Abstract
Lactic acid bacteria are nutritionally demanding bacteria which need, among other things, amino acids for optimal growth. We identified the branched-chain amino acid (BCAA) biosynthesis pathway as an essential pathway for optimal growth of Streptococcus thermophilus in milk. Through random insertional mutagenesis, we isolated and characterized two mutants for which growth in milk is affected as a consequence of ilvB and ilvC gene interruptions. This situation demonstrates that the BCAA biosynthesis pathway is active in S. thermophilus. BCAA biosynthesis is necessary but not sufficient for optimal growth of S. thermophilus and is subject to retro-inhibition processes. The specificity of the BCAA biosynthesis pathway in S. thermophilus lies in the independent transcription of the ilvC gene encoding a keto acid reductoisomerase acting on acetolactate at the junction of the BCAA and acetoin biosynthesis pathways. The possible advantages for S. thermophilus of keeping this biosynthesis pathway active could be linked either to adaptation of the organism to milk, which is different than that of other dairy bacteria, or to the role of the pathway in maintaining the internal pH.
The search for nutriments and especially for amino acids constitutes a real challenge for lactic acid bacteria. These organisms are auxotrophic for several amino acids which they cannot synthesize from simpler nitrogen sources (6, 8, 26). Only proteinase-positive lactic acid bacteria, which are capable of hydrolyzing caseins, usually grow significantly in milk, which contains only small amounts of amino acids and short peptides (39).
Most of the functions identified as being essential for optimal growth of Lactococcus lactis in milk concern the amino acid supply. Quite some time ago, Thomas and Mills (39) underlined the importance of the cell wall-anchored lactococcal proteinase (PrtP) to growth in milk. If the proteinase is absent, caseins are not hydrolyzed and L. lactis cell density reaches only 10% of that of a proteinase-positive (Prt+) strain. The cell wall proteinase liberates oligopeptides from caseins, some of which are transported into the cytoplasm by the oligopeptide transport system. This transport system is essential for optimal growth of L. lactis in milk (44). When internalized, the oligopeptides are further hydrolyzed into amino acids by intracellular peptidases. The most important peptidases in this process have been identified by using negative mutants. Suppression of these enzymes significantly reduces the L. lactis growth rate in milk (24). L. lactis can also synthesize amino acids, at least to some extent. The aspartate biosynthesis pathway is active in L. lactis, and lack of this pathway reduces the growth rate in milk to half the normal rate (45).
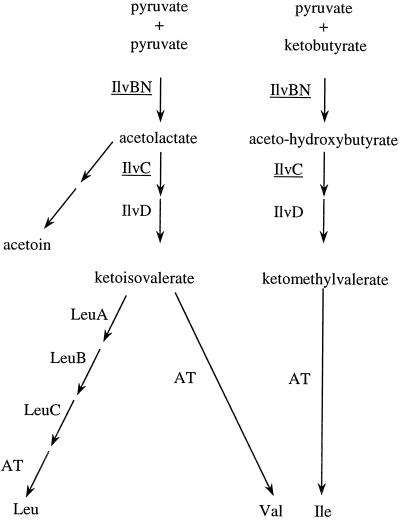
In the present work we identified, for the first time, the branched-chain amino acid (BCAA) biosynthesis pathway as a key pathway for optimal growth of Streptococcus thermophilus in milk. Since the BCAA are a quantitatively important group of amino acids in bacterial proteins (they account for 20% of the total protein amino acids in Escherichia coli and L. lactis [25, 27]), the BCAA biosynthesis pathways have been extensively studied and are well-known in several bacterial genera. They share the following features: first, they involve three enzymes common to the three amino acid synthesis pathways; and second, they are composed of steps which are specific to leucine and isoleucine-valine biosynthesis. In addition, some metabolic intermediates are linked to other metabolic pathways, such as acetoin, butanediol, and coenzyme A synthesis (Fig. 1). As a consequence, the BCAA biosynthesis pathway is under complex control, including retro-inhibition processes and regulation at the transcription level (5, 11, 14, 32, 42).
FIG. 1.
Branched-chain biosynthesis of the three BCAA. IlvBN, acetolactate synthase; IlvC, keto acid reductoisomerase; IlvD, dihydroxy acid dehydratase; LeuA, isopropyl malate synthase; LeuB, isopropyl malate dehydrogenase; LeuC, isopropyl malate dehydratase; AT, aminotransferase. The proteins whose encoding genes have been disrupted are underlined.
Investigation of the amino acid requirements of L. lactis by using a single-omission technique revealed that the dairy lactococcal strains are auxotrophic for at least six amino acids (Glu, Met, Leu, Ile, Val, and His) while lactococci from vegetal origins are prototrophic for all amino acids (5, 6). The amino acid auxotrophies, including those for BCAA, are due to minor genetic lesions that, in most cases, are reparable by single-step mutations (8, 13).
