Abstract
SIRT6 belongs to class III sirtuin family with NAD+-dependent histone deacetylase activities and controls multiple processes including aging, metabolism and inflammation. In recent years, increasing studies showed tumor suppressor role of SIRT6 in HCC development. We established a two-stage DEN followed CCl4 induced liver carcinogenesis in the hepatic-specific SIRT6 HKO mice models and found that hepatic SIRT6 deficit significantly promotes liver injury and liver cancer through inhibition of the ERK1/2 pathway. SIRT6 was compensatory upregulated in mice tumor tissues and human HCC cells and overexpressed SIRT6 inhibits tumor growth both in vitro and in vivo. Taken together, we provide a useful mouse model for delineating the molecular pathways involved in chronic liver diseases and primary liver cancer and suggest that SIRT6 can be a promising target for HCC therapies.
Keywords: ERK1/2 pathway, HCC, Liver carcinogenesis, Mouse model, SIRT6
Abbreviations: HCC, hepatocellular carcinoma; DEN, Diethylnitrosamine; SIRT, sirtuin; CCl4, carbon tetrachloride
Introduction
Chronic hepatopathy and hepatocellular carcinoma (HCC) are increasing global human problems. HCC as the most lethal and prevalent form of primary liver cancer occurs not as a sporadic event, but a slow progression from chronic liver disease, and remains one of the deadliest cancer types.1, 2, 3 More than 800,000 people worldwide are diagnosed with liver cancer annually and lack successful treatment options. The incidence and mortality rates of liver cancer in men are about three times to women almost worldwide.4 While HCC is a multifactorial disease associated with several risks, including hereditary factors, environmental, lifestyle and diet factors, the vast majority of cases were commonly progressed from cirrhosis.5 Given the increasing risk of death and limited clinical treatment options for HCC, to understand cancer-promoting mechanisms underlying HCC is urgently needed.6
Over the past decades, numerous mouse models have been established to research the pathogenesis of HCC. Diethylnitrosamine (DEN), as a well-known liver chemical carcinogen, can initiate liver cancer with human HCC similarity by producing reactive oxygen species and forming mutagenic DNA adducts.7 C57BL/6 genetic background mice injected intraperitoneally with a single dose of 25 mg/kg (body weight) DEN at day 14 and developed tumor nodules in the liver after 9 months.8 Liver cancer is usually associated with cirrhosis background secondary to chronic hepatopathy.9 Domenicali et al established advanced liver cirrhosis in mice with continuous exposure of carbon tetrachloride (CCl4) at about 12 weeks.10 As a promoter, weekly administration of CCl4, along with drinking alcohol, developed HCC after 104 weeks.11 Combination models of co-treatment with single DEN followed by repeated intraperitoneal injection of CCl4 showed a 100% incidence of liver tumors at 5 months of age.12 This two-stage DEN followed CCl4 induced liver carcinogenesis mouse model is more likely human HCC, which requires liver cirrhosis and offers an opportunity for researching the molecular events in the progression of the primary liver cancer.
Several molecular pathways and cellular events have been shown to contribute to the progression of HCC. SIRT6, a member of the sirtuin (SIRT) family of NAD+-dependent protein deacetylases, has been identified as a critical regulator in fundamental human processes including lifespan, metabolism and inflammation13 and global SIRT6 knockout mice died in a few weeks.14,15 Many molecular pathways in the aging process also contribute to tumor suppression,16 and increasing evidence showed the tumor suppressor role of SIRT6 in human cancer.17, 18, 19, 20 SIRT6 showed an important role in chronic liver disease by the evidence that hepatic specific loss of SIRT6 accelerates fatty liver and hepatic steatosis.21,22 However, the role and mechanism of SIRT6 in DEN and CCl4 induced liver tumorigenesis to remain unclear.
Given the critical role of SIRT6 in liver function, we performed a DEN and CCl4 induced liver tumorigenesis mouse model in hepatic-specific SIRT6 deletion mice. We firstly investigated that SIRT6 deficit contributes to liver injury and chemical-induced hepatocarcinogenesis in mice by ERK1/2 pathway activation.
