Abstract
A substantial number of male infertility is caused by azoospermia. However, the underlying etiology and the molecular basis remain largely unknown. Through single-cell (sc)RNA sequencing, we had analyzed testis biopsy samples from two patients with obstructive azoospermia (OA) and nonobstructive azoospermia (NOA). We found only somatic cells in the NOA samples and explored the transcriptional changes in Sertoli cells in response to a loss of interactions with germ cells. Moreover, we observed a germ cell population discrepancy between an OA (postvasectomy) patient and a healthy individual. We confirmed this observation in a secondary study with two datasets at GSM3526588 and GSE124263 for detailed analysis wherein the regulatory mechanisms at the transcriptional level were identified. These findings thus provide valuable information on human spermatogenesis, and we also identified insightful information for further research on reproduction-related diseases.
Keywords: Androgen signaling, Retinoic acid, Sertoli cell-only syndrome, Single-cell, Spermatogenesis
Introduction
Spermatogenesis comprised of a series of highly orchestrated cellular events to ensure the continuous supply of millions of spermatozoa on a daily basis since puberty in humans. Spermatogonial stem cells (SSCs) either self-renew or initiate spermatogenesis by producing undifferentiated progenitors that go through amplifying mitotic proliferation, become differentiating spermatogonia, and then form spermatocytes that ultimately develop into haploid spermatozoa through meiosis and spermiogenesis in the testis.1,2 These delicate procedures also involve complex germ cell-Sertoli cell niche interactions to ensure long-term fertility which could be disturbed by genetic disorders, leading to male infertility.3, 4, 5 A substantial proportion of male infertility (approximately 10–15%) is accompanied by azoospermia, which is characterized by the absence of sperm from the ejaculate.6 When the azoospermic symptom is caused by failure in spermatogenesis, termed non-obstructive azoospermia (NOA), or reproductive tract obstruction, termed obstructive azoospermia (OA).
Genomic studies on azoospermia have been performed to profile the mutational landscape of the two disease subtypes, and several genetic causes have since been identified.7,8 Previous bulk transcriptome analyses in OA and NOA studies have revealed distinct transcriptional signatures of these two subtypes.9,10 However, the conclusion might be of limited use due to the highly heterogeneous nature of human testicular tissue because it is composed of numerous cell types, in particular the different germ cell types, and in different functional states (e.g., Sertoli cells, Leydig cells and peritubular myoid cells) during the epithelial cycle.11 The newly developed single-cell RNA sequencing (scRNA-Seq) technique allows transcriptome profiling to be resolved at the single-cell level. This thus enables elucidation of cell type heterogeneity within a testis biopsy sample.
In this study, we performed scRNA-Seq analysis of testicular biopsy tissues from OA and NOA patients, including integrated analysis of public scRNA-Seq data on human testes from healthy individuals versus OA and NOA patients, aiming to uncover the dynamic processes and regulators involved in human spermatogenesis by assessing these disease subtypes.
Materials and methods
Testis sample collection and processing
The study was approved by the institutional review boards at the International Peace Maternity and Child Health Hospital and at Shanghai Jiao Tong University (SJTU). Informed consent with regard to testis samples was obtained from the study participants in accordance with local Institutional Review Board requirements at the time of collection at Shanghai Renji Hospital, SJTU. The fresh biopsies were stored in GEXSCOPE Tissue Preservation Solution (Singleron Biotechnologies, Jiangsu, China) at 4 °C and transported to the processing laboratory within 3 days. The testicular tissues were washed three times with phosphate-buffered saline (PBS, 10 mM sodium phosphate, 0.15 M NaCl, pH 7.4 at 22 °C) at room temperature and subjected to a standard two-step digestion procedure. First, the tissues were digested with collagenase type IV (Sigma Aldrich Catalog Number: C5138–100MG) for 15 min at room temperature. The tubules were sedimented by centrifugation at 200g for 5 min and washed with Hank's Balanced Salt Solution (HBSS). Thereafter, tissues were digested with TrypLE™ Express Enzyme (Thermo Fisher Catalog Number: 12604013), sedimented and washed as in the first step. Single testicular cells were obtained by filtering the material through strainers with a mesh size of 70 μm (Falcon Catalog Number: 352350). The cells were pelleted by centrifugation at 200g for 5 min and washed twice with PBS.
Single-cell RNA library construction and sequencing
Single-cell suspensions of 1 × 105 cells/mL in PBS were prepared. The single-cell suspensions were then loaded onto microfluidic devices and scRNA-Seq libraries were constructed according to the Singleron GEXSCOPE protocol with GEXSCOPE Single-Cell RNA Library Kit (Singleron Biotechnologies).12 The individual library of NOA.1 and OA.1 were diluted to 4 nM and pooled for sequencing. The pools were sequenced using the Illumina HiSeq X Ten platform with 150-bp paired-end reads to obtain a sequencing depth of approximately 6.5 K reads/cell and saturation levels of 70%–80%.
