Abstract
This study investigated the role of N6-methyladenosine RNA methylation in liver regeneration following partial hepatectomy in mice. We created a liver-specific knockout mouse model by the deletion of Mettl3, a key component of the N6-methyladenosine methyltransferase complex, using the albumin-Cre system. Mettl3 liver-specific knockout mice and their wild-type littermates were subjected to 2/3 partial hepatectomy. Transcriptomic changes in liver tissue at 48 h after partial hepatectomy were detected by RNA-seq. Immunohistochemistry and immunofluorescence were used to determine protein expression levels of Ki67, hepatocyte nuclear factor 4 alpha, and cytokeratin 19. Terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling was also performed. Liver weight/body weight ratios after partial hepatectomy were significantly lower in Mettl3 liver-specific knockout mice than in wild-type mice at 48 h after 2/3 partial hepatectomy (3.1% ± 0.11% vs. 2.7% ± 0.03%). Compared with wild-type littermates, Mettl3 liver-specific knockout mice showed reduced bromodeoxyuridine staining and reduced Ki-67 expression at 48 h after 2/3 partial hepatectomy. RNA-seq analysis showed that Mettl3 liver-specific knockout delayed the cell cycle progression in murine liver by downregulating the expression levels of genes encoding cyclins D1, A2, B1, and B2. Loss of Mettl3-mediated N6-methyladenosine function led to attenuated liver regeneration by altering the mRNA decay of suppressor of cytokine signaling 6, thereby inhibiting the phosphorylation of signal transducer and activator of transcription 3 during early liver regeneration. These results demonstrated the importance of N6-methyladenosine mRNA modification in liver regeneration and suggest that Mettl3 targeting might facilitate liver regeneration.
Keywords: Liver regeneration, METTL3, N6-methyladenosine, Socs6, STAT3
Abbreviations: m6A, N6-methyladenosine; WT, wild-type; LKO, liver-specific knockout; PHx, partial hepatectomy
Introduction
N6-methyladenosine (m6A) is a common and abundant RNA modification found in eukaryotes. There has been increasing attention to post-transcriptional RNA modification mechanisms in m6A research.1 m6A-specific antibodies were first reported in the 1970s by Munns et al,2 while the first demethylase fat mass and obesity-associated protein was identified in 2011 by He et al.3 Those findings demonstrated that m6A mRNA modifications are detectable, reversible, and dynamic. m6A is deposited by the m6A methyltransferase complex (i.e., the METTL3/METTL14/WTAP complex and other cofactor proteins, including RBM15 and RBM15B) and erased by m6A demethylases (e.g., fat mass and obesity-associated protein and ALKBH5). m6A is recognized by m6A reader proteins (i.e., YTHDF1/2, YTHDC1/2, YTHDF3, and IGF2BP1/2/3) that may mediate the divergent roles of m6A in RNA metabolism, including the stability, translation, and splicing of m6A-containing mRNAs.4
Recent studies have substantially contributed to the understanding of m6A post-transcriptional modification in regulating transcription, RNA processing events, splicing, RNA stability, and translation.5 Additionally, a key methyltransferase in RNA m6A modification, METTL3, has been reported to play important roles in many biological processes (e.g., spermatogenesis, T-cell homeostasis, and stem cell differentiation).6, 7, 8 A previous study showed that highly methylated genes are enriched in regulatory pathways related to growth and development, metabolic processes, and protein catabolic processes during porcine postnatal liver development.9 The liver has a unique ability to regenerate in response to injury; improved understanding of the regenerative process may aid in the treatment of liver failure and may elucidate the mechanism of cancer development within cirrhotic liver.10 However, the function of METTL3-mediated m6A mRNA methylation in liver regeneration remains obscure.
Here, we performed liver-specific ablation of METTL3 in mice, which were then subjected to 2/3 partial hepatectomy (PHx). Our results showed that METTL3-mediated m6A modification is an important epigenetic mechanism involved in regulating the cell cycle during early liver regeneration. Notably, dysregulated m6A modification caused reduction of suppressor of cytokine signaling (Socs6) mRNA decay, thus inhibiting signal transducer and activator of transcription (STAT)3 signaling during early liver regeneration.
