Abstract
The anaerobic bacterium Dehalococcoides ethenogenes is the only known organism that can completely dechlorinate tetrachloroethene or trichloroethene (TCE) to ethene via dehalorespiration. One of two corrinoid-containing enzymes responsible for this pathway, TCE reductive dehalogenase (TCE-RDase) catalyzes the dechlorination of TCE to ethene. TCE-RDase dehalogenated 1,2-dichloroethane and 1,2-dibromoethane to ethene at rates of 7.5 and 30 μmol/min/mg, respectively, similar to the rates for TCE, cis-dichloroethene (DCE), and 1,1-DCE. A variety of other haloalkanes and haloalkenes containing three to five carbon atoms were dehalogenated at lower rates. The gene encoding TCE-RDase, tceA, was cloned and sequenced via an inverse PCR approach. Sequence comparisons of tceA to proteins in the public databases revealed weak sequence similarity confined to the C-terminal region, which contains the eight-iron ferredoxin cluster binding motif, (CXXCXXCXXXCP)2. Direct N-terminal sequencing of the mature enzyme indicated that the first 42 amino acids constitute a signal sequence containing the twin-arginine motif, RRXFXK, associated with the Sec-independent membrane translocation system. This information coupled with membrane localization studies indicated that TCE-RDase is located on the exterior of the cytoplasmic membrane. Like the case for the two other RDases that have been cloned and sequenced, a small open reading frame, tceB, is proposed to be involved with membrane association of TCE-RDase and is predicted to be cotranscribed with tceA.
The toxic solvents tetrachloroethene (PCE) and trichloroethene (TCE) have been widely used as degreasers and chemical feedstocks. Because of leakage and poor disposal practices, these solvents are now among the most common groundwater contaminants (1). Fortunately, a variety of microbe-mediated processes can catalyze conversion of these chlorinated solvents to harmless products. In aerobic environments, TCE, dichloroethenes (DCEs), and vinyl chloride (VC) are cometabolized by bacteria containing oxygenases, such as methane monooxygenase, toluene monooxygenase, and toluene dioxygenase, but PCE is not transformed (8, 17, 25). In contrast, anaerobic bacterial degradation of PCE proceeds readily via reductive dehalogenation to TCE, cis-1,2-DCE, VC, and ethene. In contaminated anaerobic environments, dechlorination often terminates at DCE or VC, but complete dechlorination to ethene has been observed (34). Complete reductive dechlorination of PCE and TCE to ethene or ethane has also been observed in anaerobic enrichment cultures (6, 7). Thus far, all but one of the pure cultures of anaerobic dechlorinating bacteria that have been isolated reduce PCE or TCE only to cis-DCE (9, 28). However, one organism, Dehalococcoides ethenogenes, is able to completely dechlorinate PCE to ethene (21).
The evolutionary history of dehalorespiring organisms is of considerable interest. Many dehalorespirers are gram-positive bacteria that cluster with the Clostridium-Bacillus subphylum, while the others lie in the ɛ and γ branches of the Proteobacteria (13). On the other hand, D. ethenogenes is more phylogenetically distant from the other dehalorespiring bacteria. D. ethenogenes is a bacterium possessing a unique archaeon-like cell wall, and its precise relationship to other bacteria is uncertain (21), though a recent phylogenetic analysis suggests that it lies within the green nonsulfur division of bacteria (14). One of the most remarkable characteristics of this organism is that the only known electron acceptors that support its growth are certain chlorocarbons, i.e., PCE, TCE, cis-DCE, 1,1-DCE, and 1,2-dichloroethane (20, 21).
The dehalogenation reactions unique to dehalorespiring bacteria are catalyzed by reductive dehalogenases (RDases), many of which have been purified and characterized (3, 4, 12, 18, 19, 21, 22, 23, 26, 31). The identified substrates of these enzymes are either chlorinated ethenes or substituted chloroaromatics. Four of the five chloroethene RDases that have been characterized to date are membrane bound, and the other may be anchored to the membrane through an accessory protein (24). All five enzymes have a subunit molecular mass of 50 to 65 kDa and contain cobalamin and iron-sulfur clusters (13). Three of the four known PCE-RDases dechlorinate PCE or TCE to cis-DCE, as would be expected from the metabolism of their parent organisms, but the PCE-RDase from D. ethenogenes accepts only PCE as a substrate, converting it to TCE (19). D. ethenogenes also contains a second enzyme, TCE-RDase, which is the only known RDase that is able to effect the complete dechorination of TCE, DCEs, and VC to ethene (19).
Prior to this work, the genes for two RDases had been cloned and sequenced: the PCE-RDase from Dehalospirillum multivorans and the ortho-chlorophenol RDase (oCP-RDase) from Desulfitobacterium dehalogenans. Both organisms contain a single functional gene (pceA or cprA, respectively), which encodes the dehalogenase, and a short open reading frame (ORF) immediately downstream (pceB or cprB, respectively), which may encode a small integral membrane protein involved in the association of the dehalogenase with the cytoplasmic membrane. Both pceA and cprA contain the twin-arginine signal sequence characteristic of some redox cofactor-containing proteins that must be translocated into or across the cytoplasmic membrane in their native state (2). The two structural genes also contain the iron-sulfur cluster binding motif common to eight-iron ferredoxins. However, only seven of the eight cysteine ligands are conserved, suggesting either the presence of one Fe4S4 cluster and one Fe3S4 cluster (24, 32) or alternative ligation (24).
In this study, we report the cloning and sequencing of the gene for TCE-RDase from D. ethenogenes, tceA. Comparisons of tceA with the genes for the RDases from D. multivorans and D. dehalogenans are discussed. Information about the membrane localization of TCE-RDase is also presented. Finally, alternative substrates for the TCE-RDase are examined.
