Abstract
Dravet syndrome (DS) is a developmental and epileptic encephalopathy caused by monoallelic loss-of-function variants in the SCN1A gene. SCN1A encodes for the alpha subunit of the voltage-gated type I sodium channel (NaV1.1), the primary voltage-gated sodium channel responsible for generation of action potentials in GABAergic inhibitory interneurons. In these studies, we tested the efficacy of an adeno-associated virus serotype 9 (AAV9) SCN1A gene regulation therapy, AAV9-REGABA-eTFSCN1A, designed to target transgene expression to GABAergic inhibitory neurons and reduce off-target expression within excitatory cells, in the Scn1a+/− mouse model of DS. Biodistribution and preliminary safety were evaluated in nonhuman primates (NHPs). AAV9-REGABA-eTFSCN1A was engineered to upregulate SCN1A expression levels within GABAergic inhibitory interneurons to correct the underlying haploinsufficiency and circuit dysfunction. A single bilateral intracerebroventricular (ICV) injection of AAV9-REGABA-eTFSCN1A in Scn1a+/− postnatal day 1 mice led to increased SCN1A mRNA transcripts, specifically within GABAergic inhibitory interneurons, and NaV1.1 protein levels in the brain. This was associated with a significant decrease in the occurrence of spontaneous and hyperthermia-induced seizures, and prolonged survival for over a year. In NHPs, delivery of AAV9-REGABA-eTFSCN1A by unilateral ICV injection led to widespread vector biodistribution and transgene expression throughout the brain, including key structures involved in epilepsy and cognitive behaviors, such as hippocampus and cortex. AAV9-REGABA-eTFSCN1A was well tolerated, with no adverse events during administration, no detectable changes in clinical observations, no adverse findings in histopathology, and no dorsal root ganglion-related toxicity. Our results support the clinical development of AAV9-REGABA-eTFSCN1A (ETX101) as an effective and targeted disease-modifying approach to SCN1A+ DS.
Keywords: SCN1A, Dravet syndrome, encephalopathy, channelopathy, gene regulation therapy, preclinical models
INTRODUCTION
Dravet syndrome (DS) is a severe, early-onset developmental and epileptic encephalopathy and channelopathy that significantly impacts affected children and their families. DS is manifested by frequent prolonged seizures, status epilepticus events, significant cognitive delays, sleep abnormalities, motor impairment, and profound behavioral difficulties.1–6 Up to 20% of DS patients die before adulthood, frequently due to sudden unexpected death in epilepsy (SUDEP).7,8 DS is a rare disease with an estimated prevalence of 1 in 15,500 live births.9
Over 85% of DS cases are caused by heterozygous loss-of-function variants in a single copy of the SCN1A gene, encoding the alpha subunit of the voltage-gated type I sodium channel (NaV1.1).10 NaV1.1 is predominantly expressed within the axon initial segment of GABAergic inhibitory interneurons, where it generates and propagates action potentials.11 Genetic reduction of NaV1.1 substantially reduces the frequency and amplitude of action potentials generated by GABAergic inhibitory interneurons, thereby impairing their inhibitory function, which is dependent on high-frequency firing.12–14
Multiple lines of evidence have established impaired excitability of GABAergic inhibitory interneurons as the central driver of key DS phenotypes, including seizures, mortality, and cognitive deficits.12,13,15–21 Specific deletion of SCN1A within forebrain GABAergic inhibitory interneurons is sufficient to recapitulate sensitivity to hyperthermia-induced seizures (HTS), premature death, as well as behavioral and cognitive impairments observed in global SCN1A knockouts.13,15 Furthermore, enhancement of GABAergic activity with the allosteric GABA-A receptor agonist, clonazepam, rescues abnormal social behaviors and learning and memory deficits in DS mice.13 Taken together, this evidence suggests that a cell-specific genetic approach aimed at restoring expression levels and function of SCN1A within GABAergic inhibitory interneurons would have the potential to address the underlying cellular and molecular etiology of SCN1A+ DS while minimizing potential off-target effects of NaV1.1 expression.
Gene replacement therapy using recombinant, nonreplicating adeno-associated virus (AAV)-based vectors has demonstrated transformative clinical benefits for the treatment of severe monogenic central nervous system (CNS) disorders.22 However, as the SCN1A gene (cDNA ∼6 kb) exceeds the packaging capacity of AAV (4.7 kb DNA),23 AAV-mediated gene replacement therapy for SCN1A+ DS is not a viable approach. Therefore, we developed AAV9-REGABA-eTFSCN1A (ETX101), an investigational AAV-mediated SCN1A gene regulation therapy candidate, which expresses an engineered transcription factor (eTFSCN1A) designed to upregulate the SCN1A gene from the endogenous genome. A cell-selective regulatory element (REGABA) was incorporated to target transgene expression specifically to GABAergic inhibitory interneurons.
In these studies, we utilize two previously described DS mouse models,11,24,25 and demonstrate that AAV-mediated upregulation of SCN1A with AAV9-REGABA-eTFSCN1A increases SCN1A transcripts specifically within GABAergic inhibitory interneurons, prolongs survival, and reduces spontaneous seizures and HTS. In nonhuman primates (NHPs), a one-time unilateral intracerebroventricular (ICV) injection of AAV9-REGABA-eTFSCN1A led to widespread vector biodistribution and robust transgene expression throughout the brain, including forebrain, midbrain, and particularly the cortex and hippocampus, two key structures involved in epilepsy and cognitive symptoms in DS. ICV-administered doses of AAV9-REGABA-eTFSCN1A at up to 8E13 vector genomes (vg) per animal were well tolerated, with no detectable changes in clinical observations, no adverse findings in histopathology, and no dorsal root ganglion (DRG)-related toxicity. Our results support the clinical development of AAV9-REGABA-eTFSCN1A gene regulation therapy for the treatment of SCN1A+ DS.
EXPERIMENTAL PROCEDURES
Vector design: eTFSCN1A and REGABA
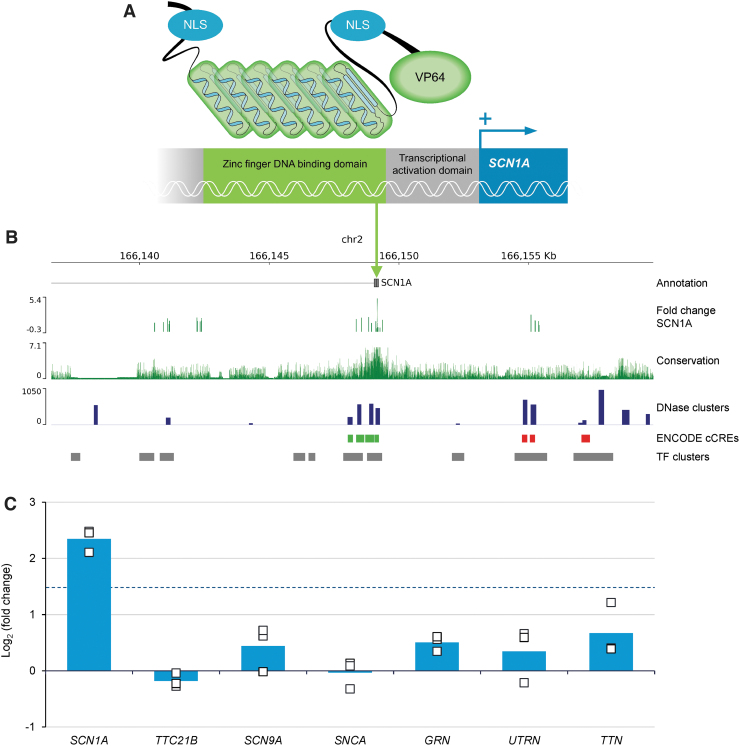
AAV9-REGABA-eTFSCN1A was developed through interrogation of the SCN1A gene and human genetic sequences surrounding the GAD1 gene, expressed specifically in GABAergic interneurons. The selectivity of the backbone of AAV9-REGABA-eTFSCN1A was determined by generating vectors that expressed visualization markers (enhanced green fluorescent protein [EGFP]). The design of eTFSCN1A was based on the Barbas zinc finger design principles,26,27 whereby 6-finger domains targeting defined 18 base-pair (bp) target sequences can confer genome-wide specificity.28 A VP64 transcriptional activation domain, as well as nuclear localization signals at the N-terminal and linker regions were included to facilitate transcriptional upregulation (Fig. 1A).27
Figure 1.
Structure of eTFSCN1A and its targeted activity at a unique and highly conserved 18-bp noncoding DNA sequence for gene-specific upregulation of SCN1A. (A) Schematic of eTFSCN1A structure. eTFSCN1A is composed of a polydactyl zinc finger DNA-binding domain, a short linker sequence, and a C-terminal VP64 transcriptional activation domain.26,28 Each of the six zinc finger domains interacts with a specific triplet nucleotide, conferring specific interaction with an 18-bp target sequence. NLS domains derived from simian vacuolating virus (SV40) and nucleoplasmin are included at the N-terminal and linker domains. (B) Genomic features of eTFSCN1A target site. Human genome track at the SCN1A locus. Tracks indicate human SCN1A gene annotation, chromosome 2 coordinates (hg38 genome assembly). FC SCN1A track shows target vector candidate screening data in HEK293cells. Mean Log2-FCs in SCN1A expression following transient transfection of eTFSCN1A candidates targeted against different genomic targets are plotted by target coordinates. The eTFSCN1A 18-bp target sequence, which induced the strongest upregulation of SCN1A within the screening window, is indicated by the green arrow. Sequence conservation track is indicated by 100-species base-wise conservation score (PhyloP).29 Regulatory element feature tracks are indicated (UCSC genome browser, ENCODE consortia31,32). DNase cluster track indicates DNase I hypersensitivity cluster score; ENCODE cCREs track marks candidate cis-regulatory element regions (green: PLS, red: dELS); TF clusters track marks transcription factor binding peak clusters. (C) eTFSCN1A upregulates SCN1A in a gene-specific manner in vitro. Change in endogenous gene expression in HEK293 cells is reported in the eTFSCN1A transfected condition relative to control condition for each replicate. Bars represent mean Log2-FC for n = 3 biological replicates (n = 2 for PAPBC1), dots indicate individual replicate measurements. Dotted line indicates Log2FC = 1.5. bp, base-pair; cCREs, candidate cis-regulatory elements; dELS, distal enhancer-like signature; FC, fold change; NLS, nuclear localization signal; PLS, promoter-like signature; TF, transcription factor.
We designed several candidate target sequences based on conservation (PhyloP)29,30 and genomic regulatory markers surrounding the distal SCN1A promoter (NM_001202435), including species sequence conservation (Fig. 1B),29 accessibility data (DNase hypersensitivity), and transcription factor binding site enrichment (chromatin immunoprecipitation sequencing [ChIP-seq] clusters) (Fig. 1B).31,32 Based on these features, candidate sequences were screened empirically for efficacy in upregulating SCN1A expression (see Fig. 1B, “fold change SCN1A” track). SCN1A upregulation and genetic selectivity were evaluated by reverse transcription–quantitative polymerase chain reaction (RT-qPCR) in vitro through transient transfection studies.
