Abstract
The BRCA1-PALB2-BRCA2 axis, or the BRCA pathway, plays key roles in genome stability maintenance and suppression of breast and several other cancers. Due to frequent p53 mutations in human BRCA1 breast cancers and mouse mammary tumors from Brca1, Brca2 and Palb2 conditional knockout models, it is often thought that p53 inactivation accelerates BRCA1/2 and PALB2-associated tumorigenesis. Here, we studied tumor development in mice with a mutation in Palb2 that disengages the PALB2-BRCA1 interaction in different Trp53 backgrounds. Rather than mammary tumors, Palb2 and Trp53 compound mutant mice developed, with greatly reduced latencies, lymphomas and sarcomas that are typically associated with germline Trp53 inactivation. Whole exome sequencing failed to identify any significant differences in genomic features between the same tumor types of Trp53 single mutant and Palb2;Trp53 compound mutant mice. These results suggest that loss of the BRCA pathway accelerates p53-associated tumor development, possibly without altering the fundamental tumorigenic processes.
Keywords: BRCA1, BRCA2, Osteosarcoma, p53, PALB2, Thymic lymphoma
Introduction
BRCA1 and BRCA2, the two major breast cancer susceptibility proteins, play key roles in the DNA damage response, especially in homologous recombination (HR)-mediated DNA double strand break repair and cell cycle checkpoint control, thereby maintaining genome integrity and suppressing tumor development.1,2 The two BRCA proteins are physically and functionally linked in HR and checkpoint control by PALB2, which was first identified as a major binding partner of BRCA2.3, 4, 5, 6 Like BRCA1/2, monoallelic germline mutations in PALB2 cause high risk of breast cancer and also increase the risks of ovarian, pancreatic and other cancers.7,8
To understand the in vivo function of the BRCA1/2 genes and the mechanisms of their associated tumorigenesis, several different mouse models were created for each gene soon after their cloning.9 The first studies found that germline, systemic knockout of each gene led to embryonic lethality,9 underscoring the importance of the genes for fundamental cellular processes and development. Conditional ablation of each gene in the mammary gland indeed led to mammary tumor development; however, the latencies were long, averaging about 1.5 years,10, 11, 12, 13 suggesting the existence of a strong barrier of tumor formation following the loss of the proteins. This barrier is widely believed to be p53, as its mutations were found in most mammary tumors arising from conditional knockout models of both genes.10,11,13 Co-deletion of Trp53 with each gene led to much more efficient mammary tumor formation,14,15 and a Trp53 heterozygous background also promoted mammary tumorigenesis in Brca1 CKO mice.12,13 Similar findings were made with Palb2 conventional and conditional KO mouse models.16, 17, 18, 19 The above findings suggest that p53 loss of function is required for BRCA- and PALB2-asociated mammary tumorigenesis, at least in mice.
To bypass the embryonic lethality of germline Palb2 knockout and to study the role of the BRCA1-PALB2 interplay in vivo, we previously created a Palb2CC6 strain with a mutation in the N-terminal coiled-coil motif of PALB2 that disrupts the binding of BRCA1.20 Homozygous mutant mice showed increased endogenous DNA damage,21 a moderate defect in spermatogenesis,20 and susceptibility to radiation-induced tumor development.21 In this study, we monitored spontaneous tumor development in the Palb2CC6 mice with different Trp53 backgrounds. Our results showed that combined mutations in the two genes led to accelerated development of tumor types typically associated with loss of Trp53 rather than Palb2.
Materials and methods
Mice
Generation of the Palb2CC6 mutant strain was described before.20 The strain was backcrossed to C57BL/6 background for 6 generations and then crossed with Trp53−/− mice22 on 129sv background; the progenies were then intercrossed to generate the different cohorts for observation. All animal work was approved by the Institutional Animal Care and Use Committee (IACUC) of the Rutgers Robert Wood Johnson Medical School.
Tumor collection and processing
Tumors were collected from mice immediately after euthanization by CO2 asphyxiation. Half of each tumor was snap frozen and the other half fixed overnight in phosphate-buffered formalin, transferred to 70% ethanol and embedded in paraffin. Paraffin-embedded tumors were sectioned at 5-µm thickness, stained with hematoxylin and eosin (H&E) for histological review.
