose solubility threshold is already elevated in a defined cell.9,19,55,57 Abnormal and misfolded proteins, comprising different structural entities (monomers, oligomers and aggregates), can thus be removed by several mechanisms, such as via intercellular bridges and by active release or secretion via non-vesicular and vesicular transports (Fig. 1).58–60 Seminal works in Caenorhabditis elegans suggest that unconventional secretion induced by cellular stress of aberrant protein aggregates prevents cellular protein overload and furthermore promotes extracellular proteolysis of proteins that are usually not secreted.61,62 More specifically in neurodegenerative diseases, prionoids such as misprocessed Aβ forms and mutant PrP escape endoplasmic reticulum (ER) quality control and are exported via the vesicular conventional secretory pathway, whereas α-synuclein, tau, TDP-43, FUS and SOD1 are cytoplasmic or nuclear leaderless proteins, and their extracellular release involves what is called unconventional secretory pathways.63–65 Special attention has been paid in recent years to the rules of different types of secreted vesicles (lysosomes, endosomes, exosomes, extracellular vesicles and exophers)66 in prionoid propagation, and there is some molecular evidence that cytoplasmic misfolded proteins are mistargeted to these different vesicles.67,68 Subsequently, absorption of prionoids by neighbouring or contacting cells allows their propagation in the CNS, an event that is called cross-seeding (Fig. 1). The cellular pathways involved in the internalization of ALS prion-like proteins in a recipient cell are still poorly characterized, but some specific receptors have been identified (Fig. 1). After cellular entry, prion-like proteins reach the cytosol by a still unknown mechanism and initiate a new cycle of seeding/nucleation (Fig. 1). The molecular mechanisms of the cross-seeding and nucleation of SOD1, TDP-43 and FUS are not yet fully characterized, as they do not behave like classical amyloids. Finally, infected cells will de novo transfer prion-like proteins to other ‘naïve’ cells. The key steps of prion-like protein propagation will be described in more detail for SOD1, TDP-43 and FUS.
Figure 1.
Cellular events involved in the generation and spread of prionoids. Seeding/nucleation: intracellular accumulation of cytoplasmic misfolded proteins induces prionoid seeding and nucleation in non-amyloid or amyloid-like structures termed ‘aggregates’. Oligomers are presumably intermediate structural protein species in the nucleation process. Propagation: mechanisms of cellular extrusion of misfolded proteins, comprising intercellular bridges like tubular nanotubes and unconventional secretion of proteins in a soluble state or contained in various secreted vesicles (lysosomes, endosomes, exosomes, extracellular vesicles, exophers).49 Cellular uptake: uptake of prionoids by macropinocytosis. This process seems to be regulated by several receptors that recognize extracellular protein aggregates.
Cellular and molecular mechanisms potentially involved in ALS prionoid propagation
Secretion pathways of SOD1, TDP-43 and FUS
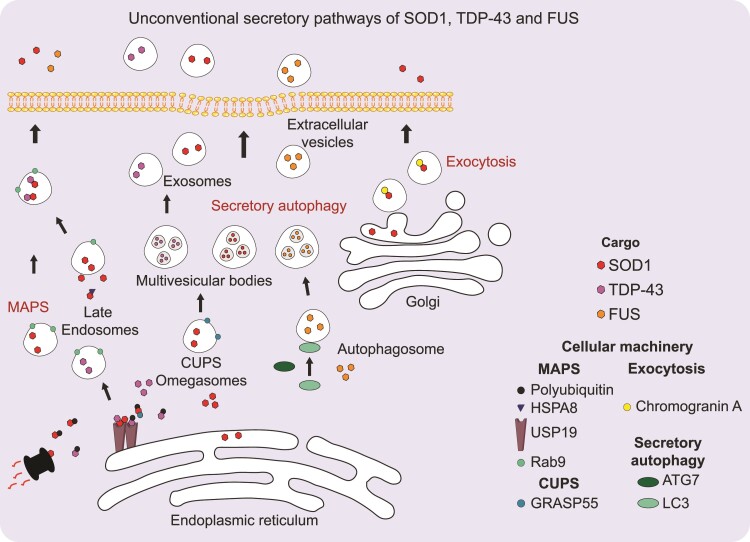
A large proportion of the prion-like proteins associated with various neurodegenerative diseases are leaderless proteins: α-synuclein, tau and huntingtin, as well as SOD1, TDP-43 and FUS. The secretion of proteins that do not bear a signal peptide has received particular attention for decades but remain poorly understood.63,69 Complex and diverse pathways of unconventional secretion, which are not linked to the canonical ER–Golgi secretory pathways, have emerged.63,70 The type of unconventional secretion varies depending on the target protein, and different pathways have been described for ALS-linked leaderless proteins (Fig. 2).
Figure 2.
Unconventional secretion pathways of SOD1, TDP-43 and FUS. The cytoplasmic cargos SOD1, FUS and TDP-43, are represented by red, orange and purple circles, respectively. SOD1 is released either by a specific secretory pathway for cytoplasmic ubiquitinated misfolded proteins called MAPS65 or by an autophagic-like secretory pathway involving a specialized organelle called CUPS.71 SOD1 mutants are also secreted via exocytosis from motoneurons.72 The MAPS pathway seems also to promote wildtype TDP-43 secretion. FUS has, for instance, only been detected in extracellular vesicles from LC3+ autophagosomes.73 MAPS involves the targeting of cargos in Rab9+ (green circles) late endosomes allowing protein secretion in a vesicles-free form.65 This process depends on the binding of HSPA8 (marine blue triangle) and the deubiquitinase USP19 (brown stick) to polyubiquitinated (black circle) proteins. The CUPS-dependent pathway involves the Golgi reassembly-stacking protein (GRASP55) that appears to shuttle between the Golgi and CUPS.71
Before the discovery of fALS-linked mutations in the SOD1 gene, studies in mouse bone marrow cell cultures had revealed regulated secretion of wildtype SOD1.74,75 Several studies have demonstrated the secretion of wildtype SOD1 by various cell types.74,76–78 SOD1 export is increased by membrane depolarization in excitable neuroendocrine cells,74 occurs through an ATP-dependent mechanism in neuroblastoma cells,79 and could be related to secretory autophagy in yeast.77 Indeed, during nutrient starvation of yeast, SOD1 is targeted to a specific cellular location called the compartment for unconventional protein secretion (CUPS), which is similar to the pre-autophagic compartment omegasome in mammalian cells.77,80 This CUPS-associated export of SOD1 is also dependent on some components of the endosomal sorting complexes required for transport. A diacidic motif, D76-E77, in SOD1 is required for this unconventional secretion, and interestingly, mutations (D77V, D77Y) in this motif are found in fALS.77,81,82 However, the SOD1G93A mutation also leads to a decreased secretion compared with wildtype SOD1 that is greater than when the mutation impacts the diacidic motif itself.77
Other studies have also described unconventional secretion of wildtype and mutant SOD1 via exosomes (Fig. 2).83–85 The contribution of wildtype SOD1 secretion to cell survival and response to oxidative stress still needs to be clarified.86,87 Interestingly, SOD1G93A mutant expression in astrocytes induces a global decrease in secreted proteins but an increase in protein content in the exosomal fraction compared with wildtype SOD1.84 This observation could be a result of an increased unfolded protein response.86,88 The abundance of SOD1G93A in comparison to wildtype SOD1 appears to paradoxically decrease in exosomes but increase in the non-exosomal fraction.84 The exosomes from SOD1G93A mouse astrocytes are toxic to motoneurons, but it is not known whether this toxicity is directly due to exosomal SOD1G93A.84 Secretion of SOD1 mutants can also occur through a more ‘conventional’ secretory pathway.72,89–91 Identified as a specific interaction partner of SOD1 mutants, the neuroendrocrine secretory protein chromogranin A recognizes only mutant SOD1 in motoneurons and targets it to exocytosis vesicles.72 How chromogranin transfers mutant SOD1 in exocytotic vesicles is still unknown but seems to be dependent on interactions with the Golgi apparatus. Furthermore, SOD1 import into the ER has been shown to be a post-translational and signal recognition particle-independent mechanism.89 Although post-translational ER targeting of proteins appears to be more frequent than expected, there are few examples of leaderless small proteins being targeted in this way, with the exception of proteins smaller than 50 aas.92,93 Another unconventional secretion pathway specifically for misfolded proteins, called misfolding-associated protein secretion (MAPS), is involved in wildtype SOD1 release.94 MAPS involves the ER-resident ubiquitin-specific protease 19 (USP19) deubiquitinase that redirects proteins destined for the proteasomal pathway to secretory endosomes (Fig. 2).94,95 Many cytosolic misfolded proteins, including mutant SOD1, might follow this unconventional secretion pathway. The USP19-dependent pathway involves the HSPA8 heat shock protein that targets misfolded proteins to USP19 (Fig. 2).94,95
The functional implications of these different unconventional secretions have been poorly exploited in SOD1-linked ALS mouse models, but some pioneering works have explored the secretion and intercellular transfer of SOD1 in vivo.96 In chimeric transgenic mice, transfer of both human mutant SOD1 and wildtype SOD1 between motoneurons and oligodendrocytes has been observed.96 It appears that transferred mutant SOD1 is not aggregated and does not co-aggregate with differently tagged human SOD1 in acceptor cells. This cell-to-cell protein transfer does not seem to be specific for mutant SOD1, as transfer of overexpressed wildtype human SOD1 and GFP between motoneurons and oligodendrocytes also occurs.96 Cellular mechanisms of SOD1 transfer in vivo are still unknown, but interestingly, motoneuron-specific deletion of autophagy related 7 (Atg7) in SOD1G93A mice has peculiar effects on ALS pathogenesis.97 ATG7 is well known to be involved in secretory autophagy, an unconventional secretory pathway that facilitates the export of leaderless proteins.98 Aggregation of the autophagy receptor p62 into skein-like inclusions is first observed in spinal motoneurons and then in interneurons in SOD1-mutant mice. Inhibition of autophagy in motoneurons delays the formation of p62 inclusions and accumulation of misfolded SOD1 in interneurons. Aggregation of p62 in interneurons occurs concomitantly with astrogliosis and microgliosis. These results highlight the non-cell-autonomous effect of motoneuron autophagy on interneurons and glial cells, maintaining the possibility that direct spreading of misfolded SOD1 from motoneurons to interneurons occurs.97 It will be of interest to explore whether this phenotype is linked to a decrease in human SOD1 propagation, as has been observed for Aβ peptides in Alzheimer mouse models crossed with ATG7 knockout mice.99
The transfer of TDP-43 oligomers from cell to cell has been shown in primary neurons using protein complementation assays.100,101 This unconventional secretion of TDP-43 is induced by oxidative stress and mediated by exosomes (Fig. 2).102,103 Moreover, secretion of wildtype and mutant TDP-43 in exosomes is also observed in basal conditions in transformed cells and might be promoted by secretory autophagy (Fig. 2).102 Mutated TDP-43 has also been found in exosomal fractions purified from the brains of patients with ALS and the spinal cords of TDP-43A315T transgenic mice.102 It is noteworthy that the inhibition of exosome biogenesis by GW4869, an inhibitor of sphingomyelinase, or the inhibition of exosome release by Rab27 silencing in neuro2a cells increases cytoplasmic and nuclear aggregation of phosphorylated TDP-43. This suggests that release and biogenesis can be coupled events.100,101 However, inhibition of exosome release with GW4869 has minor effects on the phenotypic behaviour of TDP-43A315T transgenic mice.102 Wildtype TDP-43 can also be unconventionally secreted by MAPS in an exosome-independent manner in vitro (Fig. 2).94 The interconnection between the MAPS pathway and secretory autophagy remains unknown.