Materials and methods
Generation of Mettl3 LKO mice and modeling of liver regeneration
Mettl3 conditional knockout mice were generated by inserting a loxP site into both the first and last introns of the Mettl3 gene using the CRISPR–Cas9-based genome-editing system. These mice were gifts from Pr. Huabing Li, Shanghai Jiao Tong University School of Medicine. Mettl3F/FAlbumin-Cre (liver-specific knockout [LKO]) mice and their wild-type (WT) littermates were sacrificed at several time points after 2/3 PHx; 2/3 PHx was performed as previously described.11 Remnant liver tissue was then harvested, flash-frozen, and processed for RNA and immunoblotting analyses. Liver/body weight ratios were calculated to determine the recovery of liver mass. Mice were maintained in a standard 12-h light/dark system. All procedures involving animals were performed in accordance with approval from the Animal Care and Use Committee of Shanghai Jiaotong University.
Immunoblotting and immunohistochemistry
Protein expression levels were investigated using standard sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblotting methodology. Primary polyclonal antibodies used in this study were as follows: anti-METTL3 (Abcam, Cambridge, MA, USA; cat. no. ab195352), anti-β-actin (Proteintech, Rosemont, IL, USA; cat. no. 60008-1-Ig), anti-m6A (Cell Signaling Technology, Danvers, MA, USA; cat. no. 56593), anti-STAT3 (Cell Signaling Technology; cat. no. 30835), anti-phospho-STAT3 (Tyr727) (Cell Signaling Technology; cat. no. 49081S) and anti-SOCS6 (Abcam; cat. no. ab197335). The terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling and immunohistochemistry protocols were performed in accordance with the reagent manufacturers’ instructions. Slides were probed with anti-bromodeoxyuridine (Cell Signaling Technology; cat. no. 5292S) and anti-Ki67 (Cell Signaling Technology; cat. no. 9449) antibodies, then stained with Alexa488-conjugated anti-rabbit secondary antibody. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. RNA N6-methyladenosine (m6A) dot blotting was performed as previously described,12 primary polyclonal antibody m6A antibody (Abcam; cat. no. ab151230) was used in this assay.
Quantitative polymerase chain reaction (qPCR) and RNA sequencing
Total RNA was extracted using TRIzol reagent. The relative mRNA abundances of target genes were measured using SYBR Green PCR Master Mix (Life Technologies, Carlsbad, CA, USA; cat. no. 4367659). For RNA-seq data analysis, genes were considered significantly differentially expressed when log2 [fold change] ≥ 1 or ≤ −1 and adjusted P value < 0.05. Gene set analysis was performed and enriched reactome pathways were obtained through online bioinformatics tools. Primers used for qPCR are shown in Table S1.
Methylated RNA immunoprecipitation (MeRIP)-qPCR assay
Five micrograms of m6A antibody (Abcam; cat. no. ab151230) and normal rabbit IgG (Cell Signaling Technology; cat. no. 3900) were respectively mixed with 50 μl of protein A/G-conjugated magnetic beads (Bio-Rad, Hercules, CA, USA; cat. no. 161-4023), overnight at 4 °C. Approximately 200 μg of fragmented total RNA were incubated with each antibody in immunoprecipitation buffer (50 mM Tris–HCl, 750 mM NaCl, and 0.5% nonidet-P40) supplemented with 40 IU RNase inhibitor, overnight at 4 °C. RNA was eluted from the beads by incubation with 300 μl of elution buffer (5 mM Tris–HCl, 1 mM ethylenediaminetetraacetic acid, and 0.05% sodium dodecyl sulfate) and 8 μg of proteinase K for 1.5 h at 50 °C. Following phenol extraction and ethanol precipitation, both input and m6A-enriched RNA were reverse transcribed. The extent of enrichment was determined by qPCR. The primers used for gene expression are shown in Table S1.
RNA degradation assay
For signaling-dependent degradation analysis, primary Mettl3 LKO and WT hepatocytes were treated with actinomycin-D at a concentration of 5 μg/ml for 0, 2 h, or 4 h; they were then lysed with TRIzol. The amount of a particular mRNA transcript remaining after various durations of treatment was used to calculate the mRNA decay rate for each transcript.