MATERIALS AND METHODS
Chemicals.
Inorganic chemicals of ACS reagent grade or better were obtained from Aldrich (Milwaukee, Wis.), Sigma (St. Louis, Mo.), or Fisher (Pittsburgh, Pa.). Seakem GTG agarose was from FMC Bioproducts (Rockland, Maine). Restriction enzymes were from New England Biolabs (Beverly, Mass.) or Stratagene (La Jolla, Calif.). Phenol-chloroform-isopentanol (25:24:1) and ampicillin were from Fisher. Proteinase K and kanamycin were from Roche Molecular Biochemicals (Indianapolis, Ind.). Yeast extract and Bacto-tryptone were from Difco Laboratories (Franklin Lakes, N.J.). Halogenated solvents, alkanes, alkenes, and dienes were obtained from Aldrich or Chem Service (West Chester, Pa.). Tribromoethene was prepared by dehydrohalogenation of 1,1,2,2-tetrabromoethane with sodium hydroxide in methanol and twice distilled (33). Buffers, methyl viologen, Triton X-100, and other enzymes were obtained from Sigma.
Enzyme assays.
Membrane-bound TCE-RDase was purified from an anaerobic enrichment culture containing D. ethenogenes maintained on the electron donor-acceptor pair of methanol-PCE, as previously described (19). Briefly, PCE-RDase and TCE-RDase were solubilized from the membrane fraction (20 mg [wet weight] per ml) with 0.1% Triton X-100, applied to a POROS HP/M column, and eluted with 0.25 M (NH4)2SO4. The enzymes were applied to a POROS PH/M column, and the two dehalogenases were separated by a descending gradient of (NH4)2SO4. Enzyme assays were performed as previously described with the following modifications. Halocarbons were prepared as 5 to 10% (vol/vol) solutions in ethanol. Ten microliters (2 to 10 μmol) of halocarbon solution was injected into parallel 15-ml vials containing 2.0 ml of 25 mM bis-Tris propane (pH 7), 150 mM NaCl, 2 mM methyl viologen, 2 mM titanium(III) citrate, and either 0.2 μg of TCE-RDase or no enzyme. Enzyme assay mixtures were incubated at 30°C for between 10 min and 16 h. A 100-μl aliquot of the headspace was analyzed on a Hewlett-Packard model 5890 gas chromatograph (GC) equipped with a Carbopack SP-1000 column coupled to a flame ionization detector (19) or on an Hewlett-Packard model 5890 GC equipped with a model 5971 quadrupole mass selective detector using a GSQ megabore column, or both.
Amino acid sequencing of TCE-RDase.
TCE-RDase from the POROS HP/M column was subjected to electrophoresis on a sodium dodecyl sulfate (SDS)–6% polyacrylamide gel (16). TCE-RDase was electrotransferred to a polyvinylidene difluoride membrane and subjected to Edman degradation in an ABI 477A amino acid sequencer in order to determine the N-terminal sequence (Protein Chemistry Facility, University of Florida, Gainesville). Approximately 15 μg of partially purified TCE-RDase was further purified on an SDS–7.5% polyacrylamide gel, and the resulting band was excised. The protein was digested with trypsin in the gel matrix, the peptides were separated by high-pressure liquid chromatography, and one of the internal peptides was sequenced in an ABI 477A sequencer (Protein Structure Core Facility, University of Nebraska, Omaha).
Preparation of genomic DNA.
One liter of the PCE-methanol anaerobic enrichment culture containing D. ethenogenes was harvested at 5,000 rpm in a Beckman JA-10 rotor for 10 min, resulting in 1.1 g of cell paste. The cells were suspended in 50 mM glucose–25 mM Tris-HCl (pH 8)–10 mM EDTA containing lysozyme (100 μg/ml) and lysed with 1% SDS (27). The cell lysate was centrifuged for 10 min at 5,000 rpm in a JA-20 rotor to remove cell debris. Total DNA was precipitated with ethanol, hooked with a thin glass rod, and dissolved in 4 ml of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). RNase A was added to 250 μg/ml and incubated at 37°C for 1.5 h, followed by digestion with proteinase K (250 μg/ml) at 50°C for 2 h. The digested solution was extracted one time each with phenol-chloroform-isopentanol (25:24:1) and chloroform. The DNA was precipitated and washed in cold 70% ethanol. The DNA was dissolved in 10 ml of TE buffer containing 0.1 M NaCl and 5% Triton X-100, and the genomic DNA was purified by equilibrium centrifugation in CsCl (27).
Synthesis of degenerate oligonucleotides and PCR.
The sequence of the N-terminal peptide of TCE-RDase was used to design the 128-fold-degenerate oligonucleotide TFOR (5′-GCIAAYAARGTIAAYAAYCAYCCNTGGTGGG-3′). The internal peptide sequence was used to design the 256-fold degenerate oligonucleotide TREV (5′-CCYTCCCAYTTIGGRTARTTNGTNGT-3′). These and all other oligonucleotides were synthesized by Genosys (The Woodlands, Tex.). PCR mixtures (50 μl) contained 140 ng of genomic DNA, 0.6 μM each primer, 0.2 mM each deoxynucleoside triphosphate, and 2.5 U of Taq DNA polymerase (Qiagen, Santa Clarita, Calif.) in 1× Qiagen reaction buffer (1.5 mM MgCl2). PCR was carried out with a Techne Genius thermal cycler using the following parameters: 3 min at 94°C; 28 cycles of 45 s at 94°C, 45 s at 60°C, and 60 s at 72°C; and 10 min at 72°C. The single 0.5-kb PCR product was cloned using the Zero-Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, Calif.). Both strands of the PCR product were sequenced by using an ABI Big-Dye Terminator kit (PE Applied Biosystems, Foster City, Calif.) followed by analysis of the products on an ABI 377 instrument (Amplicon Express, Pullman, Wash.).