Although FUS (as well as SOD1 and TDP-43) has been found in plasma microvesicles and exosomes from patients with ALS, the mechanism of unconventional FUS secretion remains poorly documented.104 Recently, proteome characterization of extracellular vesicles, which depend on microtubule-associated protein 1A/1B-light chain 3 (LC3) and ATG8 proteins for their secretion, has notably revealed a strong enrichment in RBP proteins, including FUS (Fig. 2).73 In basal conditions, RBPs associated with mRNAs account for a large proportion of secreted proteins.73 Whether this pathway implicating components of the autophagy machinery is increased in response to cellular stress remains an open question, but it provides a molecular basis for the unconventional secretion of FUS and other ALS-related RBPs.105(p3)
Cellular uptake of extracellular prionoids
Uptake of extracellular SOD1 aggregates has been extensively studied in cultures of neuronal and non-neuronal cells expressing different ALS-related mutant SOD1.106 Studies revealed that extracellular mutant SOD1 aggregates can enter cells and nucleate endogenous mutant but not wildtype SOD1.106,107 It was subsequently suggested that SOD1 aggregates enter acceptor cells by macropinocytosis in order to reach the cytosol.106 Macropinocytosis is a form of non-selective endocytosis generally induced by growth factors and used by cells to internalize large numbers of soluble macromolecules or particles too large for other forms of endocytosis.108,109 Because macropinosomes are characterized by the absence of a coat structure to guide their formation and by their heterogeneous size and shape, they give non-phagocytic cells the ability to take up large particles in response to an external stimulus.109 Subsequent work has revealed that SOD1 aggregates stimulate this mechanism to enter the cells.110 However, how SOD1 aggregates stimulate macropinocytosis remains unclear.110 It has been shown that the Rho-ROCK1-LIMK1 signalling pathway regulates extracellular SOD1 macropinocytosis via cofilin activation.111 In addition, by displaying heparan sulfate proteoglycans, receptor for advanced glycation end-products, CD36, integrins and receptor tyrosine kinases, neurons can trigger macropinocytosis following the binding of extracellular protein aggregates (Fig. 1).112 In microglial cells, the binding and internalization of extracellular SOD1 aggregates involve lipid raft formation, scavenger receptors and CD14.113,114
Seeding and nucleation of SOD1, TDP-43 and FUS
Self-perpetuation of SOD1 aggregation
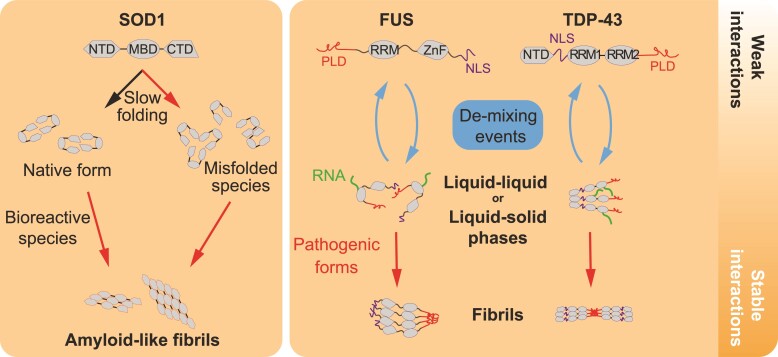
Once ALS prionoids enter the cell and reach the cytoplasmic compartment, they must escape cellular quality control and degradation to co-aggregate with their wildtype or mutant counterpart (Fig. 1). The seeding and nucleation mechanisms of ALS prion-like proteins are still not known, but there are striking fundamental structural differences between SOD1 and the two RBPs, TDP-43 and FUS. Whereas folded wildtype SOD1 is stable, TDP-43 and FUS contain intrinsic disorder domains (IDRs) that promote their self-assembly and lead to a phase transition.115 Thus, in contrast with SOD1, TDP-43 and FUS oscillate easily between different oligomeric structural states that can make them more prone to aggregation.115,116
The first studies to suggest a prion-like activity of SOD1 in vivo showed that the expression of human wildtype SOD1 aggravates the phenotype of mice expressing ALS-linked mutant SOD1.117–119 In addition, the co-expression of a truncated form of mutant SOD1 with wildtype SOD1 in mice triggered the induction of wildtype SOD1 aggregation by mutant SOD1.120 Soluble heterodimers or oligomers of wildtype and mutant SOD1 can also be detected in cell culture.121,122In vitro, several studies have shown the seeding and nucleation of wildtype SOD1 in the presence of mutant SOD1.123–129 The nucleation conditions of SOD1 in vitro necessitate reducing and denaturing agents to favour metal-deficient apoSOD1 and formation of amyloid-like fibrils. This suggests that newly synthetized and unfolded SOD1 is the species that is most likely to form amyloid-like fibrils in the presence of misfolded SOD1. The folding of SOD1 as a fully mature homodimeric metalloenzyme has been well studied in vitro and appears to be a slow process.130–134 Indeed, SOD1 in an unfolded state has a longer lifetime than most other proteins in a test tube, therefore increasing the probability of it forming aberrant oligomeric structures during folding.131,132,135 SOD1 bears several peptide sequences at both N- and C-terminus that have been implicated in fibril formation.136 Furthermore, mutations in these peptide segments found in fALS often enhance the aggregation propensity of SOD1.136 One particular sequence (aas 28–38) adopts a peculiar corkscrew-like structure that is indispensable for the formation of soluble toxic oligomers and likely leads to seeding of unfolded SOD1.137,138 In addition, nucleation of wildtype SOD1 can induce the formation of intermolecular disulfide bonds in the highly reducing environment of the cytoplasm.106,129,135,139 However, intermolecular disulfide bonds are not necessary for the initiation of seeding but could be involved in the sequestration of potentially toxic oligomeric SOD1.140 Other evidence suggests that the formation of these hyperoxidized SOD1 aggregates takes place in the ER, which provides a more suitable environment for disulfide bond formation.91
The cell-to-cell transmission of misfolded mutant SOD1 was evidenced in studies in which preformed SOD1 aggregates were added to cell cultures.106,129 While the extracellular aggregates applied were readily proteolyzable, the intracellularly induced aggregates appeared to be persistent and transferred from cell to cell.106 This transmission is more likely due to endogenous cycles of secretion, internalization and nucleation of wildtype SOD1 than to the persistence during successive cell divisions of the SOD1 aggregates that were initially added to the cultures. This suggests that self-amplification either during cell division or/and passage through the extracellular medium is a prerequisite for the cellular spreading of SOD1.106,107,129 Furthermore, in cell culture, expression of other pathogenic proteins such as FUS and TDP-43 induces wildtype SOD1 misfolding that can be transmitted to naïve cells in a prion-like fashion.85,141
Self-perpetuation of mutant TDP-43 and FUS aggregation
