Abstract
Previous studies have indicated that the yellow pigments (xanthomonadins) produced by phytopathogenic Xanthomonas bacteria are unimportant during pathogenesis but may be important for protection against photobiological damage. We used a Xanthomonas campestris pv. campestris parent strain, single-site transposon insertion mutant strains, and chromosomally restored mutant strains to define the biological role of xanthomonadins. Although xanthomonadin mutant strains were comparable to the parent strain for survival when exposed to UV light; after their exposure to the photosensitizer toluidine blue and visible light, survival was greatly reduced. Chromosomally restored mutant strains were completely restored for survival in these conditions. Likewise, epiphytic survival of a xanthomonadin mutant strain was greatly reduced in conditions of high light intensity, whereas a chromosomally restored mutant strain was comparable to the parent strain for epiphytic survival. These results are discussed with respect to previous results, and a model for epiphytic survival of X. campestris pv. campestris is presented.
Xanthomonas bacteria are the causal agents of disease on at least 124 monocot and 268 dicot plant hosts (12), and many of them can survive and multiply as epiphytes (8, 24). Most Xanthomonas bacteria produce yellow, membrane-bound, brominated aryl-polyene pigments referred to as xanthomonadins (24). Xanthomonadins are unique to Xanthomonas bacteria and serve as useful chemotaxonomic (2, 25) and diagnostic (22) markers. With methods of artificial infection, xanthomonadin-deficient strains were not affected in pathogenicity, symptomatology, or in planta growth (15). Thus, the xanthomonadins apparently are not important to the pathogen after infection of the host plant.
Xanthomonas campestris pv. campestris, the causal agent of black rot of crucifers and one of the most serious disease problems in crucifer production, naturally infects its host via hydathodes or wounds in the leaves (28). A cluster of seven transcriptional units required for xanthomonadin production (pigA to pigG) was previously identified in X. campestris pv. campestris (13, 15). In addition to a loss of xanthomonadin production, pigB mutant strains were also greatly impaired in the production of extracellular polysaccharide (EPS) and a pheromone (DF). Mutations in the other pig transcriptional units did not appear to have pleiotropic affects. When tested on the host plant, pigB mutants were significantly reduced in epiphytic survival and natural host infection via hydathodes (16). DF extracellularly restored all of these traits to a pigB mutant strain (4, 15, 16), indicating that DF is needed for xanthomonadin and EPS production, as well as for epiphytic survival and host infection. These results suggest that DF acts as a signal for the initiation of xanthomonadin and EPS production and that xanthomonadins and EPS may play a role in host infection and/or epiphytic survival.
The results of other studies suggest an association of xanthomonadins with protection against photobiological damage (11, 18). However, these studies were not conducted using modern methods of single gene mutation and chromosomal restoration, from which definitive conclusions can be drawn. In this study, we first use these techniques to demonstrate the role of xanthomonadins in the protection of X. campestris pv. campestris from photobiological damage. We then test whether xanthomonadins are needed by X. campestris pv. campestris for epiphytic survival and/or host infection.
MATERIALS AND METHODS
Bacterial strains and culture.
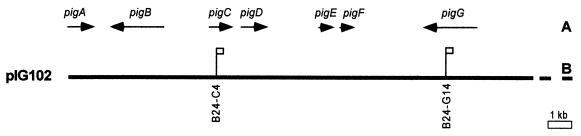
Strain B-24 is a pathogenic, cephalexin-resistant, wild-type strain of X. campestris pv. campestris which was previously used to clone and characterize the xanthomonadin-encoding region (pig) (13, 15). pIG102 (tetracycline resistant) is a pLAFR3 cosmid clone of the entire pig region which includes transcriptional units pigA to pigG (13, 15) (see Fig. 1). B24-C4 and B24-G14 are previously constructed pigC and pigG Tn3HoHo1 single insertion mutation strains (reference 15 and see Fig. 1). Chromosomally restored derivatives of mutant strains B24-C4 and B24-G14 were constructed by homologous recombination with pIG102 (Fig. 1) as previously described (16).