Statistical analysis
All data are presented as the mean ± standard deviation of at least three independent experiments. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Inc., La Jolla, CA, USA). Statistical significance was determined using a two-tailed Student's t-test or analysis of variance. Values were considered statistically significant when ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Results
Total RNA m6A level and Mettl3 expression were both upregulated during liver regeneration
To explore changes in m6A mRNA methylation during liver regeneration, we examined dynamic changes in total m6A RNA level and in eight m6A-related genes (i.e., Mettl3, Mettl14, Wtap, Fto, Alkbh5, Ythdf1, Ythdf2, and Ythdc1) during early liver regeneration. Surgeries (2/3 PHx and sham) were performed in WT C57BL6/J mice at different time points. As shown in Figure 1A, the total RNA m6A level (determined by m6A-specific antibody dot blot analysis) was significantly increased at 12 and 24 h after 2/3 PHx (P < 0.001). Among the seven m6A-related genes, the mRNA expression level of Mettl3 was upregulated after 2/3 PHx, compared with the sham operation group, beginning at 12 h postoperatively (Fig. 1B, P < 0.05). Subsequent immunoblotting analysis confirmed the upregulation of METTL3 expression in the 2/3 PHx group, beginning at 12 h postoperatively (Fig. 1C, P < 0.05). These data demonstrated the dynamic enhancement of RNA m6A modification and Mettl3 expression during liver regeneration.
Figure 1.
Mettl3 and total RNA m6A levels were upregulated during liver regeneration. (A) m6A antibody dot blot staining (left panel) and its quantification (right panel) of total RNA (100 ng) from liver lysate after PHx. Total RNA (100 ng) was stained with methylene blue as a control. (B) qPCR analysis of m6A-related genes in WT mice at different times after PHx. (C) Results of immunoblotting (left panel) and analysis of relative METTL3 protein level (right panel) in WT mice at different times after PHx. ∗P < 0.05, ∗∗∗P < 0.001. Abbreviations: WT, wild-type; N.S., not significant.
Mettl3 LKO mice were successfully constructed and used for evaluation of mid-to-late fetal liver development
To further investigate the roles of m6A modification and Mettl3 expression in murine hepatocyte proliferation, as well as liver regeneration, we obtained Mettl3 LKO mice and age-matched WT littermates. Successful Mettl3 knockout in murine hepatocytes was confirmed by qPCR and immunoblotting (Fig. 2A, B). Because METTL3 and METTL14 form a complex for m6A deposition, we examined the expression level of METTL14 in Mettl3 LKO mice. We found that METTL14 was downregulated in Mettl3 LKO hepatocytes (Fig. S1A). m6A dot blot analysis indicated that the global mRNA m6A level in Mettl3 LKO hepatocytes was also reduced (Fig. 2C).
Figure 2.
Mettl3 LKO delayed hepatocyte proliferation during liver regeneration. (A, B) qPCR and immunoblotting analyses of Mettl3 expression in purified hepatocytes from Mettl3 LKO mice and their WT littermates. (C) m6A antibody dot blot staining (left panel) and its quantification (right panel) of total RNA in hepatocytes from Mettl3 LKO mice and their WT littermates. Total RNA was stained with methylene blue as a control. (D) Ratios of liver weight to body weight (LW/BW) in Mettl3 LKO mice and their WT littermates at different times after PHx (n = 7 per genotype). (E) Representative immunofluorescence images showing bromodeoxyuridine (BrdU) staining (red) and 4′,6-diamidino-2-phenylindole (DAPI) staining (blue) in liver sections from 6–8-week-old Mettl3 LKO mice and their WT littermates at 48 and 72 h after PHx (scale bar, 25 μm). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Abbreviations: WT, wild-type F/FMETTL3F/F mice; LKO, METTL3F/FAlb-Cre+/− mice.
To explore the influence of Mettl3 LKO on murine liver development and function, we compared body and liver weights between Mettl3 LKO mice and their WT littermates; we found no differences between groups in these parameters at 3 weeks of age. Moreover, we found no differences in gross liver morphology or hematoxylin and eosin staining results between the two groups at 3 weeks of age (data not shown). Furthermore, as shown in Figure S1B, the expression patterns of hepatocyte nuclear factor 4 alpha (a marker for liver differentiation) and cytokeratin 19 (a marker for cells of epithelial origin) were comparable between 3-week-old Mettl3 LKO mice and their WT littermates. In 8-week-old Mettl3 LKO livers, the absence of Mettl3 did not lead to enhanced apoptosis, as revealed by terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling staining (Fig. S1C). These data indicated that METTL3 and its regulation of m6A RNA methylation were not essential during mid-to-late fetal liver development.