Materials and methods
Additional materials and methods are described in the Supporting Materials and Methods online. All participants were provided written informed consent to take part in the study. All samples collection and the procedures for animal studies were approved by the Ethics Committee and the Institutional Animal Care and Use Committee of the Nanjing Medical University.
DEN and CCl4-induced hepatocarcinogenesis mice model
The C57BL/6 J genetic background SIRT6loxp/loxp mice and albumin-cre mice used in this study were friendly provided by the Animal Center (Nanjing Medical University, China). Hepatic-specific SIRT6 HKO mice were generated by crossing SIRT6loxp/loxp mice with albumin-cre mice to drive Cre/loxp recombination in liver cells. The wild type (WT) mice were the SIRT6loxp/loxp mice without the Cre gene.39 All mice were housed in the Animal Research Center of Nanjing Medical University on a 12-hr light/dark cycle. All animal experiments were performed according to the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University.
To establish hepatocarcinogenesis mouse model, a single dose of 25 mg/kg DEN (N-Nitrosodiethylamine, BioChem Partner, China) was i.p injected into both female and male WT and HKO mice (N = 8 each group) at 14 days of age, followed by administration of 10% CCl4 (v/v in corn oil, 8 ml/kg of body weight, Solarbio, China) solution (twice a week) by IP starting from 8 weeks old for 14 weeks. Six weeks after the last injection, mice were sacrificed for macroscopically and microscopically liver examination, and blood and liver tissues were collected for subsequent experiments.
Xenograft mice experiments
The male BALB/c nude mice were purchased from the Animal Centre of Nanjing Medical School. Total 1 × 107 HuH7 human liver cancer cells expressed pcDNA3.1 or pcDNA3.1-SIRT6 (suspended in 200 μL 1:1 serum-free DMEM/Matrigel) were subcutaneously injected into the flanks of 5–6 weeks old nude mice (N = 5). The tumor size was monitored and measured every four days with calipers and calculated based on the formula: aⅩb2/2 (a: length, b: width). Tumor-bearing mice were sacrificed at day 34, and the tumors were removed for photographed, weighed and further study.
Statistical analysis
All statistical significance analyses in this study were used by GraphPad Prism software. Data are expressed as means ± SEM from at least three independent experiments except where otherwise indicated. Differences between groups were determined by a 2-tailed Student's t-test. P < 0.05 was considered significant.
Results
SIRT6 knockout contributes to chemical-induced hepatocarcinogenesis
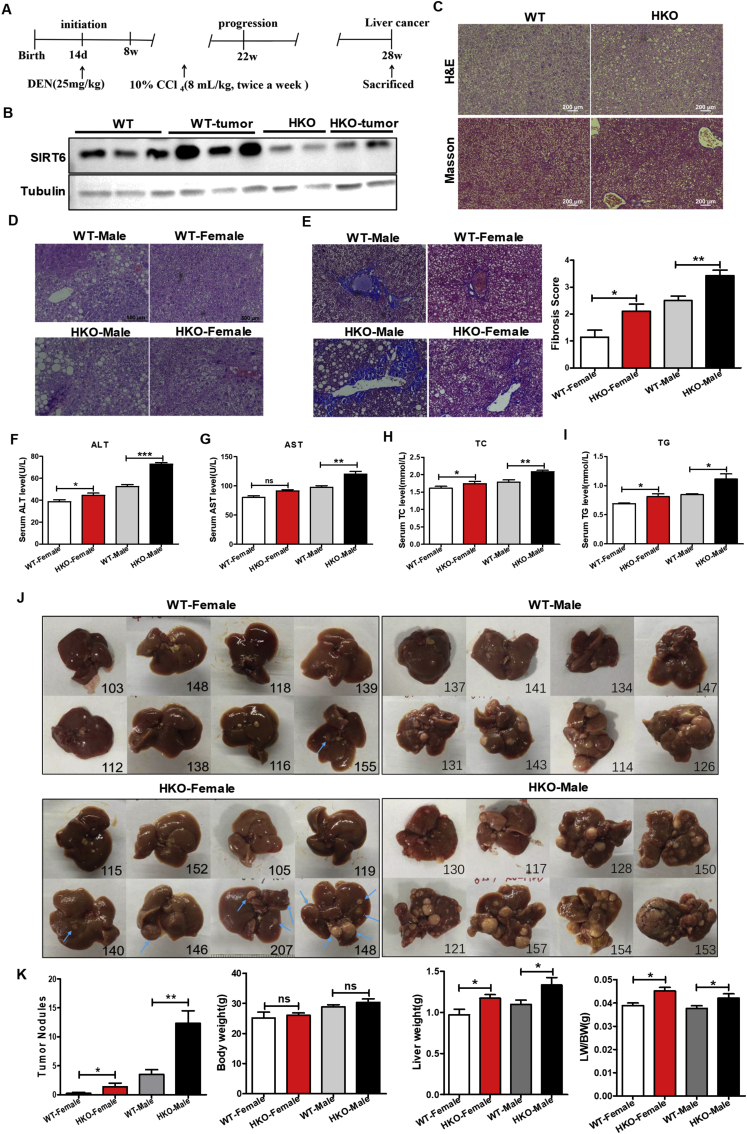