FIG. 1.
Cloned xanthomonadin encoding region (pIG102) from X. campestris pv. campestris. (A) Xanthomonadin transcriptional units. (B) Location of Tn3HoHo1 insertion mutations.
Xanthomonas strains were grown at 28°C in nutrient broth yeast extract (NBY) medium (27), nutrient starch agar (NSA) medium (21), or minimal medium (3) supplemented with 1 mg of methionine per liter (MAKC). Antibiotics were added to the medium when appropriate at the following concentrations: tetracycline (Tc), 12 mg/liter; cephalexin (Ce) and ampicillin (Ap), each at 50 mg/liter.
Biochemical and molecular techniques.
Previously described methods were used for conjugal plasmid transfer and xanthomonadin quantification (13), Southern hybridization (14), EPS quantification, and the determination of DF production (15).
Photosensitivity.
Bacterial strains were grown in MAKC broth medium and tested for sensitivity to UV light using previously described methods (5). A UV light source of 254 nm was used, and dilutions of exposed bacterial suspensions were plated on NSA agar medium. Bacterial colonies were then enumerated after 3 days of incubation at 28°C.
Bacterial strains were tested for sensitivity to visible light in the presence of the exogenous photosensitizer toluidine blue, which has an absorption maximum at 635 nm (9). Bacteria were grown in NBY broth overnight, washed in 0.01 M PO4 buffer (pH 7.0), and resuspended and diluted in PO4 buffer to a concentration of ca. 5 × 105 CFU/ml. Portions (5 ml) were then placed in 22-mm glass culture tubes, and toluidine blue was added to a final concentration of 5 μM. Tubes were shaken at 28°C and 200 rpm under a 400-W, high-pressure sodium lamp (10,000 lx) for 1.0 h. At 15-min intervals, dilutions of bacterial suspensions were plated on NSA agar medium, and bacterial colonies were subsequently enumerated after 3 days of incubation at 28°C.
Epiphytic survival and host infection.
In previous studies, we used sonication to remove bacteria from leaf surfaces (16). We found that this method provided accurate estimates of total epiphytic bacteria for up to 1 week after application. However, by 2 to 3 weeks after application, large pathogen populations developed which were not removed by sonication and were resistant to bleach treatment. This was the case even with a pigB mutant strain, which was unable to infect the host plant. However, even though not all epiphytic bacteria were removed by sonication, the sizes of the populations remaining on the leaf surfaces. Thus, these populations were referred to as easily removable epiphytic bacteria (EREB) and were taken to represent the total epiphytic population levels. We used the same methods in the present study; thus, our numbers reflect EREB population levels.
The procedures used for the growth of cauliflower seedlings, the preparation of bacterial suspensions, and the misting of plants with bacterial suspensions were as previously described (16). Misted plants were initially left in the dew chamber in the dark for 16 to 20 h, during which time continual leaf wetness was maintained. Plants were then incubated for 3 weeks in growth chambers at 25 to 30°C and 35 to 50% relative humidity or in the greenhouse at 20 to 30°C and 25 to 50% relative humidity. Plants in growth chambers were subjected to a photoperiod of 16 h with a light intensity of ca. 4,000 lx. In the greenhouse during the period of the summer solstice, plants were subjected to a natural photoperiod of ca. 16 h with an average light intensity of ca. 50,000 to 60,000 lx. There were five pots per treatment, and the pots were arranged in a completely randomized design. Upon transfer to growth chambers or the greenhouse and each week thereafter, the leaves were sampled for populations of the pathogen by plating sonicates on selective medium as previously described (16). The data were log transformed, and the means and the standard errors of the mean were then calculated and plotted. Three weeks after the spraying of the plants, the numbers of typical black-rot lesions found originating on the periphery of plant leaves were recorded.