In brief, adherent HEK293 cells (293AAV; Cell Biolabs) were cultured and transfected (FuGENE HD; Promega) with either an eTFSCN1A-expressing plasmid construct or an EGFP-expressing control 24 h postplating. RNA was isolated (RNeasy Mini kit; Qiagen) 48 h post-transfection, DNase treated, and reverse transcribed using OligoDT primers (Superscript IV; Invitrogen; Supplementary Table S1). cDNA samples were analyzed by qPCR and analysis of relative expression was performed using the ΔΔCt method. Referenced ENCODE datasets were accessed from the ENCODE portal (https://www.encodeproject.org/)33 or UCSD Genome Browser34 using the Table Browser tool (https://www.genome.ucsc.edu).35 Accession numbers for underlying data are EH38E1393970, GSE29692, GSE32970, and GSE26386.
AAV vector production
Replication-incompetent, recombinant AAV9 particles were produced in HEK293 cells via transient triple cotransfection: a transgene-containing plasmid, a packaging plasmid for Rep and Cap genes, and a plasmid containing adenoviral helper genes. Following transfection, cells were harvested, lysed to release viral particles, and treated with Benzonase. For mouse studies, AAV9 viral vector produced in the adherent HEK293 system was purified by iodixanol gradient followed by buffer exchange (video electroencephalography [EEG] studies), or generated by Vector Biolabs (Malvern, PA) using an adherent HEK293 system and purification by CsCl centrifugation (all other mouse studies). Vector was formulated in phosphate-buffered saline (PBS) and stored at −80°C. AAV vector titer for each production was originally measured by qPCR method and subsequently retentates were retitered using a qualified digital droplet polymerase chain reaction (ddPCR) assay (reported values).
For the primate study, AAV9 vector was produced at larger scale for both adherent and suspension HEK293 production platforms. Viral vector produced in the adherent platform was purified by iodixanol gradient and followed by ion exchange chromatography. AAV vector produced in the suspension platform was purified using chromatography-based methods. Vector was formulated in PBS with 0.001% pluronic and titer was determined using a qualified ddPCR method.
Animals
Mouse studies were conducted at Encoded Therapeutics, Inc. and performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Heterozygous Scn1a+/− breeders were generously provided by Dr. Jennifer Kearney at Northwestern University (Chicago, IL). F1 C57:129S hybrid Scn1a+/− mice were crossed to 129S6/SvEvTac Scn1a+/− mice to generate F1 offspring. Scn1aR1407X/+ mice (R1407X) were obtained from Riken University. Heterozygous animals were generated at Charles River Laboratories by fertilizing ova from C57BL/6N mice with sperm from Scn1aR1407X/+ mice followed by transplantation of fertilized embryo into CD-1 surrogates. Genotyping was performed by automated RT-PCR at TransnetYX, Inc. [see Supplementary Materials for primer sequences].
All mice were maintained on a 12:12-h light:dark cycle and had ad libitum access to food and water throughout the experiments. Mice were euthanized using CO2 followed by cervical dislocation. For molecular analysis, cortical brain tissues were collected, flash frozen, and stored at −80°C. For immunohistochemistry (IHC), brains were collected and preserved in 10% neutral buffered formalin for 24 h then switched to 70% ethanol and stored at 4–8°C until histopathologic processing.
NHP studies were conducted using AAV9-seronegative juvenile cynomolgus macaques (Macaca fascicularis) at Charles River (Mattawan, MI), and were performed according to local health authority guidelines. Sera were screened for preexisting AAV9 neutralizing antibodies and animals with titers <1:5 were selected. Animals were group-housed in acclimatized holding rooms with water ad libitum. Animals were given meals of balanced composition and additional food was offered to provide environmental enrichment. At the end of the study, euthanasia procedures were conducted in accordance with CR-M protocols and standard operating procedures, and brain tissues were collected.
The whole brain from rostral to caudal was cut into coronal 5-mm thick slabs, and interleaving slabs were either fixed for histology in 4% paraformaldehyde for 24–48 h then transferred and stored in 70% ethanol until processing, or 8-mm punches were taken across nine brain regions in RNAlater™ or flash-frozen for DNA/RNA/protein analysis. The spinal cord was removed, and the cervical C5, thoracic T4 and T11, and lumbar level L3 of spinal cord and associated DRGs were collected. Tissue samples were shipped under appropriate conditions from Charles River to Encoded Therapeutics, Inc.
ICV injections
At postnatal day (PND)1, mice were anesthetized on ice for ∼3 min. Three microliters of viral suspension or vehicle control were administered into each hemisphere using an insulin syringe. Lambda and bregma skull sutures were referenced to determine injection site ∼1 mm lateral, 1 mm rostral, respectively. The needle was lowered to an approximate depth of 1.5 mm ventral for injection. After injection, mice were placed in a warming pad and returned to the mother in the cage.
For unilateral ICV administration in juvenile cynomolgus macaques, presurgical MRI was performed to establish brain ventricle coordinates within the left or right hemisphere. A small burr hole was made through the frontal bone and a spinal needle (22 gauge) was introduced. The correct needle placement was verified by injecting up to 0.05 mL of contrast agent (Omnipaque™) under real-time fluoroscopy examination. Then, a controlled volume of vehicle or test article of 2.0 mL was injected at a rate of 0.1 mL/min.
Immunohistochemistry
IHC was performed on serial section slides in two independent multiplexed batches. The first batch followed the sequence mouse anticalcium/calmodulin-dependent protein kinase II alpha (MA1-048; Invitrogen), rabbit antisomatostatin (T-4102; Peninsula Laboratories), rabbit anti-GFP (ab290; Abcam), and mouse anti-PV (PV235; Swant). The second batch followed the sequence, mouse anti-glutamate decarboxylase (GAD67, ab26116; Abcam), mouse anti-fox-3 (NeuN, MAB 377; Millipore), and rabbit anti-GFP. After primary and secondary antibody incubation, slides were stained with DAPI. Whole slide images were captured at 20 × using an Akoya Biosciences Polaris instrument. Image exposures for each fluorochrome were constant for all slides. Detailed image analysis is described in the Supplementary Materials.
NHP histopathology
At necropsy, whole brain and spinal cord with associated DRGs were collected and processed for formalin-fixed paraffin embedding. Tissues were sectioned at 5 μm and stained with hematoxylin and eosin on 16–20 slides per animal. Macroscopic and microscopic morphologic observations and evaluations were obtained from three independent board-certified veterinary pathologists.
Quantification of vector copy number by ddPCR
DNA was isolated from tissue using AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer's instructions. TaqMan primers were directed against regions of transgene (eTFSCN1A) sequence (eTFSCN1A forward primer 5′-GAATGTGGGAAATCATTCAGTCGC-3′, eTFSCN1A reverse primer 5′-GCAAGTTATCCTCTCGTGAGAAGG-3′, eTFSCN1A probe 5′-GCGACAACCTGGTGAGACATCAACGCACC-3′) and mouse Tfrc (MmTfrc) gene or monkey Albumin (MfAlb) gene as an internal control for normalizing genomic DNA content for mouse tissues. Droplets were generated and templates were amplified using automated droplet generator and thermo cycler (Bio-Rad). After the PCR step, the plate was read by QX2000 Droplet Reader to quantify vector copy number (VCN) levels in tissues. VCN per microgram of DNA was converted to copy number per diploid genome.
Quantification of RNA expression by RT-ddPCR
RNA was isolated using AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer's instructions. TaqMan primers were directed against regions of transgene (eTFSCN1A) sequence and mouse GUSB (MmGUSB) gene or monkey ARFGAP2 (MfARFGAP2) gene as an internal control for normalizing genomic DNA content for mouse tissues (Supplementary Table S2). The no-RT reaction served as negative control for each sample. Droplets were generated and templates were amplified using automated droplet generator and thermo cycler (Bio-Rad). Gene expression copies per microgram RNA was calculated based on the output of QX2000 Droplet Reader (copies per microliter ddPCR reaction) multiplied by the total reaction volume and divided by the RNA input.
Single-nucleus RNA sequencing
Brain tissue was homogenized as previously described.36 The nuclei preparations were stained with DAPI, and 100,000 DAPI-positive events were sorted using a BD FACS Aria II cell sorter (UCSF Gladstone Flow Cytometry Core Facility). Single-nucleus RNAseq (snRNAseq) was performed using the Chromium Single Cell 3′ v3 kit (10 × Genomics), according to manufacturer's instructions. An enrichment PCR was utilized to specifically amplify eTF cDNA, thereby improving the sensitivity of detection of eTFSCN1A within the cDNA pool. The resulting cDNA libraries and enrichment PCR products underwent next-generation sequencing using a NextSeq 500 (Illumina). CellRanger (v5.0) was used to align reads and generate UMI (Unique Molecular Identifier) counts for each gene.
These counts were used for dimensionality reduction (Seurat 4.0 [https://satijalab.org/seurat/] and ZINB-WaVE37) and clustering using the Louvain algorithm (https://pypi.org/project/louvain/, version 0.6.1) to define neuronal subpopulations by their genetic signatures. Cells with ≥1 UMI count for the eTFSCN1A sequence were considered “infected,” whereas cells with no UMI count for the eTFSCN1A sequence were considered “uninfected.”
Quantification of membrane-associated NaV1.1 protein by Meso Scale Discovery electrochemiluminescence-based sandwich immunoassay
Mouse forebrain and midbrain tissues were homogenized using a Qiagen TissueLyser II, according to manufacturer's instructions, and plasma membranes isolated by a series of centrifugation steps. Total protein concentration was determined using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.) and total protein 1 mg/mL was loaded onto a Meso Scale Discovery (MSD) plate for measurement via an electrochemiluminescence-based sandwich immunoassay. Membrane-associated NaV1.1 was detected with a rabbit anti-NaV1.1 pAb against the C-terminus of mouse NaV1.1 as capture antibody and a mouse mAb against the same molecular region as detection. For NaV1.1 quantification, a recombinant mouse NaV1.1 C-terminus protein fragment was used as reference standard. The level of membrane-associated NaV1.1 in the sample was normalized by total protein loaded in each well and reported as percentage of that in wild-type (WT) mice.
HTS assay
Mice were placed in a TCAT 2DF (Physitemp) box for 15 min before assay initiation. A flexible rectal probe (RET-4; Physitemp) was inserted in the mice and secured with medical tape. Mice were placed in a 4-L beaker under a heat lamp and body temperature was increased by ∼0.5°C every 2 min until the onset of the first tonic-clonic seizure, defined by uncontrolled movements accompanied by loss of posture, or until 43.5°C was reached (as described previously38). Seizure events, defined as tonic-clonic seizures with loss of posture, were recorded by an experimenter blinded to treatment group and genotype. The experiment was terminated when the first tonic-clonic seizure occurred or when the mouse reached 43.5°C for 2 min.
Video-EEG seizure analysis
Scn1a+/− and WT mice were implanted with three electrodes on PND21: one each in the frontal cortex, parietal cortex, and hippocampal regions. Video-EEG recordings were performed for nine consecutive days using the Pinnacle Technology 8206 data conditioning and acquisition system (Pinnacle Technology, Inc.). Electrographic seizure events were identified with automated spike detection. Spike trains consisting of a clustering of ≥10 spikes with amplitude >3 × basal EEG background activity were considered electrographic seizure events and were manually confirmed by accompanying video recordings.