Inverse PCR and sequencing of tceA coding region.
A pair of primers for inverse PCR (30), IF1 (5′-TGCGGATCCAACCTGTAATATAG-3′) and IR1 (5′-TTGGGATCCTCATGATCACG-3′), were designed from the sequence of the 0.5-kb PCR product. Genomic DNA (140 ng) was digested with 5 U of PstI or HindIII for 30 min at 37°C (25 μl), and the restriction endonucleases were inactivated by heating to 80°C for 20 min. The digested solutions were diluted to 2.8 ng of DNA per μl (50 μl) and circularized with 2 U of T4 DNA ligase (New England Biolabs) per μl in ligase buffer containing 1 mM ATP for 16 h at 16°C. Inverse-PCR mixtures (100 μl) contained 28 ng of DNA, 0.5 μM primers, 125 μM each deoxynucleoside triphosphate, dNTP, 1× cloned Pfu polymerase buffer, and 5 U of cloned Pfu DNA polymerase (Stratagene). Reagents were mixed on ice, Pfu polymerase was added last, and the reaction mixtures were transferred directly to a thermal cycler preheated to 94°C. Thermal cycler parameters were as follows: an initial denaturation step of 2 min at 94°C; 26 cycles of 30 s at 94°C, 30 s at 58°C, and 3 min at 72°C; and a final extension step of 10 min at 72°C. Inverse-PCR products were purified by electrophoresis in a 1% agarose–Tris-borate-EDTA gel and extracted using tha Qiaex II gel extraction kit (Qiagen). PCR products were sequenced with the primers IF1 and IR1 and then with IPF1 (5′-TACTTCGGGGCTTCTTCC-3′) and IPR1 (5′-AATTAGATTGGGAGGGAC-3′) (Iowa State University DNA Sequencing Facility). Another inverse-PCR product was prepared and purified as described above using the primer set IF2 (5′-TTGCAGGCCTTGGCTATAA-3′) and IR2 (5′-CTGAATGCGTGCCTCAACC-3′). The second PstI inverse-PCR product was sequenced with IF2, IR2, and 2345F (5′-TGCACAACTTGGTCAAGTCC). A final PCR amplicon containing the entire tceA (for TCE-RDase functional gene) coding sequence was prepared using primers 797F (5′-ACGCCAAAGTGCGAAAAGC) and 2490R (5′-TAATCTATTCCATCCTTTCTC). Clustal W alignments of translated genes were performed with MacVector 6.5.1.
Membrane localization.
The following experiments were performed using anaerobic procedures. Membrane suspensions {20 mg/ml in 25 mM 1,3-bis[tris(hydroxymethyl)methylamino] propane (BTP), pH 7} were prepared from the mixed culture, as described previously (19). Microfuge tubes containing 1.0 ml of the membrane suspension were incubated in an anaerobic glove box for 1 h at 4°C with either 0.1% Triton X-100, 1.0 M NaCl, 10 mM EDTA, 1% (vol/vol) n-butanol, 100 mM potassium phosphate at pH 9, or no added reagent. Soluble protein was separated from membrane fragments by centrifugation at 16,000 × g for 30 min. The supernatant and resuspended pellet were assayed for TCE-RDase activity. The percent extraction was calculated relative to the activity in the soluble fraction of the positive control, 0.1% Triton X-100.
Nucleotide sequence accession numbers.
The coding sequence of tceA, tceB, and orf1 has been deposited in GenBank under accession number AF228507.
RESULTS
Inverse PCR and assembly of the coding sequence of tceA.
It is difficult to grow pure cultures of D. ethenogenes strain 195 due to its undefined nutritional requirements (21). Furthermore, the pure culture fails to attain high cell densities, and thus it is difficult to obtain sufficient biomass for the isolation and purification of enzymes and DNA. The anaerobic enrichment culture from which this organism was isolated is more robust and yields high quantities of biomass (>1 g/liter), approximately one-third of which is D. ethenogenes (based on 16S ribosomal DNA analysis [unpublished data]). Therefore, the mixed culture was utilized for the isolation of TCE-RDase and genomic DNA described in this work.
Analysis of the mature TCE-RDase and a peptide from the trypsin-digested enzyme yielded amino acid sequences of the N terminus (KDVDDLLSAGKALEGDHANKVNNHPWW) and an internal peptide (TTNYPKWEGTPEENLLIM). These sequences were used to design a pair of degenerate primers, TFOR and TREV, for PCR amplification of DNA extracted from the enrichment culture containing D. ethenogenes. A 491-bp PCR product resulted from this reaction. Subsequently, a small sample of D. ethenogenes genomic DNA was obtained from S. H. Zinder and A. Carroll (Cornell University) and subjected to PCR analysis using the same degenerate primer pair. A single PCR product of identical size to the product generated with DNA from the mixed culture was obtained.