The thermophilic bacterium S. thermophilus requires fewer amino acids than lactococci and lactobacilli (9). Only glutamine and glutamic acid, along with the sulfur amino acids, are essential for all of the strains that have been tested (3, 28; data not shown). This situation could be attributed to active amino acid biosynthesis pathways not yet described for this species. In the present work, we demonstrated that the S. thermophilus BCAA synthesis pathway is functional, while the BCAA synthesis pathway is not functional in L. lactis of dairy origin. Moreover, this biosynthesis pathway is essential for optimal growth of S. thermophilus in milk.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
E. coli TG1 RepA+ (TG1, which contains a chromosomal copy of the repA gene, was kindly provided by P. Renault and referenced as TIL206) and E. coli TG1 RepA+ containing the pG+h9::ISS1 plasmid (29) (= strain TIL401) were used. Both strains were grown on Luria-Bertani medium (35) at 37°C with shaking and in the presence of erythromycin (150 μg/ml) when required. S. thermophilus Prt+, plasmid-free strain St18 was provided by Rhodia-Food (Dangé Saint-Romain, France). Four media were used for cultures of S. thermophilus. Two of them were milk based and were used specifically for screening mutants. The first medium was Fast Slow Difference Agar (FSDA) (19), which contained erythromycin (5 or 3 μg/ml) when it was needed. This medium is a milk-based agar medium, which made it possible to differentiate bacteria that exhibited slow or limited growth in milk from bacteria that exhibited rapid or optimal growth after 48 h of incubation at 37°C under anaerobic conditions. The second medium was reconstituted low-heat 10% (wt/vol) skim milk (Nilac; Nederlands Instituut von Zuivelonderzoek, Ede, The Netherlands) that was autoclaved at 110°C for 12 min, buffered with 2.5% 3 M sodium glycerophosphate, and in some cases contained 3 g of Bacto Tryptone (pancreatic digest of casein; Difco Laboratories, Detroit, Mich.) per liter. Bacterial growth was monitored by measuring the optical density at 480 nm (OD480) after clarification of milk by 10-fold dilution in a solution containing 2 g of EDTA (pH 12) per liter (40). Two other media were used for general manipulation and growth rate experiments. The first of these was M17Lac medium (38), in which bacterial growth was monitored by measuring the OD600. The second was a chemically defined medium (CDM) containing nucleotides, vitamins, amino acids, salts, potassium phosphate buffer (pH 6.7), and 1% (wt/vol) lactose (31), which was sterilized by filtration. When required, in some CDM growth experiments isoleucine, leucine, or valine was omitted or the corresponding precursor keto acids, β-ketoisovalerate and ketomethylvalerate, were added at concentrations of 0.8 and 0.2 g/liter, respectively (Fig. 1). Growth rate experiments were then performed at 37°C with a Microbiology Reader Bioscreen C (Labsystems, Helsinki, Finland) in 100-well sterile covered microplates. Each well contained 200 μl of culture medium. Overnight M17Lac cultures of S. thermophilus were washed twice and resuspended in a volume of sterile phosphate buffer equal to the culture volume. Four microliters of the suspension was used to inoculate each well. The OD600 was measured every 20 min after gentle shaking. The apparent growth rate was defined as the maximum slope of the semi-logarithmic graph of growth determined by measuring optical density.
S. thermophilus mutagenesis.
The method used for insertional mutagenesis with pG+h9::ISS1 in S. thermophilus St18 was adapted from the method previously described by Maguin et al. (22). Plasmid pG+h9::ISS1 was first purified from E. coli TIL401. S. thermophilus St18 was transformed by electroporation (18) with 1 μg of purified pG+h9::ISS1, and plasmid-containing bacteria were selected on M17Lac medium containing erythromycin (5 μg/ml) at 28°C under anaerobic conditions. Integration was performed as follows. A saturated overnight culture of an erythromycin-resistant (Emr) colony containing pG+h9::ISS1 was cultivated in M17Lac medium supplemented with erythromycin (5 μg/ml) and then diluted 1:100 with fresh M17Lac medium without erythromycin and incubated at 28°C for 2.5 h. To reduce the plasmid copy number, the culture was incubated at 42°C for another 2.5 h. The culture was diluted and plated on FSDA in the presence or in the absence of erythromycin and incubated at 42°C (under anaerobic conditions) to induce chromosomal integration of the plasmid. At this step, the concentration of erythromycin was only 3 μg/ml to limit tandem insertion of pG+h9::ISS1. Emr mutants were selected after 24 h. To excise transposed pG+h9::ISS1 and obtain stable mutants, a method similar to that described previously (22) was used.