Behavioral scoring for all electrographic seizure events was evaluated based on the following criteria: Nonbehavioral seizure: no observed change in behavior; Mild seizure: mild movements with or without head twitching; Convulsive seizure: tonic-clonic seizure with or without loss of posture. Electrographic seizure events that demonstrated spiking activity across all three EEG channels were considered generalized seizure events. All assessments were performed by experimentalists blinded to treatment.
Statistical analyses
For in vitro RNA expression analysis, RT-qPCR and RT-ddPCR analyses were performed in triplicate biological samples, and one-sample z-tests were used to calculate if the mean Log2-fold change (Log2FC) was significantly different from zero. Differences in expression for each gene were considered significant if they passed differential expression thresholds of mean Log2FC ≥1.5 or mean Log2FC ≤ −1.5. Log-rank test was performed on HTS assay and survival data. Unpaired t-tests were used to compare EEG seizure frequency between groups and for IHC colocalization analysis. Chi-squared test was used to compare EEG seizure severity. p-Values are denoted as: *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant, p ≥ 0.05.
RESULTS
eTFSCN1A targets a unique and highly conserved 18-bp noncoding DNA sequence and upregulates SCN1A in a gene-specific manner
To achieve selective upregulation of SCN1A expression in GABAergic inhibitory interneurons, a GABAergic cell-selective regulatory element (REGABA) and a transgene expressing an engineered transcription factor targeted to the SCN1A gene (eTFSCN1A) were incorporated into AAV9 vector for transgene delivery, referred to as AAV9-REGABA-eTFSCN1A. eTFSCN1A comprises a synthetic DNA-binding zinc-finger protein fused to the transcriptional activator, VP64 (Fig. 1A),26,28 and was designed to selectively bind a conserved regulatory region upstream of the SCN1A transcription start site (NM_001202435) (see Experimental Procedures section). Within this promoter region, candidate target sequences were screened in vitro by transient transfection, and an 18-bp sequence element was identified that produced robust upregulation of SCN1A exceeding threshold criteria (Log2FC >1.5, p < 0.05) (Fig. 1B).
This target binding site corresponds to a peak in multispecies sequence conservation, across human, mouse, and NHP genomes (Fig. 1B), allowing the use of preclinical models to assess the safety and efficacy of transcriptional activation of SCN1A.
Next, we evaluated specificity of the eTFSCN1A transgene in vitro by transient transfection. HEK293 cells were transfected with plasmids expressing either the eTFSCN1A transgene or EGFP under the control of a strong constitutive promoter element. While transfection with eTFSCN1A resulted in upregulation of SCN1A transcript, no significant upregulation was observed in a panel of six off-target genes, including the two nearest neighboring genes to SCN1A (TTC21B, SCN9A) and four additional control genes (SNCA, GRN, UTRN, TTN) (Fig. 1C). These results indicate that upregulation by eTFSCN1A is selectively targeted to the SCN1A gene, and not a general feature of transcriptional activator protein overexpression.
To evaluate potential genomic off-target binding sites, BLASTn (NCBI) analysis and target annotation were performed against human genome on the 18-bp target sequence. Importantly, this sequence is unique within the human genome, with few predicted off-target binding sites. None of the identified homologous binding sites overlapped with identified promoter or enhancer regions (Supplementary Table S3).
REGABA selectively targets GABAergic interneurons in vivo
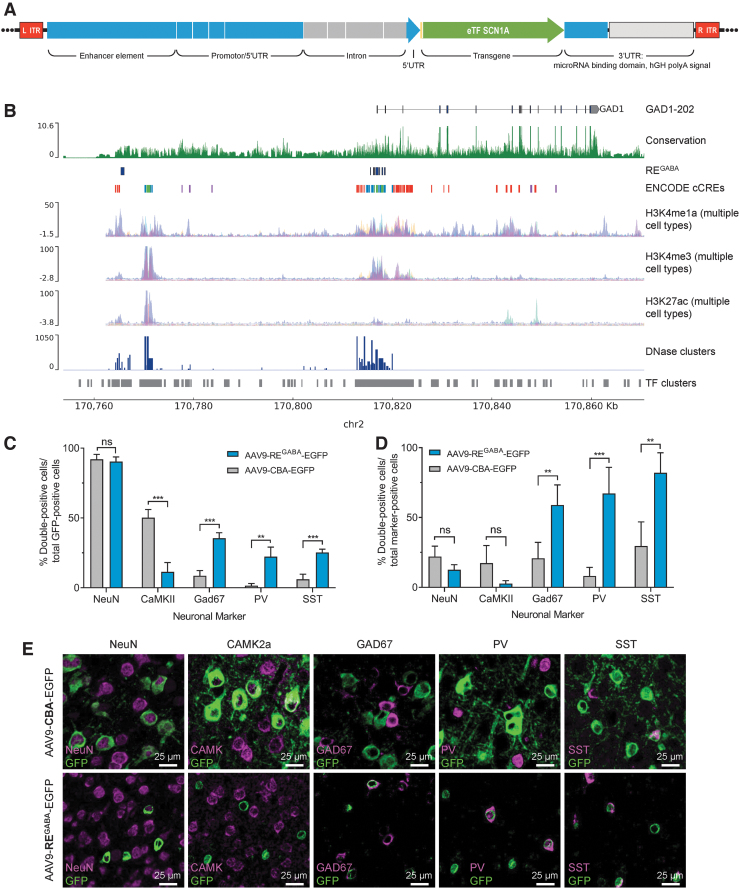
To selectively express the AAV9-REGABA-eTFSCN1A transgene in GABAergic inhibitory neurons, we developed a novel regulatory element cassette (REGABA), designed from human-derived sequence features to collectively confer specificity to GABAergic interneurons. REGABA comprised enhancer, promoter, 5′UTR (untranslated region), and intronic sequences derived from the GABA-selective hGAD1 gene locus, and a 3′UTR sequence designed to further reduce residual expression in excitatory neurons (Fig. 2A).36,39,40 These elements were designed to capture regulatory sequence features that confer endogenous cell selectivity in compact form, enabling cell-selective expression compatible with the 4.7 kb AAV packaging capacity.
Figure 2.
The REGABA regulatory element in AAV9-REGABA-eTFSCN1A selectively targets GABAergic interneurons in vivo. (A) REGABA construct design. REGABA comprised upstream and 5′UTR sequence components, and a downstream 3′UTR sequence. The upstream regulatory sequence is derived from human genomic sequence elements: an enhancer element located ∼50 kb upstream of the GAD1 gene locus, and proximal promoter and 5′UTR element sequence derived from the human GAD1 gene locus. The promoter is composed of genomic sequence segments in the proximal promoter and 5′UTR of the hGAD1 gene. To enhance gene expression40 and to capture additional regulatory features derived from intronic elements, this promoter also incorporates a truncated intron derived from the first intron of the GAD1 gene, which includes sequences flanking the splice donor and acceptor sites, and internal intronic sequences that overlap high-conservation predicted regulatory sequences. The 3′UTR element includes a collection of eight cognate target motifs derived from excitatory neuron-enriched miRNAs miR128 and mir221,36,39 and a hGH-pA. (B) Genomic features of REGABA source sequence. Human genome track at the human GAD1 locus. Tracks indicate GAD1 gene annotation, chromosome 2 coordinates (hg38 genome assembly). Sequence conservation (PhyloP29,30), and ENCODE regulatory element feature tracks are indicated, as well as additional composite epigenetic marker tracks for histone markers H3K4me1, H3K4me3, and H3K27ac, which can signal active enhancer and promoter regions (UCSC genome browser, ENCODE consortia). (C–E) Expression of EGFP in vivo 27 days post-ICV injection of AAV9-EGFP vectors driven by CBA or REGABA promoters in PND1 mice. Animals (n = 4/group) were administered 2.0E10 vg per animal of AAV-CBA-EGFP or AAV-REGABA-EGFP on PND1 by bilateral ICV injection. On PND28, brain sections were analyzed by IHC for the presence of GFP and neuron-specific markers. Quantitation of colocalization between neuron-specific markersa and EGFP: (C) among GFP-positive cells and (D) compared to the total number of neuron-marker positive cells; **p < 0.01; ***p < 0.001; ns, p ≥ 0.05 (unpaired t-tests, two-stage step-up FDR = 1%; data shown are means and error bars represent SD). (E) Representative images demonstrating colocalization of EGFP driven by each vector with each of the neuronal markers evaluated. aNeuron-specific markers: NeuN (neurons); CAMK2a (glutamatergic neurons); GAD67 (GABAergic interneurons); PV (parvalbumin-positive interneurons); SST (somatostatin-positive interneurons). AAV, adeno-associated virus; CBA, chicken β-actin promoter; chr2, chromosome 2; EGFP, enhanced green fluorescent protein; eTF, engineered transcription factor; hGH-pA, human growth hormone-derived poly-adenylation signal; IHC, immunohistochemistry; ICV, intracerebroventricular; ITR, inverted terminal repeat; ns, not significant; PND, postnatal day; REGABA, GABAergic cell-selective regulatory element; SD, standard deviation; UTR, untranslated region; vg, vector genomes.
Sequence selection was informed by genomic sequence conservation (Fig. 2B), and genomic regulatory element markers such as DNase I hypersensitivity peaks, epigenetic histone features, and transcription factor ChIP-seq clusters (Fig. 2B). The REGABA enhancer element is derived from a novel enhancer element located 50 kb upstream of the GAD1 gene (Fig. 2A, B). The 3′UTR element comprised cognate target motifs for excitatory neuron-enriched miRNAs: miR128 and mir221,36,39 upstream of a human growth hormone poly-adenylation signal (Fig. 2A).
To evaluate the cell-selectivity of REGABA promoter in vivo, PND1 mice (n = 4/group) were dosed via bilateral ICV injection with 2.0E10 vg per animal of AAV9 with a chicken β-actin (CBA) promoter driving expression of EGFP [AAV9-CBA-EGFP], or EGFP expression driven by REGABA [AAV9-REGABA-EGFP].
Quantitation of GFP colocalization with a subset of neuron-specific markers, including NeuN (neurons), CAMK2a (glutamatergic neurons), GAD67 (GABAergic interneurons), PV (parvalbumin-positive interneurons), and SST (somatostatin-positive interneurons), was then evaluated by IHC 27 days postinjection. AAV9-CBA-EGFP and AAV9-REGABA-EGFP exhibited similar selectivity for expression in neurons (NeuN, ∼90% of expressing cells in both cases). However, AAV9-CBA-EGFP expression exhibited significantly higher selectivity for excitatory neurons and AAV9-REGABA-EGFP showed higher selectivity of expression in GABAergic interneurons, as indicated by colocalization of EGFP and CAMK2a or GAD67 for excitatory neurons and GABAergic interneurons, respectively. Consistent with selectivity of AAV9-REGABA-EGFP for GABAergic interneurons, REGABA-driven EGFP had significantly higher colocalization with the GABAergic subtype markers PV and SST, compared with AAV9-CBA-EGFP (Fig. 2C).