Whole-exome sequencing analysis
DNA extraction was performed using the DNeasy Blood and Tissue Kit (Qiagen). Tumor and normal DNA samples were subjected to whole-exome sequencing (WES; SureSelect Mouse All Exon Kit, Agilent Technologies) at RUCDR Infinite Biologics (Piscataway, NJ). Paired-end sequencing data were aligned to the reference mouse genome mm10 using the Burrows-Wheeler Aligner (BWA, v0.7.15). Local realignment, duplicate removal and base quality score recalibration was performed using the Genome Analysis Toolkit (GATK, v3.1.1). After pooling the reads from each normal sample and masking repetitive regions using RepeatMasker (v4.0), somatic single nucleotide variants (SNVs) were identified using MuTect (v.1.1.4), and small insertions and deletions (indels) were detected using VarScan2 (v2.3.6) and Strelka (v3.1.1). To identify indels greater than 3 bp, Lancet, Platypus, and Scalpel were employed, and the results were combined to define a consensus call as previously described. SNVs and indels outside the WES capture were filtered out, as were SNVs and indels for which the variant allele fraction (VAF) in the tumor sample was less than 5 times the VAF of the paired normal tissue as previously described. Allele specific copy number aberrations (CNAs), tumor purity and ploidy were obtained from the WES data using FACETS.
Genomic features of HR DNA repair defects
Large-scale state transition (LST) scores were computed from the results of FACETS using the WES data according to Popova et al.23 A cut-off of ≥15 was employed to classify tumors as LST high. The NtAI score, which assesses telomeric allelic imbalance, was defined according to Birkbak et al.24 The number and length of small deletions in the tumors samples were assessed as previously described.25
Statistical analyses
P values were calculated by two-tailed unpaired t-test in GraphPad Prism 8. P values <0.05 were considered significant.
Results and discussion
To understand the importance of the BRCA1-PALB2 interaction in spontaneous tumorigenesis and the role of p53 in the process, we crossed the Palb2CC6 mice (C57BL/6 background) with Trp53 knockout mice (129sv background), in which exons 2–7 of the gene were replaced by a neomycin drug selection cassette,22 to produce Palb2CC6/CC6 mice with wt, +/− and −/− Trp53 status. These mice were aged along with wt, Trp53+/− and Trp53−/− mice generated in the same breeding process. Compared with wt mice, Palb2CC6/CC6 mice showed increased spontaneous tumor development (Fig. 1A). Overall, males showed earlier tumor development than females (Fig. 1B). Among males, the most common tumor type was liver tumor (69%), followed by lymphoma (including thymic lymphoma, splenic tumors, and other lymphomas) (16%) and soft tissue sarcoma (8%, Fig. 1C,D). Among females, lymphoma was the most common (36%), followed by tumors in the liver (24%), ovary (18%) and mammary gland (12%). The high incidence of liver tumor, especially in males, was unexpected; it could be explained by a possible chronic inflammation caused by increased oxidative stress and/or constitutive activation of NF-κB in these mice, as we reported recently,21 although this remains to be tested experimentally.
Figure 1.
Tumor development in Palb2CC6 mice with different Trp53 backgrounds. (A,B) Tumor-free survival of mice of indicated genotypes with males and females combined (A) or separated (B). (C) Summary of tumor types and numbers from mice of different genotypes. (D) Tumor spectra of mice with different genotypes. The lack of data from Palb2CC6/CC6;Trp53−/− females is due to female-specific embryonic lethality of this genotype.
Trp53+/− mice in a C57BL/6 × 129sv mixed background are known to succumb to a variety of tumors mostly between one to two years of age, with lymphomas and sarcomas being the major tumor types.26 Our observations were similar, with a male/female combined median latency of 482 days and lymphoma, soft tissue sarcoma, osteosarcoma and liver tumor being the top tumor types (Fig. 1A,D). Again, males showed faster tumor development and shorter life span than females (Fig. 1B). Additionally, males were more often affected by soft tissue sarcoma, whereas females showed higher propensity to develop osteosarcoma (Fig. 1D).