Selection of mutants whose growth in milk was affected.
Mutants whose growth in milk was affected were selected in two steps. The first selection was made on FSDA. On this medium, colonies whose growth in milk was affected remained small and translucent while the colonies with normal growth were large and white. This first selection was confirmed by comparing the growth of mutants in milk to that of the wild-type strain.
DNA manipulation and sequencing.
Plasmid DNA manipulation and transformation of E. coli TIL206 were performed as previously described (35). RNA was prepared by using S. thermophilus grown in M17Lac medium. The DNAs of mutants were digested by EcoRI or HindIII and ligated. TIL206 electrocompetent cells were transformed with ligation products, and Emr colonies were screened by PCR after 24 h of incubation at 37°C. PCR amplifications were performed with a Gene Amp 2400 PCR system (Perkin-Elmer Corp., Norwalk, Conn.) by using Taq polymerase (Appligene Oncor, Illkirch, France) and oligonucleotides from the pG+h9::ISS1 sequences (5′ ACT ACT GAC AGC TTC CAA GGA 3′ and 5′ ATA GTT CAT TGA TAT ATC CTC 3′ for EcoRI digestion and 5′ GTA AAA CGA CGG CCA GTG 3′ and 5′ TAT CTA CTG AGA TTA AGG TCT 3′ for HindIII digestion). A dye terminator kit and a 310 genetic analyzer (Applied Biosystems, Foster City, Calif.) were used for DNA sequencing; each strand was sequenced twice by using independent PCR products. DNA sequences were analyzed with Genetics Computer Group sequence analysis software from the University of Wisconsin (10) and Mail Fasta (National Center for Biotechnology Information). Southern and Northern hybridizations were performed by using a positively charged nylon membrane (Appligene Oncor) for transfer according to the instructions for the ECL detection system (Amersham, Buckinghamshire, England).
Nucleotide sequence accession number.
The GenBank, EMBL, and DDBJ nucleotide sequence accession number for a 1,955-bp partial sequence of the ilvBNC operon of S. thermophilus St18 is AF220670.
RESULTS
Set of S. thermophilus mutants resulting from random mutagenesis.
The transformation yield of S. thermophilus St18 (72 transformants/μg of pG+h9::ISS1) was low but comparable to that which has been previously described for other S. thermophilus strains (23). We obtained 1.183 × 104 Emr mutants on FSDA. The integration frequency (i.e., the ratio of the number of Emr mutants to the total number of clones) was 7.7 × 10−3, a value which is quite similar to that obtained for L. lactis (22). On the basis of their phenotypes on FSDA and their slow growth in milk, we isolated 72 integrants.
Characterization of two mutants that exhibited slower growth in milk.
After Southern analysis of digested chromosomal DNAs of the mutants that grew slowly in milk, we selected 14 clones in which pG+h9::ISS1 was integrated at only one locus, and the locus was distinct for each clone. In 12 of these clones, pG+h9::ISS1 was tandemly integrated, which gave two hybridization bands when pG+h9 was used as a probe. These two observations are illustrated in Fig. 2 for two of the mutants, mutants 1 and 2, which were characterized further in the present work.
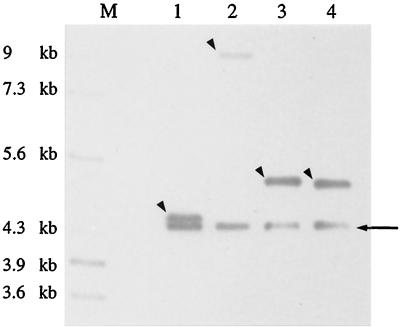
FIG. 2.
Southern analysis of pG+h9::ISS1 mutants 1 and 2. Chromosomal DNAs of mutants were digested by EcoRI (lanes 1 and 3) or HindIII (lanes 2 and 4) and probed with pG+h9. Two bands were observed for mutants 1 and 2 (lanes 1 and 2 and lanes 3 and 4, respectively), which corresponded to tandem transposition. One band corresponded to pG+h9::ISS1 (arrow), and the other band corresponded to the integrated structure of the chromosome (arrowheads). No hybridization was observed with the wild-type strain DNA (data not shown). The RAOUL marker (Appligene) was used as a size reference (lane M).
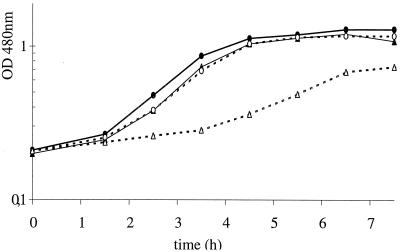
Mutants 1 and 2 had similar growth curves in both milk and milk with Bacto Tryptone (Fig. 3). Their growth rates in milk (0.37 h−1) were significantly lower than that of the wild-type strain (0.73 h−1). Rapid growth was restored by addition of Bacto Tryptone (growth rate for the mutants 0.71 h−1; growth rate for the wild-type strain, 0.85 h−1), suggesting that the affected functions were related to nitrogen nutrition.