Global SIRT6-loss mice died on day 24 after birth.14 Therefore, to understand the effect of SIRT6 deficit on liver tumorigenesis, we specifically deleted SIRT6 in the liver using alb-cre transgenic mice, which mediating Cre/loxp recombination predominantly in hepatocytes. Diethylnitrosamine (DEN) is a widely accepted model genotoxic agent to induce liver carcinogenesis, occurs 100% in males and 30% in females after 45–104 weeks,23,24 consecutive administration of carbon tetrachloride (CCl4) promotes the progression of hepatic fibrosis.25 To investigate whether knockout of SIRT6 would affect DEN and CCl4-induced liver cancer development, we injected a single dose of 25 mg/kg DEN as an initiator into fourteen-day-old female and male mice and performed repeat injection of CCl4 (in corn oil) as a promoter for up to 14 weeks (Fig. 1A). At the endpoint of 7 months, macroscopic tumor development can be seen, SIRT6 knockout mice displayed more HCC nodules both in female and male mice, however, the WT female group only 1 out of 8 mice developed one tumor nodule (Fig. 1J). The chemical-induced liver development without affecting their body weight, quantitative analyses revealed that loss of SIRT6 increased the ratio of liver weight to body weight by 16% in female HKO mice and 11% in male HKO mice compared to their WT counterparts (Fig. 1K).
Figure 1.
Liver-specific knockout of SIRT6 promotes DEN and CCl4-induced liver cancer in mice. (A) Experimental design of chemical-induced hepatocellular carcinoma mouse model. A single treatment of 25 mg/kg DEN was administered at day 14 after birth, followed by twice a week injection of CCl4 starting from 8 weeks-old for 14 consecutive weeks, and mice were sacrificed at 28 weeks. (B) The representative western blotting images show the knockdown of SIRT6 in the liver/liver tumor of the HKO mice. Tubulin as a loading control. (C) H&E and Masson's trichrome stained liver sections of normal WT and HKO mice without chemical treatment at 7 months. HKO mice showed a little more steatosis and fibrosis without inducing. Scale bar: 200 μm. (D) Representative microscope images of H&E-stained liver sections. Histopathological examination revealed that HKO mice are more prone to ballooning, steatohepatitis, and inflammation infiltrates. Scale bar: 500 μm. (E) Representative Masson's trichrome staining of liver tissues and morphometrical analysis for the fibrotic score by the method of METAVIR liver fibrosis scoring. HKO mice exhibit more severe liver fibrosis. Scale bar: 500 μm. (F–I) Mice serum levels of ALT, AST, TC and TG. N = 5 in each group values are given as mean ± SEM, ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. (J) Gross liver morphology of the four groups at the time of sacrifice. SIRT6 knockout displayed more HCC nodules both in the female and male groups even though female mice were not prone to tumor formation. The blue arrows indicate macroscopic tumor nodules in female mice. Male mice are almost full of nodules hard to show arrows. The number in each graph is the mark of each mouse for discrimination. N = 8 in each group. (K) Quantification of tumor nodules in (J), gross body weight (BW) and liver weight (LW) and the ratio of LW to BW of the four group mice at 7 months. Data are expressed as means ± SEM, and the P-value was analyzed by a 2-tailed Student's t-test. ns: not significant, ∗P < 0.05.