Palb2CC6/CC6;Trp53+/− mice showed greatly accelerated tumor development compared with mice with either mutation alone, with a combined median tumor latency of 253 days (Fig. 1A). Interestingly, although we expected Trp53 heterozygosity to shift the tumor susceptibility of the Palb2CC6/CC6 mice towards mammary and perhaps also ovarian and pancreas cancers, the tumor spectrum of the compound mutant mice instead showed a shift to osteosarcoma, a key phenotype of Trp53+/− mice,26 and thymic lymphoma, the signature phenotype of Trp53−/− mice22 (Fig. 1D). In fact, mammary and ovarian tumor incidence was substantially reduced in the compound mutant mice compared with Palb2CC6/CC6 single mutant mice (4% vs 30%), so was liver tumor incidence in males (5% vs 69%). This suggests that functional loss of the BRCA1-PALB2-BRCA2 pathway accelerates p53-associated tumorigenesis, rather than p53 loss promoting BRCA1/2- and PALB2-associated tumor development.
Consistent with results from earlier studies,22,26 all Trp53−/− mice developed tumors within 7 months of age (Fig. 1A), with thymic lymphoma being the major tumor type, especially in males (Fig. 1D). Remarkably, tumor development was still faster in Palb2CC6/CC6;Trp53−/− mice (Fig. 1A), and thymic lymphoma was the only tumor type in the double mutant mice (Fig. 1D). This finding again demonstrates that disruption of the BRCA1-PALB2-BRCA2 axis accelerates p53-associated tumor development. During the breeding, we noted a female-specific embryonic lethality of Palb2CC6/CC6;Trp53−/− mice (therefore only males were monitored). As Trp53−/− mice are known to be prone to female-specific defects in neural tube closure,27,28 it is possible that mutation of Palb2 or loss of the BRCA1-PALB2 interaction exacerbated the defect leading to embryonic lethality.
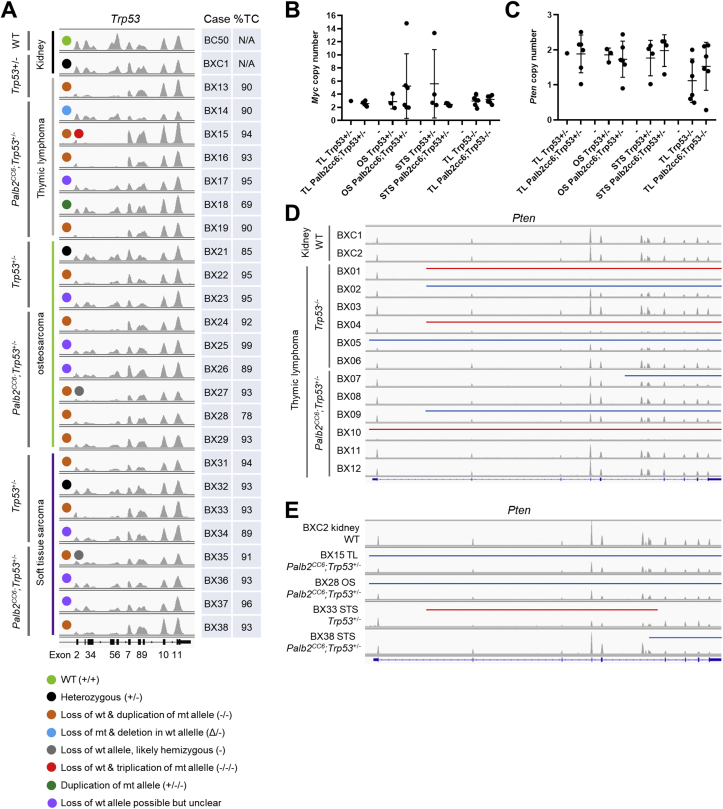
To gain insights into the genetic mechanisms of tumor development in the Palb2/Trp53 mutant mice, we conducted whole exome sequencing (WES) for a total of 36 tumors of 3 different types (thymic lymphoma, osteosarcoma and soft tissue sarcoma) from mice of 6 different genotypes (Table S1). It has been reported that the majority of tumors arising from Trp53+/− mice undergo loss of heterozygosity (LOH), losing the wt allele.22 Therefore, we first examined the Trp53 locus in tumors from Trp53+/− and Palb2CC6/CC6;Trp53+/− mice (Fig. 2A). Among thymic lymphomas, the only one from Trp53+/− mice and 3 of 6 from Palb2CC6/CC6;Trp53+/− mice clearly lost the wt allele, as evidenced by a lack of reads in exons 2–7 (Fig. 2A, orange dots). Note that the residual reads in exons 2–7 present in some cases were likely due to impurity of the tumors, i.e. the presence of normal (Trp53+/−) cells. Interestingly, among the remaining 3 thymic lymphomas from the Palb2CC6/CC6;Trp53+/− mice sequenced, one lost the mutant allele but also contained a deletion of exons 8 and 9 of the wt allele (blue dot), while another appeared to have duplicated the mutant allele while still maintaining the wt allele (dark green dot). Among osteosarcomas, 1 of 3 from Trp53+/− mice and 4 of 6 from Palb2CC6/CC6;Trp53+/− mice lost the wt allele. Among soft tissue sarcomas, loss of wt allele occurred in at least 2 of 4 from each group.