Raw data processing
Raw reads were processed to generate gene expression profiles using an in-house pipeline. Briefly, after filtering R1 reads without poly-T tails, the cell barcodes and unique molecular identifiers (UMIs) were extracted. The adapters and poly-A tails were trimmed (Cutadapt v1.8.1) before the R2 reads were mapped to reference genome hg19 (STAR v2.5.4a and featureCounts 1.6.2).13 Reads with the same cell barcode, UMI and gene were grouped together to calculate the number of UMIs per gene per cell. The UMI count tables of each cellular barcode were used for further analysis. Public scRNA-Seq expression matrixes were obtained from Gene Expression Omnibus (GEO), as noted in Table 1.
Table 1.
Information for scRNA-Seq datasets.
| ID | Conditiona | Library | Reference | Accession IDb |
|---|---|---|---|---|
| OA.1 | OA | Singleron GEXSCOPE™ | – | OEP000778 |
| NOA.1 | NOA | Singleron GEXSCOPE™ | – | OEP000778 |
| Healthy | Healthy | 10x Genomics | 19 | GSE112013 |
| NOA.a | NOA | Smart-Seq2 | 18 | GSE106487 |
| OA.a | Postvasectomy | 10x Genomics | 23 | GSM3526588 |
| OA.b | Postvasectomy | 10x Genomics | 23 | GSM3526590 |
OA: obstructive azoospermia; NOA: non-obstructive azoospermia; healthy: deceased patients without testicular pathology.
Data obtained from our scRNA-Seq were deposited in the NODE Project with Accession Number OEP000778 (https://www.biosino.org/node/project/detail/OEP000778) which are accessible to all investigators. Other public data were deposited in Gene Expression Omnibus (GEO).
Dataset integration, cell identification and clustering analysis
The R package Seurat (http://satijalab.org/seurat/, R package, v.3.1.2) was used for cell type identification and clustering analysis.14 UMI count tables were loaded into R using Read10X and read.table function, and Seurat objects were built from each experiment. Cells were retained only when they showed expression of more than 200 genes; fewer than 20% of the reads mapped to the mitochondrial genome. To mitigate batch effects, we applied the new standard integration workflow implanted in Seurat program (Version 3).15 After performing log-normalization and identification of the top 2000 variable genes in each dataset with default parameters, we identified anchors using the Seurat FindIntegrationAnchors function. The generated anchors, which represent pairwise correspondences between individual cells (one in each dataset) that we hypothesized and originated from the same biological state, were used to perform batch correction and integration via the IntegrateData function with the top 20 dimensions. Twenty statistically significant principal component (PC) dimensions were selected for uniform manifold approximation and projection (UMAP) and FindNeighbors analysis. Thereafter, we set the parameter resolution to 0.6 for the FindClusters function to perform clustering analyses. Multiple cell type-specific/enriched marker genes previously described in the literature were used to identify each cell type (Fig. 1C, S1). Testing for differential expression (DE) between different samples or consecutive clusters was performed using the function FindMarkers with the following parameters: min.pct = 0.25, logfc.threshold = 0.25, and test.use = “MAST”.16 Gene set enrichment analysis (GSEA) was performed to identify cellular pathways using the clusterProfiler (v3.14)17 package and the Gene Ontology (GO) biological process dataset as a reference. Cell–cell interaction analysis was performed by using the CCInx package at https://github.com/BaderLab/CCInx.
Figure 1.
Single-cell transcriptome analyses of the human testis. (A) UMAP projections of scRNA-Seq data (n = 10,909) and number of detected cells. (B) Proportion of cells originating from Healthy, OA.1, NOA.1 and OA.a testes. (C) Violin plot showing the distribution of expression levels of well-known representative cell-type-enriched marker genes across 13 cell types. (D) Heatmap of genes exhibiting differential expression in Sertoli cells in NOA and OA. (E, F) Selected key genes showing differential expression in distinct Sertoli cell states.
Data and code availability
Data obtained from our scRNA-Seq were deposited in the NODE Project with Accession Number OEP000778 at https://www.biosino.org/node/project/detail/OEP000778 freely accessible to all investigators. The R code to reproduce the analysis can be obtained from: https://github.com/dioncst/hTestes_scRNA-seq.
Results
Patient information
Altogether, 6 scRNA-Seq datasets for 10 individuals were utilized. Among the individuals, two (NOA.1 and OA.1) were recruited in our study and for analysis as reported here. OA.1 was a 36-year-old man who had one 8-year-old child and had a history of epididymitis approximately 4 years ago. His testicular volume(left:15 mL, right:16.4 mL), and FSH level(3.9 IU/L, normal range: 3.8–8.8) is in the normal range. NOA.1 was a 31-year-old man with small testes (left: 7.8 mL, right: 6.1 mL) and an increased FSH level (23.35 IU/L). Surgical testicular sperm extraction successfully obtained live sperm from OA.1 testes but failed to obtain sperm from NOA.1 testes.
