Abstract
We determined the compositions of bacterioplankton communities in surface waters of coastal California using clone libraries of 16S rRNA genes and fluorescence in situ hybridization (FISH) in order to compare the community structures inferred from these two culture-independent approaches. The compositions of two clone libraries were quite similar to those of clone libraries of marine bacterioplankton examined by previous studies. Clones from γ-proteobacteria comprised ca. 28% of the libraries, while approximately 55% of the clones came from α-proteobacteria, which dominated the clone libraries. The Cytophaga-Flavobacter group and three others each comprised 10% or fewer of the clone libraries. The community composition determined by FISH differed substantially from the composition implied by the clone libraries. The Cytophaga-Flavobacter group dominated 8 of the 11 communities assayed by FISH, including the two communities assayed using clone libraries. On average only 10% of DAPI (4′,6′-diamidino-2-phenylindole)-stained bacteria were detected by FISH with a probe for α-proteobacteria, but 30% of DAPI-stained bacteria appeared to be in the Cytophaga-Flavobacter group as determined by FISH. α-Proteobacteria were greatly overrepresented in clone libraries compared to their relative abundance determined by FISH, while the Cytophaga-Flavobacter group was underrepresented in clone libraries. Our data show that the Cytophaga-Flavobacter group can be a numerically dominant component of coastal marine bacterioplankton communities.
An important first step towards understanding the roles of various bacteria in the ocean is determining the numbers and relative abundances of different bacterial groups (21). Culture-independent studies are essential for determining how many different types of bacteria are present in bacterial communities, because <1% of bacteria in nature can be cultured with currently available methods (4). The most widely used approach to examine bacterial diversity is based on clone libraries of 16S rRNA genes, which are typically collected from naturally occurring bacteria using PCR with general bacterial or universal 16S rRNA gene primers. Data from the PCR-based clone library approach indicate that marine microbial communities contain novel, uncultivated species that are widespread in the major oceans of the world (12, 20, 21).
Clone libraries can deviate from the compositions of in situ communities because of biases at each step of the method, including sample collection, cell lysis, nucleic acid extraction, PCR amplification, and cloning (47). The PCR step has been studied the most extensively. Experiments using controlled mixtures of 16S ribosomal DNA show that the relative abundance of targeted DNA molecules in the final PCR product can differ substantially from that expected (16, 40, 43, 44). Several precautions have been proposed for minimizing the biases during PCR (47), but the amount of bias is not known for pelagic habitats. No study has examined the relationship between clone library composition and community composition determined without a PCR step for bacterial communities from the water columns of aquatic systems.
Fluorescence in situ hybridization (FISH) using 16S rRNA probes is one approach for determining bacterial community composition without PCR (14). Using the FISH method, cells are identified by detection with fluorescent oligonucleotide probes specific for different bacteria. In several environments results from FISH are similar to the community composition suggested by clone libraries (7, 8, 17, 41). In particular, results from both clone libraries (36) and FISH (1, 46, 49) indicate that β-proteobacteria dominate freshwater bacterioplankton communities, although the two methods have not been applied simultaneously to the same sample. In contrast, FISH results with marine bacterioplankton seem to differ from those from clone libraries. Marine α-proteobacteria typically dominate 16S rRNA gene clone libraries (21), while the limited data collected using FISH suggest that members of the Cytophaga-Flavobacter group dominate marine bacterioplankton communities (25, 42). It is not clear if these differences reflect spatial and temporal variation or methodological differences, because no study has applied both analyses to the same community.
In this study we determined the compositions of coastal California bacterioplankton communities using clone libraries and FISH to determine whether α-proteobacteria or the Cytophaga-Flavobacter group dominates marine bacterioplankton communities. It is important to know which phylogenetic groups of bacteria dominate marine bacterioplankton communities because abundant groups may be proportionally more influential in carbon cycling and other biogeochemical processes. Furthermore, understanding why particular bacteria dominate microbial communities is a fundamental ecological question. We found that α-proteobacteria dominated clone libraries of 16S rRNA genes, as shown previously for marine bacterioplankton communities, whereas FISH indicated that the Cytophaga-Flavobacter cluster was usually the most abundant group of bacterioplankton in coastal California waters.
MATERIALS AND METHODS
Sample collection and isolation of environmental DNA.
Samples were collected using a trace-metal-clean pump from a depth of 5 m at 11 stations located 5 to 40 km from the California coast between Point Sur and Point Arena in June 1999. Samples were processed promptly after collection. See reference 28 for general oceanographic information on the region.
Twenty-liter seawater samples collected at station 5 and station 9 near Point Sur and Point Reyes, respectively, were filtered sequentially through 3- and 1-μm-pore-size polycarbonate membrane filters before the bacterial size fraction was collected on 0.2-μm-pore-size Gelman Supor filters. Filtered samples were stored frozen at −20°C in a storage buffer (23). Frozen samples were thawed, and the cells were lysed using sodium dodecyl sulfate and proteinase K. The lysate was extracted sequentially with phenol-chloroform and chloroform. Extracted nucleic acids were precipitated with ethanol (6).