Mettl3 LKO suppressed hepatocyte proliferation during early liver regeneration
To investigate the role of Mettl3 and its regulation of m6A RNA methylation in liver proliferation and regeneration, we performed 2/3 PHx in 8-week-old Mettl3 LKO mice and their WT littermates. As shown in Figure 2D, the liver weight to body weight ratio was significantly lower in Mettl3 LKO mice than in WT mice at both 48 and 72 h after 2/3 PHx. Additionally, there was no significant difference in liver weight to body weight ratio between Mettl3 LKO mice and their WT littermates at 96 h after 2/3 PHx (data not shown). Intraperitoneal injection of bromodeoxyuridine (a marker for analysis of DNA synthesis and cell proliferation) revealed reduced staining at 48 and 72 h after 2/3 PHx in Mettl3 LKO mice, compared with WT mice (Fig. 2E). These results indicated that dysregulated m6A modification led to impaired hepatocyte regeneration following liver injury. Furthermore, METTL3-mediated m6A modification presumably affected early liver regeneration.
METTL3 knockout led to delayed hepatocyte cell cycle progression through the SOCS6/STAT3 pathway in mice subjected to 2/3 PHx
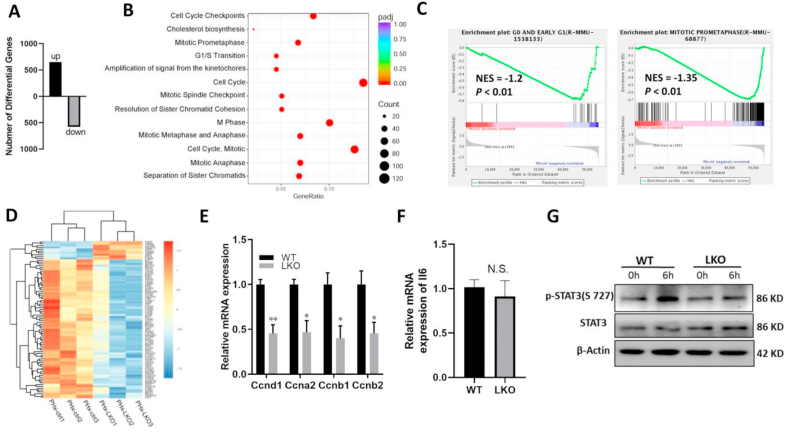
To further elucidate the molecular mechanisms underlying the functions of Mettl3 during liver proliferation and regeneration, we performed genome-wide transcriptome profiling analysis using RNA-seq in liver tissue harvested at 48 h after 2/3 PHx in 8-week-old Mettl3 LKO mice and their WT littermates (n = 3 per group). Compared with WT mice, 650 upregulated genes and 580 downregulated genes were found in Mettl3 LKO mice after 2/3 PHx (Fig. 3A, log2 [fold change] ≥ 1, adjusted P value < 0.05). Analysis of functional annotation enrichment revealed that most downregulated genes were involved in G1/S transition and mitotic phase regulation (mitotic prometaphase, mitotic anaphase and mitotic spindle checkpoint) (Fig. 3B). Gene Set Analysis Enrichment revealed that Mettl3 LKO mice significantly downregulated G0/G1 and mitotic prometaphase pathway genes after 2/3 PHx (Fig. 3C). The 86 cell cycle-related genes that were investigated in Mettl3 LKO mice after 2/3 PHx are shown in Figure 3D; approximately 88% (76 of 86 genes) were significantly reduced in Mettl3 LKO mice. qPCR analysis confirmed that genes encoding cyclins D1, A2, B1, and B2 were significantly downregulated in the livers of Mettl3 LKO mice after 2/3 PHx (Fig. 3E). Overall, the findings indicate that Mettl3 LKO delayed cell cycle progression in hepatocytes of mice subjected to 2/3 PHx.
Figure 3.