RESULTS
Strain development and characterization.
Previously constructed pigC (B24-C4) and pigG (B24-G14) Tn3HoHo1 insertion mutant strains (reference 15 and Fig. 1) were characterized with respect to xanthomonadin, EPS, and DF pheromone production. These mutant strains appeared colorless, but with our assay xanthomonadin pigment production was about 10% of that observed with the parent strain B-24 (Table 1). This low level of absorbance may have been due to the extraction and absorbance of biosynthetic intermediates. Both B24-C4 and B24-G14 were comparable to the parent strain in EPS and DF pheromone production (Table 1). No other phenotypic differences were observed between these strains and the parent strain. Strains B24-C4, B24-G14, and cosmid pIG102 (Fig. 1) were used to construct the chromosomally restored mutant strains B24-C4R and B24-G14R. Replacement of Tn3HoHo1 insertion DNA with functional DNA from pIG102 was verified by Southern hybridization (data not shown). B24-C4R and B24-G14R were fully restored for xanthomonadin production and were comparable to the parent strain for EPS and DF pheromone production (Table 1).
TABLE 1.
Phenotypic traits of the X. campestris pv. campestris strains used in this study
| Strain | Genotype | Pigmentsa | EPSb (mg/ml) | DFc | Lesions/plantd |
|---|---|---|---|---|---|
| B-24 | pig+ | 0.37 a | 1.59 a,b,c | Positive | 2.6 a |
| B24-C4 | pigC::Tn3HoHo1 | 0.04 b | 1.64 a,b | Positive | 1.4 b |
| B24-C4R | pig+ (Tn3HoHo1−) | 0.32 a | 1.52 a,b,c | Positive | 2.2 a,b |
| B24-G14 | pigG::Tn3HoHo1 | 0.05 b | 1.43 a,c | Positive | ND |
| B24-G14R | pig+ (Tn3HoHo1−) | 0.34 a | 1.47 a,b,c | Positive | ND |
As determined by absorbance measurements at 441 nm of boiled methanol extracts. Values are averages of three replicates, and values followed by the same lowercase letter were not significantly different, as determined by Fisher's protected least significant difference (LSD) (P = 0.05). The experiment was performed twice with similar results.
EPS production as determined by quantification with Anthrone reagent of CTAB (cetyltrimethylammonium bromide) and ethanol precipitates. Values are the averages of three replicates, and values followed by the same lowercase letter were not significantly different, as determined by Fisher's protected LSD (P = 0.05). The experiment was performed twice with similar results.
Production of the Xanthomonas pheromone DF, as determined by adjacent streaking with the indicator strain B24-B2. The experiment was performed twice with similar results.
Values are the averages of five replicates, and those followed by the same letter were not significantly different, as determined by Fisher's protected LSD (P = 0.05). The experiment was performed twice with similar results. ND, not determined.
Sensitivity to photobiological damage.