To clarify the effect of SIRT6 loss on the regression of DEN and CCl4 -induced liver injury, we performed H & E and Masson's trichrome staining on liver sections and found that HKO mice are more prone to ballooning, steatohepatitis and inflammation infiltrate than WT mice both in the female and male group, and male mice present a higher incidence of liver damage (Fig. 1D, E). We used the METAVIR liver fibrosis scoring to scale for fibrosis/cirrhosis ranging from 0 (no fibrosis) to 4 (cirrhosis). Quantitative analysis showed a significant severe in the fibrotic score. Serum ALT, AST, TC and TG as markers of hepatic injury were significantly increased in HKO mice after DEN and CCl4 exposure (Fig. 1F–I). Taken together, we firstly investigated that SIRT6 deficit results in DEN and CCl4-induced hepatocarcinogenesis in mice.
SIRT6 was upregulated in tumor tissues and limited HCC cell proliferation and tumor growth
It was reported that hepatic specific deletion of SIRT6 in mice developed 90% fatty liver at about 7.5–13 months.21 We performed H&E and Masson's trichrome staining and did find HKO mice exhibit more severe steatosis than normal controls without chemical treatment at 7 months (Fig. 1C). In light of the finding that SIRT6 as a tumor suppressor and deficiency of SIRT6 results in liver injury and cancer, we examined the expression of SIRT6 in tumor tissues and showed that SIRT6 was upregulated in tumors even in the HKO mice tumors compared to non-tumor tissues (Fig. 1B), and SIRT6 was also upregulated in human HCC cell lines compared to normal human liver cells analyzed by western blotting assay (Fig. 2A). SIRT6 may be compensatory increased in tumors to suppress cancer development. To evaluate the effect of SIRT6 in cell proliferation in vitro, we successfully knocked down SIRT6 in human HCC cell line HuH7 by shRNA (Fig. 2B, C) and overexpressed SIRT6 by transfection with pcDNA3.1-SIRT6(Fig. 2D, E). The cell clone formation assay showed overexpressed SIRT6 inhibits cell clone formation in HuH7 cells, and knockdown of SIRT6 significantly promotes clone formation ability (Fig. 2F, G). Taken together, these data indicate that SIRT6 was compensatorily overexpressed in tumors and played a role in limiting cell clone formation in vitro.
Figure 2.
SIRT6 was upregulated in tumor tissues and contributed to HCC cell proliferation and tumor growth inhibition. (A) The representative western blotting images show the expression of SIRT6 in the human HCC cell lines and normal human liver cell lines. Tubulin as a loading control. (B,C) RT-qPCR and western blotting assay was conducted to verify the knockdown effect of SIRT6 by shRNA in HuH7 cells. β-actin: loading control. (D,E) The overexpressed effect of SIRT6 was confirmed by RT-qPCR and western blotting analysis. β-actin: loading control. (F) SIRT6 overexpressed inhibits cell clone formation in the HuH7 cell, and knockdown of SIRT6 significantly promotes clone formation ability. (G) The quantitative analysis of the cell clone formation assay. Values represent the mean ± SEM, ∗∗P < 0.01; ∗∗∗P < 0.001. (H)In vivo antitumor efficacy of SIRT6 in xenograft tumor model the implantation of HuH7 cells expressing pcDNA3.1 or pcDNA3.1-SIRT6. Representative images of xenograft tumors dissected from mice of each group (N = 5), and the tumor weight was calculated. (I) Tumor size was measured every four days until 34 days after injection. The data represent mean ± SEM, ∗P < 0.05; ∗∗P < 0.01. (J) Immunoblotting analysis of SIRT6 in four representative xenograft tumors from each group. Tubulin as a loading control. (K) Representative images of expression of Ki-67 in xenograft tumor tissues analyzed by immunohistochemistry. Scale bars: 500 μm.