At its N-terminus, TDP-43 is composed of a dimerization domain (N-terminal domain), a nuclear-localized sequence and two RNA recognition motifs (RRMs).110,141,142 The C-terminal part has two IDRs and a glycine-rich domain involved in multivalent weak intermolecular interactions.115 Like other proteins containing IDRs, also called prion-like domains, TDP-43 is well known to self-assemble and creates a separate phase in membrane-less compartments like stress granules (Fig. 3).143,144 The two domains RRM1 and RRM2 of TDP-43 recognize GU-rich RNA sequences on several pre-mRNAs.142,144 mRNA concentrations seem to be key regulators of TDP-43 amyloid-like structures. Indeed, TDP-43 aggregates are formed when cytoplasmic mRNA levels decrease.145,146 This property explains why TDP-43 and FUS form aggregates predominantly in the cytoplasm rather than in the nucleus.145 ALS-causing mutations in TDP-43 have been shown to increase the susceptibility of TDP-43 to forming cytoplasmic insoluble oligomers (Fig. 3).147,148 Widely recognized to occur in ALS and FTD, wildtype TDP-43 labile condensates can also be transformed into insoluble aggregates.146 The circumstances that promote wildtype TDP-43 cytoplasmic aggregates, as well as their pathological relevance, are a matter of intense debate.149,150 In non-pathological conditions, cytoplasmic inclusions of TDP-43 amyloid-like structures called myo-granules have been found to be essential for myotube formation in skeletal muscle.151,152 Interestingly, myo-granules are mostly insoluble, contain sarcomere mRNAs and can seed monomeric TDP-43 in vitro.152 This means than even cytoplasmic TDP-43 amyloid-like oligomers can be formed and disassembled in some cell types and that failure of disaggregation might be the clue to cellular intoxication. Furthermore, an elegant study using real-time microscopy revealed that TDP-43 undergoes highly dynamic mixing and de-mixing into droplets in the cell nucleus (Fig. 3).40 De-mixing is a new term for the segregation and oligomerization of TDP-43 that well defines this physicochemical reaction that is based on weak protein–protein interactions.40 The functional consequence of this condensation process driven by the coordination of the two IDRs is to concentrate a subset of pre-RNAs in the nucleus.143,144 However, when TDP-43 de-mixing occurs in the cytoplasm, TDP-43 is depleted in the nucleus, leading to cell death.40 Mislocalization and aggregation of TDP-43 can be induced independently of stress granule formation by nuclear import disruption and by a broad range of cellular stresses, including proteasomal inhibition, ER misfolding and reactive oxygen species.40,153–155 Importantly, extracellular TDP-43 fibrils can induce irreversible cytoplasmic TDP-43 de-mixing reminiscent of prion-like activity as it has been suggested earlier.40,100,156 As exogenous FUS fibrils but not SOD1 fibrils also induce similar TDP-43 de-mixing, it is not clear if TDP-43 and FUS fibrils directly nucleate endogenous TDP-43.40 Interestingly, cytoplasmic TDP-43 myo-granules, similar to structures observed in injured neurons, are formed in differentiated myotubes during muscle regeneration.152 However, TDP-43 in myo-granules does not seem to be post-translationally modified as observed in cytoplasmic TDP-43 aggregates found in neuronal tissues from sALS.152 It is unknown why cytoplasmic TDP-43 de-mixing in myotubes is reversible and safe, whereas it induces cell death when induced by RBP fibrils in neurons.152 Though the properties of TDP-43 aggregation in cells are becoming better understood, the structural and biophysical determinants involved in the seeding and nucleation of TDP-43 remain largely uncharacterized (Fig. 3).
Figure 3.
Pathological aggregations of SOD1, FUS and TDP-43. Schematic view of pathways leading to the aggregation of SOD1 and the two RBPs, FUS and TDP-43. SOD1 aggregation results in the accumulation of misfolded oligomeric or monomeric species during the folding process that can be due to intrinsically disordered structure (fALS mutations) and defects of the protein quality control and folding machineries. Alternatively, the properly folded SOD1 dimer might be also destabilized due to deleterious physicochemical perturbations (e.g. reactive oxygen species). FUS and TDP-43 aggregation might be the result of an imbalance in their oscillations between disordered monomers and highly ordered oligomeric structures. These structural properties give rise to labile phase transitions from liquid to liquid (liquid droplets) or to liquid to solid (hydrogel). ALS-causing mutations, cellular dysregulation (RNA metabolism, trafficking defects…) favour aberrant and irreversible phase transitions (liquid/solid, solid/solid) producing fibrils (seeding). NTD (N-terminal domain), MBD (metal binding domain), RRM (RNA recognition motif), ZnF (zing finger domain) and NLS (nuclear-localization sequence).
Similar to TDP-43, FUS has an IDR located at the N-terminus that promotes liquid–solid phase transition.157–159 The prion-like IDR domain can self-oligomerize and may promote amyloid-like structures of FUS in the nucleus as well as in the cytoplasm.115,159–161 These structural properties allow FUS to undergo liquid–solid phase transition in the event of increased FUS concentrations.158,161,162 fALS FUS mutations, mainly localized in the IDR, have the tendency to increase the speed of the liquid–solid transition.159,160,163 However, it is not known if the amyloid-like oligomer formation of FUS might be linked to the spread of disease progression in fALS or sALS.
Physiopathological evidence of SOD1 and TDP-43 prion-like propagation in mice
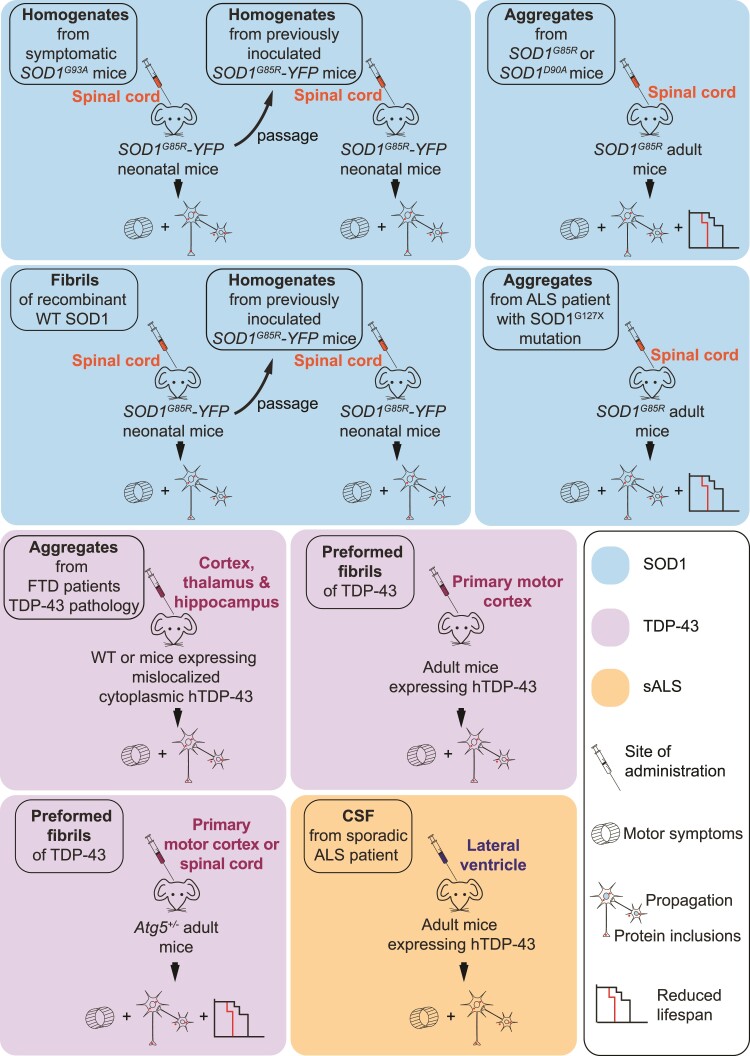
Initial evidence of prion-like propagation of human mutant SOD1 was revealed in in vivo experiments in which protein homogenates obtained from symptomatic SOD1G93A mice were injected into the spinal cords of non-symptomatic SOD1G85R-YFP mice (Fig. 4).164 Injected mice displayed motor symptoms and reduced lifespan, but the observed penetrance was about 60%. In a second passage, injection of homogenates of the spinal cords of SOD1G85R-YFP mice that had been inoculated previously and become paralyzed produced the same symptoms in mice but with full penetrance.164 The same process of passages has been used with different strains of SOD1, resulting in distinct pathologies.31 Another study showed that transgenic mice expressing different SOD1 mutants could produce distinct types of SOD1 aggregate strains.165 Two of these SOD1 strains (G85R and D90A) were then injected into the lumbar spinal cords of asymptomatic animals, resulting in SOD1 aggregation and rostral spreading of the pathology (Fig. 4).166 A rapid induction of motor deficits and premature motoneuron death were then observed.166 This work suggests the existence of multiple mutant SOD1 strains that are able to seed and spread in mutant SOD1 transgenic mice. In another study, protein homogenates from SOD1G127X patients were injected into the spinal cords of SOD1G85R-expressing mice.167 The animals then developed fatal ALS-like pathology and exhibited spreading of SOD1 aggregation.167 Additionally, injection of mutant SOD1 seeds into the sciatic nerve induced disease spread in the peripheral nervous system as well as in the spinal cord.167 However, other types of peripheral administrations (intraperitoneal and intramuscular) of highly potent SOD1 seeds do not seem to allow the spread of ALS pathology in the CNS.168 In contrast with other prion-like pathogenic proteins found in Alzheimer’s disease or Parkinson’s disease, no evidence of an ALS pathology supported by wildtype human SOD1 spread have been revealed.169 Indeed, recent work reveals that transgenic mice overexpressing wildtype SOD1 appear to be resistant to misfolded mutant SOD1 seeds.169 Together, these results show that mutant SOD1 aggregates can transmit and propagate ALS-like dysfunctions when they reach the CNS.