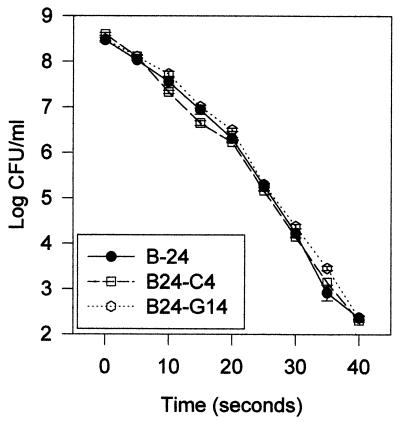
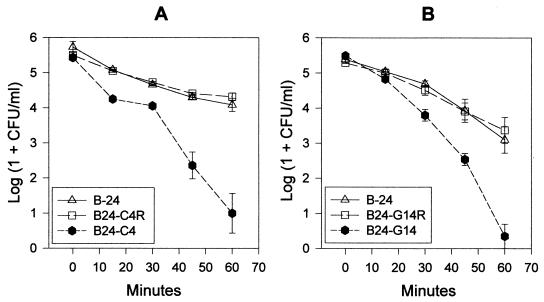
When exposed to UV irradiation, populations of parent strain B-24 decreased from ca. 8.5 to 2.4 log CFU/ml over a period of 40 s (Fig. 2). The survival of xanthomonadin mutant strains B24-C4 and B24-G14 was nearly identical to that of parent strain B-24 (Fig. 2). However, when exposed to visible light for 60 min in the presence of the photosensitizing agent toluidine blue, both xanthomonadin mutant strains were reduced ca. 1,000-fold in survival compared to the parent strain (Fig. 3). Chromosomally restored mutant strains B24-C4R and B24-G14R were fully restored to parent strain levels for survival under these conditions. Neither the mutant nor the parent strains were sensitive to toluidine blue in the absence of light or sensitive to light (10,000 or 80,000 lx) in the absence of toluidine blue (data not shown).
FIG. 2.
Survival of X. campestris pv. campestris parent (B-24) and xanthomonadin mutant (B24-C4 and B24-G14) strains in the presence of UV irradiation. Values are the means of three replicates, and the bar at each datum point shows the standard error of the mean. The experiment was repeated twice with similar results.
FIG. 3.
Survival of X. campestris pv. campestris strains in the presence of visible light and toluidine blue. (A) Parent (B-24), xanthomonadin pigC mutant (B24-C4), and restored mutant (B24-C4R) strains. (B) Parent (B-24), xanthomonadin pigG mutant (B24-G14), and restored mutant (B24-G14R) strains. Values are the means of five replicates, and the bar at each datum point shows the standard error of the mean. The experiment was repeated twice with similar results.
Epiphytic survival.
Since the two-xanthomonadin mutant strains appeared to be similar in sensitivity to photobiological damage, mutant strain B24-C4 and its chromosomally restored derivative B24-C4R were selected for further experiments. Replicated growth chamber experiments were conducted to compare strain B24-C4 to parent strain B-24 for epiphytic survival and infection frequency. In all cases, the survival and infection frequency of strain B24-C4 was not significantly different from that of the parent strain (data not shown). However, when these experiments were conducted in the greenhouse environment, different results were observed. The epiphytic survival of strains B-24 and B24-C4R was similar, but the epiphytic survival of the xanthomonadin mutant strain B24-C4 was reduced ca. 100- to 1,000-fold at 1 and 2 weeks after application (Fig. 4). The host infection frequency of strain B24-C4 was ca. 60% of that observed with the other two strains, and this difference was significant (Table 1).
FIG. 4.
Epiphytic survival of X. campestris pv. campestris parent (B-24), xanthomonadin mutant (B24-C4), and restored mutant (B24-C4R) strains in the greenhouse. Values are the means of five replicates, and the bar at each datum point shows the standard error of the mean. The experiment was repeated twice with similar results.
DISCUSSION
We used a X. campestris pv. campestris parent strain, single-site transposon insertion mutant strains, and chromosomal restoration of mutant strains to test the biological role of xanthomonadin pigments. pigC and pigG transposon insertion mutant strains were affected only in xanthomonadin production and not in EPS or DF pheromone production. Our methods of transposon mutagenesis result in a single mutation per strain (15, 16, 23), whereas chemical mutagenesis commonly results in multiple mutations per strain. This latter situation can confuse the assignment of phenotypic changes to particular mutational events. The restoration of mutant strains by plasmid-borne genes can also cause problems in interpretation. Most plasmids are multicopy; thus, the complementing gene will be present in an unnaturally high copy number and could have gene dosage affects. In addition, many plasmid vectors are unstably maintained in their bacterial hosts, resulting in only a small segment of the population harboring the plasmid. This can result in incomplete restoration of the phenotype and thus inability to determine whether the problem is due to plasmid loss or incomplete gene complementation.