Cell types and distributions in the adult human testes of healthy, OA and NOA individuals
We integrated the scRNA-Seq data generated in this study with results from public datasets18,19 which also served as references. Unsupervised hierarchical clustering analysis of the scRNA-Seq data revealed different subtypes of testicular cells between OA and NOA patients and healthy individuals (Fig. 1A). We obtained 2,632 testicular cells from one OA donor and 1,212 testicular cells from one NOA donor. Using known markers, we identified cell clusters corresponding to germ cells and somatic cells in the testis, including undifferentiated spermatogonia (Undiff S.gonia), differentiating spermatogonia (Diff S.gonia), spermatocytes (S.cytes), round spermatids (RS), elongated spermatids I(ES I), elongated spermatids II (ES II), Sertoli cells (SC), Leydig cells (LC), Mast cells, T cells, peritubular myoid cells (PTMC), testicular macrophages (tMΦ) and endothelial cells (EC). Part of the marker genes we used to identify these cell subsets are shown in Figure 1B, and Figure S1.
Only somatic cells were found in NOA.1; a few differentiating spermatogonia and spermatocytes were present in NOA.a (Fig. 1C). Therefore, it is likely that NOA.a exhibited maturation arrest (MA) and that NOA.1 exhibited Sertoli cell-only syndrome (SCOS), suggesting that scRNA-Seq might be as useful as histopathology for NOA classification. For NOA.1, the main testicular somatic cells were macrophages (257), myoid cells (272), Sertoli cells (234) and Leydig cells (266); for NOA.a, over 90% (169/183) of the cells were Sertoli cells. The OA patients had the same testicular cell subtypes as healthy individuals, confirming the functional status of spermatogenesis in these patients. However, the proportions of each germ cell subtype differed between the patients of OA and healthy individuals (Fig. 1C). Compared to the testes of healthy individuals, the proportions of sperm in OA patient testes were much lower and the proportions of early primary spermatocytes were much higher, illustrating spermatogenesis in these patients was impaired.
Sertoli cells from NOA testes exhibit distinct transcriptomic features
For Sertoli cells processed without highly abundant mitochondrial DNA-encoded genes and ribosomal protein genes, differential expression (DE) analysis revealed that 13 genes were upregulated and 21 downregulated in NOA.1 compared with OA.1 (Fig. 1D). Interestingly, several genes involved in cell junctions and focal adhesion (SYNE2, ATP2B1 and MTDH) were downregulated in NOA.1. Additionally, FATE1, which plays a significant role in antiapoptotic processes and might promote germ cell development, was upregulated in patients with NOA (Fig. 1E).20,21 In addition, expression of INHBB, which encodes a protein that participates in the formation of inhibin B (INHB), was extremely low in both NOA.1 and NOA.a (Fig. 1F), consistent with previous observations and our clinical finding that INHB levels were decreased but FSH levels increased in patients with spermatogenic failure.22
Germ cell repertoire of the OA testis is markedly altered with considerable transcriptomic changes
As mentioned above, our initial clustering analysis revealed that the proportion of spermatids markedly decreased in the OA testis. To investigate this phenomenon further, two 10x Genomics scRNA-seq datasets of testes from human males who underwent vasectomy published by Sohni A et al23 were added into our analysis. Also, only scRNA-seq datasets produced by the 10x Genomics platform were used to compare the expression profiles to avoid introducing technical bias. Two OA and one healthy control scRNA-seq datasets are integrated ideally (Fig. 2A) via Seurat v3 standard workflow (as described in methods), with little variation based on batch effect or donor origin (Fig. S3). Cluster identity was assigned based on the same cell-type marker mentioned above (Fig. 1C). Consistent with our earlier findings, the proportions of RS, ESI and ESII in OA patient testes were much lower than those in testes of healthy individuals, whereas the proportions of undifferentiated spermatogonia increased significantly (Fig. 2A, B). However, unlike the OA case in our cohort, the developmental steps in the OA testes appeared to be arrested before the early spermatid stage. To further characterize the possible mechanism, we performed DE analysis on seven major cell populations with cell counts greater than 100, including germ cells (undifferentiated spermatogonia, differentiating spermatogonia and spermatocytes) and niche cells (Leydig cells, PTMC, endothelial cells and macrophages), between the two conditions. DE analysis was not performed on Sertoli cells due to the compromised quality of the Sertoli cell data mentioned by the original study. Two OA samples were compared to a healthy sample, and expression changes that were not common between the OA samples were filtered out (for detailed gene lists, see Table S1). Thousands of genes were differentially expressed between the two conditions, displaying dramatic transcriptomic changes (Fig. 2C).
Figure 2.
Single-cell transcriptome analyses in healthy versus obstructive (postvasectomy) testes samples. (A) UMAP projections of scRNA-Seq data (n = 16,721). (left) Split by sample origin. (right) Split by cell type. (B) Each of the 7 cell types and the fraction of cells originating from each of the 4 patients. (C) Strip chart showing the average logarithmic fold-changes (avgLogFC) of genes that significantly changed between OA and healthy individuals (FDR < 0.05 and LogFC >0.25, as determined by MAST analysis, expression changes that were not common between the OA samples were filtered out) across 7 cell types. The avgLogFC values are the average of OA.a and OA.b.