Mettl3 LKO deregulated cell cycle-related genes and STAT3 signaling after 2/3 PHx. (A) Differentially expressed genes (DEGs) between liver tissue from 8-week-old Mettl3 LKO mice and their WT littermates at 48 h after PHx (log2 [fold change] > 1, adjusted P value < 0.05). (B) Bubble Diagram of Reactome pathway analysis based on downregulated genes from among the DEGs. (C) Gene set enrichment analysis (GSEA) of differentially expressed genes between liver tissue from Mettl3 LKO mice and their WT littermates at 48 h after PHx. (D) Heatmap of differentially expressed genes enriched in cell cycle function, derived from list of overall DEGs. (E) qPCR verification of cell cycle-related genes among DEGs. (F) qPCR analysis of interleukin-6 expression in liver tissue from Mettl3 LKO mice and their WT littermates at 48 h after PHx. (G) Immunoblotting analysis of STAT3 and p-STAT3 (Tyr727) protein levels in hepatocytes from 8-week-old Mettl3 LKO mice and their WT littermates at 6 h after PHx. ∗P < 0.05, ∗∗P < 0.01.
Interleukin-6 is considered a pleiotropic cytokine, which regulates hepatocyte proliferation and liver regeneration by activating the STAT3 signaling pathway.13 In this study, the interleukin-6 mRNA expression level did not differ between Mettl3 LKO mice and their WT littermates after 2/3 PHx (Fig. 3F). However, compared with WT mice, the livers of Mettl3 LKO mice exhibited significantly reduced levels of phospho-STAT3 (Tyr727) at 6 h after 2/3 PHx (Fig. 3G), suggesting that the phosphorylation of STAT3 may be inhibited by other molecules in Mettl3 LKO mice. In physiological conditions, STAT3 signaling is carefully controlled by a cohort of negative regulators, mainly comprising the SOCS family.14 Thus, we measured the mRNA expression levels of SOCS members that could inhibit STAT signaling activation in the livers of Mettl3 LKO mice and their WT littermates before and after 2/3 PHx. The results showed that the mRNA and protein expression levels of SOCS6 were upregulated in the livers of Mettl3 LKO mice before and after 2/3 PHx (Fig. 4A–C). These data indicated that Mettl3 LKO may inhibit liver regeneration after liver injury through the SOCS6/STAT3 signaling pathway.
Figure 4.
Mettl3-mediated m6A activity regulated Socs6 mRNA stability in hepatocytes. (A) Analysis of Socs6 mRNA expression in hepatocytes from 8-week-old Mettl3 LKO mice and their WT littermates before and after PHx. (B, C) Immunoblotting analysis of SOCS6 in hepatocytes from 8-week-old Mettl3 LKO mice and their WT littermates before and after PHx. (D) Socs6 mRNA decay analysis in hepatocytes from Mettl3 LKO mice and their WT littermates, following culture for 24 h and subsequent treatment with actinomycin D for 0, 2, and 4 h. (E) MeRIP qPCR confirmation that Socs6 was an METTL3-dependent m6A target in hepatocytes from Mettl3 LKO mice and their WT littermates. (F) Mutation of the m6A consensus sequence increased luciferase activity in 293T cells. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Mettl3 knockout reduced Socs6 mRNA decay during liver regeneration
m6A has been demonstrated to negatively regulate target mRNA stability in many cell types. As noted above, we confirmed that Socs6 was upregulated in Mettl3 LKO liver tissue, compared with WT liver tissue, during liver proliferation and regeneration. Primary hepatocytes isolated from both Mettl3 LKO mice and their WT littermates were pre-treated with actinomycin-D for different durations to measure their mRNA stabilities; Socs6 mRNA was degraded more slowly in Mettl3 LKO hepatocytes than in WT hepatocytes (Fig. 4D). Importantly, Socs6 mRNA was previously identified as a direct target of m6A modification; in this study, MeRIP qPCR confirmed that Socs6 mRNA was a METTL3-dependent m6A target in purified hepatocytes from 8-week-old mice (Fig. 4E).