Solar UV radiation consists of UVC (190- to 290-nm), UVB (290- to 320-nm), and UVA (320- to 400-nm) wavelengths (10). The biological mechanisms of action of UVC and UVA wavelengths are quite different, whereas those of UVB represent a mixture of UVC and UVA mechanisms (10). The carotenoid pigments are well known for their ability to protect various organisms against photo-oxidative damage resulting from exposure to UV radiation (1). In bacteria, cloned genes for the production of various carotenoids have been used to transform Escherichia coli to tolerance to UVA or UVB light damage in the presence or absence of photosensitizers (20, 26). When we compared our two X. campestris pv. campestris xanthomonadin mutant strains to their parent strain for sensitivity to UVC light damage, the three strains were almost identical. One xanthomonadin mutant strain was also compared to the parent for susceptibility to UVA light damage, and again they were not significantly different (G. Sundin, personal communication). Thus, xanthomonadins didn't protect the bacterium against any of the mechanisms of UV radiation damage, and it is likely that they don't play a role in protection against direct UV damage. However, these results do not preclude the possibility that xanthomonadins may protect against indirect UV damage, mediated by an unknown photosensitizing agent on the leaf surface.
Previous studies have indicated that xanthomonadins may protect against visible light damage in the presence of photosensitizers (11, 18). In the first study, a single, chemically induced mutant strain of X. juglandis was more sensitive than its parent strain to photosensitizer-mediated photobiological damage. However, restoration of the mutant strain with a cloned xanthomonadin gene was not attempted in this study. In the second study, chemically induced, xanthomonadin mutant strains of X. oryzae pv. oryzae were decreased ca. 100-fold in survival compared to the parent strain in the presence of light and toluidine blue. Restoration of these strains for tolerance to light damage was attempted with xanthomonadin genes cloned in a plasmid vector; however, this complemented mutant strain was still ca. 10-fold reduced in survival compared to the parent strain. Multiple mutations, complementing gene copy number effects, plasmid instability, or the use for complementation of a gene that did not correspond to the original mutated gene, could explain the incomplete restoration of this phenotype. Thus, we repeated these experiments with X. campestris pv. campestris and methods from which definitive conclusions could be drawn. In the presence of toluidine blue and light, our Tn3HoHo1 xanthomonadin mutant strains were ca. 1,000-fold decreased in survival compared to the parent strain, and our chromosomally restored mutant strains were indistinguishable from the parent strain for survival. Thus, xanthomonadins do provide protection from visible light damage in the presence of the exogenous photosensitizer, toluidine blue. Since toluidine blue does not penetrate past the cell membrane and since its damaging effects are due to the generation of reactive oxygen species in the presence of visible light, its targets are thought to be lipid components of the cell membrane (9). In addition, the membrane-bound xanthomonadins have been shown to protect lipids from peroxidation (18). Thus, it is likely that xanthomonadins protected the bacterial membrane from reactive oxygen species generated by exposure to visible light and toluidine blue.
The X. campestris pv. campestris pigC xanthomonadin mutant strain was negative for xanthomonadin production but positive for both EPS and DF pheromone production. Under conditions of high light intensity, this strain was impaired in epiphytic survival, whereas its chromosomally restored derivative was similar to the parent strain. Under conditions of low light intensity, the pigC xanthomonadin mutant strain was similar to the parent strain for epiphytic survival. These results indicate that xanthomonadins are needed for epiphytic survival, but only under high, natural light conditions. Plants produce a wide variety of photosensitizing agents (6). Thus, on the leaf surface, xanthomonadins could protect the pathogen from photobiological damage in the presence of an unknown, host-produced photosensitizer, which has a mode of action similar to that of toluidine blue. If this were the case, it is unlikely that the unknown photosensitizer would be activated by UV radiation, since the Polygal polycarbonate material of the greenhouse allows only 5 to 10% transmission of wavelengths below 385 nm (Spec-Data Sheet; Polymark, Inc.).