FIG. 3.
Comparison of the growth curves of wild-type strain St18 in milk (○) and in milk containing 3 g of Bacto Tryptone per liter (●) with the growth curves of mutant 1 in milk (▵) and in milk containing 3 g of Bacto Tryptone per liter (▴).
Identification of disrupted genes as BCAA biosynthesis genes.
Sequences of the interrupted genes were determined by PCR with oligonucleotides from pG+h9::ISS1. We obtained 116- and 1,419-bp sequences for mutants 1 and 2, respectively. A search for homologues in databases revealed that the two mutants were affected in the same BCAA biosynthesis pathway. The ilvB and ilvC genes, coding for the large subunit of acetolactate synthase and keto acid reductoisomerase, were interrupted in mutants 1 and 2, respectively (Fig. 1). Using oligonucleotides corresponding to the extremities of the two sequences obtained from mutants 1 and 2, we performed additional PCRs. We obtained a unique 1,955-bp DNA fragment that included the sequences from mutants 1 and 2 and contained three open reading frames (ORFs) (partial ilvB gene, whole ilvN gene encoding the small subunit of acetolactate synthase, and whole ilvC gene). Protein sequences deduced from the whole DNA sequence showed the highest homology with the sequences of similar proteins from L. lactis (46% identity for IlvN to 78% identity for IlvC) (14), Bacillus subtilis (44% identity for IlvN to 58% identity for IlvC) (30), and Leuconostoc mesenteroides subsp. cremoris (50% identity for IlvB to 52% identity for IlvC) (4).
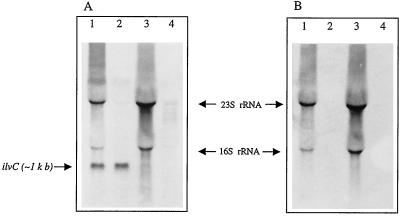
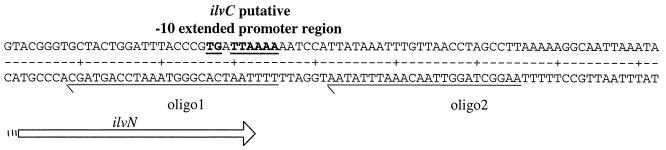
Analysis of the sequence revealed the presence of a putative −10 extended promoter sequence and two short inverted repeat sequences that were 72 and 50 bp, respectively, upstream of the ATG start codon of ilvC and the presence of a putative terminator 19 bp downstream of the stop codon of the same sequence (Fig. 4). Downstream of the ilvC gene, we found a partial ORF encoding a potential protein with 51% identity to tyrosyl tRNA synthetase of B. subtilis (17). No trace of leu genes was found in a 417-bp sequence located 3.5 kb upstream of ilvB from S. thermophilus. No ORF encoding proteins with similarity to either the IlvY regulator from E. coli (46) or the α-acetolactate decarboxylase present in L. lactis (15) were found in the vicinity of ilvC. After RNA was prepared from strain St18 grown in M17Lac medium or CDM containing all of the amino acids, Northern blot analysis revealed the presence of a 1,070-bp transcript that hybridized with the 1,955-bp ilvBNC sequence and, more precisely, with the 1,070-bp ilvC sequence (Fig. 5A). As expected, the 1,070-bp transcript was not visible when the same experiment was done with the IlvC-negative mutant (Fig. 5A). In this case, only the two bands with unusual shapes, probably resulting from larger degraded ilv transcripts stacked with 23S and 16S rRNAs, were visible. Similar bands have been observed previously for other large transcripts (21). An ilvBN probe ending at the putative ilvC promoter (oligo1) (Fig. 6) did not hybridize with the 1,070-bp transcript (Fig. 5B), while a 30-bp-longer probe obtained with oligo2 (Fig. 6) gave a slight hybridization signal (data not shown). Consequently, the start of transcription is most probably located not far downstream of the ilvC promoter. This result demonstrates that the potential promoter and terminator sequences identified upstream and downstream of ilvC are functional (Fig. 4). In all cases, larger transcripts containing at least the ilvBN sequence were not visualized, probably due to their instability.