Bacterial growth rate.
Bacterial production was estimated by the leucine incorporation method (31) in triplicate. The added leucine concentration was 20 nM, and the incubation time was 1 h. An index of the bacterial growth rate was obtained by dividing leucine incorporation rates (bacterial production) by bacterial abundance, which was measured by fluorescence microscopy of DAPI (4′,6′-diamidino-2-phenylindole)-stained samples (37).
Clone libraries.
16S rRNA genes were amplified from extracted DNA using oligonucleotide primers EubA (AAG GAG GTG ATC CAN CCR CA) and EubB (AGA GTT TGA TCM TGG CTC AG) (22). The 25-μl PCR mixtures contained 4 ng of template DNA per μl, a 0.2 mM concentration of each of the four deoxynucleoside triphosphates (dTTP, dCTP, dGTP, and dATP), 1.5 mM MgCl2, 1 μM (each) primer, and 2.5 U of Taq DNA polymerase (Promega). Thermocycling conditions included 1 min of denaturation at 94°C, 1 min of primer annealing at 50°C, and 3 min of primer extension at 72°C. This cycle was repeated 25 times. PCR products were cloned by using the TOPO-TA cloning kit with the pCR 2.1 vector (Invitrogen) according to the manufacturer's protocol.
Clone libraries were screened by dot blot hybridization of plasmids prepared using alkaline lysis of recombinant clones grown for 48 h in 96-well microtiter plates containing 200 μl of TYGPN medium per well (6). TYGPN medium contains 20 g of tryptone, 10 g of yeast extract, 10 g of glycerol, 5 g of Na2HPO4 and 10 g of KNO3 per liter of deionized water. One-third of the plasmid preparation from each well was suspended in 6× SSC (1× SSC is 150 mM NaCl plus 15 mM trisodium citrate [pH 7.7]), denatured in a boiling water bath for 10 min, and blotted by vacuum onto a Hybond-N membrane (Pharmacia). The membrane was removed from the vacuum manifold and incubated for 10 min on absorbent paper saturated with a denaturing solution containing 0.5 M NaOH and 1.5 M NaCl. The membrane was transferred to absorbent paper saturated with a neutralizing solution containing 1.5 N NaCl and 0.5 M Tris-HCl (pH 7.4) for 5 min, and the plasmids were bound to the membrane using UV light.
Clone libraries were screened using oligonucleotide probe Alf968 (25) for the α-subclass of the proteobacteria and probe CF319a (34) for the Cytophaga-Flavobacter group of the Cytophagales division. Probes were labeled with digoxigenin using oligonucleotide tailing reagents (Boehringer Mannheim) and detected colorometrically using nitroblue tetrazolium, using the protocol supplied by the manufacturer. The hybridization and washing stringency was set using the established formamide and NaCl concentration for each probe (25, 34). Dot blot hybridization specificity was confirmed using cloned 16S rRNA genes of cultured bacteria in the α-, β- and γ-subclasses of the proteobacteria and the Cytophaga-Flavobacter cluster and by nucleotide sequencing of selected clones having positive hybridization with the oligonucleotide probes. Clones that did not bind the probes for α-proteobacteria and the Cytophaga-Flavobacter cluster were classified by partial nucleotide sequencing and BLAST analysis (version 2.0; National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/BLAST/]) (2).
Nucleotide sequencing.
Nucleotide sequencing was performed using an ABI PRISM 310 (Perkin-Elmer) genetic analyzer with ABI PRISM Big Dye terminator cycle sequencing reagent and oligonucleotide primers EubB and 519R (GWA TTA CCG CGG CKG CTG) (32). Double-stranded DNA templates were prepared using an alkaline lysis procedure recommended by Perkin-Elmer.
FISH.
Bacterioplankton community compositions were determined by FISH using probe Eub338 (3) for eubacteria, Alf968 (25) for the α-subclass of the proteobacteria, Bet42a (35) for the β-subclass of the proteobacteria, Gam42a (35) for the γ-subclass of the proteobacteria, CF319a (34) for the Cytophaga-Flavobacter group, SAR11A1 (18) for the SAR11 cluster (21), SAR86/1249 (15) for the SAR86 cluster (21), and a negative control probe (29) for nonspecific probe binding. Bacterioplankton samples from 11 stations between Point Sur and Point Arena were prepared for FISH using a modification of the method described by Glöckner et al. (24). Seawater samples were prefiltered through 1.0-μm-pore-size polycarbonate membranes and mixed with 3 volumes of freshly prepared 4% formaldehyde. After 16 to 48 h in fixative at 5°C, the bacteria were filtered onto a 0.2-μm-pore-size polycarbonate membrane (Poretics), rinsed with 0.2-μm-pore-size-filtered seawater and stored at −20°C. A piece of the filter was placed on a Parafilm-covered glass slide, overlaid with 30 μl of hybridization solution containing 75 ng of Cy3-labeled oligonucleotide probe, and incubated in a sealed container for 90 min at 46°C. The hybridization solution contains 0.9 M NaCl, 20 mM Tris-HCl (pH 7.4), 0.01% sodium dodecyl sulfate, and the concentration of formamide determined to achieve specificity for the targeted group of bacteria (15, 48). The one exception was probe SAR11 A1 (18), which was hybridized at 37°C using a hybridization solution containing 0.25 M Na2HPO4 and adjusted to pH 7.2 with 85% H3PO4 (26). After hybridization, the sample was transferred to a wash solution containing 20 mM Tris-HCl (pH 7.4), 5 mM EDTA, 0.01% sodium dodecyl sulfate, and a concentration of NaCl appropriate for the probe. Probe SAR11 A1 was washed in 0.2× SSPE (1× SSPE is 180 mM NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) at 37°C, using the conditions described by Field et al. (18).