We next evaluated the percentage of neuron marker-positive cells coexpressing EGFP. At a dose level of 2.0E10 vg per animal, AAV9-REGABA-EGFP drove EGFP expression in 58.9% ± 14.4% GAD67-positive cells, 67.2% ± 18.7% PV-positive cells, and 82.0% ± 14.3% SST-positive cells, whereas AAV9-CBA-EGFP exhibited significantly lower expression coverage (Fig. 2D). Expression of GFP under the control of REGABA was limited to cells that coexpressed the GABAergic neuronal markers—GAD67, PV, and SST—while expression of GFP under the control of the constitutive CBA promoter was detected throughout the brain in multiple cell types (Fig. 2E). Minimal staining was observed in microglia and astrocytes (data not shown).
Taken together, these data demonstrate that the REGABA promoter element can significantly enhance selectivity of AAV9-driven transgene expression in GABAergic inhibitory interneurons via both a reduction in off-target expression within excitatory neurons and enhanced expression within GABAergic interneurons, compared with CBA.
AAV9-REGABA-eTFSCN1A upregulates SCN1A in a cell-type selective manner in vivo with a corresponding increase in membrane-associated NaV1.1 protein
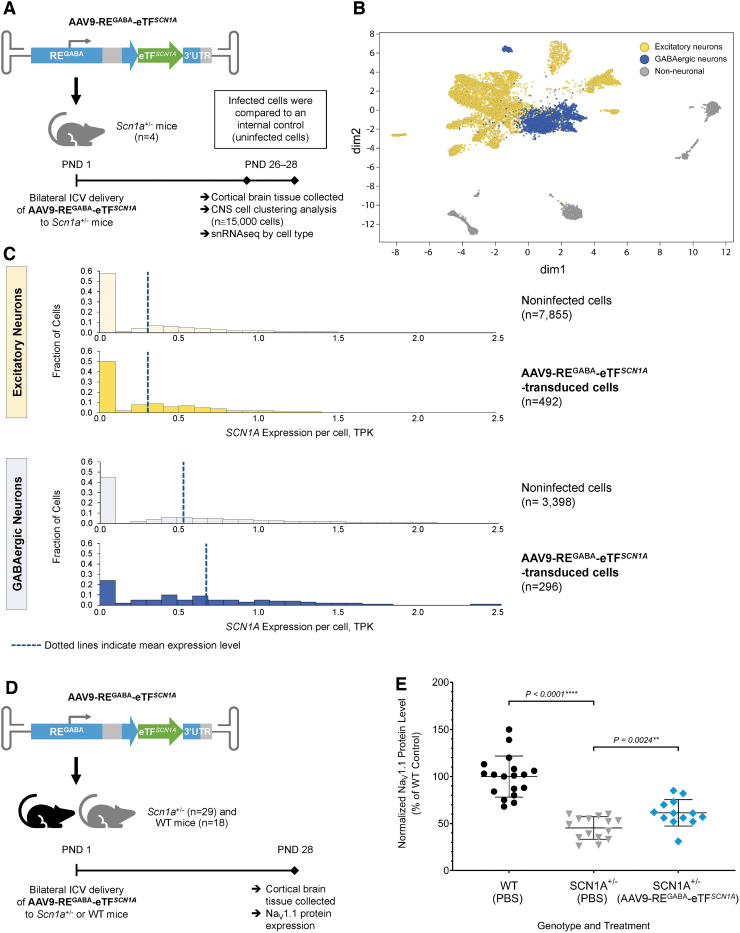
To determine SCN1A upregulation within GABAergic inhibitory interneurons, Scn1a+/− mice (n = 4) received AAV9-REGABA-eTFSCN1A (1.7E10 vg/animal) via bilateral ICV injection on PND1 (Fig. 3A). On PND28, cortical brain tissue was collected and snRNAseq was performed from isolated cortical nuclei. Clustering analysis was performed to classify over 15,000 cells into excitatory/inhibitory neurons and non-neuronal cell types based on transcriptome profiles (Fig. 3B). Cell types were annotated based on canonical markers, as shown in the heat map (Supplementary Fig. S1).
Figure 3.
AAV9-REGABA-eTFSCN1A upregulates SCN1A within GABAergic inhibitory interneurons in vivo. (A) Experimental design to evaluate SCN1A expression in excitatory and GABAergic inhibitory neurons. Four Scn1a+/− mice were treated with 1.7E10 vg per animal of AAV9-REGABA-eTFSCN1A on PND1 via bilateral ICV injection. On PND28, cortical brain tissue was collected, and clustering analysis was performed to classify over 15,000 cells into excitatory and inhibitory cell types based on transcriptome profiles. (B) Clustering of CNS cell types in cortical brain tissue. Gene expression visualization using tSNE to compute a low-dimensional representation of snRNAseq data; dim2 and dim1 are the outputs of tSNE, the 2D representation of the gene expression profiles of each cell. Yellow dots denote excitatory neurons; blue dots denote GABAergic inhibitory neurons; gray dots denote non-neuronal cells. (C) SCN1A mRNA transcript levels in excitatory and GABAergic inhibitory neurons. AAV9-REGABA-eTFSCN1A upregulates SCN1A mRNA specifically in GABAergic inhibitory neurons, with no upregulation in excitatory neurons. Dotted lines indicate mean SCN1A TPK per cell. (D) Experimental design to determine NaV1.1 protein expression. One-day-old (PND1) Scn1a+/− mice were administered PBS (n = 16) or 5.1E10 vg per animal AAV9-REGABA-eTFSCN1A (n = 13) and 18 WT littermates were administered PBS via a bilateral ICV injection. Animals were sacrificed on PND28 and brain tissue was evaluated by MSD electrochemiluminescence-based sandwich immunoassay. (E) NaV1.1 protein expression. Membrane-associated NaV1.1 protein levels from PBS-treated WT (6 males/12 females; black circle) or Scn1a+/− mice (7 males/9 females; gray triangle) and AAV9-REGABA-eTFSCN1A-treated Scn1a+/− mice (9 males/4 females; blue diamond). Mean levels ± SD are indicated; ****p < 0.0001 and **p = 0.0024, Mann–Whitney test. CNS, central nervous system; dim, dimension; MSD, Meso Scale Discovery; NaV1.1, voltage-gated type I sodium channel; PBS, phosphate-buffered saline; snRNAseq, single-nucleus RNA sequencing analysis; TPK, transcripts per 1000 total transcripts; tSNE, t-distributed stochastic neighbor embedding; WT, wild-type.
SCN1A expression was detected in both excitatory and inhibitory cells (Fig. 3C, noninfected cells, pale yellow and pale blue histograms), similar to what has been reported by other groups.41 Overall, SCN1A transcript levels in cortical brain were ∼2-fold higher in GABAergic cells (dark blue histogram; average transcripts per 1000 total transcripts [TPK] 0.55) than in excitatory cells (dark yellow histogram; average TPK 0.31). GABAergic neurons containing eTFSCN1A transcript expressed ∼30% more SCN1A than GABAergic cells that lacked eTFSCN1A (Fig. 3C; blue panel, average TPK 0.69 vs. 0.54; p < 0.001; one-sided rank-sum test). Importantly, treatment with AAV9-REGABA-eTFSCN1A did not elevate SCN1A expression in excitatory cells (average TPK 0.31) over basal levels (yellow panel, p > 0.01) (Fig. 3C). These results indicate that AAV9-REGABA-eTFSCN1A results in selective upregulation of SCN1A in GABAergic inhibitory interneurons.
Upregulation of SCN1A led to a corresponding increase in NaV1.1 protein levels in brain tissue. In a separate experiment, Scn1a+/− mice (n = 13) were administered AAV9-REGABA-eTFSCN1A (5.1E10 vg/animal) and as controls, Scn1a+/− mice (n = 16) and WT littermates (n = 18) received PBS vehicle via ICV injection, and membrane NaV1.1 protein expression was measured by MSD electrochemiluminescence-based sandwich immunoassay (Fig. 3D). As expected, NaV1.1 protein expression in vehicle-treated Scn1a+/− mice was significantly lower when compared with WT mice (45.7% ± 11.7% vs. 99.9% ± 21.8%, respectively; p < 0.0001). AAV9-REGABA-eTFSCN1A resulted in the upregulation of membrane-associated NaV1.1 protein in the CNS tissue of Scn1a+/− mice by ∼30% compared with vehicle-treated Scn1a+/− animals (61.4% ± 14.1% vs. 45.7% ± 11.7%, respectively; p = 0.0024) (Fig. 3E).
AAV9-REGABA-eTFSCN1A reduces the frequency, duration, and severity of spontaneous seizures and rescues HTS sensitivity in Dravet mice
We next tested the potential for AAV9-REGABA-eTFSCN1A treatment to reduce seizures in the DS mouse model. The optimal interventional timepoint for dosing was determined based on the kinetics of AAV9 transgene expression which requires ∼30 days to reach stable expression levels (Supplementary Fig. S2). Considering that the onset of seizures and mortality in Scn1a+/− mice occurs between the third and fourth weeks of life,17,24,42 PND1 was determined to be the optimal time for dosing as the stabilization of transgene expression at approximately PND30 occurs following onset of seizure symptoms but before high rates of animal attrition due to SUDEP.
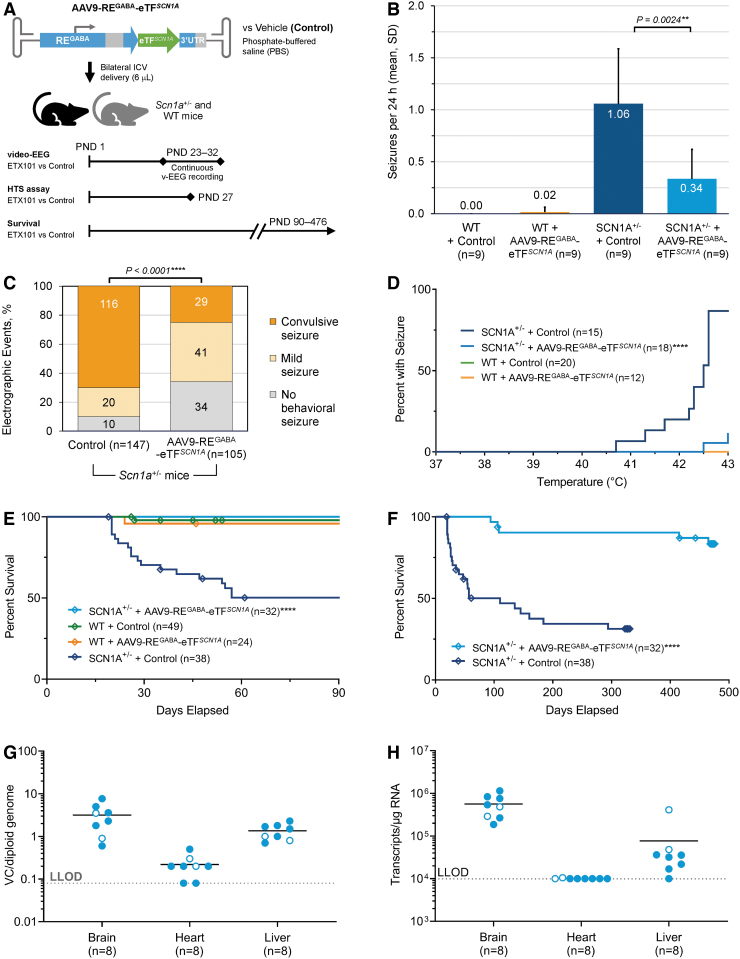
To evaluate spontaneous seizures, we dosed WT (n = 10 vehicle, n = 7 AAV9-REGABA-eTFSCN1A) and Scn1a+/− (n = 17 vehicle, n = 18 AAV9-REGABA-eTFSCN1A) mice at PND1 via bilateral ICV injection (1.2–5.4E11 vg/animal; 3 μL/hemisphere). Animals were monitored continuously by video-EEG for occurrence of electrographic seizures from PND21 for 9 days. For HTS, we dosed WT (n = 20 vehicle, n = 12 AAV9-REGABA-eTFSCN1A) and Scn1a+/− (n = 15 vehicle, n = 18 AAV9-REGABA-eTFSCN1A) mice at PND1 with a total 3.7E10 vg per animal. HTS assays were performed on PND 27 (±1 day) (Fig. 4A).