After demonstrating the impact of the absence of SIRT6 in liver cancer development both in mice and human cell lines, overexpression of SIRT6 also showed cell proliferation limitation. Next, we aimed to investigate whether SIRT6 overexpression influences the biological behavior of HCC tumors in vivo. Therefore, we used stably transfected HuH7-pcDNA3.1 or HuH7-pcDNA3 -SIRT6 cells to establish a xenograft mouse model. The animal experiments showed the limitary effect of SIRT6 in the xenograft formation and slowed tumor growth (Fig. 2H, I). Meanwhile, the SIRT6 protein level was examined and confirmed retaining a significantly high expression in the pcDNA3 -SIRT6 xenografts compared with pcDNA3.1-HuH7 negative control (Fig. 2J). We also stained with the proliferation marker Ki67 in xenograft tumor tissues and showed that Ki-67 was downregulated in SIRT6 overexpressed group (Fig. 2K). Altogether, these data indicate that SIRT6 plays a tumor suppressor role in HCC tumor growth in vivo.
SIRT6 plays a tumor-suppressive role by ERK1/2 pathway inhibition
Next, we aimed to investigate how SIRT6 inhibited liver tumorigenesis in mice and human HCC cell lines. Serine and threonine kinase ERK1/2 (extracellular signal-regulated kinase 1 and 2) as a member of Mitogen-activated protein kinases (MAPKs) family, its deregulation contributes to many cancers development, including HCC.26 It was reported that MAPK signaling is a crucial pathway linked to SIRT6 and cancer.13 We found that the SIRT6 knockdown activated ERK1/2 pathway and overexpression of SIRT6 or activation by selective SIRT6 activator MDL-800 significantly inhibited the phosphorylation of ERK1/2 in HuH7 cells (Fig. 3A–D). We also investigated that the abnormal activation status of ERK1/2 in HKO mice and tumor tissues (Fig. 3E, F). We, therefore, hypothesized that SIRT6 might inhibit liver cancer development by ERK1/2 pathway inactivation.
Figure 3.
Deficiency of SIRT6 induces ERK1/2 pathway activation. (A–D) Quantitative Western blot analysis of p-ERK and ERK expression in the indicated group. β-actin as a loading control. Group1: HuH7 cells were stable expression with negative-control or SIRT6 shRNA. Group2: HuH7 cells were transfected with pcDNA3.1 or pcDNA3.1-SIRT6 for 72 h then harvested for Western blot. Group3: HuH7 cells were harvested for Western blot after 48 h treating with 25 μM MDL-800 or vehicle control. (E,F) Western blot analysis using liver extracts from normal WT and HKO mice, and chemical-induced hepatocellular carcinoma mouse model (isolated tumor nodules). Values represent the mean ± SEM from three independent experiments, ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Discussion
In recent years, increasing evidence has shown that SIRT6, which is widely expressed in the brain, heart, kidney, liver, lung and other organ tissues of mice is closely related to the initiation and progression of different types of cancers.27 However, due to the diversiform functions and the related complicated signaling pathways, SIRT6 may play a dual role in tumors. Feng and colleagues indicated that SIRT6 as a tumor suppressor. Its overexpression significantly suppresses glioma cell growth by inactivating the JAK2/STAT3 signaling pathway.28 A recent study has found that SIRT6 overexpression can be used as a marker for malignant colorectal cancer, suggesting that SIRT6 may be a new target for the treatment of colorectal cancer.29 And a study including 86 patients with non-small cell lung cancer showed that the expression of SIRT6 was significantly correlated with cell differentiation degree, metastasis and patient survival.30 But overall, SIRT6 is widely recognized as a tumor suppressor.