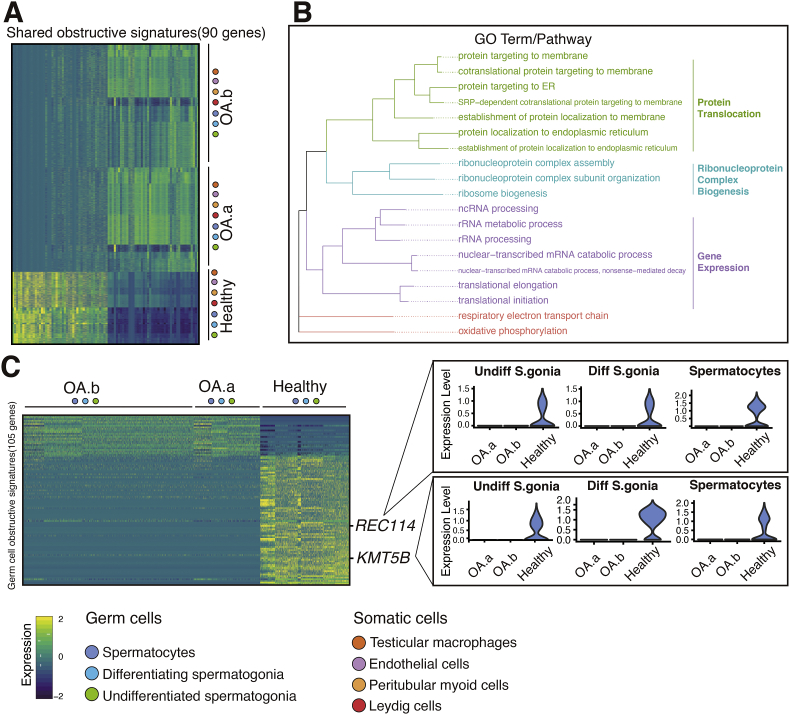
Identification of shared obstructive signatures
We noticed that several differentially expressed genes (DEGs) were shared across multiple cell types. First, to investigate the general influence of the obstructive condition, we distinguished 90 DEGs with consistent trends across all seven abovementioned cell types (see Fig. 3A and Table S2). Among these genes, the majority were found to be ribosomal protein (RBP) genes (such as RPS17), the functions of many of the genes remain to be explored. GO terms such as “Protein targeting to ER” and ‘‘nuclear-transcribed mRNA catabolic process’’ were enriched (Fig. 3B). Next, we identified 105 genes that were altered only in the three populations of germ cells to examine the effect of obstruction on the early spermatogenic process (Fig. 3C). Interestingly, we found downregulation of REC114, which acts as a partner of Spo11 in double-strand break (DSB) formation in mice,24 suggesting impaired spermatogenic dynamics. Additionally, we observed DE of several epigenetic modification genes, including KMT5B, PRMT9 and HNRNPA2B1. Notably, mutations in KMT5B have been reported to be associated with neurodevelopmental disorders and autism spectrum disorder.25
Figure 3.
Identification of shared obstructive signatures. (A) Heatmap of obstructive signatures shared (90 genes) across seven cell types. (B) Enriched GO terms of obstructive signatures in A and pathway summary. GO and pathway categories are grouped according to functional theme. (C) Heatmap of germ cell obstructive signatures (105 genes) and violin plot of selected genes showing differential expression.
OA testes exhibit disease-related archetypes among germ cells
To further explain the reduction of spermatids in OA testes, we next explored the dynamic and regulatory changes in germ cell development in response to obstruction. We began by examining undifferentiated spermatogonia, which are known to undergo self-renewal to maintain a constant cell pool and generate differentiation-committed progenitor that proceed through spermatogenesis to form sperm. As anticipated, we observed downregulation of progenitor marker NANOS326; in contrast, ID4, the key regulator to maintain the undifferentiated stem cell state,27 was considerably upregulated (Fig. 4A). This finding thus suggested that undifferentiated spermatogonia exhibited less progenitor capacity while more regenerative capacity, thereby providing an explanation for the increased population of undifferentiated spermatogonia in the testes of OA patients. In differentiating spermatogonia, we found meiosis initiation markers (MEIOB, MEIOC, SYCP3, SYCE1, SYCE3, TDRD9, TDRD12, REC114, HORMAD1 and TEX11) to be downregulated (Fig. 4B). Loss of any one of these genes would cause complete meiotic arrest, leading to azoospermia and subsequent infertility. These might be downstream targets of the abovementioned effects on multiple pathways responding to obstruction and might partially explain the reduced spermatogenesis of OA patients. In spermatocytes, in which the numbers of late-stage germ cell genes were considerably downregulated, we observed that genes involved in the apoptotic process and positive regulation of the intrinsic apoptotic signaling pathway were significantly upregulated according to GSEA analysis (Fig. 4C). Taken together, this finding indicated that spermatocytes in individuals with OA might undergo aberrant meiotic exit and apoptotic accumulation, ultimately leading to arrest before early spermatids were formed.
Figure 4.