Figure 4.
AAV9-REGABA-eTFSCN1A reduces frequency and severity of spontaneous seizures, protects against febrile seizures, and demonstrates durable survival efficacy and persistent activity for up to 470 days postdosing in Scn1a+/− mice. (A) Study design. AAV9-REGABA-eTFSCN1A or vehicle alone (PBS) were administered by bilateral ICV injection (3 μL/hemisphere) to heterozygous Scn1a+/− or WT littermates at PND1. Separate studies assessed electrographic seizure monitoring of spontaneous seizures (following doses of 1.2–5.4E11 vg/animala vs. vehicle); susceptibility to HTS (3.7E10 vg/animala vs. vehicle); and survival and long-term persistence of AAV9-REGABA-eTFSCN1A (3.7E10 vg/animala vs. vehicle). At 470 days, the animals were sacrificed, and brain, heart, and liver tissues were evaluated for AAV9-REGABA-eTFSCN1A VCN by ddPCR and eTF transgene mRNA transcript levels by RT-ddPCR. (B) EEG seizure frequency. Mice treated with AAV9-REGABA-eTFSCN1A experienced a 68% reduction in the mean daily generalized seizure frequency per animal compared with Scn1a+/− controls (**p = 0.0024, unpaired t-test). No changes in seizure frequency were detected in WT mice treated with AAV9-REGABA-eTFSCN1A. (C) EEG seizure severity. Video characterization of all recorded electrographic seizure events revealed that treatment of Scn1a+/− mice with AAV9-REGABA-eTFSCN1A significantly reduced convulsive (tonic-clonic) seizures compared with Scn1a+/− mice treated with vehicle (****p < 0.0001; chi-square test). Most events detected in AAV9-REGABA-eTFSCN1A-treated Scn1a+/– mice were mild (characterized by mild movements and/or head twitching, but without convulsions) or nonbehavioral seizures. (D) HTS assay. Percentage of AAV9-REGABA-eTFSCN1A PND1-treated Scn1a+/− mice experiencing seizures at a given temperature at PND27. WT groups overlap along the zero line (green and orange lines; only the orange trace is shown). p-Values calculated using a Log-rank test, ****p < 0.0001. (E) Ninety-day survival. The 90-day survival rate in AAV9-REGABA-eTFSCN1A-treated Scn1a+/– mice was 100% compared with 50% in control-treated Scn1a+/− mice (****p < 0.0001, Log-rank test). No difference in survival was observed between WT mice ± AAV9-REGABA-eTFSCN1A and Scn1a+/− mice + AAV9-REGABA-eTFSCN1A. (F) Long-term Survival. Long-term follow-up showed that survival benefit of AAV9-REGABA-eTFSCN1A was sustained over ∼470 days after dosing (****p < 0.0001, Log-rank test). Diamond indicates humane endpoint or end of study euthanasia at approximately day 330 for vehicle and day 470 for AAV9-REGABA-eTFSCN1A-treated animals. (G) VCN biodistribution in Scn1a+/− mice administered AAV9-REGABA-eTFSCN1A. At 470 days, the animals were sacrificed, and brain, heart, and liver tissues were evaluated for AAV9-REGABA-eTFSCN1A VCN by ddPCR and eTFSCN1A transgene mRNA transcripts levels by RT-ddPCR. (H) eTFSCN1A mRNA expression levels in Scn1a+/− mice administered AAV9-REGABA-eTFSCN1A. eTFSCN1A mRNA transcripts per microgram of RNA analyzed. Seven of eight animals had no measurable eTFSCN1A mRNA transcripts in the heart. Open circle indicates female gender. Dotted line indicates the limit of detection in both assays. aDoses in the EEG study were determined by a qPCR titering method while the HTS and survival studies were titered via ddPCR; thus the doses used across studies are not directly comparable. EEG, electroencephalography; HTS, hyperthermia-induced seizure; LLOD, lower limit of detection; qPCR, quantitative polymerase chain reaction; RT-ddPCR, reverse transcription digital droplet polymerase chain reaction; VCN, vector copy number.
Treatment of Scn1a+/− mice with AAV9-REGABA-eTFSCN1A at PND1 significantly reduced the frequency and severity of spontaneous seizures and increased the number of seizure-free mice over a 9-day assessment period. AAV9-REGABA-eTFSCN1A treatment reduced average number of daily seizures by 68% (p = 0.0024, unpaired t-test), whereas no differences in seizure frequency were detected in WT mice treated with vehicle or AAV9-REGABA-eTFSCN1A (Fig. 4B). In addition, blinded video-EEG assessment revealed that electrographic seizure events experienced by Scn1a+/− mice treated with AAV9-REGABA-eTFSCN1A were less likely to manifest as convulsive seizure events, compared with Scn1a+/− mice treated with vehicle (p < 0.0001, chi-square test, Fig. 4C). Finally, the percentage of seizure-free mice increased from 20% in vehicle-treated mice to 67% in AAV9-REGABA-eTFSCN1A-treated mice.
In the HTS assay (Fig. 4D), vehicle-treated Scn1a+/− mice exhibited temperature-induced seizures, with ∼87% exhibiting seizures at an internal body temperature of 43.0°C. Treatment of Scn1a+/− mice at PND1 with 3.7E10 vg per animal AAV9-REGABA-eTFSCN1A significantly increased the temperature threshold for developing HTS (p < 0.0001, Log-rank test). Approximately 88% of AAV9-REGABA-eTFSCN1A-treated Scn1a+/− mice remained seizure free at 43.0°C. No seizures were observed in WT mice treated with vehicle or AAV9-REGABA-eTFSCN1A. Similarly, treatment with AAV9-REGABA-eTFSCN1A also significantly increased resistance to HTS in an alternative mouse model of DS,11,25 where a nonsense mutation (R1407X) was introduced in the reading frame of one copy of SCN1A (Supplementary Fig. S3A).
AAV9-REGABA-eTFSCN1A extends survival in Dravet mice up to 470 days postdosing and persists preferentially in the brain
The ability of AAV9-REGABA-eTFSCN1A to prolong short-term and long-term survival was assessed in Scn1a+/− mice injected at PND1 with vehicle or AAV9-REGABA-eTFSCN1A (3.7E10 vg/animal; n = 70) and followed for 90 days or 470 days. As controls, WT littermates were administered PBS or AAV9-REGABA-eTFSCN1A (3.7E10 vg/animal; n = 73 at PND1) (Fig. 4A).
After 90 days, 100% of Scn1a+/− mice treated with AAV9-REGABA-eTFSCN1A were alive compared with 50% of vehicle-treated Scn1a+/− mice (Fig. 4E; p < 0.0001, Log-rank test). There were no differences in survival between WT mice treated with AAV9-REGABA-eTFSCN1A or vehicle. At 470 days, AAV9-REGABA-eTFSCN1A-treated Scn1a+/− mice showed extended survival (n = 32; 83.2%) when compared with vehicle-treated Scn1a+/− mice (n = 38; 31.4%; Fig. 4F; p < 0.0001, Log-rank test). Similarly, treatment with AAV9-REGABA-eTFSCN1A also significantly prolonged survival in the alternative Scn1a+/R1407X mouse model of DS (Supplementary Fig. S3B).
AAV9-REGABA-eTFSCN1A VCN and eTFSCN1A transcript levels were measured in brain, heart, and liver of Scn1a+/− mice (n = 8) at day 470 postdosing. We found vector persistence in the brain, with a mean of 3.2 vector copies/diploid genome (Fig. 4G), while lower levels were detected in liver and heart (mean 1.3 and 0.2 vector copies/diploid genome, respectively). Similarly, eTFSCN1A transcript levels were measured in the brain at a mean of 5.6E5 transcripts per microgram of RNA (Fig. 4H), indicating persistent mRNA expression. eTFSCN1A mRNA in the liver was 7.5-fold lower (mean 7.5E4 transcripts/μg RNA); and seven of eight animals had no measurable eTFSCN1A transcripts in the heart. One animal with detectable mRNA in the heart had a level of 1.05E4 transcripts per microgram RNA (Fig. 4H).
Collectively, these results show AAV9-REGABA-eTFSCN1A persistently and preferentially targets brain tissue in Scn1a+/− mice and is associated with extended survival up to 470 days.
Unilateral ICV administration of AAV9-REGABA-eTFSCN1A in NHPs resulted in widespread and selective CNS biodistribution and was well tolerated over 28 days
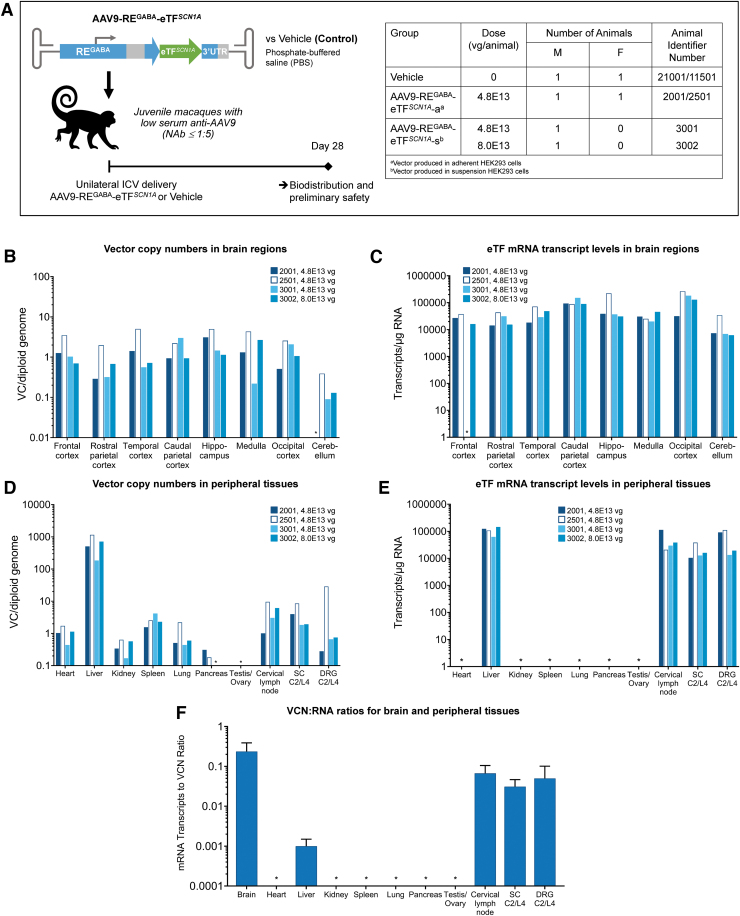
Safety, biodistribution, and activity of AAV9-REGABA-eTFSCN1A using unilateral ICV injection were evaluated in a pilot, non-GLP, 28-day study in juvenile NHPs (24–27 months of age) (Fig. 5A). Previously, we have demonstrated that unilateral ICV administration of AAV9 in NHPs results in widespread and symmetrical distribution throughout the brain.43 Animals received infusions of vehicle (n = 2), or AAV9-REGABA-eTFSCN1A at 4.8E13 vg per animal (n = 3) and 8.0E13 vg per animal (n = 1); no steroids or other immunosuppressant agents were administered. Because the difference between the two doses (4.8E13 and 8.0E13) of AAV9-REGABA-eTFSCN1A tested was less than twofold and led to no systematic differences in VCN or RNA transcript levels in the brain, the VCN and RNA expression data were combined for analysis (n = 4 animals).