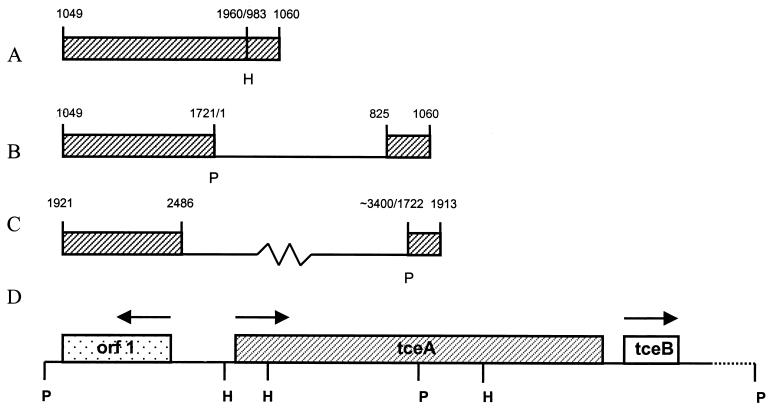
A new pair of primers, IF1 and IR1, were designed from the sequence of the PCR product. These were used in inverse PCRs with PstI and HindIII-digested, ligated DNA as templates, resulting in 1.7-kb (Fig. 1B) and 1.0-kb (Fig. 1A) products, respectively. Both PCR products were sequenced directly with IF1 and IR1. The PstI inverse-PCR product was subjected to a second round of sequencing with the primer pair IPF1 and IPR1 to complete the DNA sequence. The first round of inverse PCR resulted in sequence extending from position 1 (824 bp upstream of the start codon) to position 1960, thus covering about two-thirds of the coding sequence of tceA. A second pair of inverse PCR primers, IF2 and IR2, were designed to complement the sequence between the 3′ HindIII site at position 1721 and the 3′ PstI site at position 1960. A 2.7-kb inverse-PCR product (Fig. 1C) was generated with the PstI-digested, ligated DNA serving as the template. Sequencing of the PCR product with IF2 completed the 3′ end of the tceA coding sequence. Primers 797F and 2490R were used to amplify the entire tceA coding sequence by PCR. This final PCR amplicon served as the template for a primer-walking strategy to complete the sequencing of the coding region (Fig. 1D).
FIG. 1.
Inverse-PCR products and arrangement of sequenced genes. (A to C) Amplicons prepared using HindIII-digested ligated genomic DNA as the template with primers IF1 and IR1 (A), using PstI-digested ligated genomic DNA as the template with primers IF1 and IR1 (B), and using PstI-digested ligated genomic DNA as the template with primers IF2 and IR2. Numbers indicate the nucleotide position starting from the first PstI site 5′ to tceA; the tceA start position is 825. The position of the tceA coding sequence is indicated by the boxed areas. (D) Arrangement of the orf1, tceA, and tceB genes. Arrows indicate the direction of transcription. The dotted line represents DNA from position 2831 to approximately 3400, for which the sequence is incomplete. P: PstI site; H: HindIII site.
Sequence analysis.
The general organization of the region of chromosomal DNA containing tceA is shown in Fig. 1D. Potential ς70 promoter regions upstream of the tceA coding sequence were identified by the neural network promoter prediction tool (M. Reese, Promoter Prediction by Neural Network [http://www.fruitfly.org/seq_tools/promoter.html]). A putative ribosome binding site was identified 12 nucleotides upstream of the second in-frame ATG in the tceA sequence, suggesting that it is the start codon. The proposed coding sequence of tceA is 1,662 nucleotides long, which translates into a 554-amino-acid protein with a calculated molecular mass of 62,128 Da. A second ORF was found 24 nucleotides downstream of the tceA stop codon. This ORF, tentatively named tceB, encodes a hypothetical protein of 94 amino acids with a calculated molecular mass of 10,905 Da. No termination sites were found in the region between tceA and tceB, suggesting they could be cotranscribed. A third ORF, orf1, is found on the opposite strand upstream of the transcription start site of tceA. orf1 encodes a hypothetical 177-amino-acid protein with a calculated molecular mass of 20,604 Da.
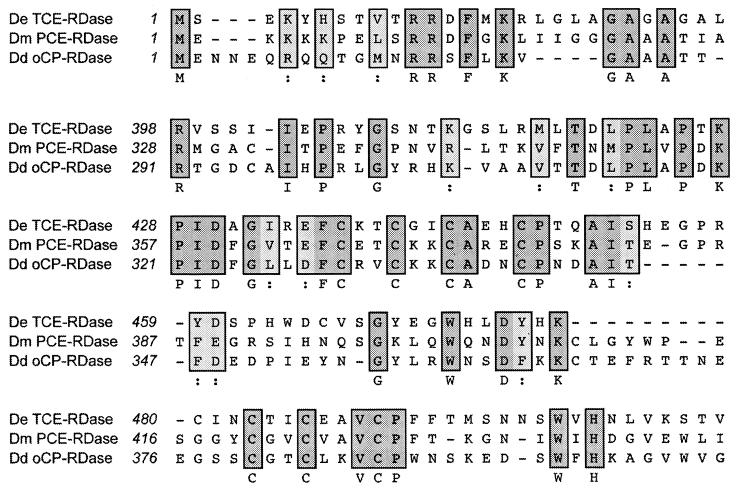
BLASTP analysis revealed that the PCE-RDase of D. multivorans (AF022812) and the oCP-RDase of Desulfitobacterium dehalogenans (AF115542) share limited homology with the TCE-RDase of D. ethenogenes (AF228507). Clustal W pairwise alignments of the complete amino acid sequences of TCE-RDase, PCE-RDase, and oCP-RDase (Fig. 2) showed that TCE-RDase shared 24% identity with PCE-RDase and 22% identity with oCP-RDase. The regions of similarity are confined to the C-terminal domains of these proteins, which contain the twin Fe4S4 cluster binding motif, (CXXCXXCXXXCP)2, characteristic of Fe8S8 ferredoxins (29), and the N- terminal leader sequence (discussed below).
FIG. 2.
Clustal W alignment of the N-terminal leader sequence and C-terminal regions of D. ethenogenes (De) TCE-RDase (AF228507), D. multivorans (Dm) PCE-RDase (AF022812), and D. dehalogenans (Dd) oCP-RDase (AF115542). Numbers correspond to the amino acid positions of the preproteins. Predicted Fe4S4 cluster binding motifs are underlined.