After hybridization, cells were stained with 2 μg of DAPI per ml and counted in 10 fields of view using a Nikon FXA microscope fitted with Cy3 filter 41007a (Chroma) and DAPI filter 31000 (Chroma).
RESULTS
Clone libraries.
We sampled surface waters at several stations from Point Arena to Point Sur, California, in order to compare the community compositions of bacterial assemblages as revealed by clone libraries and by FISH. Eighty-two and 87 clones were screened in the station 5 and station 9 libraries, respectively. BLAST analysis of nucleotide sequences indicated that most of the clones in our two libraries were highly similar (mean = 0.97) to clones representing uncultured bacteria described in previous studies of marine bacterioplankton (Table 1). The portion of the 16S rRNA gene that we sequenced should be adequate for classification to the proteobacterial subclass level, because it includes approximately 500 bp spanning the three variable regions V1, V2, and V3.
TABLE 1.
GenBank entries identified by BLAST as having the highest similarity to the station 5 and station 9 clones that were assayed by sequence analysis
| Group and GenBank entry | No. of clones | Similarity
|
|
|---|---|---|---|
| Minimum | Maximum | ||
| α-Proteobacteria | |||
| Roseobacter sp. strain PRLIST06 | 4 | 0.94 | 0.97 |
| Roseobacter sp. strain Shippagan | 2 | 0.98 | 0.99 |
| Strain HRV3# HpaAS1 | 1 | 0.99 | 0.99 |
| Strain KAT6 | 2 | 0.95 | 0.95 |
| Strain KAT8 | 1 | 0.93 | 0.93 |
| Strain GAI-36 | 1 | 0.93 | 0.93 |
| Uncultured α-proteobacteria | |||
| OCS154 (SAR11 cluster) | 1 | 0.97 | 0.97 |
| OCS53 (SAR11 cluster) | 1 | 1.00 | 1.00 |
| OM25 | 1 | 0.94 | 0.94 |
| OM42 | 9 | 0.98 | 0.99 |
| OM65 | 2 | 0.99 | 0.99 |
| Uncultured β-proteobacteria | |||
| OM43 | 1 | 0.98 | 0.98 |
| OM58 | 1 | 0.97 | 0.97 |
| OCS178 | 1 | 0.97 | 0.97 |
| γ-Proteobacteria | |||
| Pseudomonas sp. strain BAL18 | 3 | 0.96 | 0.99 |
| Acinetobacter sp. strain 79 | 2 | 0.98 | 0.99 |
| Strain HTB082 | 3 | 0.95 | 0.97 |
| Strain DPT1.2 | 1 | 0.98 | 0.98 |
| Strain NKB4 | 3 | 0.89 | 0.92 |
| Uncultured γ-proteobacteria | |||
| OCS44 (SAR86 cluster) | 12 | 0.91 | 1.00 |
| OM10 (SAR86 cluster) | 7 | 0.90 | 0.99 |
| 400m-FREE-40 | 3 | 0.95 | 0.97 |
| CRE-PA14 | 1 | 0.99 | 0.99 |
| CRE-PA50 | 3 | 0.99 | 0.99 |
| CRO-FL8 | 6 | 0.95 | 0.97 |
| OM23 | 1 | 0.98 | 0.98 |
| OM182 | 1 | 0.93 | 0.93 |
| Cytophaga-Flavobacter group | |||
| Polaribacter sp. strain MED18 | 1 | 0.90 | 0.90 |
| Strain SCB49 | 3 | 0.93 | 0.94 |
| Uncultured CRE-PA37 | 1 | 0.90 | 0.90 |
| Other groups | |||
| Uncultured Planctomyces strain CRE-FL31 | 1 | 0.89 | 0.89 |
| Uncultured Verrucomicrobiales strain HstpL11 | 2 | 0.89 | 0.89 |
| Uncultured Firmicutes CR-PA26 | 1 | 0.89 | 0.89 |
| Uncultured Actinomycetes OCS155 | 4 | 0.99 | 0.99 |
| Uncultured algal chloroplast | |||
| OCS182 | 4 | 0.98 | 0.99 |
| OCS20 | 1 | 0.99 | 0.99 |
| Prasinophyte OM39 | 2 | 0.98 | 0.99 |
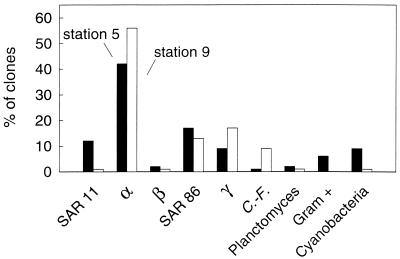
The bulk of the clones (82 to 88%) in the libraries came from proteobacteria (Fig. 1), and most of the proteobacteria in both libraries were from the α-subclass (54 to 57% of all clones). A large fraction of the α-proteobacteria were Roseobacter spp. Forty-two percent of the α-proteobacterial clones in the station 5 library and 56% in the station 9 library were similar (>94%) to the 16S rRNA genes in cultured Roseobacter spp. The α-proteobacterial clones were analyzed further to determine the fraction affiliated with the SAR11 cluster, which typically accounts for a large fraction of the clones in libraries from aquatic systems (21). Clones affiliated with the SAR11 cluster comprised 12% of the station 5 library but only 1% of the station 9 library.