Figure 5.
Unilateral ICV delivery of AAV9-REGABA-eTFSCN1A to NHPs leads to widespread vector biodistribution and increased eTFSCN1A mRNA transcript levels in the brain with low off-target vector expression in peripheral tissues of NHPs. (A) Study design. AAV9-REGABA-eTFSCN1A (4.8–8.0 E13 vg/animal) or vehicle alone (PBS) was administered by unilateral ICV injection to four juvenile cynomolgus macaques (3M and 1F). All animals were sacrificed 28 ± 2 days after injection. Biodistribution of AAV9-REGABA-eTFSCN1A vector copies and expression of transgene mRNA were measured in target neuronal and peripheral tissues using ddPCR (B–E). (B) AAV9-REGABA-eTFSCN1A vector biodistribution in brain regions. (C) eTFSCN1A mRNA expression levels in brain regions. (D) AAV9-REGABA-eTFSCN1A vector biodistribution and (E) eTFSCN1A mRNA expression levels in peripheral tissues. (F) VCN:RNA ratios for brain and peripheral tissues (N = 4, mean values; error bars indicate SD). Starred organs (*) indicate VCN or RNA levels below the limit of detection of the assay. DRG, dorsal root ganglia; NAb, neutralizing antibodies; NHP, nonhuman primate; SC, spinal cord.
ddPCR analysis of target neuronal and peripheral tissues showed that one-time administration of AAV9-REGABA-eTFSCN1A led to widespread vector biodistribution and robust transgene expression throughout the brain (Fig. 5B, C), including key structures involved in epilepsy and cognitive deficits in DS, such as cerebral cortex and hippocampus.44 The mean VCN and eTFSCN1A mRNA transcripts in the forebrain regions, including frontal cortex, temporal cortex, parietal cortex, occipital cortex and hippocampus, were measured at 1.73 copies per diploid genome and 1.71E4 transcripts per microgram RNA, respectively.
VCN and RNA expression in the medulla were similar to that of forebrain regions, with mean values of 2.14 copies/diploid genome and 3.04E4 transcripts per microgram RNA, respectively. Vector distribution was ∼10-fold lower in the cerebellum than other brain regions tested (0.16 copies/diploid genome). However, RNA expression levels were similar (1.38E4 transcripts/microgram RNA), likely due to the abundance of GABAergic Purkinje cells and inhibitory interneurons present in cerebellum.45
Vector distribution was detected in most peripheral tissues examined (Fig. 5D, E), the liver showing the highest levels. However, eTFSCN1A transgene expression in non-neuronal tissues was detected only in liver and cervical lymph nodes. Relative to VCN, average eTF mRNA levels were lower in the liver, DRG, spinal cord, and cervical lymph nodes by ∼200-, 5-, 8-, and 3.6-fold, respectively, compared with CNS tissues (Fig. 5F), indicating the selectivity of REGABA for CNS tissues.
AAV9-REGABA-eTFSCN1A was well tolerated with no adverse events occurring during administration and no detectable changes in clinical observations, body weight, or body temperature during the 28-day study. All animals survived until necropsy at 28 days. Histopathological evaluation revealed no adverse macroscopic or microscopic findings in the tissues examined, including brain, DRG, spinal cord, and non-neuronal tissues (heart, lungs, spleen, liver, and gonads). No DRG-related toxicity was observed. Blood chemistry was unchanged, except for minimum to mild transient elevations in alanine aminotransferase and aspartate aminotransferase on day 8 in AAV9-REGABA-eTFSCN1A-treated animals, which was fully resolved without treatment by day 15 or day 22. No corresponding microscopic finding was noted in the liver for any animal.
All animals had a measurable amount of serum anti-AAV9 capsid neutralizing antibodies 2 weeks after vector administration, determined by methods described in the Supplementary Materials. Titers ranged from 1:135 to 1:405 and were sustained through the end of the study (Supplementary Table S4). One animal had a positive anti-eTFSCN1A-binding antibody titer (1:400) in serum at 4 weeks postdosing (Supplementary Table S5). None of the animals had quantifiable levels of anti-AAV9 or anti-eTFSCN1A antibody response in the cerebrospinal fluid (CSF). No correlation was observed between the presence of serum antibody and AAV9-REGABA-eTFSCN1A VCN or eTFSCN1A transcript levels in the brain or peripheral organs.
DISCUSSION
Key limitations of conventional AAV-mediated gene replacement therapy have precluded DS from treatment by this modality, notably (1) transgene size restrictions due to the relatively small packaging capacity of AAV particles and (2) lack of cell type specificity to restrict transgene expression to target neurons following local CNS delivery.46,47
To overcome these key limitations, we developed AAV9-REGABA-eTFSCN1A, a novel gene regulation therapy, to selectively drive upregulation of endogenous WT SCN1A in GABAergic inhibitory interneurons to rescue haploinsufficiency of SCN1A in DS (Fig. 6). Using a compact engineered transcription factor (eTFSCN1A) to selectively target a unique and conserved genomic SCN1A regulatory sequence, we generate potent and specific upregulation in SCN1A mRNA expression in vitro and in vivo. Dynamic changes in NaV1.1 expression and localization can be influenced by intrinsic regulatory mechanisms—alternative splicing, trafficking, and post-translational modifications—that provide control mechanisms to regulate the appropriate balance of neuronal excitability.48,49
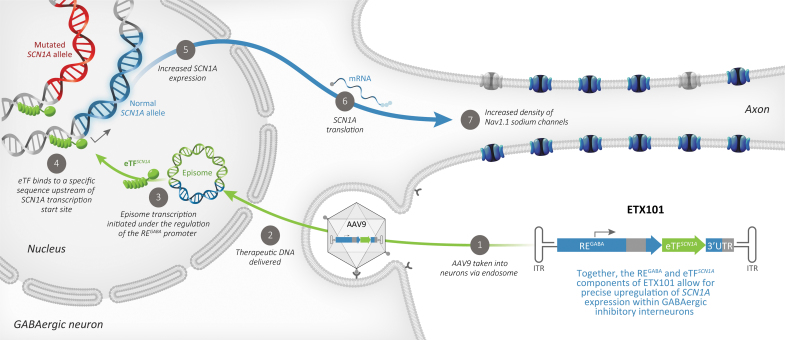
Figure 6.
Mechanism of action of AAV9-REGABA-eTFSCN1A (ETX101), a cell-selective AAV-mediated SCN1A gene regulation therapy that upregulates expression of WT SCN1A selectively in GABAergic inhibitory interneurons to compensate for the loss-of-function mutant alleles in individuals with DS. As previously described,46 AAV vector enters the cell by endocytosis in a receptor-mediated manner. Vector escapes from the endosome, enters the nucleus through the nuclear pore complex, and predominantly exists as a nonreplicating episome. Episome transcription is then initiated under the regulation of the REGABA promoter to produce eTFSCN1A preferably in GABAergic neurons. eTFSCN1A binds to a conserved regulatory region upstream of the SCN1A transcription start site, promoting increased SCN1A expression and protein translation, thereby increasing the density of membrane-associated NaV1.1 sodium channels and restoring function. While eTFSCN1A also binds the mutated SCN1A allele, it does not produce any stable protein capable of functioning at the neuronal membrane. This approach leverages natural patterns of gene expression to increase production of NaV1.1 protein and restore inhibitory function while minimizing potential off target effects. DS, Dravet syndrome; RE, regulatory element.
Our strategy regulates endogenous SCN1A transcription upstream of these intrinsic regulatory mechanisms, which may provide additional physiological control, and safeguards against overexpression. Our results show that a one-time ICV administration of AAV9-REGABA-eTFSCN1A is well tolerated in NHPs and significantly prolongs survival, and significantly reduces both spontaneous seizures and HTS in a mouse model of DS.
In the brain, NaV1.1 is predominantly expressed in GABAergic inhibitory interneurons, where it maintains the appropriate balance of inhibitory neurotransmission. Multiple lines of evidence have established impaired excitability of GABAergic inhibitory interneurons due to NaV1.1 haploinsufficiency as fundamental in driving key DS phenotypes, and suggest that enhancement of GABA signaling may improve seizure control and other manifestations associated with DS.12,13,15–21 On the contrary, pan-neuronal overexpression of Nav1.1 in both GABAergic inhibitory interneurons and excitatory neurons would lead to an opposing physiological effect, increasing the excitability of both cell types and further disrupting the balance of excitation and inhibition.50–53
Accordingly, we designed an expression cassette (collectively referred to as REGABA) to drive potent expression in multiple subtypes of GABAergic interneurons, with minimal expression in excitatory neurons, by utilizing optimized human genomic regulatory sequences surrounding the GAD1 gene locus, combined with a 3′ excitatory detargeting UTR sequences.39
When compared with the constitutive CBA promoter, we show that expression of eTFSCN1A under the control of REGABA upregulates SCN1A expression specifically within GABAergic neurons in vivo, resulting in ∼30% elevation in NaV1.1 protein levels. This level of increase was sufficient to rescue multiple disease phenotypes in a DS mouse model, highlighting the importance of cell-type selectivity. In addition, we detected low levels of transgene expression in peripheral tissues of NHPs, including the liver. This is in contrast to the finding that intrathecal delivery of AAV vectors to the CSF still results in peripheral transgene expression.54 By emulating endogenous expression patterns and gene regulation mechanisms, we can avoid supraphysiological protein expression and improve safety and potency of gene therapies. Beyond DS, GABAergic dysfunction has been implicated in other autism spectrum disorder-related syndromes, including Angelman syndrome, Rett syndrome, and others.55–59
Thus, targeted gene delivery to GABAergic inhibitory interneurons may have the potential to address the neurocircuitry deficits observed across multiple neurodevelopmental and epileptic encephalopathies. Our results provide proof-of-concept for future targeted gene delivery strategies for other interneuronopathies.