To further investigate the effect of m6A modification on the expression of Socs6, we constructed a reporter system in which the WT or mutant 3′-untranslated region of Socs6 was tethered to the 3′-untranslated region of the firefly luciferase coding sequence. The adenosine bases in the m6A consensus sequence were changed to cytosine bases, thus abolishing m6A modification. Relative luciferase activities of the WT or mutant Socs6 3′-untranslated region-fused reporter were compared in 293T cells. Compared with the WT Socs6 sequence, mutation of m6A consensus sequences enhanced luciferase activity in 293T cells (Fig. 4F). These data indicated that Socs6 is a major functional target of m6A in hepatocytes, and that m6A may primarily regulate Socs6 mRNA decay.
Discussion
RNA m6A modification is reportedly involved in many physiological processes. This study demonstrated the following properties of m6A modification in liver proliferation and regeneration: 1) METTL3 and its regulation of m6A RNA methylation are not essential during mid-to-late fetal liver development; 2) METTL3-mediated m6A modification promotes liver regeneration by activating Socs 6-STAT signaling during early liver regeneration; and 3) deletion of METTL3 in hepatocytes led to elevated Socs6 expression and may regulate mRNA stability in an m6A-dependent manner.
Recent studies showed that Mettl3 is required for differentiation of embryonic stem cells and hematopoietic stem cells, as well as differentiation of spermatogonia and initiation of meiosis.15,16 Notably, the albumin gene begins expression during embryogenesis shortly after the appearance of the liver bud (9.5 days post coitum) and weak mRNA levels increase following liver development.17 During mid-to-late fetal liver development, hepatic progenitor cells differentiate into hepatocytes and cholangiocytes.18 Previous studies have shown that m6A RNA modification controls cell fate transition in mammalian embryonic stem cells and specification in hematopoietic progenitor cells.19 Our data showed that METTL3 LKO mice were generally normal and did not become acutely ill, suggesting that METTL3 was not required for early fetal liver development and hepatic maturation during mid-to-late fetal development.
The liver regeneration process after PHx or serious liver damage is an important clinical consideration, especially because of the growing population of patients who either require liver transplantation or have toxicity/virus-related liver injuries.20 Liver regeneration is largely driven by hepatocyte proliferation, which relies on robust cell cycle function. Our study showed that Mettl3 LKO led to delayed hepatocyte cell cycle progression during liver regeneration (i.e., G1/S transition and S phase). Thus, hepatocyte proliferation was delayed and liver regeneration was impaired; these findings imply that METTL3-mediated m6A modification has an important role in promoting early liver regenerative activity after PHx. Hepatocytes reportedly require rapid proliferation and regeneration after small-for-size liver transplantation.21 In a previous study of a mouse model of this transplantation approach, METTL3-mediated m6A modification led to significant improvement in survival rate after hepatectomy.
A prior study showed that the SOCS protein family is modified by m6A and is a direct downstream target of METTL3 in T cells; moreover, METTL3-mediated m6A modifications of Socs 1, Socs 3, and Cish mRNAs promote their decay. These inhibitors of the STAT signaling axis may be involved in T-cell homeostasis and differentiation.6 METTL3 expression was enhanced in hepatocellular carcinoma, which promoted liver cancer cell line tumorigenicity and metastasis by repressing the expression of Socs genes in an m6A-YTHDF2-dependent manner.22 In our study, we found that Mettl3 LKO caused upregulation of Socs6 proteins and reduction of Socs6 mRNA decay in hepatocytes, thus inhibiting the STAT3 activation necessary for early liver regeneration.
Previous studies showed that some inhibitors of m6A methylation-related proteins may have promising effects on cancer development and metabolic disease.23 Fat mass and obesity-associated protein is known to negatively regulate m6A; several fat mass and obesity-associated protein inhibitors (i.e., rhein, radicicol, and epigallocatechin gallate) have been shown to treat both cancer and metabolic disease.4 In our study, METTL3 and its regulation of m6A modification promoted early liver regeneration; however, no specific inhibitors or activators of METTL3 have been found thus far. Further studies are needed to explore inhibitors of m6A methylation-related enzymes.
Previous studies have shown that m6A has regulatory roles in modification of histones, lncRNAs, microRNAs, piRNAs, and mRNAs24,25; it may also participate in regulation of DNA replication and the cell cycle. More research is needed to elucidate how METTL3 and its regulation of m6A modification may directly affect hepatocyte proliferation and liver regeneration.