Cell type-specific responses to obstruction in germ cells. (A) Violin plots of two key genes reflecting the state of undifferentiated spermatogonia. (B) Dot plot of genes that played crucial roles in meiosis initiation and were downregulated in differentiating spermatogonia of both OA individuals. (C) GSEA results indicated that the obstructive condition correlated significantly and positively with apoptotic signaling. On the left panel, the “Intrinsic apoptotic signaling pathway by p53 class mediator” is at the top (overexpressed) of the list corresponding to differences in expression between case OA.a and healthy. On the right panel, the “positive regulation of intrinsic apoptotic signaling pathway” and “positive regulation of apoptotic signaling pathway” is at the top (overexpressed) of the list corresponding to differences in expression between case OA.b and healthy controls.
PTMC (peritubular myoid cell) responses to reproductive tract obstruction
Smooth muscle-like PTMC are the main cellular components of the wall of seminiferous tubules and could modulate spermatogenesis directly and indirectly through collaborating with other somatic cells. Interestingly, GATA4, a negative regulator of contractility,28 was downregulated, whereas MYH11, a gene encoding a major contractile protein, was upregulated, suggesting that PTMC in individuals with OA tend to be in a contraction state to support seminiferous tubule fluid movement to the rete testis. This finding thus confirms a previous observation that obstruction leads to retention of fluid in seminiferous tubules and causes pressure-mediated spermatogenic damage,29 suggesting that PTMC might act as a pressure “sensor” and thus induce changes in intercellular communication and pathways.
OA testis gene regulatory network deviates markedly from controls
Next, we sought to explore how obstruction-driven changes in gene expression might affect intercellular communication among somatic cells in the testis and consequently influence germ cell development indirectly. We highlighted four major pathways that were most affected and that play important roles in the development, regulation, and maintenance of the testis (Fig. 5). These pathways may exert modulatory effects either synergistically or independently.
Figure 5.
Violin plot of key genes from four major pathways. (A–D) Violin plots the showing the cell-type-specific transcriptomic changes of key genes in (A) PDGF (B) Hedgehog (C) Androgen (D) Retinoic acid signaling pathway between the OA samples and the healthy samples.
Platelet-derived growth factors (PDGFs) are known to be essential for Leydig cell development, increasing LH-stimulated testosterone production and subsequently modulating spermatogenesis in rodents.30, 31, 32 In the present human data, we found that PDGFB and PDGFC localized in endothelial cells might interact with PDGFRA and PDGFRB in Leydig and myoid cells (Fig. S2). Moreover, expression of both PDGF ligands and their receptors was decreased under obstructive conditions (Fig. S2).
Hedgehog (Hh) signaling influences germ cell ontogenesis and controls fetal myoid and Leydig cell development in mice.33,34 We observed downregulation of Hh pathway components, including PTCH1 in Leydig cells, PTCH2 and GLI1 in PTMC and PTCH2 in macrophages. Although we lack data for Sertoli cells, we can infer from the downregulation of receptors (PTCH1 and PTCH2) and effectors (GLI1) in the present data that their upstream ligand DHH, which is expressed by Sertoli cells and regulates spermatogenesis,35 might also be downregulated under obstructive conditions.
Androgen signaling is required for germ cell survival and maturation.36 Androgen receptor (AR) has been indicated to be present in niche cells but absent from postnatal germ cells; in addition, it can promote the differentiation of undifferentiated progenitors via an indirect regulatory pattern. We found that AR was downregulated in Leydig cells and PTMC; reduced expression of genes encoding steroidogenic enzymes was also observed in Leydig cells (HSD17B3 and HSD17B11), PTMC (HSD17B4), endothelial cells (HSD17B4) and macrophages (HSD17B11). HSD17B3 is a key gene for testosterone biosynthesis that converts androstenedione to testosterone.37 As revealed by the CCInx analysis results, AR might act as a mediator of crosstalk with Notch signaling, which can repress GDNF and promote SSC differentiation in mice,38 by interacting with JAG1 (Fig. S2).
Retinoic acid (RA) is known to profoundly affect the induction of differentiation in the male germline. Thus, we explored the possible regulatory pattern of RA signaling in response to obstruction and observed that STRA8 was downregulated in the transition of differentiating spermatogonia to spermatocytes under obstructive condition. Additionally, KIT was downregulated, though its fold change was slightly under our cutoff value (logFC = 0.23; cutoff > 0.25). Moreover, PRAME, which encodes a protein that acts as a repressor of retinoic acid receptor (RAR),39 was upregulated. This evidence indicates that obstruction not only reduces differentiation commitment evoked by RA stimuli but also promotes inhibition of RAR. In addition, we observed downregulation of many genes encoding RA response-related transcription factors (HOXB4 and PBX1)40,41 and retinol metabolic enzymes (RDH14, RDH11, ALDH1A3 and CYP26B1). Overall, data reported here have indicated that these changes may modulate, either synergistically or separately, obstructive-related processes that take place in the testis.