Figure 4.
Experimental paradigms demonstrating the prion-like properties of SOD1 and TDP-43 in mice. This schema is a compilation of several published results obtained by the administration of misfolded SOD1 and TDP-43 aggregates in different mouse models. The nature of protein extracts (crude homogenates, purified proteins, recombinant proteins), the route of delivery and the genetic background of injected mice are indicated. For the observed physiopathological outcome, three criteria are mentioned: the levels of protein inclusions/aggregates in the CNS, the motor symptoms evaluated by behaviour tests and the lifespan of injected mice.
The potential transmissibility and spreading of TDP-43 proteinopathy has been revealed by a study in which insoluble protein extracts from the brains of patients with sporadic FTD were injected into the CNS of mice (Fig. 4).170 Pathological TDP-43 spreading was progressively observed in a spatio-temporal manner in different brain areas, supporting cell-to-cell transmission throughout the CNS.170 The CSF as a route for spreading of pathology has been explored by a study in which the CSF of patients with sALS was intracerebroventricularly injected into mice expressing wildtype human TDP-43.171 Motor, cognitive and pathological changes were observed in the transgenic mice that received ALS CSF. These ALS-like alterations also suggest a threshold effect for seeding of TDP-43. Indeed, the motor phenotype in response to ALS CSF infusion was more pronounced in mice expressing human TDP-43 than in wildtype mice, and cytoplasmic aggregation of TDP-43 was observed only in transgenic mice and not in wildtype mice. Until recently, it was still unknown whether different strains of TDP-43 might support multiple TDP-43 proteinopathies in the CNS, but recent studies have provided new evidence about the spreading of TDP-43 preformed fibrils in human TDP-43 and in Atg5+/− mice. These studies observed spreading all along the corticospinal tract in a time-dependent manner, which supports Braak’s model.172–174 In the case of FUS, although it can behave like a prionoid in vitro, no in vivo experimental evidence currently exists that demonstrates the spread of FUS proteinopathy.
Limits and caveats of prion theory in ALS
Despite the evidence presented above, some points continue to cast doubt on the importance of prion-like mechanisms for transmission and spreading in ALS. A fundamental difference between ALS prionoids and PrP or prionoids including Aβ, α-synuclein and tau is that the latter can form amyloid fibrils in human CNS. Amyloid fibrils are extracellular deposits of a fibrillary protein with a β-sheet secondary structure.175 Biochemical properties of amyloids such as seeding and protease resistance obviously facilitate their dissemination. In addition, the natural secretion of PrP and Aβ peptides that occurs facilitates their dissemination even further. For now, no kind of disease transmission between individuals has been documented in the case of ALS, whereas an observation of this kind has been reported for Alzheimer’s disease.176 Furthermore, inoculation of brain homogenates from patients with ALS into the brain of non-human primates did not reveal any disease transmission.177 In this large-scale study, similar conclusions were made for other non-spongiform neurological diseases, including Alzheimer’s disease and Parkinson’s disease. The phylogenetic distance between human and non-human primates might explain the failure to transmit non-spongiform diseases. Recently, inoculation of brain homogenates from patients with Alzheimer’s disease into mouse lemurs led to the spreading of Aβ and tau pathology in different brain regions associated with cognitive defects.178 It should also be noted that a more detailed histopathological study and follow-up of the motor capacity of the primates could have helped to reveal potential transmission. Another study reported that intramuscular or intraperitoneal inoculation of spinal cord homogenates from endstage SOD1G85R mice that contained SOD1 aggregates did not transmit SOD1 aggregation to the CNS or peripheral tissues of recipient mice.168 It is noteworthy that the SOD1 aggregates were found to be highly sensitive to protease degradation, which is not the case for highly transmissible species, such as the scrapie isoform of PrP. This observation could explain the reduced ability to transmit the disease-associated aggregation from the peripheral tissues to the CNS. Finally, a recent study showed that genetic ablation of the SOD1G37R transgene in corticofugal projection neurons, which prevent corticospinal neuron degeneration, does not influence disease onset, progression, life expectancy or spinal motoneuron survival. Interestingly, the corticofugal projection neuron-selective excision of mutant SOD1 did not influence the levels of misfolded mutant SOD1 in the spinal cord.179 These results led the authors to conclude that corticofugal disease propagation is not mediated by a prion-like mechanism but rather by cortical hyperexcitability.179 Their findings do not seem to be consistent with Braak’s staging scheme,174 where mutant SOD1 in corticofugal neurons may not be essential for disease propagation, although a prion-like mechanism may still operate in the spinal cord.
Clinical and therapeutic considerations
The eventual progression of ALS from an initial site to distant regions in the CNS is well described. The disease starts in one part of the body, in the vast majority of cases in one hand, one foot or as a speech disturbance. Within months or years, the disease spreads to other parts of the body and gradually becomes more debilitating. It has been noted that contiguous areas are more rapidly involved in the disease process of ALS than non-contiguous areas.180 Importantly, pathological studies in humans have confirmed this progressive and stepwise spreading of ALS, following a model of corticofugal spread.174 Although neuronal involvement progresses at different rates from one patient to another, it nevertheless follows a similar sequence between patients. In animal models, and particularly in mutant SOD1 mice, the same phenomenon has been described and follows anatomical pathways.181 To date, in clinical neurology, few other potential explanations have been proposed to interpret the progression of ALS. One interesting suggestion was that a progressive and stepwise activation of microglia might also occur, secondary to initial prionoid spreading.182 A study evaluated this microglial hypothesis by use of selective Cre recombinase-mediated SOD1 deletion in microglial cells via targeting of the CD11b promoter. The impact on disease progression was highly significant; the lifespan of the mice improved by 75 days.182 In another study, the addition of SOD1 aggregates to microglial cells in culture triggered their activation.114 This study suggests that mutant SOD1 aggregates can contribute to disease progression by promoting microglial activation and subsequent release of toxic factors to motoneurons.114 Targeting both extracellular and intracellular misfolded SOD1, anti-misfolded SOD1 antibodies have been successfully used as a potential immunotherapeutic approach in mice.183,184 More recently, peripheral administration of an antibody specifically recognizing misfolded SOD1 delayed disease onset, improved the decline of motor function, and increased the lifespan of SOD1G37R and SOD1G93A mice.185 An alternative approach has been the development of an ALS vaccine targeting a conformation-specific epitope. This aimed to reduce the burden of extracellular SOD1 and has provided therapeutic benefits in ALS mice.186,187 From a therapeutic point of view, the discovery of prionoid spreading in people with ALS opens perspectives that cover a large number of pathways. Proteostasis is a particularly important mechanism to consider. It has been shown that riluzole, the only marketed drug for ALS that has a positive effect on ALS patients’ survival, enhances the expression of cytosolic chaperons HSP70 and HSP90 by increasing the heat shock protein response.188 Clinical trials are underway to evaluate a series of drugs acting on proteostasis, such as arimoclomol,189 fasudil190 and memantine.191
Other promising approaches that tackle prionoid activity at the root consist of silencing SOD1. The proof-of-principle of SOD1 gene silencing in mice with virally delivered shRNA has already been established192,193; more recently it was shown that subpial delivery of adeno-associated adenovirus serotype 9 prevented disease onset when injected before ALS onset, and blocked disease progression when delivered after onset.194 One promising therapeutic approach is gene modulation by antisense oligonucleotides (ASOs). ASOs have been given intrathecally to patients with ALS; treatment has been shown to be well tolerated and result in significantly lowered SOD1 levels in the CSF.195 A phase I/II trial with torfersen ASO in 50 patients with SOD1 mutations not only revealed a good tolerance profile, but also suggested that the clinical impact of this therapy is worth exploring in a phase III trial.196 In October 2021, results of the tofersen phase III study (VALOR) were presented at the American Neurological Association annual meeting (https://investors.biogen.com/). Although the primary efficacy endpoint as determined by the revised ALS functional rating scale was not met, secondary measures, including SOD1 CSF and plasma neurofilament light chain (a neuronal degeneration marker) levels, reached the goal of change from baseline. Trends towards a benefit for respiratory function, muscle strength and quality of life are further promising signs. One of the possible directions that could lead to a better outcome with this treatment is earlier and longer intervention, and Biogen is currently running a phase III trial (ATLAS) in presymptomatic SOD1 mutation carriers (https://clinicaltrials.gov/ct2/show/NCT04856982).
Conclusion
Perhaps more so than in other neurodegenerative diseases, the field of ALS research is incredibly dynamic, as functionally and structurally unrelated new genes involved in fALS are being discovered each year and illuminating novel molecular and cellular fundamental mechanisms. A challenge for the scientific community remains as to how all these diverse pathological genetic dysfunctions lead to a disease that is quite homogenous. In this context, the study of the spread of prionoids in ALS remains in its infancy and might offer new therapeutic approaches. Preventing disease progression in ALS is an important therapeutic strategy that will be boosted by our increased knowledge about the molecular and cellular mechanisms that lead to the dissemination of neurotoxic prionoids.