In conclusion, we demonstrated an important role for RNA m6A modification in hepatocyte regeneration. Our data illustrated that alterations of METTL3 and m6A levels led to impaired hepatocyte proliferation and early liver regeneration. Thus, targeting m6A modification by means of METTL3 alterations may provide a novel therapeutic strategy for preventing hepatic regeneration failure due to PHx or serious liver damage.
Conflict of interests
The authors declare no conflicts of interest in this work.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2017YFC0908100 [Q.X.]) and the Key Clinical Subject Construction Project of Shanghai (No. shslczdzk05801).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.11.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Frye M., Harada B.T., Behm M., He C. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munns T.W., Sims H.F., Liszewski M.K. Immunospecific retention of oligonucleotides possessing N6-methyladenosine and 7-methylguanosine. J Biol Chem. 1977;252(9):3102–3104. [PubMed] [Google Scholar]
- 3.Jia G., Fu Y., Zhao X., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Z., Meng J., Su R., et al. Epitranscriptomics in liver disease: basic concepts and therapeutic potential. J Hepatol. 2020;73(3):664–679. doi: 10.1016/j.jhep.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H.B., Tong J., Zhu S., et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batista P.J., Molinie B., Wang J., et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Z., Hsu P.J., Xing X., et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27(10):1216–1230. doi: 10.1038/cr.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He S., Wang H., Liu R., et al. mRNA N6-methyladenosine methylation of postnatal liver development in pig. PLoS One. 2017;12(3):e0173421. doi: 10.1371/journal.pone.0173421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forbes S.J., Newsome P.N. Liver regeneration - mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol. 2016;13(8):473–485. doi: 10.1038/nrgastro.2016.97. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell C., Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3(7):1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 12.Nagarajan A., Janostiak R., Wajapeyee N. Dot blot analysis for measuring global N(6)-methyladenosine modification of RNA. Methods Mol Biol. 2019;1870:263–271. doi: 10.1007/978-1-4939-8808-2_20. [DOI] [PubMed] [Google Scholar]
- 13.Ozaki M. Cellular and molecular mechanisms of liver regeneration: proliferation, growth, death and protection of hepatocytes. Semin Cell Dev Biol. 2020;100:62–73. doi: 10.1016/j.semcdb.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Bi J., Sun K., Wu H., Chen X., Tang H., Mao J. PPARgamma alleviated hepatocyte steatosis through reducing SOCS3 by inhibiting JAK2/STAT3 pathway. Biochem Biophys Res Commun. 2018;498(4):1037–1044. doi: 10.1016/j.bbrc.2018.03.110. [DOI] [PubMed] [Google Scholar]
- 15.Lee H., Bao S., Qian Y., et al. Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol. 2019;21(6):700–709. doi: 10.1038/s41556-019-0318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu K., Yang Y., Feng G.H., et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27(9):1100–1114. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellendonk C., Opherk C., Anlag K., Schütz G., Tronche F. Hepatocyte-specific expression of Cre recombinase. Genesis. 2000;26(2):151–153. doi: 10.1002/(sici)1526-968x(200002)26:2<151::aid-gene17>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 18.Carpentier R., Suñer R.E., van Hul N., et al. Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology. 2011;141(4):1432–1438. doi: 10.1053/j.gastro.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng H., Huang H., Wu H., et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22(2):191–205. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vollmar B., Menger M.D. The hepatic microcirculation: mechanistic contributions and therapeutic targets in liver injury and repair. Physiol Rev. 2009;89(4):1269–1339. doi: 10.1152/physrev.00027.2008. [DOI] [PubMed] [Google Scholar]
- 21.Dahm F., Georgiev P., Clavien P.A. Small-for-size syndrome after partial liver transplantation: definition, mechanisms of disease and clinical implications. Am J Transplant. 2005;5(11):2605–2610. doi: 10.1111/j.1600-6143.2005.01081.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen M., Wei L., Law C.T., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 23.Dai D., Wang H., Zhu L., Jin H., Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9(2):124. doi: 10.1038/s41419-017-0129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu N., Parisien M., Dai Q., Zheng G., He C., Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19(12):1848–1856. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T., Hao Y.J., Zhang Y., et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16(3):289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.