FIG. 1.
Percentages of clones represented by the major phylogenetic groups of bacteria in libraries of 16S rRNA genes in samples collected near Point Sur (station 5) (black bars) and Point Reyes (station 9) (white bars). Clones corresponding to the SAR11 cluster (SAR11), α-proteobacteria (α), β-proteobacteria (β), SAR86 cluster (SAR86), γ-proteobacteria (γ), Cytophaga-Flavobacter group (C.-F.), Planctomyces, gram-positive group (Gram +), and cyanobacteria were identified using oligonucleotide probing and nucleic acid sequence analysis. Eighty-two and 87 clones were screened in the Big Sur and Point Reyes libraries, respectively.
Clones from γ-proteobacteria were a substantial fraction of the two clone libraries, comprising 26 and 30% of the station 5 and station 9 libraries, respectively (Fig. 1). Nucleotide sequence analysis together with the dot blot hybridizations with probe SAR86/1249 (15) indicated that a modest fraction of the clones was affiliated with the SAR86 cluster, which is a group of uncultured γ-proteobacteria commonly found in clone libraries (21). Seventeen percent of the clones in the station 5 library came from the SAR86 cluster, while 13% of the clones in the station 9 library were affiliated with this group. The remaining γ-proteobacterial clones, comprising 9 and 17% of the clones in the station 5 and station 9 libraries, respectively, were similar (>91%) to 16S rRNA genes in different cultured and uncultured γ-proteobacteria (Table 1).
Clones from the Cytophaga-Flavobacter group, Planctomyces, Verrucomicrobiales, Actinobacteria, and cyanobacteria were present in both libraries but were not abundant (Fig. 1). One percent of the clones in the station 5 library and 9% of the clones in the station 9 library came from the Cytophaga-Flavobacter group. Cyanobacteria were represented by a similarly small fraction (9% or less) of the clones. Clones closely related to Planctomyces accounted for 2% or less of the libraries. Six percent of the clones in the station 5 library came from Actinobacteria, while none of the clones in the station 9 library were affiliated with this group. As is typical for marine bacterioplankton libraries (21), β-proteobacteria were not abundant in the two clone libraries. Only 2% or fewer of the clones in the two libraries belonged to this subclass of the proteobacteria.
FISH.
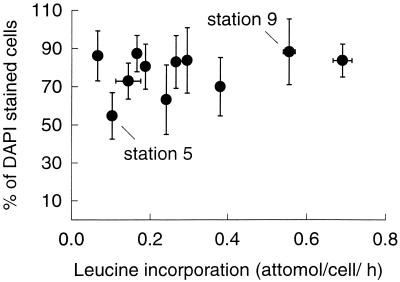
The percentage of DAPI-stained cells detected with the eubacterial probe was usually quite high, greater than 70% at 9 of the 11 stations (Fig. 2). This detection by FISH was consistently high even though bacterial growth varied by 1 order of magnitude (0.07 to 0.7 amol of leucine/cell/h). The percentage of bacteria detected by probe Eub338 varied from 55% (standard deviation [SD] = 12%) to 88% (SD = 17%) of the cells detected with DAPI. The percentage of bacteria detected with a negative control probe (29), which has at least three mismatches with the 16S rRNA genes in the Ribosomal Database Project (release 8.0, 1 June, 2000), varied from 0 to 2% of the DAPI-stained bacteria. Results with this negative control probe, which accounts for autofluorescence of cells and nonspecific probe binding, were subtracted from the percentages detected with probes for the bacterial groups.
FIG. 2.
Percentages of DAPI-stained bacteria detected with the eubacterial probe Eub338 and 3H-leucine incorporation off the coast of California. Error bars are ± 1 SD. Clone libraries were constructed at stations 5 and 9.