Data availability
Data sharing is not applicable to this article as no new data were created or analysed in this study.
Abbreviations
- aas =
amino acids
- ATG7 =
autophagy related 7
- CUPS =
compartment for unconventional protein secretion
- ER =
endoplasmic reticulum
- fALS =
familial ALS
- FTD =
frontotemporal dementia
- FUS =
fused in sarcoma
- IDR =
intrinsic disorder domain
- LC3 =
microtubule-associated protein 1A/1B-light chain 3
- MAPS =
misfolding-associated protein secretion
- PrP =
prion protein
- RBP =
RNA-binding protein
- RRM =
RNA recognition motif
- sALS =
sporadic ALS
- SOD1 =
Cu/Zn superoxide dismutase 1
- TDP-43 =
TAR DNA-binding protein of 43 kDa
- USP19 =
ubiquitin-specific protease 19
Contributor Information
Philippe Gosset, INM, Univ Montpellier, INSERM, CNRS, Montpellier 34095, France.
William Camu, INM, Univ Montpellier, INSERM, CNRS, Montpellier 34095, France; ALS Center, Univ Montpellier, CHU Montpellier, Montpellier, France.
Cedric Raoul, INM, Univ Montpellier, INSERM, CNRS, Montpellier 34095, France; Laboratory of Neurobiology, Kazan Federal University, Kazan, Russia.
Alexandre Mezghrani, INM, Univ Montpellier, INSERM, CNRS, Montpellier 34095, France.
Funding
This work was supported by a grant from the French national research agency (ANR SPREADALS); the association Française pour la recherche sur la SLA (ARSLA); Marie Sklodowska-Curie Actions Individual fellowship ADELE (CR) and the national institute of health and medical research (INSERM).
Competing interests
The authors report no competing interests.
References
- 1. Jucker M, Walker LC. Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat Neurosci. 2018;21(10):1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vaquer-Alicea J, Diamond MI. Propagation of protein aggregation in neurodegenerative diseases. Annu Rev Biochem. 2019;88(1):785–810. [DOI] [PubMed] [Google Scholar]
- 3. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. [DOI] [PubMed] [Google Scholar]
- 4. Goedert M. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled A, tau, and -synuclein. Science. 2015;349(6248):1255555. [DOI] [PubMed] [Google Scholar]
- 5. Prusiner SB. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet. 2013;47:601–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlson GA, Prusiner SB. How an infection of sheep revealed prion mechanisms in Alzheimer’s disease and other neurodegenerative disorders. Int J Mol Sci. 2021;22(9):4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kara E, Marks JD, Aguzzi A. Toxic protein spread in neurodegeneration: Reality versus fantasy. Trends Mol Med. 2018;24(12):1007–1020. [DOI] [PubMed] [Google Scholar]
- 8. Wong YC, Krainc D. α-Synuclein toxicity in neurodegeneration: Mechanism and therapeutic strategies. Nat Med. 2017;23(2):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. [DOI] [PubMed] [Google Scholar]
- 10. McAlary L, Plotkin SS, Yerbury JJ, Cashman NR. Prion-like propagation of protein misfolding and aggregation in amyotrophic lateral sclerosis. Front Mol Neurosci. 2019;12(262):262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Polymenidou M, Cleveland DW. The seeds of neurodegeneration: Prion-like spreading in ALS. Cell. 2011;147(3):498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bertolotti A. Importance of the subcellular location of protein deposits in neurodegenerative diseases. Curr Opin Neurobiol. 2018;51:127–133. [DOI] [PubMed] [Google Scholar]
- 13. Taylor JP, Brown RH, Cleveland DW. Decoding ALS: From genes to mechanism. Nature. 2016;539(7628):197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shatunov A, Al-Chalabi A. The genetic architecture of ALS. Neurobiol Dis. 2021;147:105156. [DOI] [PubMed] [Google Scholar]
- 15. Lattante S, Ciura S, Rouleau GA, Kabashi E. Defining the genetic connection linking amyotrophic lateral sclerosis (ALS) with frontotemporal dementia (FTD). Trends Genet. 2015;31(5):263–273. [DOI] [PubMed] [Google Scholar]
- 16. Mathis S, Goizet C, Soulages A, Vallat JM, Masson GL. Genetics of amyotrophic lateral sclerosis: A review. J Neurol Sci. 2019;399:217–226. [DOI] [PubMed] [Google Scholar]
- 17. Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Al-Chalabi A, Jones A, Troakes C, King A, Al-Sarraj S, van den Berg LH. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012;124(3):339–352. [DOI] [PubMed] [Google Scholar]
- 19. Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5(3):781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ciryam P, Kundra R, Morimoto RI, Dobson CM, Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol Sci. 2015;36(2):72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakashima-Yasuda H, Uryu K, Robinson J, et al. . Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114(3):221–229. [DOI] [PubMed] [Google Scholar]
- 22. Amador-Ortiz C, Lin WL, Ahmed Z, et al. . TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol. 2007;61(5):435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tziortzouda P, Van Den Bosch L, Hirth F. Triad of TDP43 control in neurodegeneration: Autoregulation, localization and aggregation. Nat Rev Neurosci. 2021;22(4):197–208. [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Branicky R, Noë A, Hekimi S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J Cell Biol. 2018;217(6):1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364(6435):362. [DOI] [PubMed] [Google Scholar]
- 26. Reaume AG, Elliott JL, Hoffman EK, et al. . Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13(1):43–47. [DOI] [PubMed] [Google Scholar]
- 27. Fischer LR, Li Y, Asress SA, Jones DP, Glass JD. Absence of SOD1 leads to oxidative stress in peripheral nerve and causes a progressive distal motor axonopathy. Exp Neurol. 2012;233(1):163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bruijn LI, Houseweart MK, Kato S, et al. . Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281(5384):1851–1854. [DOI] [PubMed] [Google Scholar]
- 29. Bowling AC, Barkowski EE, McKenna-Yasek D, et al. . Superoxide dismutase concentration and activity in familial amyotrophic lateral sclerosis. J Neurochem. 1995;64(5):2366–2369. [DOI] [PubMed] [Google Scholar]
- 30. Cleveland DW, Laing N, Hurse PV, Brown RH. Toxic mutants in Charcot’s sclerosis. Nature. 1995;378(6555):342–343. [DOI] [PubMed] [Google Scholar]
- 31. Ayers JI, Diamond J, Sari A, et al. . Distinct conformers of transmissible misfolded SOD1 distinguish human SOD1-FALS from other forms of familial and sporadic ALS. Acta Neuropathol. 2016;132(6):827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paré B, Lehmann M, Beaudin M, et al. . Misfolded SOD1 pathology in sporadic amyotrophic lateral sclerosis. Sci Rep. 2018;8(1):14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Forsberg K, Jonsson PA, Andersen PM, et al. . Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS One. 2010;5(7):e11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Forsberg K, Graffmo K, Pakkenberg B, et al. . Misfolded SOD1 inclusions in patients with mutations in C9orf72 and other ALS/FTD-associated genes. J Neurol Neurosurg Psychiatry. 2019;90(8):861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pokrishevsky E, Grad LI, Yousefi M, Wang J, Mackenzie IR, Cashman NR. Aberrant localization of FUS and TDP43 is associated with misfolding of SOD1 in amyotrophic lateral sclerosis. PLoS One. 2012;7(4):e35050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Da Cruz S, Bui A, Saberi S, et al. . Misfolded SOD1 is not a primary component of sporadic ALS. Acta Neuropathol. 2017;134(1):97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ratti A, Buratti E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J Neurochem. 2016;138:95–111. [DOI] [PubMed] [Google Scholar]
- 38. Panza F, Lozupone M, Seripa D, et al. . Development of disease-modifying drugs for frontotemporal dementia spectrum disorders. Nat Rev Neurol. 2020;16(4):213–228. [DOI] [PubMed] [Google Scholar]
- 39. Sreedharan J, Blair IP, Tripathi VB, et al. . TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gasset-Rosa F, Lu S, Yu H, et al. . Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron. 2019;102(2):339–357.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Purice MD, Taylor JP. Linking hnRNP function to ALS and FTD pathology. Front Neurosci. 2018;12:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arai T, Hasegawa M, Akiyama H, et al. . TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351(3):602–611. [DOI] [PubMed] [Google Scholar]
- 43. Neumann M, Sampathu DM, Kwong LK, et al. . Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. [DOI] [PubMed] [Google Scholar]
- 44. Hasegawa M, Arai T, Nonaka T, et al. . Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol. 2008;64(1):60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim G, Gautier O, Tassoni-Tsuchida E, Ma XR, Gitler AD. ALS genetics: Gains, losses, and implications for future therapies. Neuron. 2020;108(5):822–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kwiatkowski TJ, Bosco DA, Leclerc AL, et al. . Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. [DOI] [PubMed] [Google Scholar]
- 47. Vance C, Rogelj B, Hortobágyi T, et al. . Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tyzack GE, Luisier R, Taha DM, et al. . Widespread FUS mislocalization is a molecular hallmark of amyotrophic lateral sclerosis. Brain. 2019;142(9):2572–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Korenkova O, Pepe A, Zurzolo C. Fine intercellular connections in development: TNTs, cytonemes, or intercellular bridges? Cell Stress. 2020;4(2):30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Asai H, Ikezu S, Tsunoda S, et al. . Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci. 2015;18(11):1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Donnelly KM, DeLorenzo OR, Zaya AD, et al. . Phagocytic glia are obligatory intermediates in transmission of mutant huntingtin aggregates across neuronal synapses. Elife. 2020;9:e58499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wood H. Evidence for trans-synaptic and exo-synaptic tau propagation in Alzheimer disease. Nat Rev Neurol. 2015;11(12):665–665. [DOI] [PubMed] [Google Scholar]