Figure 2.
Genomic analyses of tumors from Palb2CC6 mutant mice in different Trp53 backgrounds. (A) Status of the Trp53 locus in tumors from Trp53+/− and Palb2CC6;Trp53+/− mice. Images were generated with IGV based on WES data. Data range for each track is normalized using the size of the corresponding BAM file, which reflects the number of total reads obtained for the tumor. (B,C) Dot plots of computed copy numbers of Myc (B) and Pten (C) in all tumors sequenced. (D,E) Status of the Pten locus in all 12 sequenced tumors from Trp53−/− and Palb2CC6;Trp53+/− mice (D) and selected tumors from Palb2CC6;Trp53+/− mice (E). Data range for each track is normalized as in A. Regions of monoallelic and biallelic deletion are indicated by blue and red bars, respectively.
For all 3 tumor types, when the wt allele was lost, the mutant Trp53 allele was often duplicated (copy-neutral LOH); however, in one (thymic lymphoma) case the mutant allele was triplicated (red dot), while a few other cases appeared to be hemizygous (with deletion of the wt allele, grey dots). Therefore, although all tumors from the Palb2CC6/CC6;Trp53+/− mice did not lose the wt Trp53 allele, overall LOH appeared to be accelerated due to the disruption of the BRCA1-PALB2 interaction, given the faster tumor development. The remaining wt allele could be disrupted by rearrangements, which is not detectable by WES.
We next analyzed the copy number of key cancer genes known to be involved in relevant tumor types studied here. Using computed copy number of 3 as a cutoff, 8/24 (33%) of the tumors arising from Trp53+/− and Palb2CC6/CC6;Trp53+/− mice contained a gain or amplification of the Myc oncogene. In particular, 3/6 (50%) of osteosarcomas from the Palb2CC6/CC6;Trp53+/− mice contained 5 or more copies of the gene (Fig. 2B). Gain of Myc (3–4 copies) was also observed in 4/6 (67%) and 3/6 (50%) of thymic lymphomas from Trp53−/− and Palb2CC6/CC6;Trp53−/− mice, respectively. Among tumor suppressor genes, frequent deletions in Pten have been reported to occur in thymic lymphomas arising from Trp53−/− mice.29 This was observed in the current study, as 4/6 (67%) of such tumors showed some forms of deletion in the gene (Fig. 2C, D). In comparison, partial or complete loss of Pten were observed in 3/6 (50%) of thymic lymphomas from the Palb2CC6/CC6;Trp53−/− mice. Additionally, partial deletions of Pten were also seen in a small number of tumors from Palb2CC6/CC6;Trp53+/− mice (Fig. 2E). With respect to the overall copy numbers of the 2 genes, no statistically significant difference was found between the Palb2 wt and mutant subgroups of each tumor type.