The difference between the putative start codon and the N-terminal sequence of the mature protein determined by Edman degradation (KDVDD…) indicated the presence of a 42-amino-acid leader sequence. It contains the twin-arginine motif RRXFXK followed by a stretch of predominantly hydrophobic amino acid residues, from position 17 to 37, predicted by the dense-alignment surface (DAS) method to form a membrane-spanning region (5). These primary and secondary protein structural features are characteristic of periplasmic or membrane-bound enzymes containing complex redox cofactors that are translocated across the membrane in their native state by the Sec-independent transport system (2). The identification of this leader sequence in TceA is consistent with TCE-RDase being a membrane-bound enzyme containing a corrinoid cofactor and Fe4S4 clusters. The calculated molecular mass for the 512-amino-acid processed protein is 57,658 Da, which is in reasonable agreement with the size of the mature enzyme, 61 kDa, determined by SDS-polyacrylamide gel electrophoresis (19).
BLAST analyses of tceB and orf1 against GenBank failed to reveal any homologous sequences. A DAS analysis of the amino acid sequence encoded by tceB indicated the existence of three potential membrane-spanning regions from amino acid residues 6 to 22, 39 to 50, and 70 to 86 in the 94-amino-acid protein. This suggests that tceB encodes an integral membrane protein. DAS analysis of orf1 predicts one membrane-spanning helix from residue 28 to 42. With the exception of this 15-amino-acid helix, the predicted gene product of orf1 is very hydrophilic, rich in polar and charged residues.
Location of TCE-RDase in the membrane.
It was previously suggested that TCE-RDase is a membrane protein, as all of the activity was associated with the membrane fraction of lysed cells (19). The sequence analysis and experiments presented below indicate that TCE-RDase is a peripheral membrane protein. A DAS analysis of the TCE-RDase sequence predicted only one transmembrane helix with certainty, located in the signal sequence, as noted above (5). Similarly, the Kyte-Doolittle hydrophilicity profile of TCE-RDase showed only short hydrophobic regions (fewer than 10 amino acids), with the exception of the signal sequence (15). This is unlike the case for an integral membrane protein, which would be expected to possess membrane-spanning regions. Solubilization of a protein without the use of detergent is also diagnostic of a peripheral membrane protein (11). Membrane suspensions were extracted with 0.1% Triton X-100, 1.0 M NaCl, 10 mM EDTA, 1% (vol/vol) n-butanol, 100 mM potassium phosphate at pH 9, or no added reagent (negative control). The treatment with 1.0 M NaCl released 66% of the TCE-RDase activity into the soluble fraction, and 0.1 M potassium phosphate (pH 9) released 12% of the activity, relative to the positive control containing 0.1% Triton X-100. The negative control and all other reagents contained less than 2% of the total TCE-RDase activity in the supernatant. Furthermore, TCE-RDase remained soluble in the absence of detergent after dissociation from the membrane and throughout the liquid chromatography steps during the routine purification procedure. The cumulative data are consistent with the classification of TCE-RDase as a peripheral membrane protein. The presence of the twin-arginine motif in the translated sequence of tceA suggests that TCE-RDase is located on the outer face of the cytoplasmic membrane.
Substrate range of TCE-RDase.
A suite of halocarbons were used to qualitatively assess the substrate range of TCE-RDase in order to elicit information about the mechanism of the enzymatic reductive dehalogenation reaction and to assess the utility of TCE-RDase for the dehalogenation of other problematic environmental pollutants. Parallel assays of each halocarbon were conducted with and without TCE-RDase to control for chemical reduction of the substrate by titanium(III) citrate and methyl viologen. Vials containing pentachloroethane or 1,1,2,2-tetrachloroethane produced PCE or TCE, respectively, regardless of whether or not TCE-RDase was included. All other controls were negative with regard to chemical alteration of the substrate. Table 1 shows the results with substrates categorized by the number of carbons or branching of the carbon chain. Quantitative rates were not determined for all of the halocarbons due to the large number of substrates and products and the fact that some of the products were not commercially available. However, qualitative rates based on comparison of peak areas of slow substrates versus peak areas obtained with TCE as the substrate are included, as they are useful for assessing trends in reactivity. Potential products that were commercially available were acquired and used to identify unknown products by comparison of elution times on the GC flame ionization detector. If any ambiguities existed, the unknown was subjected to GC-mass spectrometry to obtain a clear identification. The identities of products that were unavailable in neat form were determined by comparison of their mass spectra to a library of mass spectra. Exceptions were the cis-trans isomers, many of which were unavailable as pure conformers and were indistinguishable by GC-mass spectrometry.
TABLE 1.