DS has been successfully recapitulated in well-characterized mouse models using several genetic strategies, including heterozygous Scn1a+/− models, the R1407X knock-in model resulting in a truncated α-subunit, and others.11,12,24 Importantly, the Scn1a+/− model recapitulates seizure-modifying efficacy for several treatments clinically indicated for DS patients. Hawkins et al.60 used the Scn1a+/− model to evaluate the efficacy of multiple antiseizure medications approved for DS, including clobazam, topiramate, stiripentol, and valproic acid. In this study, the authors found no correlation between seizure reduction (as measured by the HTS assay) and extended survival, even at supratherapeutic doses of these compounds.60
In contrast, we demonstrate that a single bilateral ICV administration of AAV9-REGABA-eTFSCN1A at PND1 leads to both a significant reduction in HTS sensitivity and extended survival in two distinct genetic mouse models of DS (Scn1a+/− and Scn1a+/R1407X). We also show that severity of spontaneous seizures as measured by EEG activity is significantly reduced in Scn1a+/− mice.
Importantly, reduction of sensitivity to HTS was also observed when mice were dosed as late as PND5, indicating a potentially broad effective treatment window of AAV9-REGABA-eTFSCN1A (Supplementary Fig. S4). In support of this, a recent study showed that reactivation of Scn1a expression in a Scn1a conditional knock-in mouse model can rescue seizure activity and behavioral abnormalities, and normalize interneuron excitability, even months after symptom onset.61 Together, these data suggest that disease phenotype may be reversed and strongly support continued development of AAV9-REGABA-eTFSCN1A as a novel gene therapy with the potential to durably rescue multiple phenotypes in DS patients.
In AAV-mediated gene therapy, the careful selection of the proper route of administration is important as it can influence biodistribution, efficacy, and safety. Intravenous administration for systemic exposure to multiple organs usually requires larger doses, while local administration to smaller organs, such as the eye and brain, requires smaller doses for transduction of relevant cells and may limit systemic exposure. CNS-administered investigational AAV-mediated gene therapies are in development for multiple CNS disorders and use different local routes, including intrathecal, intraparenchymal, intracisterna magna, and ICV. Because these routes show different AAV biodistribution profiles in the brain, the decision on the optimal one is intimately related to the underlying biology of the specific disease and affected areas.62
Previously, we demonstrated that unilateral ICV administration of AAV9 in NHPs results in widespread and symmetrical distribution throughout the brain, with significantly improved transduction of forebrain structures compared to intrathecal-lumbar administration.43 ICV is a well-established and widely used method of drug delivery for the treatment of pediatric and adult patients with a broad range of diseases.63 Here, we show that a one-time, unilateral ICV injection of AAV9-REGABA-eTFSCN1A in NHPs was well tolerated and led to widespread vector biodistribution and robust transgene expression throughout the brain. These results confirm ICV as an efficient and safe delivery route for AAV9-REGABA-eTFSCN1A, reaching areas of the brain associated with DS manifestations that are not currently addressed by antiseizure medications.
Several potential disease-modifying therapies for DS are under various stages of development,64–69 which differ in their approaches and neuronal cell types targeted. For example, a non-GABA selective, antisense oligonucleotide-based therapy has been reported to be well tolerated in DS patients at multiple monthly doses of up to 20 mg.66,69 The safety, pharmacokinetic profile, and efficacy of higher doses under placebo-controlled conditions will determine the frequency of intrathecal injections required to maintain therapeutic effect.70,71
Importantly, AAV9-mediated gene therapies have shown transgene persistence after a single administration.72 AAV-delivered transgenes form stable, nonreplicating circular episomal DNA, which is gradually lost in dividing cells, but persists in postmitotic cells, such as neurons, due to the very low cellular turnover.46 Recently published AAV-mediated gene therapies demonstrate compelling proof of concept, but are limited in their potential for clinical development. A GABA-targeted gene therapy strategy for DS utilizing a NaV1.1 channel modulator (AAV-Navβ1) reported only partial phenotypic rescue, with no effect on febrile seizure susceptibility, and limited GABA specificity.65
Conditional Cas9-based transcriptional activators, combined with GABA-specific Cre mouse lines or codelivery of a targeting vector emphasize the potential for a GABA-targeted SCN1A gene regulation in treating cognitive and behavioral effects, improving survival rate, and reducing seizure susceptibility.64,68 However, GABA-selective expression of these described transgenes is required for therapeutic efficacy, and the size of these effector transgenes precludes packaging within a single AAV particle.49,53 AAV9-REGABA-eTFSCN1A achieves GABA-specific expression of an SCN1A transcriptional activator using a single AAV vector, enabling a clinically tractable path for an AAV-mediated gene therapy for DS.
CONCLUSIONS
Our preclinical data demonstrate the efficacy and preliminary safety of AAV9-REGABA-eTFSCN1A for selectively increasing SCN1A gene activity and NaV1.1 protein expression in the brain. This resulted in reduced frequency of spontaneous seizures and HTS and increased long-term survival in a robust and translatable mouse model of SCN1A+ DS. To our knowledge, this is the longest follow-up reported in the DS mouse model, with unprecedented survival achieved after a single treatment administration. The duration of AAV9-REGABA-eTFSCN1A expression in CNS in humans is not yet quantified, but early results on the durability of AAV-mediated gene therapy in other indications have been positive, with a persistence of treatment effect reported in patients with SMA for over 6 years.73 Preliminary safety findings in juvenile NHPs support the feasibility of AAV9-REGABA-eTFSCN1A delivery by one-time unilateral ICV administration.
Collectively, these preclinical findings establish strong proof-of-concept for an AAV9-mediated gene regulation therapy that targets GABAergic inhibitory neurons to address the underlying cellular and genetic mechanism of SCN1A+ DS. Current treatments for this devastating condition provide only partial control of select aspects of the DS phenotype, underscoring the need for disease-modifying therapies. This unmet need supports the ongoing clinical development of AAV9-REGABA-eTFSCN1A (ETX101) to address the full spectrum of SCN1A+ DS manifestations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Barak Gunter and the team at Charles River (Mattawan, MI) for their invaluable technical assistance in delivering AAV vectors in nonhuman primates; Stephanie White-Hunt, BA, BVSc, MRCVS, DACVP (Charles River Laboratories, Inc.), Nicola M.A. Parry BSc, MSc, BVSc, DACVP, FRCVS (Midwest Veterinary Pathology, LLC), and Sarah Cramer, DVM, PhD, DACVP (StageBio) [Tox Path Specialists] for NHP histopathology evaluations; Psychogenics for their expertise and assistance in performing the EEG experiments and data analysis; and the ENCODE Consortium and UCSD Genome Browser group for access to genomic datasets. We also thank the team at Encoded Therapeutics, in particular Keith Place and David Rodriguez, and all our partners for their support of these studies. Finally, we thank Joanne Fitz-Gerald, BPharm and Jonathan A.C. Lee, PhD at FourWave Medical Communications for writing assistance and preparation of artwork files.
AUTHORs' CONTRIBUTIONS
A.T.: Conceptualization, methodology, investigation, and writing—original draft. T.S.: Validation, formal analysis, investigation, data curation, and writing—original draft. A.Y.: Conceptualization, methodology, formal analysis, investigation, and writing—review and editing. J.M.: Methodology, software, validation, formal analysis, investigation, data curation, and writing—review and editing, visualization. R.A.: Validation, formal analysis, investigation, data curation, and writing—review and editing. I.W.L.: Methodology, validation, formal analysis, investigation, data curation, and writing—review and editing. J.L.: Methodology, validation, investigation, and writing—review and editing. R.H.: Methodology, software, formal analysis, investigation, data curation, and writing—review and editing, visualization. M.C.: Methodology, validation, investigation, and writing—review and editing. J.L.: Methodology and writing—review and editing. T.C.: Investigation and validation. S.P.: Methodology, validation, investigation, and writing—review and editing. M.C.V.: Writing—original draft, writing—review and editing, and visualization. J.A.K.: Methodology, resources, and writing—review and editing. M.M.: Conceptualization, methodology, supervision, and writing—review and editing. A.B.: Conceptualization, methodology, supervision, and writing—review and editing. S.T.: Conceptualization, methodology, writing—review and editing, supervision, and funding acquisition.
AUTHOR DISCLOSURE
A.T., J.M., R.A., I.W.L., R.H., M.C., J. Li, S.P., M.C.V., M.M., A.B., and S.T. are current employees of and hold stock options in Encoded Therapeutics. A.Y., J.L., T.S., and T.C. are former employees of Encoded Therapeutics. J.A.K. serves as a consultant to Encoded Therapeutics, Praxis Precision Medicines, and NeuroCycle Therapeutics, and serves on the Scientific Advisory Boards of the Dravet Syndrome Foundation and FamilieSCN2A Foundation.
FUNDING INFORMATION
This work was supported by Encoded Therapeutics, Inc., South San Francisco, CA, USA.