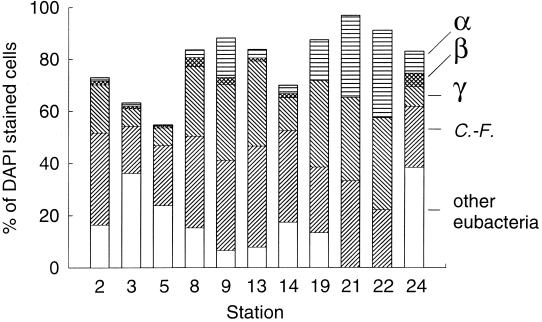
The FISH results indicate that the Cytophaga-Flavobacter group was more abundant than the proteobacteria. On average 30% of the bacteria in communities from the 11 stations were detected with the probe for the Cytophaga-Flavobacter group (Fig. 3). The relative abundance of the Cytophaga-Flavobacter group varied approximately twofold, from 18 to 39% of the cells detected by DAPI staining. The relative abundance of γ-proteobacteria varied from 7 to 42% of the bacterial community. A small fraction of the bacteria was detected with a probe for the SAR86 cluster of the γ-proteobacteria (undetectable to 9%) (Table 2).
FIG. 3.
Percentages of DAPI-stained bacteria detected by FISH with probes for α-proteobacteria (α), β-proteobacteria (β), γ-proteobacteria (γ), and the Cytophaga-Flavobacter group (C.-F.). The percentage of DAPI-stained bacteria detected with the probe for eubacteria (Eub338) corresponds to the maximum bar height. The white portions of the bars indicate cells detected with probe Eub338 but not with any group-specific probe (other eubacteria).
TABLE 2.
Relative abundances of the SAR11 and SAR86 clusters and total α-proteobacteria and γ-proteobacteria detected by FISH in bacterioplankton communities of the coastal Pacific Oceana
| Station | Location | % of DAPI-stained cellsb
|
|||
|---|---|---|---|---|---|
| SAR11 cluster | Total α-proteobacteria | SAR86 cluster | Total γ-proteobacteria | ||
| 2 | Monterey Bay | <1 (1) | 1 (2) | 0 | 19 (6) |
| 3 | 4 km south of Point Sur | 0 | 1 (1) | <1 (1) | 7 (3) |
| 5 | Point Sur | <1 (1) | <1 (1) | 0 | 7 (3) |
| 8 | Point Reyes | <1 (1) | 3 (2) | 9 (9) | 27 (7) |
| 9 | 5 km north of Point Reyes | 0 | 15 (6) | 3 (4) | 29 (8) |
| 13 | Point Arena | 0 | 3 (2) | 4 (4) | 33 (14) |
| 14 | 20 km south of Point Arena | <1 (1) | 3 (2) | 3 (3) | 13 (9) |
| 19 | Point Año Nuevo | 0 | 15 (7) | 0 | 33 (11) |
| 21 | Point Sur | 0 | 30 (8) | 0 | 32 (13) |
| 22 | 18 km south of Point Sur | 0 | 34 (17) | <1 (1) | 35 (10) |
| 24 | Point Sur, 37 km offshore | 0 | 8 (5) | 2 (2) | 8 (3) |
The SAR11 cluster is a group of α-proteobacteria and the SAR86 cluster is a group of γ-proteobacteria. The SAR11 cluster was detected with probe SAR11A (18), and the SAR86 cluster was detected with probe SAR86/1249 (15). Total α-proteobacteria and γ-proteobacteria were detected with probes Alf968 (25) and Gam42a (35), respectively.
The values in parentheses are SDs from 10 microscopic fields of view.
On average α-proteobacteria comprised 10% of the bacteria detected by DAPI staining (Fig. 3). Less than 3% of the community at 6 of 11 stations was detected with the probe for α-proteobacteria. At the other five stations α-proteobacteria comprised 8 to 34% of the bacterial community. Fewer than 1% of the bacteria at all stations were detected with a probe for the SAR11 cluster of the α-proteobacteria (Table 2).
β-Proteobacteria were a small fraction of the bacteria in coastal Pacific Ocean communities, comprising at most 5% (SD = 3%) of the community (Fig. 3).
Usually, the number of bacteria detected by the eubacterial probe exceeded or equaled the sum of the numbers of bacteria detected by probes Alf968, Bet42a, Gam42a, and CF319a (Fig. 3). At nine stations, however, 6 to 38% of the cells detected with probe Eub338 were not detected by these probes. The results were quite different at station 21 and station 22, where the percentages of cells detected with the group-specific probes were 137 and 116%, respectively, of the cells detected with the eubacterial probe Eub338, suggesting that some cells bound more than one group-specific probe.
Clone libraries versus FISH.
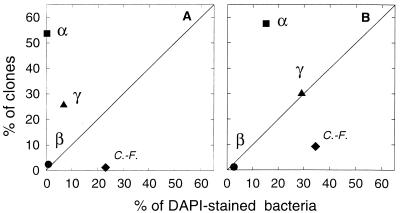
The bacterioplankton community compositions determined by FISH and clone libraries of 16S rRNA genes were compared graphically by plotting percentages from one approach against the other (Fig. 4). Data points above the 1:1 line indicate phylogenetic groups that are overrepresented in clone libraries compared to their relative abundances determined by FISH. Similarly, points below the 1:1 line indicate phylogenetic groups that are underrepresented in clone libraries compared to their relative abundances determined by FISH. The percentage of clones representing α-proteobacteria was far greater than the relative abundance of α-proteobacteria determined by FISH. Clones representing α-proteobacteria dominated the two libraries, comprising 54 and 57% of the station 5 and station 9 libraries, respectively (Fig. 4). In contrast, fewer than 1 and 15% of the DAPI-stained bacteria in surface seawater at station 5 and station 9, respectively, were detected in the FISH assay by probe Alf968 for α-proteobacteria (Fig. 4).