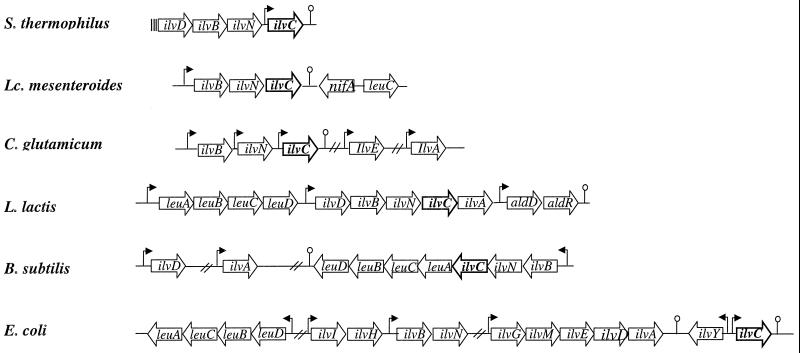
FIG. 4.
Organization of the ilv and leu genes in different bacteria. , putative promoter; , putative terminator; |||, partial ORF; ⫽, interrupted sequence. Data for S. thermophilus were obtained from this study; data for L. mesenteroides, C. glutamicum, L. lactis, B. subtilis, and E. coli were obtained from references 4, 7, 14, 30, and 2, respectively.
FIG. 5.
Northern analysis of the S. thermophilus wild-type (lanes 1 and 2) and IlvC-negative mutant (lanes 3 and 4) RNAs. RNAs were prepared from bacteria grown in M17lac medium (lanes 1 and 3) or CDM containing all of the amino acids (lanes 2 and 4). ilvC and ilvBN probes were used in the experiments whose results are shown in panels A and B, respectively. RNA Marker 0.2-10 kb (Sigma) was used as a size reference.
FIG. 6.
Schematic representation of the positions of the oligonucleotides (oligo1 and oligo2) used to amplify the ilvBN fragments used as probes for the Northern analysis.
Working of the ilv biosynthesis pathway in S. thermophilus.
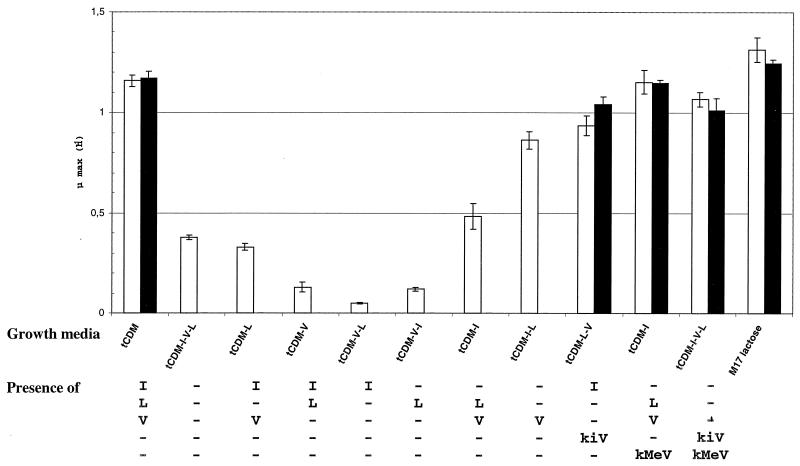
To understand the general working of the biosynthesis pathway in S. thermophilus, we performed growth experiments with CDM lacking Ile, Leu, and Val or containing the corresponding precursor keto acids and compared the growth rates of mutant 1 or 2 and the wild-type strain (Fig. 7). The wild-type strain grew in all media, which demonstrated that the BCAA biosynthesis pathway works in S. thermophilus. However, growth of the wild-type strain was limited when the three BCAA were absent, showing that the BCAA biosynthesis pathway is necessary but insufficient to ensure optimal growth of S. thermophilus in the absence of BCAA. In contrast, mutant 1 or 2 did not grow if one of the BCAA was missing.
FIG. 7.
Growth rate (μ max) obtained with the wild-type strain (open bars) and mutant 1 or 2 (solid bars) in CDM in which the nitrogen source was variable. The data are averages obtained from four independent experiments. tCDM is CDM which contained all of the amino acids. I, isoleucine; L, leucine; V, valine; kiV, ketoisovalerate; kMeV, ketoisomethylvalerate.
Variations in the BCAA content of the medium changed the growth rate of S. thermophilus. The presence of Ile (without Val and Leu) or Leu (without Val and Ile) in CDM decreased the growth rate of the wild-type strain compared to that in CDM without any BCAA. Similarly, addition of Ile or Leu to CDM containing Val as the sole BCAA decreased the growth rate of the strain. The presence of valine did not inhibit growth of the wild-type strain. No differences in the growth rates of the wild-type and mutant strains were observed in rich media or in CDM without BCAA but with intermediate precursors. These results confirm that the second part of the BCAA biosynthesis pathway involving leu genes products and aminotransferases is functional in the mutants (Fig. 1).