Previously, pigB mutant strains exhibited an infection frequency which was only 2.6% that of the parent strain (16), whereas in this study, the pigC mutant strain infection frequency was ca. 60% that of the parent or restored mutant strains. Thus, although the loss of xanthomonadins appears to have a measurable effect on infection frequency, it cannot account for the large reduction observed with pigB mutant strains. This suggests that some other pigB-DF-regulated trait must be associated with host infection.
The time course of xanthomonadin production and the effect of exogenous DF on this time course (17) is consistent with DF regulating xanthomonadin production in a cell density-dependent manner, referred to as autoinduction or quorum sensing (see references 7 and 19 for reviews). During growth of the bacterial population the autoinducer accumulates in the surrounding environment until a specific concentration is achieved which triggers expression of the structural genes for the regulated trait. Thus, we propose the following model for the epiphytic survival of X. campestris pv. campestris. When single cells or small aggregates of the pathogen arrive on the plant surface, low cell densities result in a level of DF which is insufficient to trigger xanthomonadin production. These cells are susceptible to lethal damage from the combination of high light conditions and host-produced photosensitizers. Under favorable conditions (low light), some cells survive, and pathogen populations now begin to expand. Subsequently, the extracellular concentrations of DF reach a level sufficient to induce xanthomonadin production. The pathogen populations are now able to withstand conditions of high light intensity and survive for long periods to provide a source of pathogen propagules when host infection conditions become favorable. Future studies will test this model and possibly identify other epiphytic fitness traits that are controlled by pigB and DF.
ACKNOWLEDGMENTS
This work was supported in part by grant 9304168 from USDA/NRICGP.
We acknowledge the technical assistance of Cole Bryngelson and helpful discussions with George Sundin.
REFERENCES
- 1.Armstrong G A. Eubacteria show their true colors: genetics of carotenoid pigment biosynthesis from microbes to plants. J Bacteriol. 1994;176:4795–4802. doi: 10.1128/jb.176.16.4795-4802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradbury J F. Genus II. Xanthomonas Dowson 1939, 187.AL. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. I. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 199–210. [Google Scholar]
- 3.Chatterjee A K. Acceptance by Erwinia spp. of R plasmid R68.45 and its ability to mobilize the chromosome of Erwinia chrysanthemi. J Bacteriol. 1980;142:111–119. doi: 10.1128/jb.142.1.111-119.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun W, Cui J, Poplawsky A. Purification, characterization, and biological role of a pheromone produced by Xanthomonas campestris pv. campestris. Physiol Mol Plant Pathol. 1997;51:1–14. [Google Scholar]
- 5.Clowes R C, Hayes W, editors. Experiments in microbial genetics. Oxford, England: Blackwell Scientific Publications; 1968. pp. 13–16. [Google Scholar]
- 6.Downum K R, Wen J. The occurrence of photosensitizers among higher plants. In: Heitz J R, Downum K R, editors. Light-activated pest control. ACS Symposium series 616. Washington, D.C.: The American Chemical Society; 1995. pp. 135–143. [Google Scholar]
- 7.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirano S S, Upper C D. Ecology and epidemiology of foliar bacterial plant pathogens. Annu Rev Phytopathol. 1983;21:243–269. [Google Scholar]
- 9.Itoh T. Toluidine blue: the mode of photodynamic action in yeast cells. Photochem Photobiol. 1977;25:47–53. doi: 10.1111/j.1751-1097.1977.tb07423.x. [DOI] [PubMed] [Google Scholar]