In HCC, SIRT6 acts as both suppressor and oncogene. According to the literature, Ran et al demonstrated that SIRT6 was frequently upregulated in clinical HCC samples and act as a protumorigenic factor.31 In contrast, SIRT6 was reported downregulated in human hepatocellular carcinomas from the Oncomine Cancer Microarray database, and loss of the aging-related gene SIRT6 might lead to the development HCC formation.31 The pathways of SIRT6 involved in cancer are very complicated, which can be specific to tumor type, certain context, or stage. In our study, we established a chemical-induced liver cancer model and found that SIRT6 was upregulated in tumors and human HCC cell lines, overexpressed SIRT6 has protective roles in liver cancer. We hypothesized that SIRT6 was compensatory overexpression in HCC to inhibit tumor growth, or perhaps some more important tumor suppressors lead to the upregulation of SIRT6 to inhibit HCC progression. Due to the complex and critical functions of SIRT6 in cancer development, we need further study to understand the detailed mechanisms of SIRT6 in liver cancer development.
In this study, we established a DEN and CCl4-induced liver cancer mouse model and shed light on that SIRT6 acts as a potential cancer suppressor since a hepatic deficit of SIRT6 promotes liver injury and the development of liver cancer in mice. Overexpression of SIRT6 significantly inhibits tumor growth of HCC cells in the xenograft mice model. Our findings demonstrate that SIRT6 plays a critical role in the development of human and mouse liver cancer. In vitro experiments demonstrated that overexpression of SIRT6 or activation by a selective SIRT6 activator MDL-800 significantly inhibited the ERK1/2 signaling pathway, consistent with the finding that upregulated SIRT6 suppressed HCC cell growth by blocking the ERK1/2 signaling.32 Previous studies have reported that SIRT6 bound to the promoters of ERK1/2 and deacetylated histone 3 at Lys9 (H3K9), thereby inhibiting ERK1/2 expression.33 Cea et al showed that SIRT6 interacts with the transcription factor ELK1 and with the ERK signaling-related gene. By binding to their promoters and deacetylating H3K9, SIRT6 downregulates the expression of MAPK pathway genes and proliferation.34 Kim et al showed that cyclic AMP signaling reduces SIRT6 expression by promoting ubiquitin-proteasomal degradation via inhibition of the Raf/MEK/ERK Pathway.35 Many other signaling pathways may also be included in this study to better understand the mechanism associated with SIRT6 in liver cancer development, like the NF-κB pathway, which can be attenuated by SIRT6,36 Twist-related protein 1 (Twist1) signaling which has been demonstrated regulating tumor progression.37,38
In conclusion, our study demonstrated that liver-specific knockout of SIRT6 in mice promotes liver tumorigenesis and antitumor effects of SIRT6 through inhibition of the ERK1/2 signaling pathway. Considering this, combined activation of SIRT6 and inhibition of ERK1/2 activities may profoundly improve liver injury and HCC therapies.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Author contributions
M.W., H.X. designed the study, performed experiments and analyzed the data. L.L., F.Y, S.J., H.X., C.Z., G.Z., J.X. participated in discussion of related experiments, acquired and analyzed the clinical and experimental data. All authors reviewed the manuscript.
Conflict of interests
All authors of this paper declare no potential conflicts of interest.
Funding
This study was supported grants from the National Natural Science Foundation of China (No. 81902803, 81972233), the Overseas Young Talents Project of China, “Innovative and Entrepreneurial Team” (No. (2018)2015), Science and Technology Grant (No. BE2019758) and the Natural Science Foundation (No. BK20190657) of Jiangsu Province, Southeast University-Nanjing Medical University Cooperative Research Project (No. 2242018K3DN33), Fund of Nanjing Medical University and the China Scholarship Council (No. 201906090247).
Acknowledgements
We thanks Prof.Jian Zhang from the Shanghai Jiao Tong University School of Medicine for kindly providing the MDL-800.40
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.08.007.
Contributor Information
Guoren Zhou, Email: zhouguoren888@126.com.
Hongping Xia, Email: xiahongping@njmu.edu.cn.