Substrates and products of TCE-RDase
| Halocarbon substratea | Product(s) | Rate (μmol/min/mg or relativeb) |
|---|---|---|
| Two carbons | ||
| TCE | DCEs, VC, ethene | 5.2 |
| Tribromoethene | Dibromoethenes, vinyl bromide, ethene | NDc |
| cis-1,2-DCE | VC, ethene | 12.1 |
| trans-1,2-DCE | VC, ethene | 0.45 |
| 1,1-DCE | VC, ethene | 8.7 |
| cis- and trans-1,2-Dibromoethene | Vinyl bromide, ethene | ND |
| VC | Ethene | 0.036 |
| Vinyl bromide | Ethene | 0.18 |
| 1,2-Dichloroethane | Ethene, VC (<1%) | 7.5 |
| 1,2-Dibromoethane | Ethene, vinyl bromide (<1%) | 30 |
| Three carbons | ||
| 3-Chloro-1-propene | Propene | Moderate |
| 1,3-Dichloropropene | An isomer of 1-chloropropene, propene | Moderate |
| cis-1-Bromo-1-propene | Propene | Low |
| trans-1-Bromo-1-propene | Propene | Very low |
| 1,1-Dichloropropene | cis- and trans-1-Chloropropene, propene (trace) | Very low |
| 1,2-Dibromopropane | Propene | Moderate |
| 1,2-Dichloropropane | Propene | Very low |
| 1,2,3-Trichloropropane | 2-Chloropropene, propene | Low |
| 1,3-Dichloropropane | Propene | Very low |
| Four or five carbons | ||
| 1,4-Dichlorobutane | 4-Chloro-1-butene, 1,3-butadiene | Very low |
| 1,4-Dibromobutane | 4-Bromo-1-butene, 1,3-butadiene | Very low |
| 1,5-Dichloropentane | 5-Chloro-1-pentene, 1,4-pentadiene | Very low |
| 1,5-Dibromopentane | 5-Bromo-1-pentene, 1,4-pentadiene | Very low |
| 5-Bromo-1-pentene | 1,4-Pentadiene | Very low |
| Branched | ||
| 1-Bromo-2-methylpropane | 2-Methylpropene | Low |
| 2-Bromo-2-methylpropane | None | |
| 1-Bromo-3-methylbutane | 3-Methyl-1-butene | Very low |
| 2-Chloro-2-methylbutane | None | |
| 3-Chloro-2-methylpropene | 2-Methylpropene | Moderate |
No reaction was observed with carbon tetrachloride, chloroform, bromoform, dichloromethane, 1,1,1-trichloroethane, 1,1,2,2-tetrachloroethane, pentachloroethane, or PCE.
Relative rates are based on qualitative comparison of peak areas of products to product peak areas obtained with TCE as the substrate. Due to expected variation in the response of different compounds in the flame ionization detector and differences in partitioning between the gas and liquid phases, the data are presented qualitatively.
ND, not determined.
DISCUSSION
TCE-RDase is a peripheral membrane-bound protein of the cytoplasmic membrane which serves as one of two identified terminal reductases in the dehalorespiratory electron transport chain of D. ethenogenes. TCE-RDase accepts electrons from an unknown electron donor or donors (probably located in the membrane), which in turn receive electrons from a membrane-bound hydrogenase. Previous characterization of TCE-RDase coupled with sequence analysis of tceA from D. ethenogenes suggests that TCE-RDase is a member of the recently discovered class of halocarbon RDases, which contain cobalamin and iron-sulfur clusters.
The sequence of the translated TCE-RDase gene (tceA) is unique, sharing limited amino acid homology with the PCE-RDase of D. multivorans (pceA) and the oCP-RDase of D. dehalogenans (cprA). The homology in the N-terminal regions of the three RDases is confined to the short twin-arginine motif, a signal sequence found in other periplasmic or cytoplasmic membrane proteins containing complex redox cofactors (2). The region of greatest homology between the three RDases is in the C-terminal domain of the protein, which contains the twin Fe4S4 cluster binding motif, (CXXCXXCXXXCP)2, characteristic of eight-iron ferredoxins (Fig. 2). Both PCE-RDase and oCP-RDase are missing the first cysteine of the second group in the eight-iron ferredoxin motif, suggesting either alternative ligation to one of the Fe4S4 clusters or the presence of one Fe4S4 cluster and one Fe3S4 cluster. Electron paramagnetic resonance (EPR) spectroscopy of oCP-RDase demonstrated that it contains an Fe4S4 cluster and an Fe3S4 cluster (32). Based on sequence similarity with oCP-RDase, the PCE-RDase of D. multivorans probably has the same complement of iron-sulfur clusters, although no spectroscopic information is available (24). In contrast, EPR studies of the PCE-RDase from Dehalobacter restrictus indicated the presence of two Fe4S4 clusters. Therefore, this enzyme probably has eight cysteine ligands, but this cannot be confirmed, as the sequence has not been reported. Since the sequence of TCE-RDase has all eight cysteines of the motif, it probably contains two Fe4S4 clusters, although in this case no EPR spectroscopy has been performed, due to difficulties in purifying a sufficient mass of the enzyme.
The two other ORFs near tceA (tceB and orf1) showed no homology to proteins in GenBank. tceB, like pceB of D. multivorans and cprB of D. dehalogenans, encodes a small protein with predicted membrane-spanning helices. In analogy to the proposed functions of the other genes, the tceB gene product may serve as a membrane anchor for the TCE-RDase. If the membrane anchor function of these proteins is correct, they would likely tolerate a high degree of divergence in primary sequence, as long as the secondary structure homology and an affinity for their respective dehalogenase were retained.
The extent of divergence of the sequences of RDase genes may indicate that they are ancient enzymes. Perhaps the natural substrates of the various RDases simply have not been discovered. On the other hand, since many halocarbons, including PCE and TCE, are produced naturally (10), it is possible that higher concentrations of halocarbons were present on earth during previous biogeochemical conditions, e.g., during periods of high volcanism. These conditions could have driven the evolution of dehalorespiration and the associated dehalogenases. Alternatively, the ability to detoxify halocarbons may have been a sufficient selective advantage to direct the evolution of the dehalogenases, while dehalorespiration may have evolved recently as the substrates became plentiful due to production by humans. Another possibility is that the ability to dehalogenate halocarbons evolved recently based on an ancient reductive-type enzyme scaffold, which originally did not act upon halocarbons. If this is the case, the appearance of RDases may be an instance of convergent evolution, given the wide species distribution of these enzymes and the observed sequence dissimilarity.