Discussion
Azoospermia is among the most severe forms of male infertility, affecting approximately 1% of the male population worldwide.42 Interestingly, the underlying mechanisms that lead to azoospermia remain poorly characterized. In the clinic, the cause of NOA remains elusive in up to 72% of cases.43 Through scRNA-Seq analysis, we attempted to provide more information regarding the etiology of this disease. In the present study, we used a more efficient scRNA-Seq libraries construction method on NOA and OA sample, which captured more cells in NOA sample than the previous study18 at 1212 cells vs. 183. Herein, we captured macrophages, myoid cells, and Leydig cells in the NOA testis which the earlier report18 failed to do. Interestingly, only somatic cells were found in the NOA.1 sample, indicating the existence of SCOS. Sertoli cells are considered the “nurse cells” of the testis and play an essential role in sex determination during embryogenesis as well as in male gametogenesis during adulthood.44 However, we did not identify any evidence supporting that changes in Sertoli cells lead to the loss of germ cells. Instead, these changes might result from weakening interactions with germ cells. We observed that some of these secondary responses tended to promote the survival and antagonize the apoptosis of germ cells, especially undifferentiated spermatogonia. For example, Sertoli cells stimulated FSH, which acts as an antiapoptotic survival factor, by repressing inhibin B. Additionally, other antiapoptotic factors, including FATE1, were upregulated. These novel data provide evidence for future functional investigations to understand these Sertoli cell signals.
Moreover, we observed a germ cell population discrepancy between OA individuals and healthy individuals in the first run of the analysis. We confirmed this phenomenon and performed further analyses using published datasets. OA is defined as the absence of spermatozoa in the ejaculate due to occlusion of the reproductive tract despite normal spermatogenesis. Vasectomy and infection are frequent causes of OA.45 Previous studies have reported that significant morphologic changes occur in the human testis after vasectomy and show positive relationships with the obstructive interval.46,47 However, no previous study has profiled and compared the transcriptomes of OA and healthy testes due to the unsatisfactory resolution of bulk RNA-Seq data. As such, despite the successful identification of morphologic changes, previous studies have not resolved the underlying mechanism(s) that lead to OA. By applying computational analysis, we compared the two conditions (OA and healthy) and observed noticeable cell-to-cell transcriptional variation. Our findings offer new insight into the regulation of spermatogenesis in humans and will help in the design of effective therapeutics that improve male infertility. Additionally, we identified expression changes shared across multiple cell types, which might be used as obstructive signatures for clinical diagnosis in the future. Nevertheless, detailed functional studies are needed to comprehensively uncover and confirm the pathways of regulation.
As a cautious note, owing to the low quantity and quantity of Sertoli cells sequenced in the OA.a, OA.b and healthy samples, Sertoli cells were not considered in the analyses. Sertoli cells are the largest cell type in the testis, and their diameter can exceed 40 μm, even in suspension when these cells were freshly isolated. Therefore, the use of a 40-μm filter during digestion procedures could largely reduce the number of Sertoli cells that could be harvested for scRNA-Seq, and might even induce Sertoli cell fragmentation. Additionally, the width of the microfluidic channel of the 10x Genomics controller is less than ~50–60 μm. Therefore, it might not be possible to partition Sertoli cells into Gel Beads in Emulsion (GEMs), or they might clog microfluidic channels during GEM generation. For future studies, in order to avoid fragmentizing Sertoli cells and to increase the number of Sertoli cells to be examined, the 40 μm cell strainers mesh used in size filtering should be replaced with 70 μm. Also, alternative library constructs methods, such as smart-seq2 (used in NOA.a) and microwell-based GEXSCOPE (used in NOA.1 and OA.1), may produce more ideal results which should be carefully evaluated in future studies.
Author contributions
F.S. and C. Yan Cheng conceived and designed the study. G.A. and P.P. collect the samples. F.S. acquired funding. Y.M.J., J.F., C.T.S., H.S.W. and X.L.W., performed the testis sample preparation, single cell RNA-seq performance and library preparation and sequencing. S.T.C. and L.F.H. performed data analysis and visualization. S.T.C., F.S. and C. Yan Cheng drafted, reviewed, and edited the manuscript. All authors approved the final manuscript.
Conflict of interests
Authors have nothing to declare.
Funding
This work was supported by the National Key R&D Program of China (No. 2018YFC1003500 to F.S.), the National Natural Science Foundation of China (No. 81671510 to F.S.) and the Science and Technology Program of Guangdong Province (No. 2019A1515011439 to G.A.).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.09.004.
Contributor Information
C. Yan Cheng, Email: ccheng@rockefeller.edu.
Fei Sun, Email: sunfei@shsmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Cell type-specific results of differential expression testing between OA and healthy controls.
Obstructive signatures.