Jinglin Xia, Email: xiajinglin@wzhospital.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA: Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Muller M., Bird T.G., Nault J.C. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol. 2020;72(5):990–1002. doi: 10.1016/j.jhep.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs B.C., Hoshida Y., Fujii T., et al. Epidermal growth factor receptor inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology. 2014;59(4):1577–1590. doi: 10.1002/hep.26898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H.J., Hu M.H., Xu F.G., Xu H.J., She J.J., Xia H.P. Understanding the inflammation-cancer transformation in the development of primary liver cancer. Hepatoma Res. 2018;4(7):29. [Google Scholar]
- 7.Wang Z., Lin H., Hua F., Hu Z.W. Repairing DNA damage by XRCC6/KU70 reverses TLR4-deficiency-worsened HCC development via restoring senescence and autophagic flux. Autophagy. 2013;9(6):925–927. doi: 10.4161/auto.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabachini T., Fernandez-Marrero Y., Montani M., et al. BOK promotes chemical-induced hepatocarcinogenesis in mice. Cell Death Differ. 2018;25(4):708–720. doi: 10.1038/s41418-017-0008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llovet J.M., Zucman-Rossi J., Pikarsky E., et al. Hepatocellular carcinoma. Nat Rev Dis Prim. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 10.Uehara T., Pogribny I.P., Rusyn I. The DEN and CCl4 -induced mouse model of fibrosis and inflammation-associated hepatocellular carcinoma. Curr Protoc Pharmacol. 2014;66:14 30 11–10. doi: 10.1002/0471141755.ph1430s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farazi P.A., Glickman J., Horner J., Depinho R.A. Cooperative interactions of p53 mutation, telomere dysfunction, and chronic liver damage in hepatocellular carcinoma progression. Canc Res. 2006;66(9):4766–4773. doi: 10.1158/0008-5472.CAN-05-4608. [DOI] [PubMed] [Google Scholar]
- 12.Uehara T., Ainslie G.R., Kutanzi K., et al. Molecular mechanisms of fibrosis-associated promotion of liver carcinogenesis. Toxicol Sci : Off J Soc Toxicol. 2013;132(1):53–63. doi: 10.1093/toxsci/kfs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitiello M., Zullo A., Servillo L., et al. Multiple pathways of SIRT6 at the crossroads in the control of longevity, cancer, and cardiovascular diseases. Ageing Res Rev. 2017;35:301–311. doi: 10.1016/j.arr.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Mostoslavsky R., Chua K.F., Lombard D.B., et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 15.Kanfi Y., Naiman S., Amir G., et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 16.Irminger-Finger I. Science of cancer and aging. J Clin Oncol : Off J Am Soc Cli Oncol. 2007;25(14):1844–1851. doi: 10.1200/JCO.2007.10.8928. [DOI] [PubMed] [Google Scholar]
- 17.Sebastian C., Zwaans B.M., Silberman D.M., et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151(6):1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kugel S., Feldman J.L., Klein M.A., et al. Identification of and molecular basis for SIRT6 loss-of-function point mutations in cancer. Cell Rep. 2015;13(3):479–488. doi: 10.1016/j.celrep.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kugel S., Sebastian C., Fitamant J., et al. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell. 2016;165(6):1401–1415. doi: 10.1016/j.cell.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerrer B., Cohen H.Y. The guardian: metabolic and tumour-suppressive effects of SIRT6. EMBO J. 2013;32(1):7–8. doi: 10.1038/emboj.2012.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H.S., Xiao C., Wang R.H., et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metabol. 2010;12(3):224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ka S.O., Bang I.H., Bae E.J., Park B.H. Hepatocyte-specific sirtuin 6 deletion predisposes to nonalcoholic steatohepatitis by up-regulation of Bach1, an Nrf2 repressor. FASEB J: Off Publ Fed Am Soc Exp Biol. 2017;31(9):3999–4010. doi: 10.1096/fj.201700098RR. [DOI] [PubMed] [Google Scholar]