- 10.Jagger J. Solar-UV actions on living cells. New York, N.Y: Praeger Publishers; 1985. [Google Scholar]
- 11.Jenkins C L, Starr M P. The brominated aryl-polyene (xanthomonadin) pigments of Xanthomonas juglandis protect against photobiological damage. Curr Microbiol. 1982;7:323–326. [Google Scholar]
- 12.Leyns F, DeCleene M, Swings J G, Deley J. The host range of the genus Xanthomonas. Bot Rev. 1984;50:308–356. [Google Scholar]
- 13.Poplawsky A R, Kawalek M D, Schaad N W. A xanthomonadin-encoding gene cluster for the identification of pathovars of Xanthomonas campestris. Mol Plant-Microbe Interact. 1993;6:545–552. [Google Scholar]
- 14.Poplawsky A R, Chun W. Strains of Xanthomonas campestris pv. campestris with atypical pigmentation isolated from commercial crucifer seeds. Plant Dis. 1995;79:1021–1024. [Google Scholar]
- 15.Poplawsky A R, Chun W. pigB determines a diffusible factor needed for EPS and xanthomonadin production in Xanthomonas campestris pv. campestris. J Bacteriol. 1997;179:439–444. doi: 10.1128/jb.179.2.439-444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poplawsky A R, Chun W. Xanthomonas campestris pv. campestris requires a functional pigB for epiphytic survival and host infection. Mol Plant-Microbe Interact. 1998;11:466–475. doi: 10.1094/MPMI.1998.11.6.466. [DOI] [PubMed] [Google Scholar]
- 17.Poplawsky A R, Chun W. The Xanthomonas campestris pv. campestris DF pheromone and additional regulatory functions of the pig gene cluster. Phytopathology. 1999;89:S61. [Google Scholar]
- 18.Rajagopal L, Sundari C S, Balasubramanian D, Sonti R V. The bacterial pigment xanthomonadin offers protection against photodamage. FEBS Lett. 1997;415:125–128. doi: 10.1016/s0014-5793(97)01109-5. [DOI] [PubMed] [Google Scholar]
- 19.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. The bacterial 'enigma': cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 20.Sandmann G, Kuhn S, Boger P. Evaluation of different carotenoids in Escherichia coli transformants as protectants against UV-B radiation. Appl Environ Microbiol. 1998;64:1972–1974. doi: 10.1128/aem.64.5.1972-1974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaad N W, Kendrick R. A qualitative method for detecting Xanthomonas campestris in crucifer seed. Phytopathology. 1975;65:1034–1036. [Google Scholar]
- 22.Schaad N W, Stall R E. Xanthomonas. In: Schaad N W, editor. Laboratory guide for identification of plant pathogenic bacteria. 2nd ed. St. Paul, Minn: The American Phytopathological Society; 1988. pp. 81–94. [Google Scholar]
- 23.Stachel S E, An G, Flores C, Nester E W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starr M P. The genus Xanthomonas. In: Starr M P, Stolp H, Truper H G, Balows A, Schlegel H G, editors. The prokaryotes. I. Berlin, Germany: Springer-Verlag; 1981. pp. 742–763. [Google Scholar]
- 25.Starr M P, Jenkins C L, Bussey L B, Andrewes A G. Chemotaxonomic significance of the xanthomonadins, novel brominated aryl-polyene pigments produced by bacteria of the genus Xanthomonas. Arch Microbiol. 1977;113:1–9. doi: 10.1007/BF00428572. [DOI] [PubMed] [Google Scholar]
- 26.Tuveson R W, Larson R A, Kagan J. Role of cloned carotenoid genes expressed in Escherichia coli in protecting against inactivation by near-UV light and specific phototoxic molecules. J Bacteriol. 1988;170:4675–4680. doi: 10.1128/jb.170.10.4675-4680.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidaver A K. Synthetic and complex media for rapid detection of fluorescence of phytopathogenic pseudomonads: effect of carbon source. Appl Microbiol. 1967;15:1523–1524. doi: 10.1128/am.15.6.1523-1524.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams P H. Black rot: a continuing threat to world crucifers. Plant Dis. 1980;64:736–742. [Google Scholar]