- 53. Cicardi ME, Marrone L, Azzouz M, Trotti D. Proteostatic imbalance and protein spreading in amyotrophic lateral sclerosis. EMBO J. 2021;40(10):e106389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hipp MS, Kasturi P, Hartl FU. The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol. 2019;20(7):421–435. [DOI] [PubMed] [Google Scholar]
- 55. Ruegsegger C, Saxena S. Proteostasis impairment in ALS. Brain Res. 2016;1648:571–579. [DOI] [PubMed] [Google Scholar]
- 56. Douglas PM, Dillin A. Protein homeostasis and aging in neurodegeneration. J Cell Biol. 2010;190(5):719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hetz C. Adapting the proteostasis capacity to sustain brain healthspan. Cell. 2021;184(6):1545–1560. [DOI] [PubMed] [Google Scholar]
- 58. Peng C, Trojanowski JQ, Lee VMY. Protein transmission in neurodegenerative disease. Nat Rev Neurol. 2020;16(4):199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Victoria GS, Zurzolo C. The spread of prion-like proteins by lysosomes and tunneling nanotubes: Implications for neurodegenerative diseases. J Cell Biol. 2017;216(9):2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vilette D, Courte J, Peyrin JM, et al. . Cellular mechanisms responsible for cell-to-cell spreading of prions. Cell Mol Life Sci. 2018;75(14):2557–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gallotta I, Sandhu A, Peters M, et al. . Extracellular proteostasis prevents aggregation during pathogenic attack. Nature. 2020;584(7821):410–414. [DOI] [PubMed] [Google Scholar]
- 62. Melentijevic I, Toth ML, Arnold ML, et al. . C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature. 2017;542(7641):367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27(3):230–240. [DOI] [PubMed] [Google Scholar]
- 64. Sitia R, Rubartelli A. Evolution, role in inflammation, and redox control of leaderless secretory proteins. J Biol Chem. 2020;295(22):7799–7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Volkmar N, Fenech E, Christianson JC. New MAPS for misfolded proteins. Nat Cell Biol. 2016;18(7):724–726. [DOI] [PubMed] [Google Scholar]
- 66. Shiina Y, Arima K, Tabunoki H, Satoh JI. TDP-43 dimerizes in human cells in culture. Cell Mol Neurobiol. 2010;30(4):641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Holm MM, Kaiser J, Schwab ME. Extracellular vesicles: Multimodal envoys in neural maintenance and repair. Trends Neurosci. 2018;41(6):360–372. [DOI] [PubMed] [Google Scholar]
- 68. Lim YJ. Lee SJ. Are exosomes the vehicle for protein aggregate propagation in neurodegenerative diseases? Acta Neuropathol Commun. 2017;5(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nickel W. The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur J Biochem. 2003;270(10):2109–2119. [DOI] [PubMed] [Google Scholar]
- 70. Pallotta MT, Nickel W. FGF2 and IL-1β—explorers of unconventional secretory pathways at a glance. J Cell Sci. 2020;133(21):jcs250449. [DOI] [PubMed] [Google Scholar]
- 71. Cruz-Garcia D, Malhotra V, Curwin AJ. Unconventional protein secretion triggered by nutrient starvation. Semin Cell Dev Biol. 2018;83:22–28. [DOI] [PubMed] [Google Scholar]
- 72. Urushitani M, Sik A, Sakurai T, Nukina N, Takahashi R, Julien JP. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat Neurosci. 2006;9(1):108–118. [DOI] [PubMed] [Google Scholar]
- 73. Leidal AM, Huang HH, Marsh T, et al. . The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol. 2020;22(2):187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Santillo M, Secondo A, Serù R, et al. . Evidence of calcium- and SNARE-dependent release of CuZn superoxide dismutase from rat pituitary GH3 cells and synaptosomes in response to depolarization. J Neurochem. 2007;102(3):679–685. [DOI] [PubMed] [Google Scholar]
- 75. Pluthero FG, Shreeve M, Eskinazi D, et al. . Purification of an inhibitor of erythroid progenitor cell cycling and antagonist to interleukin 3 from mouse marrow cell supernatants and its identification as cytosolic superoxide dismutase. J Cell Biol. 1990;111(3):1217–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cimini V, Ruggiero G, Buonomo T, et al. . CuZn-superoxide dismutase in human thymus: Immunocytochemical localisation and secretion in thymus-derived epithelial and fibroblast cell lines. Histochem Cell Biol. 2002;118(2):163–169. [DOI] [PubMed] [Google Scholar]
- 77. Cruz-Garcia D, Brouwers N, Duran JM, Mora G, Curwin AJ, Malhotra V. A diacidic motif determines unconventional secretion of wild-type and ALS-linked mutant SOD1. J Cell Biol. 2017;216(9):2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mondola P, Annella T, Santillo M, Santangelo F. Evidence for secretion of cytosolic CuZn superoxide dismutase by Hep G2 cells and human fibroblasts. Int J Biochem Cell Biol. 1996;28(6):677–681. [DOI] [PubMed] [Google Scholar]
- 79. Mondola P, Ruggiero G, Serù R, et al. . The Cu, Zn superoxide dismutase in neuroblastoma SK-N-BE cells is exported by a microvesicles dependent pathway. Brain Res Mol Brain Res. 2003;110(1):45–51. [DOI] [PubMed] [Google Scholar]
- 80. Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14(2):207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Andersen PM, Nilsson P, Keränen ML, et al. . Phenotypic heterogeneity in motor neuron disease patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain. 1997;120(10):1723–1737. [DOI] [PubMed] [Google Scholar]
- 82. Segovia-Silvestre T, Andreu AL, Vives-Bauza C, Garcia-Arumi E, Cervera C, Gamez J. A novel exon 3 mutation (D76V) in the SOD1 gene associated with slowly progressive ALS. Amyotroph Lateral Scler Other Motor Neuron Disord. 2002;3(2):69–74. [DOI] [PubMed] [Google Scholar]
- 83. Gomes C, Keller S, Altevogt P, Costa J. Evidence for secretion of Cu, Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci Lett. 2007;428(1):43–46. [DOI] [PubMed] [Google Scholar]
- 84. Basso M, Pozzi S, Tortarolo M, et al. . Mutant copper-zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: Implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J Biol Chem. 2013;288(22):15699–15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Grad LI, Yerbury JJ, Turner BJ, et al. . Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc Natl Acad Sci U S A. 2014;111(9):3620–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Petrozziello T, Secondo A, Tedeschi V, et al. . ApoSOD1 lacking dismutase activity neuroprotects motor neurons exposed to beta-methylamino-L-alanine through the Ca2+/Akt/ERK1/2 prosurvival pathway. Cell Death Differ. 2017;24(3):511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cruz-Garcia D, Brouwers N, Malhotra V, Curwin AJ. Reactive oxygen species triggers unconventional secretion of antioxidants and Acb1. J Cell Biol. 2020;219(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Smith HL, Freeman OJ, Butcher AJ, et al. . Astrocyte unfolded protein response induces a specific reactivity state that causes non-cell-autonomous neuronal degeneration. Neuron. 2020;105(5):855–866.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Urushitani M, Ezzi SA, Matsuo A, Tooyama I, Julien JP. The endoplasmic reticulum–Golgi pathway is a target for translocation and aggregation of mutant superoxide dismutase linked to ALS. FASEB J. 2008;22(7):2476–2487. [DOI] [PubMed] [Google Scholar]
- 90. Chang RC, Parakh S, Coates JR, Long S, Atkin JD. Protein disulphide isomerase is associated with mutant SOD1 in canine degenerative myelopathy. Neuroreport. 2019;30(1):8–13. [DOI] [PubMed] [Google Scholar]
- 91. Medinas DB, Rozas P, Martínez Traub F, et al. . Endoplasmic reticulum stress leads to accumulation of wild-type SOD1 aggregates associated with sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2018;115(32):8209–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Allen WJ, Collinson I, Römisch K. Post-translational protein transport by the Sec complex. Trends Biochem Sci. 2019;44(6):481–483. [DOI] [PubMed] [Google Scholar]
- 93. Haßdenteufel S, Nguyen D, Helms V, Lang S, Zimmermann R. ER import of small human presecretory proteins: Components and mechanisms. FEBS Lett. 2019;593(18):2506–2524. [DOI] [PubMed] [Google Scholar]
- 94. Xu Y, Cui L, Dibello A, et al. . DNAJC5 facilitates USP19-dependent unconventional secretion of misfolded cytosolic proteins. Cell Discov. 2018;4(11):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lee JG, Takahama S, Zhang G, Tomarev SI, Ye Y. Unconventional secretion of misfolded proteins promotes adaptation to proteasome dysfunction in mammalian cells. Nat Cell Biol. 2016;18(7):765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Thomas EV, Fenton WA, McGrath J, Horwich AL. Transfer of pathogenic and nonpathogenic cytosolic proteins between spinal cord motor neurons in vivo in chimeric mice. Proc Natl Acad Sci U S A. 2017;114(15):E3139–E3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rudnick ND, Griffey CJ, Guarnieri P, et al. . Distinct roles for motor neuron autophagy early and late in the SOD1G93A mouse model of ALS. Proc Natl Acad Sci U S A. 2017;114(39):E8294–E8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V. Secretory autophagy. Curr Opin Cell Biol. 2015;35:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nilsson P, Loganathan K, Sekiguchi M, et al. . Aβ secretion and plaque formation depend on autophagy. Cell Rep. 2013;5(1):61–69. [DOI] [PubMed] [Google Scholar]