Finally, we compared the number of mutations, large-scale state transition (LST) scores, and the number and length of indels in the different groups of tumors (Fig. S1). A larger number of mutations was found in osteosarcomas from Palb2CC6/CC6;Trp53+/− mice than the same tumor type from Trp53+/− mice. Other than that, no significant differences in any parameter were found in other tumor types and groups. Therefore, in most cases, disruption of the BRCA1-PALB2 axis appeared to accelerate rather than alter the genetic changes that lead to tumor development in p53 mutant mice.
In summary, our finding that the Palb2CC6 mutation accelerates p53-associated tumor development (Fig. 1A, B) suggests that p53 is the more dominant factor that determines tumor spectrum, i.e. tissue specificity. Given the much shorter tumor latency in Palb2CC6/CC6;Trp53+/− than Trp53+/− mice and a lack of clear differences in genomic alterations in tumors arising from the 2 groups of mice, our data suggest that loss of the BRCA1-PALB2-BRCA2 axis accelerates p53-associated tumor development without fundamentally altering the tumorigenic process. As for the mechanism, WES of tumors from Palb2CC6/CC6 mice with a Trp53 heterozygous background (Fig. 2) suggest that loss of the BRCA1-PALB2 interaction accelerates loss of heterozygosity (LOH) at Trp53. In almost all tumors with full Trp53 inactivation, it occurred by losing, rather than mutating, the wt allele and duplicating the mutant allele, with the one exception being a deletion within the wt allele. Notably, the Palb2CC6 mutation also accelerated p53-associated tumor formation (mostly thymic lymphomas) in mice with biallelic knockout of Trp53, indicating that accelerating Trp53 LOH is only part of the mechanism and that accelerated alterations in other cancer genes, such as Myc, also contributes to accelerated tumor development.
Our findings also imply that at least a subset of BRCA- and PALB2-associated breast cancers may be, by nature, a malignancy caused by somatic p53 loss of function or dysfunction that is accelerated by a defect in the BRCA1-PALB2-BRCA2 axis. This hypothesis is also supported by the fact that virtually all BRCA1 mutant human breast cancer harbor TP53 mutations.30 However, TP53 mutations only occur in a minority of BRCA2 and PALB2 mutant breast cancers.31 Therefore, it would be important to determine if p53 or its pathway is inactivated in BRCA2 and PALB2 mutant cancers at levels other than primary gene sequence. It should also be noted that as human BRCA and PALB2 mutation carriers are mostly heterozygous, cancer development in humans generally requires somatic, spontaneous inactivation of the wt allele by LOH or mutations. This is not only an extra step compared with biallelic ablations or mutations in mouse models but also a major variable, in terms of timing and frequency, among the 3 genes due to their different genomic loci and chromatin structures. As such, the different requirements of TP53 inactivation may stem from differences in the timing, frequency, or nature of the inactivation of the wt BRCA or PALB2 alleles in the breast epithelial cells of human mutation carriers.