DISCUSSION
Regulation and physiological significance of the BCAA biosynthesis pathway.
We showed that the BCAA biosynthesis pathway is active in S. thermophilus, just as it is in nondairy L. lactis strains, as shown previously (14). This phenotype seems to be widespread in S. thermophilus since 12 strains whose nutritional requirements were tested were all capable of growing in the absence of BCAA (data not shown). Regulation of BCAA biosynthesis in S. thermophilus apparently involves end product inhibition effects similar to those observed in L. lactis (14). Addition of Ile to a medium containing Leu as the only BCAA or addition of Leu to a medium containing Ile as the only BCAA decreased the growth rate of the wild-type strain, which suggests that synthesis of at least one of the missing BCAA is reduced by Ile or Leu. For L. lactis (14), E. coli (37), L. mesenteroides subsp. cremoris (4), and B. subtilis (16) such regulation occurs at the transcription level. The decrease in the growth rate of S. thermophilus when Ile or Leu was added to CDM containing Val was probably not due to competition of BCAA for a putative common transport system, as described previously for L. lactis (31). The concentrations of individual amino acids used were identical to those used in complete CDM.
In S. thermophilus St18, the expected large transcript corresponding to ilv genes was not visualized, while a smaller transcript corresponding to ilvC was clearly visible. We cannot be sure whether this was because the genes were poorly transcribed during growth in rich medium or because the transcript was very unstable. The specificity of the BCAA biosynthesis pathway in S. thermophilus is reflected by the independent transcription of the ilvC gene visualized on the Northern blot (Fig. 5). Independent transcription of ilvC has also been observed in E. coli (46) and Corynebacterium glutamicum (20) but not in dairy bacteria (Fig. 4). It is probably very important since the ilvC gene product works at the junction between the BCAA biosynthesis pathway and acetolactate-acetoin metabolism (Fig. 1). The substrate of IlvC, acetolactate, is also an intermediate of pyruvate transformation into acetoin and 2,3-butanediol. The acetoin pathway is generally thought to assist in internal pH maintenance by changing the metabolism from acid to neutral compounds and to participate in the regeneration of NAD+ (34, 41). Independent transcription of ilvC could inhibit the accumulation of acetolactate and control its partition between the BCAA and acetoin pathways. A similar role has been assigned to acetolactate decarboxylase, which is the enzyme acting on acetolactate towards acetoin in L. lactis (15).
Evolution and conservation of active amino acid biosynthesis pathways.
The fact that dairy starter bacteria very frequently possess inactive amino acid biosynthesis pathways raises the question whether auxotrophies are beneficial to the bacteria (43, 47). The BCAA biosynthesis pathway, which is not functional in the dairy starter bacteria L. lactis and L. mesenteroides subsp. cremoris but is active in lactococci from vegetal origins (4, 14), reflects the different evolutionary pathways of these organisms. Godon et al. (13) have suggested that auxotrophy of dairy L. lactis strains could be a consequence of an adaptation to milk. In the present work, we demonstrated that the same BCAA biosynthesis pathway is functional in another dairy starter species, S. thermophilus. This finding contradicts what was previously suggested and can be explained in two ways. First, if we look at a cell wall proteinase capable of providing bacteria with peptides containing BCAA, we see that it appears more frequently in L. lactis than in S. thermophilus (36). L. lactis does not, therefore, really need to maintain a functional biosynthesis pathway. Mutants 1 and 2 grew rapidly again after addition of either Bacto Tryptone (Fig. 3) or an amino acid mixture (data not shown) to milk. This observation strongly suggests that the main role of the S. thermophilus BCAA biosynthesis pathway is to supply BCAA, a role complementary to the BCAA transport role already described for S. thermophilus (1). The difficulty in finding BCAA in milk encountered by S. thermophilus has been observed, to a much lesser extent, with L. lactis (12). In most cases, this difficulty is probably overcome by coculture of S. thermophilus with Lactobacillus delbrueckii subsp. bulgaricus, which produces amino acids and peptides and stimulates S. thermophilus growth (33). The second possible explanation is linked to the different genetic organizations of the BCAA biosynthesis pathways in L. lactis (14) or L. mesenteroides (4) and S. thermophilus (this study). In this case, independent translation of ilvC probably allows finer regulation of the pathway, which is sufficiently beneficial to S. thermophilus to keep the BCAA biosynthesis pathway active.
ACKNOWLEDGMENTS
This work was financed by Danone, Rhodia-Food, and Sodiaal in the framework of the contract “Substrates of fermentation.”
We thank Annie Sepulchre, Patricia Ramos, Jérôme Mengaud, Françoise Rul, and Donald White for critically reading the manuscript and Pierre Renault for the gift of strain TIL206.