References
- 1.Soraggi S., Riera M., Rajpert-De Meyts E., Schierup M.H., Almstrup K. Evaluating genetic causes of azoospermia: what can we learn from a complex cellular structure and single-cell transcriptomics of the human testis? Hum Genet. 2021;140(1):183–201. doi: 10.1007/s00439-020-02116-8. [DOI] [PubMed] [Google Scholar]
- 2.Muciaccia B., Boitani C., Berloco B.P., et al. Novel stage classification of human spermatogenesis based on acrosome development. Biol Reprod. 2013;89(3):60. doi: 10.1095/biolreprod.113.111682. [DOI] [PubMed] [Google Scholar]
- 3.Wu S., Yan M., Ge R., Cheng C.Y. Crosstalk between sertoli and germ cells in male fertility. Trends Mol Med. 2020;26(2):215–231. doi: 10.1016/j.molmed.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 4.McLachlan R.I., O'Bryan M.K. Clinical review#: state of the art for genetic testing of infertile men. J Clin Endocrinol Metab. 2010;95(3):1013–1024. doi: 10.1210/jc.2009-1925. [DOI] [PubMed] [Google Scholar]
- 5.Oud M.S., Volozonoka L., Smits R.M., Vissers L., Ramos L., Veltman J.A. A systematic review and standardized clinical validity assessment of male infertility genes. Hum Reprod. 2019;34(5):932–941. doi: 10.1093/humrep/dez022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olesen I.A., Andersson A.M., Aksglaede L., et al. Clinical, genetic, biochemical, and testicular biopsy findings among 1,213 men evaluated for infertility. Fertil Steril. 2017;107(1):74–82. doi: 10.1016/j.fertnstert.2016.09.015. e7. [DOI] [PubMed] [Google Scholar]
- 7.Arafat M., Har-Vardi I., Harlev A., et al. Mutation in TDRD9 causes non-obstructive azoospermia in infertile men. J Med Genet. 2017;54(9):633–639. doi: 10.1136/jmedgenet-2017-104514. [DOI] [PubMed] [Google Scholar]
- 8.Riera-Escamilla A., Enguita-Marruedo A., Moreno-Mendoza D., et al. Sequencing of a ‘mouse azoospermia’gene panel in azoospermic men: identification of RNF212 and STAG3 mutations as novel genetic causes of meiotic arrest. Hum Reprod. 2019;34(6):978–988. doi: 10.1093/humrep/dez042. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Z., Li C., Yang S., et al. Dynamics of the transcriptome during human spermatogenesis: predicting the potential key genes regulating male gametes generation. Sci Rep. 2016;6:19069. doi: 10.1038/srep19069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang T., Wang Y., Zhu M., et al. Transcriptome-wide association study revealed two novel genes associated with nonobstructive azoospermia in a Chinese population. Fertil Steril. 2017;108(6):1056–1062. doi: 10.1016/j.fertnstert.2017.09.023. e4. [DOI] [PubMed] [Google Scholar]
- 11.Gille A.S., Lapoujade C., Wolf J.P., Fouchet P., Barraud-Lange V. Contribution of single-cell transcriptomics to the characterization of human spermatogonial stem cells: toward an application in male fertility regenerative medicine? Int J Mol Sci. 2019;20(22):5773. doi: 10.3390/ijms20225773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dura B., Choi J.Y., Zhang K., et al. scFTD-seq: freeze-thaw lysis based, portable approach toward highly distributed single-cell 3' mRNA profiling. Nucleic Acids Res. 2019;47(3):e16. doi: 10.1093/nar/gky1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 14.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chothani S., Adami E., Ouyang J.F., et al. deltaTE: detection of translationally regulated genes by integrative analysis of Ribo-seq and RNA-seq data. Curr Protoc Mol Biol. 2019;129(1):e108. doi: 10.1002/cpmb.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finak G., McDavid A., Yajima M., et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M., Liu X., Chang G., et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018;23(4):599–614. doi: 10.1016/j.stem.2018.08.007. e4. [DOI] [PubMed] [Google Scholar]
- 19.Guo J., Grow E.J., Mlcochova H., et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28(12):1141–1157. doi: 10.1038/s41422-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olesen C., Larsen N.J., Byskov A.G., Harboe T.L., Tommerup N. Human FATE is a novel X-linked gene expressed in fetal and adult testis. Mol Cell Endocrinol. 2001;184(1–2):25–32. doi: 10.1016/s0303-7207(01)00666-9. [DOI] [PubMed] [Google Scholar]
- 21.Maxfield K.E., Taus P.J., Corcoran K., et al. Comprehensive functional characterization of cancer-testis antigens defines obligate participation in multiple hallmarks of cancer. Nat Commun. 2015;6:8840. doi: 10.1038/ncomms9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luisi S., Florio P., Reis F.M., Petraglia F. Inhibins in female and male reproductive physiology: role in gametogenesis, conception, implantation and early pregnancy. Hum Reprod Update. 2005;11(2):123–135. doi: 10.1093/humupd/dmh057. [DOI] [PubMed] [Google Scholar]