FIG. 4.
Relationship between compositions of 16S rRNA gene clone libraries and bacterial community compositions in the coastal Pacific Ocean at Point Sur (station 5) (A) and Point Reyes (station 9) (B). Clones and DAPI-stained bacteria were classified as α-proteobacteria (α), β-proteobacteria (β), γ-proteobacteria (γ) and members of the Cytophaga-Flavobacter group (C.-F.).
Bacteria in the Cytophaga-Flavobacter group were greatly underrepresented in clone libraries of 16S rRNA genes relative to their abundances in bacterioplankton communities determined by FISH. Only 1 and 9% of the clones in the station 5 and station 9 libraries, respectively, came from the Cytophaga-Flavobacter group (Fig. 4). In contrast, FISH suggested that the Cytophaga-Flavobacter group dominated the bacterial communities, comprising 23 and 35% of the DAPI-stained bacteria in the station 5 and station 9 communities, respectively.
Estimates of the relative abundances of γ-proteobacteria determined by FISH and clone libraries differed as well at one of the two stations. Clone libraries overestimated the relative abundance of γ-proteobacteria in the station 5 sample. Twenty-six percent of the clones in the station 5 library came from γ-proteobacteria, while only 7% of the bacteria in the station 5 community were detected with the Gam42a probe for γ-proteobacteria (Fig. 4A). In contrast, FISH and clone libraries yielded similar estimates of the relative abundance of γ-proteobacteria in the station 9 sample. Both approaches indicated that γ-proteobacteria made up 30% of the community at this station (Fig. 4B).
β-Proteobacteria comprised equally small fractions of the clone libraries and of the bacterial communities as determined by FISH (Fig. 4).
DISCUSSION
Our study revealed substantial differences between the community compositions of marine bacterioplankton communities determined by FISH and clone libraries of 16S rRNA genes. Similar to marine bacterioplankton clone libraries from previous studies (21), our coastal California Pacific Ocean clone libraries were dominated by α-proteobacteria. In contrast, direct microscopic analysis of the same samples by FISH showed that the Cytophaga-Flavobacter group was far more abundant than the α-proteobacteria group. Previous studies using FISH have shown that the Cytophaga-Flavobacter group also dominates marine bacterioplankton communities in the North Sea and Antarctic Ocean (25, 42).
Our data suggest that clone libraries of 16S rRNA genes amplified using general bacterial primers overestimate the relative abundance of α-proteobacteria and underestimate the Cytophaga-Flavobacter group. Although additional data from more aquatic environments are clearly needed, the data so far collected with FISH suggest that the Cytophaga-Flavobacter group may comprise a larger fraction of marine bacterioplankton communities than clone libraries have indicated. Studies using clone libraries of 16S rRNA genes suggest that the Cytophaga-Flavobacter group is enriched on particles but comprises a much smaller fraction of free-living communities (11, 12, 38). For example, the Cytophaga-Flavobacter group accounted for 75% of the cloned 16S rRNA genes amplified from the community of particle-associated bacteria in the Columbia River estuary (11). Enrichment of the Cytophaga-Flavobacter group on particles is probably real, because bias associated with the PCR-based clone library approach should influence results for both the free-living and particle-associated communities.
The PCR primers used to determine diversity in microbial communities using clone libraries are critical for obtaining an accurate determination of community composition. Primers that are ineffective with the Cytophaga-Flavobacter group would obviously lead to underestimates of the relative abundance of this group. However, there are no obvious mismatches between the general bacterial primers we used and 16S rRNA gene sequences for the Cytophaga-Flavobacter group now available in GenBank. The forward primer EubB matches all of the 11 Cytophaga-Flavobacter sequences that have been determined for the binding site of this primer. Seven of the nine Cytophaga-Flavobacter sequences match the reverse EubA primer exactly; one sequence has a single mismatch, while another sequence has three mismatches. However, the mismatches in this reverse primer are probably not responsible for clone libraries underestimating the relative abundance of the Cytophaga-Flavobacter group. The commonly used universal reverse primer (1492R) (32) matches all 22 of the Cytophaga-Flavobacter genes that have been completely sequenced in the region where this probe binds. Libraries made with universal primers also have low representation by the Cytophaga-Flavobacter group (21).
However, lack of amplification by the general bacterial primers is still a likely explanation for the difference between the FISH and clone library results. Although the GenBank database indicates that the bacterial and universal PCR primers should be effective for the Cytophaga-Flavobacter group, the current sequence data may not yield the most robust test. GenBank sequences come largely from cultured bacteria, but we know that these bacteria represent a small subset of total bacterial diversity. In addition, sequences in the database from uncultured bacteria obviously would not include any sequences that do not match currently used primers, because they would not be retrieved by PCR. In short, our data suggest that a dominant group of marine bacteria, i.e., the Cytophaga-Flavobacter cluster, is underrepresented in the GenBank database. We suspect that further examination of uncultured bacteria in the Cytophaga-Flavobacter cluster will reveal differences in the binding sites for general bacterial and universal primers.