- 100. Feiler MS, Strobel B, Freischmidt A, et al. . TDP-43 is intercellularly transmitted across axon terminals. J Cell Biol. 2015;211(4):897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ishii T, Kawakami E, Endo K, Misawa H, Watabe K. Formation and spreading of TDP-43 aggregates in cultured neuronal and glial cells demonstrated by time-lapse imaging. PLoS One. 2017;12(6):e0179375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Iguchi Y, Eid L, Parent M, et al. . Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain. 2016;139:3187–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nonaka T, Masuda-Suzukake M, Arai T, et al. . Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 2013;4(1):124–134. [DOI] [PubMed] [Google Scholar]
- 104. Sproviero D, La Salvia S, Giannini M, et al. . Pathological proteins are transported by extracellular vesicles of sporadic amyotrophic lateral sclerosis patients. Front Neurosci. 2018;12:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Delorme-Axford E, Klionsky DJ. The LC3-conjugation machinery specifies cargo loading and secretion of extracellular vesicles. Autophagy. 2020;16:1169–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Münch C, O’Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc Natl Acad Sci U S A. 2011;108(9):3548–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Grad LI, Pokrishevsky E, Silverman JM, Cashman NR. Exosome-dependent and independent mechanisms are involved in prion-like transmission of propagated Cu/Zn superoxide dismutase misfolding. Prion. 2014;8(5):331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mercer J, Helenius A. Gulping rather than sipping: Macropinocytosis as a way of virus entry. Curr Opin Microbiol. 2012;15(4):490–499. [DOI] [PubMed] [Google Scholar]
- 109. Swanson JA. Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol. 2008;9(8):639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zeineddine R, Pundavela JF, Corcoran L, et al. . SOD1 protein aggregates stimulate macropinocytosis in neurons to facilitate their propagation. Mol Neurodegener. 2015;10:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhong Z, Grasso L, Sibilla C, Stevens TJ, Barry N, Bertolotti A. Prion-like protein aggregates exploit the RHO GTPase to cofilin-1 signaling pathway to enter cells. EMBO J. 2018;37(6):e97822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yerbury JJ. Protein aggregates stimulate macropinocytosis facilitating their propagation. Prion. 2016;10(2):119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhao W, Beers DR, Henkel JS, et al. . Extracellular mutant SOD1 induces microglial-mediated motoneuron injury. Glia. 2010;58(2):231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Roberts K, Zeineddine R, Corcoran L, Li W, Campbell IL, Yerbury JJ. Extracellular aggregated Cu/Zn superoxide dismutase activates microglia to give a cytotoxic phenotype. Glia. 2013;61(3):409–419. [DOI] [PubMed] [Google Scholar]
- 115. Loughlin FE, Wilce JA. TDP-43 and FUS-structural insights into RNA recognition and self-association. Curr Opin Struct Biol. 2019;59:134–142. [DOI] [PubMed] [Google Scholar]
- 116. Mompean M, Laurents DV. Intrinsically disordered domains, amyloids and protein liquid phases: Evolving concepts and open questions. Protein Pept Lett. 2017;24(4):281–293. [DOI] [PubMed] [Google Scholar]
- 117. Deng HX, Shi Y, Furukawa Y, et al. . Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc Natl Acad Sci U S A. 2006;103(18):7142–7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Prudencio M, Durazo A, Whitelegge JP, Borchelt DR. An examination of wild-type SOD1 in modulating the toxicity and aggregation of ALS-associated mutant SOD1. Hum Mol Genet. 2010;19(24):4774–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang L, Deng HX, Grisotti G, Zhai H, Siddique T, Roos RP. Wild-type SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. Hum Mol Genet. 2009;18(9):1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Han-Xiang D, Hujun J, Ronggen F, et al. . Molecular dissection of ALS-associated toxicity of SOD1 in transgenic mice using an exon-fusion approach. Hum Mol Genet. 2008;17(15):2310–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Witan H, Kern A, Koziollek-Drechsler I, Wade R, Behl C, Clement AM. Heterodimer formation of wild-type and amyotrophic lateral sclerosis-causing mutant Cu/Zn-superoxide dismutase induces toxicity independent of protein aggregation. Hum Mol Genet. 2008;17(10):1373–1385. [DOI] [PubMed] [Google Scholar]
- 122. Witan H, Gorlovoy P, Kaya AM, et al. . Wild-type Cu/Zn superoxide dismutase (SOD1) does not facilitate, but impedes the formation of protein aggregates of amyotrophic lateral sclerosis causing mutant SOD1. Neurobiol Dis. 2009;36(2):331–342. [DOI] [PubMed] [Google Scholar]
- 123. Banci L, Bertini I, Durazo A, et al. . Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc Natl Acad Sci U S A. 2007;104(27):11263–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Banci L, Bertini I, Boca M, et al. . SOD1 and amyotrophic lateral sclerosis: Mutations and oligomerization. PLoS One. 2008;3(2):e1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chan PK, Chattopadhyay M, Sharma S, et al. . Structural similarity of wild-type and ALS-mutant superoxide dismutase-1 fibrils using limited proteolysis and atomic force microscopy. Proc Natl Acad Sci U S A. 2013;110(27):10934–10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Chattopadhyay M, Valentine JS. Aggregation of copper-zinc superoxide dismutase in familial and sporadic ALS. Antioxid Redox Signal. 2009;11(7):1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chia R, Tattum MH, Jones S, Collinge J, Fisher EMC, Jackson GS. Superoxide dismutase 1 and tgSOD1 mouse spinal cord seed fibrils, suggesting a propagative cell death mechanism in amyotrophic lateral sclerosis. PLoS One. 2010;5(5):e10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Furukawa Y, Kaneko K, Yamanaka K, O’Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2008;283(35):24167–24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Grad LI, Guest WC, Yanai A, et al. . Intermolecular transmission of superoxide dismutase 1 misfolding in living cells. Proc Natl Acad Sci U S A. 2011;108(39):16398–16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. Solution structure of Apo Cu, Zn superoxide dismutase: Role of metal ions in protein folding. Biochemistry. 2003;42(32):9543–9553. [DOI] [PubMed] [Google Scholar]
- 131. Cohen NR, Kayatekin C, Zitzewitz JA, Bilsel O, Matthews CR. Friction-limited folding of disulfide-reduced monomeric SOD1. Biophys J. 2020;118(8):1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ding F, Furukawa Y, Nukina N, Dokholyan NV. Local unfolding of Cu, Zn superoxide dismutase monomer determines the morphology of fibrillar aggregates. J Mol Biol. 2012;421(4–5):548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: Disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci U S A. 2004;101(45):15893–15898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Mojumdar S S, Scholl Z N, Dee DR, et al. . Partially native intermediates mediate misfolding of SOD1 in single-molecule folding trajectories. Nat Commun. 2017;8(1):1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kayatekin C, Zitzewitz JA, Matthews CR. Disulfide-reduced ALS Variants of Cu, Zn superoxide dismutase exhibit increased populations of unfolded species. J Mol Biol. 2010;398(2):320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ivanova MI, Sievers SA, Guenther EL, et al. . Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proc Natl Acad Sci U S A. 2014;111(1):197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sangwan S, Zhao A, Adams KL, et al. . Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc Natl Acad Sci U S A. 2017;114(33):8770–8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Sangwan S, Sawaya MR, Murray KA, Hughes MP, Eisenberg DS. Atomic structures of corkscrew-forming segments of SOD1 reveal varied oligomer conformations. Protein Sci. 2018;27(7):1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Furukawa Y, Fu R, Deng HX, Siddique T, O’Halloran TV. Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu, Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc Natl Acad Sci U S A. 2006;103(18):7148–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Karch CM, Borchelt DR. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283(20):13528–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Grad LI, Pokrishevsky E, Cashman NR. Intercellular prion-like conversion and transmission of Cu/Zn superoxide dismutase (SOD1) in cell culture. Methods Mol Biol. 2017;1658:357–367. [DOI] [PubMed] [Google Scholar]
- 142. Afroz T, Hock EM, Ernst P, et al. . Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat Commun. 2017;8(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hallegger M, Chakrabarti AM, Lee FCY, et al. . TDP-43 condensation properties specify its RNA-binding and regulatory repertoire. Cell. 2021;184(18):4680–4696.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Mann JR, Donnelly CJ. RNA modulates physiological and neuropathological protein phase transitions. Neuron. 2021;109(17):2663–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Maharana S, Wang J, Papadopoulos DK, et al. . RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360(6391):918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Mann JR, Gleixner AM, Mauna JC, et al. . RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 2019;102(2):321–338.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. French RL, Grese ZR, Aligireddy H, et al. . Detection of TAR DNA-binding protein 43 (TDP-43) oligomers as initial intermediate species during aggregate formation. J Biol Chem. 2019;294(17):6696–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Ling SC, Albuquerque CP, Han JS, et al. . ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A. 2010;107(30):13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Hergesheimer RC, Chami AA, de Assis DR, et al. . The debated toxic role of aggregated TDP-43 in amyotrophic lateral sclerosis: A resolution in sight? Brain. 