REFERENCES
- 1.Akpemado K M, Bracquart P A. Uptake of branched-chain amino acids by Streptococcus thermophilus. Appl Environ Microbiol. 1983;45:136–140. doi: 10.1128/aem.45.1.136-140.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B J. Linkage map of Escherichia coli K12. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 7th ed. Washington, D.C.: American Society for Microbiology; 1987. pp. 807–876. [Google Scholar]
- 3.Bracquart P, Lorient D. Effet des acides aminés et peptides sur la croissance de Streptococcus thermophilus. III Peptides comportant Glu, His et Met. Milchwissenschaft. 1979;34:676–679. [Google Scholar]
- 4.Cavin J F, Dartois V, Labarre C, Divies C. Cloning of branched-chain amino acid biosynthesis genes and assays of α-acetolactate synthase activities in Leuconostoc mesenteroides subsp. cremoris. Res Microbiol. 1999;150:189–198. doi: 10.1016/s0923-2508(99)80035-7. [DOI] [PubMed] [Google Scholar]
- 5.Chopin A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:21–38. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 6.Cogain-Bousquet M, Garrigues C, Novak L, Lindley N D, Loubiere P. Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J Appl Bacteriol. 1995;79:108–116. [Google Scholar]
- 7.Cordes C, Mockel B, Eggeling L, Sahm H. Cloning, organization and functional analysis of ilvA, ilvB and ilvC genes from Corynebacterium glutamicum. Gene. 1990;112:113–116. doi: 10.1016/0378-1119(92)90311-c. [DOI] [PubMed] [Google Scholar]
- 8.Deguchi Y, Morishita T. Nutritional requirements in multiple auxotrophic lactic acid bacteria: genetic lesions affecting amino acid biosynthetic pathways in Lactococcus lactis, Enterococcus faecium, and Pediococcus acidilactici. Biosci Biotechnol Biochem. 1992;56:913–918. doi: 10.1271/bbb.56.913. [DOI] [PubMed] [Google Scholar]
- 9.Desmazeaud M. L'état des connaissances en matière de nutrition des bactéries lactiques. Lait. 1983;63:267–316. [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epelbaum S, La Rossa R A, Van Dyk T, Elkayam T, Chipman D M, Barak Z. Branched-chain amino acid biosynthesis in Salmonella typhimurium: a quantitative analysis. J Bacteriol. 1998;180:4056–4067. doi: 10.1128/jb.180.16.4056-4067.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flambard B, Helinck S, Richard J, Juillard V. The contribution of caseins to the amino acid supply for Lactococcus lactisdepends on the type of cell envelope proteinase. Appl Environ Microbiol. 1998;64:1991–1996. doi: 10.1128/aem.64.6.1991-1996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godon J J, Delorme C, Bardowski J, Chopin M C, Ehrlich S D, Renault P. Gene inactivation in Lactococcus lactis: branched-chain amino acid biosynthesis. J Bacteriol. 1993;175:4383–4390. doi: 10.1128/jb.175.14.4383-4390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godon J J, Chopin M C, Ehrlich S D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992;174:6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goupil-Feuillerat N, Cogain-Bousquet M, Godon J J, Ehrlich S D, Renault P. Dual role of α-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J Bacteriol. 1997;179:6285–6293. doi: 10.1128/jb.179.20.6285-6293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandoni J A, Zahler S A, Calvo J M. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J Bacteriol. 1992;174:3212–3219. doi: 10.1128/jb.174.10.3212-3219.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henkin T M, Glass B L, Grundy F J. Analysis of the Bacillus subtilis tyrSgene: conservation of a regulatory sequence in multiple tRNA synthetase genes. J Bacteriol. 1992;174:1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremorisgrown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huggins A M, Sandine W E. Differentiation of fast and slow milk coagulating isolates in strains of streptococci. J Dairy Sci. 1984;67:1674–1679. [Google Scholar]
- 20.Keilhauer C, Eggleling L, Sahn H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvCoperon. J Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llanos R M, Hillier A J, Davidson B. Cloning, nucleotide sequence, expression, and chromosomal location of ldh, the gene encoding l-(+)-lactate dehydrogenase from Lactococcus lactis. J Bacteriol. 1992;174:6956–6964. doi: 10.1128/jb.174.21.6956-6964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maguin E, Prévost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marciset O, Mollet B. Multifactorial experimental designs for optimizing transformation: electroporation of Streptococcus thermophilus. Biotechnol Bioeng. 1994;43:490–496. doi: 10.1002/bit.260430609. [DOI] [PubMed] [Google Scholar]