SUPPLEMENTARY MATERIAL
REFERENCES
- 1. Dravet C, Guerrini R. Dravet Syndrome. Topics in Epilepsy, 3rd ed. Paris: John Libbey Eurotext, 2011. [Google Scholar]
- 2. Nabbout R, Chemaly N, Chipaux M, et al. . Encephalopathy in children with Dravet syndrome is not a pure consequence of epilepsy. Orphanet J Rare Dis 2013;8:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brunklaus A, Ellis R, Reavey E, et al. . Prognostic, clinical and demographic features in SCN1A mutation-positive Dravet syndrome. Brain 2012;135(Pt 8):2329–2336. [DOI] [PubMed] [Google Scholar]
- 4. Gataullina S, Dulac O. From genotype to phenotype in Dravet disease. Seizure 2017;44:58–64. [DOI] [PubMed] [Google Scholar]
- 5. Wirrell EC, Laux L, Donner E, et al. . Optimizing the diagnosis and management of Dravet syndrome: recommendations from a North American Consensus Panel. Pediatr Neurol 2017;68:18–34.e3. [DOI] [PubMed] [Google Scholar]
- 6. Villas N, Meskis MA, Goodliffe S. Dravet syndrome: characteristics, comorbidities, and caregiver concerns. Epilepsy Behav 2017;74:81–86. [DOI] [PubMed] [Google Scholar]
- 7. Skluzacek JV, Watts KP, Parsy O, et al. . Dravet syndrome and parent associations: the IDEA League experience with comorbid conditions, mortality, management, adaptation, and grief. Epilepsia 2011;52(Suppl 2):95–101. [DOI] [PubMed] [Google Scholar]
- 8. Cooper MS, McIntosh A, Crompton DE, et al. . Mortality in Dravet syndrome. Epilepsy Res 2016;128:43–47. [DOI] [PubMed] [Google Scholar]
- 9. Symonds JD, Zuberi SM, Stewart K, et al. . Incidence and phenotypes of childhood-onset genetic epilepsies: a prospective population-based national cohort. Brain 2019;142:2303–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu YW, Sullivan J, McDaniel SS, et al. . Incidence of Dravet syndrome in a US population. Pediatrics 2015;136:e1310–e1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogiwara I, Miyamoto H, Morita N, et al. . Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci 2007;27:5903–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu FH, Mantegazza M, Westenbroek RE, et al. . Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci 2006;9:1142–1149. [DOI] [PubMed] [Google Scholar]
- 13. Han S, Tai C, Westenbroek RE, et al. . Autistic-like behaviour in Scn1a+/− mice and rescue by enhanced GABA-mediated neurotransmission. Nature 2012;489:385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tai C, Abe Y, Westenbroek RE, et al. . Impaired excitability of somatostatin- and parvalbumin-expressing cortical interneurons in a mouse model of Dravet syndrome. Proc Natl Acad Sci U S A 2014;111:E3139–E3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheah CS, Yu FH, Westenbroek RE, et al. . Specific deletion of NaV1.1 sodium channels in inhibitory interneurons causes seizures and premature death in a mouse model of Dravet syndrome. Proc Natl Acad Sci U S A 2012;109:14646–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohno Y, Sofue N, Ishihara S, et al. . Scn1a missense mutation impairs GABAA receptor-mediated synaptic transmission in the rat hippocampus. Biochem Biophys Res Commun 2010;400:117–122. [DOI] [PubMed] [Google Scholar]
- 17. Kalume F, Westenbroek RE, Cheah CS, et al. . Sudden unexpected death in a mouse model of Dravet syndrome. J Clin Invest 2013;123:1798–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tatsukawa T, Ogiwara I, Mazaki E, et al. . Impairments in social novelty recognition and spatial memory in mice with conditional deletion of Scn1a in parvalbumin-expressing cells. Neurobiol Dis 2018;112:24–34. [DOI] [PubMed] [Google Scholar]
- 19. Higurashi N, Uchida T, Lossin C, et al. . A human Dravet syndrome model from patient induced pluripotent stem cells. Mol Brain 2013;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun Y, Paşca SP, Portmann T, et al. . A deleterious Nav1.1 mutation selectively impairs telencephalic inhibitory neurons derived from Dravet Syndrome patients. Elife 2016;5:e13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oakley JC, Cho AR, Cheah CS, et al. . Synergistic GABA-enhancing therapy against seizures in a mouse model of Dravet syndrome. J Pharmacol Exp Ther 2013;345:215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kantor B, McCown T, Leone P, et al. . Clinical applications involving CNS gene transfer. Adv Genet 2014;87:71–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chamberlain K, Riyad JM, Weber T. Expressing transgenes that exceed the packaging capacity of adeno-associated virus capsids. Hum Gene Ther Methods 2016;27:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller AR, Hawkins NA, McCollom CE, et al. . Mapping genetic modifiers of survival in a mouse model of Dravet syndrome. Genes Brain Behav 2014;13:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito S, Ogiwara I, Yamada K, et al. . Mouse with Nav1.1 haploinsufficiency, a model for Dravet syndrome, exhibits lowered sociability and learning impairment. Neurobiol Dis 2013;49:29–40. [DOI] [PubMed] [Google Scholar]
- 26. Gersbach CA, Gaj T, Barbas CF, 3rd. Synthetic zinc finger proteins: the advent of targeted gene regulation and genome modification technologies. Acc Chem Res 2014;47:2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mandell JG, Barbas CF, 3rd. Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res 2006;34:W516–W523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Q, Segal DJ, Ghiara JB, et al. . Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci U S A 1997;94:5525–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pollard KS, Hubisz MJ, Rosenbloom KR, et al. . Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 2010;20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Siepel A, Bejerano G, Pedersen JS, et al. . Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005;15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davis CA, Hitz BC, Sloan CA, et al. . The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 2018;46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sloan CA, Chan ET, Davidson JM, et al. . ENCODE data at the ENCODE portal. Nucleic Acids Res 2016;44:D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kent WJ, Sugnet CW, Furey TS, et al. . The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karolchik D, Hinrichs AS, Furey TS, et al. . The UCSC Table Browser data retrieval tool. Nucleic Acids Res 2004;32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mo A, Mukamel EA, Davis FP, et al. . Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 2015;86:1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Risso D, Perraudeau F, Gribkova S, et al. . A general and flexible method for signal extraction from single-cell RNA-seq data. Nat Commun 2018;9:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hawkins NA, Lewis M, Hammond RS, et al. . The synthetic neuroactive steroid SGE-516 reduces seizure burden and improves survival in a Dravet syndrome mouse model. Sci Rep 2017;7:15327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keaveney MK, Tseng HA, Ta TL, et al. . A microRNA-based gene-targeting tool for virally labeling interneurons in the rodent cortex. Cell Rep 2018;24:294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shaul O. How introns enhance gene expression. Int J Biochem Cell Biol 2017;91(Pt B):145–155. [DOI] [PubMed] [Google Scholar]
- 41. Goff KM, Goldberg EM. Vasoactive intestinal peptide-expressing interneurons are impaired in a mouse model of Dravet syndrome. Elife 2019;8:e46846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oakley JC, Kalume F, Yu FH, et al. . Temperature- and age-dependent seizures in a mouse model of severe myoclonic epilepsy in infancy. Proc Natl Acad Sci U S A 2009;106:3994–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Belle A, Lin W, Li J, et al. . Efficient delivery of clinically validated adeno-associated viral vectors to the central nervous system: a systematic evaluation of multiple routes of administration and viral serotypes in cynomolgus macaques. Mol Ther 2019;27(Suppl 1):257–258. [Google Scholar]
- 44. Stein RE, Kaplan JS, Li J, et al. . Hippocampal deletion of Na(V)1.1 channels in mice causes thermal seizures and cognitive deficit characteristic of Dravet Syndrome. Proc Natl Acad Sci U S A 2019;116:16571–16576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cerminara NL, Lang EJ, Sillitoe RV, et al. . Redefining the cerebellar cortex as an assembly of non-uniform Purkinje cell microcircuits. Nat Rev Neurosci 2015;16:79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 2019;18:358–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuzmin DA, Shutova MV, Johnston NR, et al. . The clinical landscape for AAV gene therapies. Nat Rev Drug Discov 2021;20:173–174. [DOI] [PubMed] [Google Scholar]
- 48. Chen C, Bharucha V, Chen Y, et al. . Reduced sodium channel density, altered voltage dependence of inactivation, and increased susceptibility to seizures in mice lacking sodium channel beta 2-subunits. Proc Natl Acad Sci U S A 2002;99:17072–17077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vacher H, Trimmer JS. Trafficking mechanisms underlying neuronal voltage-gated ion channel localization at the axon initial segment. Epilepsia 2012;53(Suppl 9):21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huberfeld G, Menendez de la Prida L, Pallud J, et al. . Glutamatergic pre-ictal discharges emerge at the transition to seizure in human epilepsy. Nat Neurosci 2011;14:627–634. [DOI] [PubMed] [Google Scholar]
- 51. Ogiwara I, Iwasato T, Miyamoto H, et al. . Nav1.1 haploinsufficiency in excitatory neurons ameliorates seizure-associated sudden death in a mouse model of Dravet syndrome. Hum Mol Genet 2013;22:4784–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liautard C, Scalmani P, Carriero G, et al. . Hippocampal hyperexcitability and specific epileptiform activity in a mouse model of Dravet syndrome. Epilepsia 2013;54:1251–1261. [DOI] [PubMed] [Google Scholar]
- 53. Berecki G, Bryson A, Terhag J, et al. . SCN1A gain of function in early infantile encephalopathy. Ann Neurol 2019;85:514–525. [DOI] [PubMed] [Google Scholar]
- 54. Schuster DJ, Dykstra JA, Riedl MS, et al. . Biodistribution of adeno-associated virus serotype 9 (AAV9) vector after intrathecal and intravenous delivery in mouse. Front Neuroanat 2014;8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yizhar O, Fenno LE, Prigge M, et al. . Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 2011;477:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rossignol E. Genetics and function of neocortical GABAergic interneurons in neurodevelopmental disorders. Neural Plast 2011;2011:649325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hussman JP. Suppressed GABAergic inhibition as a common factor in suspected etiologies of autism. J Autism Dev Disord 2001;31:247–248. [DOI] [PubMed] [Google Scholar]
- 58. Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav 2003;2:255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ramamoorthi K, Lin Y. The contribution of GABAergic dysfunction to neurodevelopmental disorders. Trends Mol Med 2011;17:452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hawkins NA, Anderson LL, Gertler TS, et al. . Screening of conventional anticonvulsants in a genetic mouse model of epilepsy. Ann Clin Transl Neurol 2017;4:326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Valassina N, Brusco S, Salamone A, et al. . Scn1a gene reactivation after symptom onset rescues pathological phenotypes in a mouse model of Dravet syndrome. Nat Commun 2022;13:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Naso MF, Tomkowicz B, Perry WL, 3rd, et al. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 2017;31:317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cohen-Pfeffer JL, Gururangan S, Lester T, et al. . Intracerebroventricular delivery as a safe, long-term route of drug administration. Pediatr Neurol 2017;67:23–35. [DOI] [PubMed] [Google Scholar]
- 64. Colasante G, Lignani G, Brusco S, et al. . dCas9-based Scn1a gene activation restores inhibitory interneuron excitability and attenuates seizures in Dravet syndrome mice. Mol Ther 2020;28:235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Niibori Y, Lee SJ, Minassian BA, et al. . Sexually divergent mortality and partial phenotypic rescue after gene therapy in a mouse model of Dravet syndrome. Hum Gene Ther 2020;31:339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Han Z, Chen C, Christiansen A, et al. . Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome. Sci Transl Med 2020;12:eaaz6100. [DOI] [PubMed] [Google Scholar]
- 67. Lenk GM, Jafar-Nejad P, Hill SF, et al. . Scn8a antisense oligonucleotide is protective in mouse models of SCN8A encephalopathy and Dravet syndrome. Ann Neurol 2020;87:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yamagata T, Raveau M, Kobayashi K, et al. . CRISPR/dCas9-based Scn1a gene activation in inhibitory neurons ameliorates epileptic and behavioral phenotypes of Dravet syndrome model mice. Neurobiol Dis 2020;141:104954. [DOI] [PubMed] [Google Scholar]
- 69. Wengert ER, Wagley PK, Strohm SM, et al. . Targeted Augmentation of Nuclear Gene Output (TANGO) of Scn1a rescues parvalbumin interneuron excitability and reduces seizures in a mouse model of Dravet Syndrome. Brain Res 2022;1775:147743. [DOI] [PubMed] [Google Scholar]
- 70. Turner TJ, Zourray C, Schorge S, et al. . Recent advances in gene therapy for neurodevelopmental disorders with epilepsy. J Neurochem 2021;157:229–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. ClinicalTrials.gov. NCT04442295. An Open-Label Study to Investigate the Safety of Single and Multiple Ascending Doses of STK-001 in Children and Adolescents with Dravet Syndrome. Bethesda, MD, USA: U.S. National Library of Medicine, 2022. Updated March 4, 2022. https://www.clinicaltrials.gov/ct2/show/record/NCT04442295 (accessed June 1, 2022). [Google Scholar]
- 72. Al-Zaidy SA, Mendell JR. From clinical trials to clinical practice: practical considerations for gene replacement therapy in SMA Type 1. Pediatr Neurol 2019;100:3–11. [DOI] [PubMed] [Google Scholar]
- 73. Mendell JR, Al-Zaidy SA, Lehman KJ, et al. . Five-year extension results of the Phase 1 START trial of onasemnogene abeparvovec in spinal muscular atrophy. JAMA Neurol 2021;78:834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.