- 23.Vesselinovitch S.D., Mihailovich N. Kinetics of diethylnitrosamine hepatocarcinogenesis in the infant mouse. Canc Res. 1983;43(9):4253–4259. [PubMed] [Google Scholar]
- 24.Park T.J., Kim J.Y., Oh S.P., et al. TIS21 negatively regulates hepatocarcinogenesis by disruption of cyclin B1-Forkhead box M1 regulation loop. Hepatology. 2008;47(5):1533–1543. doi: 10.1002/hep.22212. [DOI] [PubMed] [Google Scholar]
- 25.Stowell R.E., Lee C.S., Tsuboi K.K., Villasana A. Histochemical and microchemical changes in experimental cirrhosis and hepatoma formation in mice by carbon tetrachloride. Canc Res. 1951;11(5):345–354. [PubMed] [Google Scholar]
- 26.Min L., He B., Hui L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin Canc Biol. 2011;21(1):10–20. doi: 10.1016/j.semcancer.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Liszt G., Ford E., Kurtev M., Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280(22):21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 28.Feng J., Yan P.F., Zhao H.Y., Zhang F.C., Zhao W.H., Feng M. SIRT6 suppresses glioma cell growth via induction of apoptosis, inhibition of oxidative stress and suppression of JAK2/STAT3 signaling pathway activation. Oncol Rep. 2016;35(3):1395–1402. doi: 10.3892/or.2015.4477. [DOI] [PubMed] [Google Scholar]
- 29.Geng C.H., Zhang C.L., Zhang J.Y., Gao P., He M., Li Y.L. Overexpression of Sirt6 is a novel biomarker of malignant human colon carcinoma. J Cell Biochem. 2018;119(5):3957–3967. doi: 10.1002/jcb.26539. [DOI] [PubMed] [Google Scholar]
- 30.Zhu B., Yan Y., Shao B., Tian L., Zhou W. Downregulation of SIRT6 is associated with poor prognosis in patients with non-small cell lung cancer. J Int Med Res. 2018;46(4):1517–1527. doi: 10.1177/0300060517750298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran L.K., Chen Y., Zhang Z.Z., et al. SIRT6 overexpression potentiates apoptosis evasion in hepatocellular carcinoma via BCL2-associated X protein-dependent apoptotic pathway. Clin Canc Res : Off J Am Assoc Cancer Res. 2016;22(13):3372–3382. doi: 10.1158/1078-0432.CCR-15-1638. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z.G., Qin C.Y. Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signal regulated kinase signaling pathway. Mol Med Rep. 2014;9(3):882–888. doi: 10.3892/mmr.2013.1879. [DOI] [PubMed] [Google Scholar]
- 33.Li Z., Xu K., Zhang N., et al. Overexpressed SIRT6 attenuates cisplatin-induced acute kidney injury by inhibiting ERK1/2 signaling. Kidney Int. 2018;93(4):881–892. doi: 10.1016/j.kint.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 34.Cea M., Cagnetta A., Adamia S., et al. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood - J Am Soc Hematol. 2016;127(9):1138–1150. doi: 10.1182/blood-2015-06-649970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim E.-J., Juhnn Y.-S. Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase) pathway. J Biol Chem. 2015;290(15):9604–9613. doi: 10.1074/jbc.M114.633198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos-Barriopedro I., Bosch-Presegue L., Marazuela-Duque A., et al. SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-kappaB pathway. Nat Commun. 2018;9(1):101. doi: 10.1038/s41467-017-02586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han Z., Liu L., Liu Y., Li S. Sirtuin SIRT6 suppresses cell proliferation through inhibition of Twist1 expression in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(8):4774–4781. [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J., Mani S.A., Donaher J.L., et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Tang C., Liu P., Zhou Y., Jiang B., Song Y., Sheng L. Sirt6 deletion in hepatocytes increases insulin sensitivity of female mice by enhancing ERalpha expression. J Cell Physiol. 2019;234(10):18615–18625. doi: 10.1002/jcp.28500. [DOI] [PubMed] [Google Scholar]
- 40.Huang Z., Zhao J., Deng W., et al. Identification of a cellularly active SIRT6 allosteric activator. Nature Chemical Biology. 2018;14(12):1118–1126. doi: 10.1038/s41589-018-0150-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.