The low degree of homology between the TCE-RDase from D. ethenogenes and the PCE-RDase from D. multivorans is also surprising given the similarity of the reactions they catalyze. Both enzymes dechlorinate TCE to cis-DCE, but this is the only known substrate that they have in common. Although the RDases lack homology in their primary sequences, they may share greater homology at the secondary and tertiary levels that will not become apparent until their three-dimensional structures are analyzed. The reactions that the RDases catalyze fail to fit into either the rearrangement or methyl transfer categories, thus, they appear to represent a new type of cobalamin-dependent enzyme in the oxidoreductase class of enzymes. In addition, the RDases are responsible for the terminal step in the recently discovered energy-generating process termed dehalorespiration. Given these novel and notable features, determining the mechanism of the RDases is an important task.
The study of various substrates of TCE-RDase has led to an increased understanding of the relationship between structural features of the substrates and their reactivity. No reaction was detected with one-carbon compounds. Two-carbon compounds are the preferred substrates of TCE-RDase, and the reaction rate decreases dramatically as the number of carbons in the substrate increases from two to five. This effect may be due in part to steric hindrance; in other words, the active site is unable to accommodate the longer molecules. The decreased reactivity is also likely related to the lack of halide substituents on the intervening carbons. Note that halocarbons containing a single halogen atom, e.g., VC or iodoethane, are rather slow substrates or reversible inhibitors of the enzyme. The four- and five-carbon α-ω-dihaloalkanes may react in a similar manner; in other words, the second halogen substituent is too distant from the first to affect reactivity. Thus, the ends of these substrates behave as if they are two independent monohaloalkanes. The release of the monohalogenated product (e.g., 5-bromo-1-pentene from 1,5-dibromopentane) is consistent with this assessment.
The number, arrangement, and type of halogen substituents are also very important. Geminal or vicinal dihalides exhibit high reaction rates, but the cis conformation is greatly preferred to the trans conformation for the vicinal dihaloalkenes. The failure of TCE-RDase to effect observable dechlorination of 1,1,1-trichloroethane, 1,1,2,2-tetrachloroethane, pentachloroethane, and PCE may reflect steric constraints within the active site; in other words, there may be a limit to the number of bulky halogen atoms which can be accommodated, or the compounds with multiple halogen atoms on sp3 carbons may fit poorly into the active site. Among homologous series of halocarbons, the brominated substrate is dehalogenated faster than the chlorinated homologue (compare vinyl bromide to VC and 1,2-dibromoethane to 1,2-dichloroethane), indicating that carbon halide bond cleavage is rate limiting. The sum of these observations can be used to construct preferred substrate models for TCE-RDase. The ideal haloalkene substrate of TCE-RDase has two carbons and two to three halogen atoms, preferably arranged in a gem or cis conformation. Similarly, for haloalkanes, the preferred substrate has two carbons and two vicinal halogen atoms but no more than one halogen atom per carbon.
This information about the substrates of TCE-RDase may be used to predict the degree of reactivity and the products of other halocarbons of environmental or industrial significance. Some of the compounds that were examined are problematic pollutants. For example, 1,2-dichloroethane and 1,2-dibromoethane are common pollutants, due to their use as solvents, soil fumigants, and components of leaded gasoline and the major role of 1,2-dichloroethane as a chemical feedstock. The structure-activity relationships could be used to predict the bioremediation potential of TCE-RDase (or the dehalogenating organisms expressing it) for different halocarbon pollutants, although the predictions would still need to be empirically verified. An important consideration is whether alternative substrates will be used to support dehalorespiration by D. ethenogenes or simply be cometabolically degraded. Such predictions cannot be made from the current data set, although chlororespiration of 1,2-dichloroethane has already been demonstrated (20). Industrially, the TCE-RDase may have utility in chemical syntheses, given its preference for utilizing and producing the thermodynamically less favored cis conformer of dihaloalkenes.
TCE-RDase is the first enzyme isolated which can completely dechlorinate TCE to the environmentally benign product ethene. This membrane-bound enzyme catalyzes the terminal step in the electron transport pathway for dehalorespiration of TCE by D. ethenogenes. TCE-RDase, along with PCE-RDases and oCP-RDase, appears to be a member of a new subclass of oxidoreductases containing cobalamin and iron-sulfur clusters as cofactors. Systematic classification of these enzymes awaits characterization of their physiological electron donors.
The substrate specificity of TCE-RDase is broad, and the range of rates is similarly broad. This suggests that TCE-RDase might be amenable to a directed evolution approach for increasing the rate of degradation of problematic halocarbons that are currently poor substrates of the enzyme, for example, VC. Future kinetic and structural studies will provide additional insights into the enzyme mechanism that would facilitate a rational enzyme design approach to the same problem.
ACKNOWLEDGMENTS
This work was partially supported by a grant from the Strategic Environmental Research and Development Program of the Department of Defense, Department of Energy, and the U.S. Environmental Protection Agency to D.R.B. Additional support was provided by a PNNL Initiative in Microbial Biotechnology grant under Department of Energy contract DE-AC06-76RL0 1830.
We thank Stephen H. Zinder and Amy Carroll for providing a sample of pure D. ethenogenes strain 195 genomic DNA. We acknowledge the analytical protein chemistry services of the Protein Structure Core Facility, University of Nebraska, Omaha, and the Protein Chemistry Facility, University of Florida, Gainesville. We also acknowledge the DNA sequencing services of the DNA Sequencing Facility, Iowa State University, and Amplicon Express, Pullman, Wash.