2019;142(5):1176–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Loganathan S, Lehmkuhl EM, Eck RJ, Zarnescu DC. To be or not to be…toxic—is RNA association with TDP-43 complexes deleterious or protective in neurodegeneration? Front Mol Biosci. 2019;6:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Cutler AA, Ewachiw TE, Corbet GA, Parker R, Olwin BB. Myo-granules connect physiology and pathophysiology. J Exp Neurosci. 2019;13:117906951984215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Vogler TO, Wheeler JR, Nguyen ED, et al. . TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature. 2018;563(7732):508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Dewey CM, Cenik B, Sephton CF, et al. . TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31(5):1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Mateju D, Franzmann TM, Patel A, et al. . An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36(12):1669–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Walker AK, Soo KY, Sundaramoorthy V, et al. . ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS One. 2013;8(11):e81170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Zhu L, Xu M, Yang M, et al. . An ALS-mutant TDP-43 neurotoxic peptide adopts an anti-parallel β-structure and induces TDP-43 redistribution. Hum Mol Genet. 2014;23(25):6863–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Bogaert E, Boeynaems S, Kato M, et al. . Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 2018;24(3):529–537.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Holehouse AS, Pappu RV. FUS zigzags its way to cross beta. Cell. 2017;171(3):499–500. [DOI] [PubMed] [Google Scholar]
- 159. Patel A, Lee HO, Jawerth L, et al. . A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162(5):1066–1077. [DOI] [PubMed] [Google Scholar]
- 160. Murray DT, Kato M, Lin Y, et al. . Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171(3):615–627.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Murthy AC, Dignon GL, Kan Y, et al. . Molecular interactions underlying liquid–liquid phase separation of the FUS low-complexity domain. Nat Struct Mol Biol. 2019;26(7):637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Niaki AG, Sarkar J, Cai X, et al. . Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-linked FUS mutations. Mol Cell. 2020;77(1):82–94.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Murakami T, Qamar S, Lin JQ, et al. . ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88(4):678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Ayers JI, Fromholt S, Koch M, et al. . Experimental transmissibility of mutant SOD1 motor neuron disease. Acta Neuropathol. 2014;128(6):791–803. [DOI] [PubMed] [Google Scholar]
- 165. Bergh J, Zetterström P, Andersen PM, et al. . Structural and kinetic analysis of protein-aggregate strains in vivo using binary epitope mapping. Proc Natl Acad Sci U S A. 2015;112(14):4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Bidhendi EE, Bergh J, Zetterström P, Andersen PM, Marklund SL, Brännström T. Two superoxide dismutase prion strains transmit amyotrophic lateral sclerosis-like disease. J Clin Invest. 2016;126(6):2249–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Bidhendi E E, Bergh J, Zetterström P, et al. . Mutant superoxide dismutase aggregates from human spinal cord transmit amyotrophic lateral sclerosis. Acta Neuropathol. 2018;136(6):939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Keskin I, Ekhtiari Bidhendi E, Marklund M, et al. . Peripheral administration of SOD1 aggregates does not transmit pathogenic aggregation to the CNS of SOD1 transgenic mice. Acta Neuropathol Commun. 2021;9(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Ayers JI, Xu G, Dillon K, et al. . Variation in the vulnerability of mice expressing human superoxide dismutase 1 to prion-like seeding: A study of the influence of primary amino acid sequence. Acta Neuropathol Commun. 2021;9(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Porta S, Xu Y, Restrepo CR, et al. . Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat Commun. 2018;9(1):4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Mishra PS, Boutej H, Soucy G, et al. . Transmission of ALS pathogenesis by the cerebrospinal fluid. Acta Neuropathol Commun. 2020;8(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Ding X, Xiang Z, Qin C, et al. . Spreading of TDP-43 pathology via pyramidal tract induces ALS-like phenotypes in TDP-43 transgenic mice. Acta Neuropathol Commun. 2021;9(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zhang R, Chen Y, Wang X, et al. . Spreading of pathological TDP-43 along corticospinal tract axons induces ALS-like phenotypes in Atg5+/− mice. Int J Biol Sci. 2021;17(2):390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Braak H, Brettschneider J, Ludolph AC, Lee VM, Trojanowski JQ, Del Tredici K. Amyotrophic lateral sclerosis—a model of corticofugal axonal spread. Nat Rev Neurol. 2013;9(12):708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Benson MD, Buxbaum JN, Eisenberg DS, et al. . Amyloid nomenclature 2018: Recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid. 2018;25(4):215–219. [DOI] [PubMed] [Google Scholar]
- 176. Purro SA, Farrow MA, Linehan J, et al. . Transmission of amyloid-β protein pathology from cadaveric pituitary growth hormone. Nature. 2018;564(7736):415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Brown P, Gibbs CJ, Rodgers-Johnson P, et al. . Human spongiform encephalopathy: The National Institutes of Health series of 300 cases of experimentally transmitted disease. Ann Neurol. 1994;35(5):513–529. [DOI] [PubMed] [Google Scholar]
- 178. Lam S, Petit F, Hérard AS, et al. . Transmission of amyloid-beta and tau pathologies is associated with cognitive impairments in a primate. Acta Neuropathol Commun. 2021;9:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Scekic-Zahirovic J, Fischer M, Stuart-Lopez G, et al. . Evidence that corticofugal propagation of ALS pathology is not mediated by prion-like mechanism. Prog Neurobiol. 2021;200:101972. [DOI] [PubMed] [Google Scholar]
- 180. Brooks BR. The role of axonal transport in neurodegenerative disease spread: A meta-analysis of experimental and clinical poliomyelitis compares with amyotrophic lateral sclerosis. Can J Neurol Sci. 1991;18(S3):435–438. [DOI] [PubMed] [Google Scholar]
- 181. Ayers JI, Fromholt SE, O’Neal VM, Diamond JH, Borchelt DR. Prion-like propagation of mutant SOD1 misfolding and motor neuron disease spread along neuroanatomical pathways. Acta Neuropathol. 2016;131(1):103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Boillée S, Yamanaka K, Lobsiger CS, et al. . Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312(5778):1389–1392. [DOI] [PubMed] [Google Scholar]
- 183. Gros-Louis F, Soucy G, Larivière R, Julien JP. Intracerebroventricular infusion of monoclonal antibody or its derived Fab fragment against misfolded forms of SOD1 mutant delays mortality in a mouse model of ALS. J Neurochem. 2010;113(5):1188–1199. [DOI] [PubMed] [Google Scholar]
- 184. Patel P, Kriz J, Gravel M, et al. . Adeno-associated virus-mediated delivery of a recombinant single-chain antibody against misfolded superoxide dismutase for treatment of amyotrophic lateral sclerosis. Mol Ther. 2014;22(3):498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Maier M, Welt T, Wirth F, et al. . A human-derived antibody targets misfolded SOD1 and ameliorates motor symptoms in mouse models of amyotrophic lateral sclerosis. Sci Transl Med. 2018;10(470):eaah3924. [DOI] [PubMed] [Google Scholar]
- 186. Urushitani M, Ezzi SA, Julien JP. Therapeutic effects of immunization with mutant superoxide dismutase in mice models of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2007;104(7):2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Zhao B, Marciniuk K, Gibbs E, Yousefi M, Napper S, Cashman NR. Therapeutic vaccines for amyotrophic lateral sclerosis directed against disease specific epitopes of superoxide dismutase 1. Vaccine. 2019;37(35):4920–4927. [DOI] [PubMed] [Google Scholar]
- 188. Liu AYC, Mathur R, Mei N, Langhammer CG, Babiarz B, Firestein BL. Neuroprotective drug riluzole amplifies the heat shock factor 1 (HSF1)- and glutamate transporter 1 (GLT1)-dependent cytoprotective mechanisms for neuronal survival. J Biol Chem. 2011;286(4):2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Kalmar B, Greensmith L. Cellular chaperones as therapeutic targets in ALS to restore protein homeostasis and improve cellular function. Front Mol Neurosci. 2017;10:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Koch JC, Tatenhorst L, Roser AE, Saal KA, Tönges L, Lingor P. ROCK inhibition in models of neurodegeneration and its potential for clinical translation. Pharmacol Ther. 2018;189:1–21. [DOI] [PubMed] [Google Scholar]
- 191. Kataura T, Tashiro E, Nishikawa S, et al. . A chemical genomics-aggrephagy integrated method studying functional analysis of autophagy inducers. Autophagy. 2021;17(8):1856–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Ralph GS, Radcliffe PA, Day DM, et al. . Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Med. 2005;11(4):429–433. [DOI] [PubMed] [Google Scholar]
- 193. Raoul C, Abbas-Terki T, Bensadoun JC, et al. . Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med. 2005;11(4):423–428. [DOI] [PubMed] [Google Scholar]
- 194. Bravo-Hernandez M, Tadokoro T, Navarro MR, et al. . Spinal subpial delivery of AAV9 enables widespread gene silencing and blocks motoneuron degeneration in ALS. Nat Med. 2020;26(1):118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Miller TM, Pestronk A, David W, et al. . An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12(5):435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Miller T, Cudkowicz M, Shaw PJ, et al. . Phase 1–2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N Engl J Med. 2020;383(2):109–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.