- 23.Sohni A., Tan K., Song H.W., et al. The neonatal and adult human testis defined at the single-cell level. Cell Rep. 2019;26(6):1501–1517. doi: 10.1016/j.celrep.2019.01.045. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boekhout M., Karasu M.E., Wang J., et al. REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol Cell. 2019;74(5):1053–1068. doi: 10.1016/j.molcel.2019.03.023. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stessman H.A., Xiong B., Coe B.P., et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49(4):515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La H.M., Mäkelä J.A., Chan A.L., et al. Identification of dynamic undifferentiated cell states within the male germline. Nat Commun. 2018;9(1):2819. doi: 10.1038/s41467-018-04827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mäkelä J.A., Hobbs R.M. Molecular regulation of spermatogonial stem cell renewal and differentiation. Reproduction. 2019;158(5):R169–R187. doi: 10.1530/REP-18-0476. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y.Q., Batool A., Chen S.R., Liu Y.X. GATA4 is a negative regulator of contractility in mouse testicular peritubular myoid cells. Reproduction. 2018;156(4):343–351. doi: 10.1530/REP-18-0148. [DOI] [PubMed] [Google Scholar]
- 29.Ma L., Guo Y., Yuan Y., Li Y.G., Deng X.Z., Yang Z.W. Morphometric study of the testis and reproductive tract (including sperm granuloma) after vasectomy in mature rats. Asian J Androl. 2016;18(1):66–73. doi: 10.4103/1008-682X.150038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basciani S., Mariani S., Spera G., Gnessi L. Role of platelet-derived growth factors in the testis. Endocr Rev. 2010;31(6):916–939. doi: 10.1210/er.2010-0004. [DOI] [PubMed] [Google Scholar]
- 31.Gnessi L., Basciani S., Mariani S., et al. Leydig cell loss and spermatogenic arrest in platelet-derived growth factor (PDGF)-A-deficient mice. J Cell Biol. 2000;149(5):1019–1026. doi: 10.1083/jcb.149.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brennan J., Tilmann C., Capel B. Pdgfr-alpha mediates testis cord organization and fetal Leydig cell development in the XY gonad. Genes Dev. 2003;17(6):800–810. doi: 10.1101/gad.1052503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franco H.L., Yao H.H. Sex and hedgehog: roles of genes in the hedgehog signaling pathway in mammalian sexual differentiation. Chromosome Res. 2012;20(1):247–258. doi: 10.1007/s10577-011-9254-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahin Z., Szczepny A., McLaughlin E.A., et al. Dynamic Hedgehog signalling pathway activity in germline stem cells. Andrology. 2014;2(2):267–274. doi: 10.1111/j.2047-2927.2014.00187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bitgood M.J., Shen L., McMahon A.P. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996;6(3):298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 36.O'Hara L., Smith L.B. Androgen receptor roles in spermatogenesis and infertility. Best Pract Res Clin Endocrinol Metab. 2015;29(4):595–605. doi: 10.1016/j.beem.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Geissler W.M., Davis D.L., Wu L., et al. Male pseudohermaphroditism caused by mutations of testicular 17 beta-hydroxysteroid dehydrogenase 3. Nat Genet. 1994;7(1):34–39. doi: 10.1038/ng0594-34. [DOI] [PubMed] [Google Scholar]
- 38.Garcia T.X., Parekh P., Gandhi P., Sinha K., Hofmann M.C. The NOTCH ligand JAG1 regulates GDNF expression in Sertoli cells. Stem Cells Dev. 2017;26(8):585–598. doi: 10.1089/scd.2016.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Epping M.T., Wang L., Edel M.J., Carlée L., Hernandez M., Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122(6):835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Qin P., Haberbusch J.M., Soprano K.J., Soprano D.R. Retinoic acid regulates the expression of PBX1, PBX2, and PBX3 in P19 cells both transcriptionally and post-translationally. J Cell Biochem. 2004;92(1):147–163. doi: 10.1002/jcb.20057. [DOI] [PubMed] [Google Scholar]
- 41.Gould A., Itasaki N., Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21(1):39–51. doi: 10.1016/s0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 42.Agarwal A., Mulgund A., Hamada A., Chyatte M.R. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cannarella R., Condorelli R.A., Duca Y., La Vignera S., Calogero A.E. New insights into the genetics of spermatogenic failure: a review of the literature. Hum Genet. 2019;138(2):125–140. doi: 10.1007/s00439-019-01974-1. [DOI] [PubMed] [Google Scholar]
- 44.Griswold M.D. 50 years of spermatogenesis: Sertoli cells and their interactions with germ cells. Biol Reprod. 2018;99(1):87–100. doi: 10.1093/biolre/ioy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker K., Sabanegh E. Jr. Obstructive azoospermia: reconstructive techniques and results. Clinics (Sao Paulo) 2013;68(Suppl 1):61–73. doi: 10.6061/clinics/2013(Sup01)07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jarow J.P., Budin R.E., Dym M., Zirkin B.R., Noren S., Marshall F.F. Quantitative pathologic changes in the human testis after vasectomy. A controlled study. N Engl J Med. 1985;313(20):1252–1256. doi: 10.1056/NEJM198511143132003. [DOI] [PubMed] [Google Scholar]
- 47.Raleigh D., O'Donnell L., Southwick G.J., de Kretser D.M., McLachlan R.I. Stereological analysis of the human testis after vasectomy indicates impairment of spermatogenic efficiency with increasing obstructive interval. Fertil Steril. 2004;81(6):1595–1603. doi: 10.1016/j.fertnstert.2003.10.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell type-specific results of differential expression testing between OA and healthy controls.
Obstructive signatures.
Data Availability Statement
Data obtained from our scRNA-Seq were deposited in the NODE Project with Accession Number OEP000778 at https://www.biosino.org/node/project/detail/OEP000778 freely accessible to all investigators. The R code to reproduce the analysis can be obtained from: https://github.com/dioncst/hTestes_scRNA-seq.