The number of 16S rRNA genes per genome could be another reason why we found differences between the clone library composition and the actual community composition determined by FISH (16). A group having more copies would be more abundant in the library. However, α-proteobacteria which are more abundant in clone libraries seem to have fewer copies of the rRNA operon than the Cytophaga-Flavobacter group. The 8 members of the Cytophaga-Flavobacter group that have been examined average five copies of the rRNA operon, while the 17 α-proteobacteria (including Roseobacter spp.) have only three copies on average (Ribosomal RNA Operon Copy Number Database [http://rdp.cme.msu.edu/rrn]). Based on these data, we would expect the Cytophaga-Flavobacter group to be overrepresented, not underrepresented, in clone libraries. Of course, operon copy number has been determined only for cultured bacteria, and there may be problems extrapolating the results to uncultured bacteria.
A final possible explanation for our results is that the FISH method may not be detecting all bacteria. In our study the percentage of cells detected by FISH with the eubacterial probe Eub338 was generally high (>70%) (Fig. 2), but at some stations almost 45% of bacteria were not detected by FISH. The factors limiting detection of microbial cells by FISH are the abundance of ribosomes per cell (33), accessibility of the rRNA (19), and cell wall permeability (48). The percentage of cells detected by FISH did not increase with higher growth rates and presumably more ribosomes per cell (30, 33), suggesting that the growth rate did not limit detection by FISH (Fig. 2). There may be other explanations for why some cells were not detected by FISH. It is possible that gram-positive cells were not permeable to oligonucleotide probes under the conditions we used. In addition, archaea which would not be detected by the eubacterial probe, may comprise part of the community that was not detected by FISH (13).
However, detection problems with FISH cannot account for the difference we found between the clone library and FISH results. The greatest difference between the clone library and FISH data was the relative abundance of α-proteobacteria (Fig. 4). Even if all cells not detected by FISH were α-proteobacteria, the clone library composition would still differ from the community composition determined by FISH. In the station 5 sample, if we assume that the 45% of the DAPI-stained bacteria not detected by FISH are all α-proteobacteria, they would be 45% of the community (versus <1% now). This percentage (45%) is still less than the percentage of α-proteobacteria in the clone library (53%). The same argument can be made for station 9, where only 15% of the DAPI-stained bacteria were not detected by FISH. Assuming that all of these undetected cells are α-proteobacteria increases their representation to 30%, which is less than the percentage of α-proteobacteria in the clone library (58%) and still less than fraction of the community determined by FISH to be in the Cytophaga-Flavobacter group (35%).
Several studies have found a close correspondence between clone library composition and in situ community composition determined by FISH. The community of bacteria associated with the deep-sea hydrothermal vent polychaete Alvinella pompejana is one example from a marine environment (7). Similarly, clone libraries and FISH confirmed that populations of Nitrosospira and Nitrospira spp. dominated bacterial populations in a nitrifying fluidized bed reactor (41). The same phylogenetic groups dominated clone libraries of 16S rRNA genes and the natural microbial communities in a rice field soil inoculum and cellulose enrichment cultures (8). In a stable toluene-degrading consortium, clone libraries and FISH detected the same dominant bacteria and archaea as well (17). However, clone library and FISH results are not always the same. One example is the bacterial community in batch reactors modeling activated-sludge processes. Clone libraries of 16S rRNA genes amplified from activated-sludge communities were dominated by the Cytophaga-Flavobacter group, but the in situ community composition determined by FISH was dominated by β-proteobacteria (10). Although in many environments the same phylogenetic groups are detected by FISH and clone libraries, the activated-sludge study and our study of marine bacterioplankton indicate that these two approaches can yield different estimates of numerical dominance.
The hypothesis that easily cultured bacteria are not representative of most bacteria in natural bacterioplankton communities is reinforced by the large difference between community compositions determined by culture-dependent and culture-independent (e.g., clone libraries of 16S rRNA genes) approaches. However, the Cytophaga-Flavobacter group may be a noteworthy exception if this group is numerically dominant in marine bacterioplankton communities, as suggested by our study and others (25, 42). Representatives of this group typically show up in culture collections, although they are not abundant in clone libraries (45). Ecophysiological studies based on cultured bacteria (27) could be quite useful for studying the ecology of marine bacteria, even given that certain aspects of their metabolism in culture likely differ from that in nature. Bacteria from the Cytophaga-Flavobacter group seem to be good candidates for further laboratory study. More work is needed to determine if the particular types of bacteria in the Cytophaga-Flavobacter group that can be cultured are in fact abundant in natural communities.