REFERENCES
- 1.Agency for Toxic Substances and Disease Registry. Agency for Toxic Substances and Disease Registry HazDat Database. Vol. 2000. Atlanta, Ga: U.S. Department of Health and Human Services; 1999. [Google Scholar]
- 2.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 3.Christiansen N, Ahring B K. Desulfitobacterium hafniense sp. nov., an anaerobic reductively dechlorinating bacterium. Int J Syst Bacteriol. 1996;46:442–448. [Google Scholar]
- 4.Cole J R, Fathepure B Z, Tiedje J M. Tetrachloroethene and 3-chlorobenzoate dechlorination activities are co-induced in Desulfomonile tiedjei DCB-1. Biodegradation. 1995;6:167–172. doi: 10.1007/BF00695347. [DOI] [PubMed] [Google Scholar]
- 5.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the Dense Alignment Surface method. Prot Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 6.deBruin W P, Kotterman M J J, Posthumus M A, Gosse Schraa, Zehnder A J B. Complete biological reductive transformation of tetrachloroethene to ethane. Appl Environ Microbiol. 1992;58:1996–2000. doi: 10.1128/aem.58.6.1996-2000.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiStefano T D, Gossett J M, Zinder S H. Reductive dechlorination of high concentrations of tetrachloroethene to ethene by an anaerobic enrichment culture in the absence of methanogenesis. Appl Environ Microbiol. 1991;57:2287–2292. doi: 10.1128/aem.57.8.2287-2292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox B G, Borneman J G, Wackett L P, Lipscomb J D. Haloalkene oxidation by the soluble methane monooxygenase from Methylosinus trichosporium OB3b: mechanistic and environmental implications. Biochemistry. 1990;29:6419–6427. doi: 10.1021/bi00479a013. [DOI] [PubMed] [Google Scholar]
- 9.Gerritse J, Renard V, Gomes T M P, Lawson P A, Collins M D, Gottschal J C. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch Microbiol. 1996;165:132–140. doi: 10.1007/s002030050308. [DOI] [PubMed] [Google Scholar]
- 10.Gribble G W. The natural production of chlorinated compounds. Environ Sci Technol. 1994;28:310A–319A. doi: 10.1021/es00056a712. [DOI] [PubMed] [Google Scholar]
- 11.Helenius A, Simons K. Solubilization of membranes by detergents. Biochim Biophys Acta. 1975;415:29–79. doi: 10.1016/0304-4157(75)90016-7. [DOI] [PubMed] [Google Scholar]
- 12.Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder A J. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch Microbiol. 1998;169:313–321. doi: 10.1007/s002030050577. [DOI] [PubMed] [Google Scholar]
- 13.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- 14.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Wackett L P. Trichloroethylene oxidation by toluene dioxygenase. Biochem Biophys Res Commun. 1992;185:443–451. doi: 10.1016/s0006-291x(05)81005-8. [DOI] [PubMed] [Google Scholar]
- 18.Löffler F E, Sanford R A, Tiedje J M. Initial characterization of a reductive dehalogenase from Desulfitobacterium chlororespirans Co23. Appl Environ Microbiol. 1996;62:3809–3813. doi: 10.1128/aem.62.10.3809-3813.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnuson J K, Stern R V, Gossett J M, Zinder S H, Burris D R. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl Environ Microbiol. 1998;64:1270–1275. doi: 10.1128/aem.64.4.1270-1275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maymó-Gatell X, Anguish T, Zinder S H. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl Environ Microbiol. 1999;65:3108–3113. doi: 10.1128/aem.65.7.3108-3113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maymó-Gatell X, Chien Y-T, Gossett J M, Zinder S H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 22.Miller E, Wohlfarth G, Diekert G. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch Microbiol. 1998;169:497–502. doi: 10.1007/s002030050602. [DOI] [PubMed] [Google Scholar]
- 23.Neumann A, Scholz-Muramatsu H, Diekert G. Tetrachloroethene metabolism of Dehalospirillum multivorans. Arch Microbiol. 1994;162:295–301. doi: 10.1007/BF00301854. [DOI] [PubMed] [Google Scholar]
- 24.Neumann A, Wohlfarth G, Diekert G. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J Bacteriol. 1998;180:4140–4145. doi: 10.1128/jb.180.16.4140-4145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman L M, Wackett L P. Trichloroethylene oxidation by purified toluene 2-monooxygenase: products, kinetics, and turnover-dependent inactivation. J Bacteriol. 1997;179:90–96. doi: 10.1128/jb.179.1.90-96.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni S, Fredrickson J K, Xun L. Purification and characterization of a novel 3-chlorobenzoate-reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB-1. J Bacteriol. 1995;177:5135–5139. doi: 10.1128/jb.177.17.5135-5139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Scholz-Muramatsu H, Neumann A, Meßmer M, Moore E, Diekert G. Isolation and characterization of Dehalospirillum mulltivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch Microbiol. 1995;163:48–56. [Google Scholar]
- 29.Sticht H, Rösch P. The structure of iron-sulfur proteins. Prog Biophys Mol Biol. 1998;70:95–136. doi: 10.1016/s0079-6107(98)00027-3. [DOI] [PubMed] [Google Scholar]
- 30.Triglia T, Peterson M G, Kemp D J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1998;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 32.van de Pas B A, Smidt H, Hagen W R, van der Oost J, Schraa G, Stams A J, de Vos W M. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J Biol Chem. 1999;274:20287–20292. doi: 10.1074/jbc.274.29.20287. [DOI] [PubMed] [Google Scholar]
- 33.Ward A M. Investigations on the bivalency of carbon. IV. Halogen displacements from s-tetra-bromo- and -chloro-ethane and tri-bromo- and -chloro-ethylene. J Chem Soc. 1930;1930:2143–2148. [Google Scholar]
- 34.Wiedemeier T H, Swanson M A, Moutoux D E, Gordon E K, Wilson J T, Wilson B H, Kampbell D H, Haas P E, Miller R N, Hansen J E, Chapelle F H. Technical protocol for evaluating natural attenuation of chlorinated solvents in ground water. EPA/600/R-98/128. U.S. Washington, D.C.: Environmental Protection Agency; 1998. [Google Scholar]