- 24.Mierau I, Kunji E R S, Leenhouts K J, Hellendorn M A, Haandrikman A J, Poolman B, Konings W N, Venema G, Kok J. Multiple-peptidase mutants of Lactococcus lactisare severely impaired in their ability to grow in milk. J Bacteriol. 1996;178:2794–2803. doi: 10.1128/jb.178.10.2794-2803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills O E, Thomas T D. Nitrogen sources for growth of lactic streptococci in milk. N Z J Dairy Sci Technol. 1981;15:43–55. [Google Scholar]
- 26.Morishita T, Deguchi Y, Yajima M, Sakurai T, Yura T. Multiple nutritional requirements of lactobacilli: genetic lesions affecting amino acid biosynthetic pathways. J Bacteriol. 1981;148:64–71. doi: 10.1128/jb.148.1.64-71.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidhart F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology Press; 1996. pp. 13–28. [Google Scholar]
- 28.Neviani E, Giraffa G, Brizzi A, Carminati D. Amino acid requirements and peptidase activities of Streptococcus salivarius subsp. thermophilus. J Appl Bacteriol. 1995;79:302–307. doi: 10.1111/j.1365-2672.1995.tb03141.x. [DOI] [PubMed] [Google Scholar]
- 29.Obis D, Guillot A, Gripon J C, Renault P, Bolotin A, Mistou M Y. Genetic and biochemical characterization of a high-affinity betaine uptake system (BusA) in Lactococcus lactisreveals a new functional organization within bacterial ABC transporters. J Bacteriol. 1999;181:6238–6246. doi: 10.1128/jb.181.20.6238-6246.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piggot P J, Hoch J A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985;49:158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poolman B, Konings W N. Relation of growth of Streptococcus lactis and Streptococcus cremoristo amino acid transport. J Bacteriol. 1988;170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potter C A, Baumberg S. End-product control of enzymes of branched-chain amino acid biosynthesis in Streptomyces coelicolor. Microbiology. 1996;142:1945–1952. doi: 10.1099/13500872-142-8-1945. [DOI] [PubMed] [Google Scholar]
- 33.Radke-Mitchell L, Sandine W E. Associative growth and differential enumeration of Streptococcus thermophilus and Lactobacillus bulgaricus: a review. J Food Prot. 1984;47:245–248. doi: 10.4315/0362-028X-47.3.245. [DOI] [PubMed] [Google Scholar]
- 34.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsRgenes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Shahbal S, Hemme D, Renault P. Characterization of a cell envelope-associated proteinase activity from Streptococcus thermophilusH strains. Appl Environ Microbiol. 1993;59:177–172. doi: 10.1128/aem.59.1.177-182.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coligrowing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terzaghi B T, Sandine W E. Improved medium for lactic streptococci and their bacteriophage. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas T D, Mills O E. Proteolytic enzymes of starter bacteria. Neth Milk Dairy J. 1981;35:255–273. [Google Scholar]
- 40.Thomas T D, Turner K W. Preparation of skim milk to allow harvesting of starter cells from milk cultures. N Z J Dairy Sci Technol. 1977;12:15–21. [Google Scholar]
- 41.Tsau J L, Guffanti A A, Montville T J. Conversion of pyruvate to acetoin helps to maintain pH homeostasis in Lactobacillus plantarum. Appl Environ Microbiol. 1992;58:891–894. doi: 10.1128/aem.58.3.891-894.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umbarger H E. Biosynthesis of the branched-chain amino acids. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology Press; 1996. pp. 442–457. [Google Scholar]
- 43.Van Dyk T K, La Rossa R A. Prevention of endogenous 2-ketobutyrate toxicity in Salmonella typhimurium. In: Barak Z, Chipman D M, Schloss J V, editors. Biosynthesis of branched-chain amino acids. Weinheim, Germany: VCH; 1990. pp. 123–130. [Google Scholar]
- 44.Von Wright A, Tynkkynen S, Souminen M. Cloning of a Streptococcus lactis subsp. lactischromosomal fragment associated with the ability to grow in milk. Appl Environ Microbiol. 1987;53:1584–1588. doi: 10.1128/aem.53.7.1584-1588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Yu W, Coolbear T, O'Sullivan D, McKay L. A deficiency in aspartate biosynthesis in Lactococcus lactis subsp. lactisC2 causes slow milk coagulation. Appl Environ Microbiol. 1998;64:1673–1679. doi: 10.1128/aem.64.5.1673-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wek R C, Hatfield G W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in E. coliK12. J Biol Chem. 1986;261:2441–2550. [PubMed] [Google Scholar]
- 47.Zamenhof S, Eichhorm H H. Study of microbial evolution through loss of biosynthetic functions: establishment of “defective” mutants. Nature. 1967;216:456–458. doi: 10.1038/216456a0. [DOI] [PubMed] [Google Scholar]