Dominance of the Cytophaga-Flavobacter group in bacterioplankton communities has important implications for our understanding of organic matter cycling in the ocean. Cultured strains of bacteria in the Cytophaga-Flavobacter group are well known for their capacity to degrade high-molecular-weight organic compounds (39), and the same appears to be true for uncultured members of this group (9). An abundant Cytophaga-Flavobacter group using high-molecular-weight organic compounds would be consistent with work showing that this size class of organic material is a large, biologically labile pool in the ocean (5). Information on the different types of marine bacteria in the Cytophaga-Flavobacter group, as well as their capacity for organic matter consumption, should lead to a better understanding of carbon cycling by bacteria in the ocean.
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Energy and the National Science Foundation.
We thank Benedikt Meon for his assistance and David Hutchins for inviting us to participate in the Circus '99 cruise.
REFERENCES
- 1.Alfreider A, Pernthaler J, Amann R, Sattler B, Glöckner F O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amon R M W, Benner R. Bacterial utilization of different size classes of dissolved organic matter. Limnol Oceanogr. 1996;41:41–51. [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struh K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1992. [Google Scholar]
- 7.Cary S C, Cottrell M T, Stein J L, Camacho F, Desbruyeres D. Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl Environ Microbiol. 1997;63:1124–1130. doi: 10.1128/aem.63.3.1124-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin K J, Lukow T, Stubner S, Conrad R. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15 and 30°C) FEMS Microbiol Ecol. 1999;30:313–326. doi: 10.1111/j.1574-6941.1999.tb00659.x. [DOI] [PubMed] [Google Scholar]
- 9.Cottrell M T, Kirchman D L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crocetti G R, Hugenholtz P, Bond P L, Schuler A, Keller J, Jenkins D, Blackall L L. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl Environ Microbiol. 2000;66:1175–1182. doi: 10.1128/aem.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crump B C, Armbrust E V, Baross J A. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached versus free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 13.DeLong E F, Taylor L T, Marsh T L, Preston C M. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLong E F, Wickham G, Pace N. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 15.Eilers H, Pernthaler J, Glöckner F O, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ficker M, Krastel K, Orlicky S, Edwards E. Molecular characterization of a toluene-degrading methanogenic consortium. Appl Environ Microbiol. 1999;65:5576–5585. doi: 10.1128/aem.65.12.5576-5585.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field K G, Gordon D, Wright T, Rappé M, Urback E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuhrman J A, McCallum K, Davis A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giovannoni S, Rappé M. Evolution, diversity and molecular ecology of marine prokaryotes. In: Kirchman D L, editor. Microbial ecology of the oceans. New York, N.Y: Wiley-Liss; 2000. pp. 47–84. [Google Scholar]
- 22.Giovannoni S J. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: Wiley; 1991. pp. 177–203. [Google Scholar]
- 23.Giovannoni S J, DeLong E F, Schmidt T M, Pace N R. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 25.Glöckner F O, Fuchs B M, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagström Å, Pinhassi J, Zweifel U L. Biogeographical diversity among marine bacterioplankton. Aquat Microb Ecol. 2000;21:231–244. [Google Scholar]
- 28.Hutchins D A, DiTullio G R, Zhang Y, Bruland K W. An iron limitation mosaic in the California upwelling regime. Limnol Oceanogr. 1998;43:1037–1054. [Google Scholar]
- 29.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerkhof L, Kemp P. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol Ecol. 1999;30:253–260. doi: 10.1111/j.1574-6941.1999.tb00653.x. [DOI] [PubMed] [Google Scholar]
- 31.Kirchman D L. Leucine incorporation as a measure of biomass production by heterotrophic bacteria. In: Kemp P, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis; 1993. pp. 509–512. [Google Scholar]
- 32.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: Wiley; 1991. pp. 115–175. [Google Scholar]
- 33.Lee S, Malone C, Kemp P. Use of multiple 16S rRNA-targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria. Mar Ecol Prog Ser. 1993;101:193–201. [Google Scholar]
- 34.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 35.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 36.Methé B A, Hiorns W D, Zehr J P. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol Oceanogr. 1998;43:368–374. [Google Scholar]
- 37.Porter K, Feig Y. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 38.Rath J, Wu K Y, Herndl G J, DeLong E F. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 39.Reichenbach H. The order Cytophagales. In: Balows A, editor. The prokaryotes. New York, N.Y: Springer-Verlag; 1991. pp. 3631–3675. [Google Scholar]
- 40.Reysenbach A L, Giver L J, Wickham G S, Pace N R. Differential amplification of ribosomal RNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schramm A, de Beer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon M, Glöckner F O, Amann R. Different community structure and temperature optima of heterotrophic picoplankton in various regions of the Southern Ocean. Aquat Microb Ecol. 1999;18:275–284. [Google Scholar]
- 43.Suzuki M, Rappé M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Strobel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss P, Schweitzer B, Amann R, Simon M. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow) Appl Environ Microbiol. 1996;62:1998–2005. doi: 10.1128/aem.62.6.1998-2005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wintzingerode F V, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 48.Zarda B, Hahn D, Chatzinotas A, Schonhuber W, Neef A, Amann R I, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]
- 49.Zwart G, Hiorns W D, Methe B A, Van Agterveld M P, Huismans R, Nold S C, Zehr J P, Laanbroek H J. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1998;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]