Conflict of interests
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the National Cancer Institute (R01CA138804 to BX, R01GM129066 to SD, P30CA072720 to Rutgers Cancer Institute of New Jersey, and P30CA008748 to MSKCC). AHM was supported by Higher Committee for Education Development (HCED) of Iraq.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.08.012.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Chen C.C., Feng W., Lim P.X., Kass E.M., Jasin M. Homology-directed repair and the role of BRCA1, BRCA2, and related proteins in genome integrity and cancer. Annu Rev Cell Biol. 2018;2:313–336. doi: 10.1146/annurev-cancerbio-030617-050502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkitaraman A.R. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343:1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- 3.Xia B., Sheng Q., Nakanishi K., et al. Control of BRCA2 cellular and clinical functions by a nuclear partner. PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Sy S.M., Huen M.S., Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F., Ma J., Wu J., et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simhadri S., Vincelli G., Huo Y., et al. PALB2 connects BRCA1 and BRCA2 in the G2/M checkpoint response. Oncogene. 2019;38(10):1585–1596. doi: 10.1038/s41388-018-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couch F.J., Shimelis H., Hu C., et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X., Leslie G., Doroszuk A., et al. Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol. 2020;38(7):674–685. doi: 10.1200/JCO.19.01907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evers B., Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 10.Ludwig T., Fisher P., Murty V., Efstratiadis A. Development of mammary adenocarcinomas by tissue-specific knockout of Brca2 in mice. Oncogene. 2001;20:3937–3948. doi: 10.1038/sj.onc.1204512. [DOI] [PubMed] [Google Scholar]
- 11.Shakya R., Szabolcs M., McCarthy E., et al. The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. Proc Natl Acad Sci U S A. 2008;105:7040–7045. doi: 10.1073/pnas.0711032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Wagner K., Larson D., et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 13.McCarthy A., Savage K., Gabriel A., et al. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol. 2007;211(4):389–398. doi: 10.1002/path.2124. [DOI] [PubMed] [Google Scholar]
- 14.Liu X., Holstege H., van der Gulden H., et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104(29):12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonkers J., Meuwissen R., van der Gulden H., et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 16.Rantakari P., Nikkila J., Jokela H., et al. Inactivation of Palb2 gene leads to mesoderm differentiation defect and early embryonic lethality in mice. Hum Mol Genet. 2010;19(15):3021–3029. doi: 10.1093/hmg/ddq207. [DOI] [PubMed] [Google Scholar]
- 17.Bouwman P., Drost R., Klijin C., et al. Loss of p53 partially rescues embryonic development of Palb2 knockout mice but does not foster haploinsufficiency of Palb2 in tumour suppression. J Pathol. 2011;224(1):10–21. doi: 10.1002/path.2861. [DOI] [PubMed] [Google Scholar]
- 18.Bowman-Colin C., Xia B., Bunting S., et al. Palb2 synergizes with Trp53 to suppress mammary tumor formation in a model of inherited breast cancer. Proc Natl Acad Sci U S A. 2013;110(21):8632–8637. doi: 10.1073/pnas.1305362110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huo Y., Cai H., Teplova I., et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Canc Discov. 2013;3(8):894–907. doi: 10.1158/2159-8290.CD-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simhadri S., Peterson S., Patel D.S., et al. Male fertility defect associated with disrupted BRCA1-PALB2 interaction in mice. J Biol Chem. 2014;289(35):24617–24629. doi: 10.1074/jbc.M114.566141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahdi A.H., Huo Y., Tan Y., et al. Evidence of intertissue differences in the DNA damage response and the pro-oncogenic role of NF-kappaB in mice with disengaged BRCA1-PALB2 interaction. Can Res. 2018;78(14):3969–3981. doi: 10.1158/0008-5472.CAN-18-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacks T., et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 23.Popova T., Manié E., Rieunier G., et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 24.Birkbak N.J., et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2(4):366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashley C.W., et al. Analysis of mutational signatures in primary and metastatic endometrial cancer reveals distinct patterns of DNA repair defects and shifts during tumor progression. Gynecol Oncol. 2019;152(1):11–19. doi: 10.1016/j.ygyno.2018.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donehower L.A., Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009;9(11):831–841. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong J.F., Kaufman M.H., Harrison D.J., Clarke A.R. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5(8):931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 28.Delbridge A.R.D., Kueh A.J., Ke F., et al. Loss of p53 causes stochastic aberrant X-chromosome inactivation and female-specific neural tube defects. Cell Rep. 2019;27(2):442–454. doi: 10.1016/j.celrep.2019.03.048. [DOI] [PubMed] [Google Scholar]
- 29.Dudgeon C., Chan C., Kang W., et al. The evolution of thymic lymphomas in p53 knockout mice. Gene Dev. 2014;28:2613–2620. doi: 10.1101/gad.252148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holstege H., Joosse S.A., van Oostrom C.T., et al. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Can Res. 2009;69(8):3625–3633. doi: 10.1158/0008-5472.CAN-08-3426. [DOI] [PubMed] [Google Scholar]
- 31.Li A., Geyer F.C., Blecua P., et al. Homologous recombination DNA repair defects in PALB2-associated breast cancers. Npj Breast Cancer. 2019;5:23. doi: 10.1038/s41523-019-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.