Abstract
Macrophages are important for host defense against intracellular pathogens like Salmonella and can be differentiated into two major subtypes. M1 macrophages, which are pro-inflammatory and induce antimicrobial immune effector mechanisms, including the expression of inducible nitric oxide synthase (iNOS), and M2 macrophages, which exert anti-inflammatory functions and express arginase 1 (ARG1). Through the process of phagocytosis, macrophages contain, engulf, and eliminate bacteria. Therefore, they are one of the first lines of defense against Salmonella. Infection with Salmonella leads to gastrointestinal disorders and systemic infection, termed typhoid fever. For further characterization of infection pathways, we established an in vitro model where macrophages are infected with the mouse Salmonella typhi correlate Salmonella enterica serovar Typhimurium ( S. tm), which additionally expresses red fluorescent protein (RFP). This allows us to clearly characterize macrophages that phagocytosed the bacteria, using multi-color flow cytometry.
In this protocol, we focus on the in vitro characterization of pro- and anti-inflammatory macrophages displaying red fluorescent protein-expressing Salmonella enterica serovar Typhimurium, by multi-color flow cytometry.
Keywords: Salmonella Typhimurium , Macrophages, Infection Control, Arginase 1, Inducible Nitric Oxide Synthase
Background
The intracellular Gram-negative bacterium Salmonella typhi can cause severe and often life-threatening disease in humans. Globally, approximately 200,000 deaths are caused by the bacterium every year. The intracellular pathogen is ingested through contaminated food, and transmitted from person to person. The mouse correlate of human Salmonella typhi is Salmonella enterica serovar Typhimurium ( S. tm). The intracellular bacteria are phagocytosed by macrophages and are able to evade antimicrobial defense by inhibiting fusion of lysosome and phagosome. Therefore, S. tm is able to survive and replicate inside the host ( Buchmeier and Heffron, 1991 ; Navarre et al. , 2010 ; Lahiri et al. , 2010 ; Mastroeni and Grant, 2011 ; Bhutta et al. , 2018 ).
In general, the immune system is divided into cells of the innate immune system (monocytes, macrophages, dendritic cells, and natural killer cells), and the acquired immune system (B lymphocytes, and T lymphocytes). Macrophages are phagocytic cells of the innate immune system, and one of the body's first defense mechanisms, when a pathogen crosses the host’s skin barrier. They can be classified into two types: (I) M1 pro-inflammatory macrophages, which are responsible for killing bacterial and viral pathogens, and express inducible nitric oxide synthase (iNOS), and (II) M2 macrophages, which support the wound healing process, and have an anti-inflammatory effect. Importantly, M2 macrophages express the enzyme arginase 1 (ARG1) ( Mosser and Edwards, 2008 ; Mills, 2012 ; Weiss and Schaible, 2015 ; Gordon and Martinez- Pomares, 2017 ; Murray, 2017 ; Hannemann et al. , 2019 ).
The cytosolic enzyme ARG1 is primarily expressed in liver tissue. During infection, ARG1 upregulation promotes pathogen survival in macrophages, by hydroxylating l-arginine to urea and ornithine, which therefore lowers l-arginine levels for the synthesis of nitric oxide (NO) by iNOS. A fully functional iNOS needs an adequate l-arginine supply to produce NO that kills pathogens. Interferon gamma (IFNγ) and tumor necrosis factor alpha (TNFα) are potent inducers of iNOS ( Nairz et al. , 2013 ; Bogdan, 2015 ; Brigo et al. , 2021 ). ARG1 is induced by interleukin 4 (IL-4), which is produced by type 2 T helper cells. TNFα and IFNγ inhibit the transcription of ARG1, by interfering with IL-4–induced chromatin remodeling ( Ostuni and Natoli, 2011 ; Schleicher et al. , 2016 ; Piccolo et al. , 2017 ). Additionally, several studies show that increased ARG1 activity is associated with an increased concentration of various pathogens, such as Streptococcus pneumoniae, Mycobacterium bovis, Mycobacterium tuberculosis , or Toxoplasma gondii ( Iniesta et al. , 2005 ; El Kasmi et al. , 2008 ; De Muylder et al. , 2013 ; Schleicher et al. , 2016 ; Paduch et al. , 2019 ). However, it has been reported that deletion or pharmacological inhibition of ARG1 does not lead to a better control of Salmonella infection in macrophages, or in mice, in a septicemia model ( Brigo et al. , 2021 ).
Current treatment against invasive salmonellosis with antibiotics has become more difficult, due to resistance against conventional antibiotics. Therefore, the identification of new mechanisms to better understand the host-pathogen interplay in Salmonella infection, and the detailed characterization of the cells involved in the defense against this infection are urgently needed. Herein, we describe a method that identifies pro- and anti-inflammatory bone marrow-derived macrophage subtypes, during an infection with Salmonella enterica serovar Typhimurium. Furthermore, Salmonella expressing red fluorescent protein allows the visualization of phagocytosed Salmonella in these subtypes. This method supports further research on the involvement of pro- and anti-inflammatory macrophages in the defense against Salmonella .
Materials and Reagents
250 mL Erlenmeyer flask (Stoelzle Medical, catalog number: 21226368000)
0.5 mL Eppendorf tubes (Eppendorf, catalog number: 0030121.023)
Disposable cuvette (BRAND, catalog number: 759015)
1.5 mL Eppendorf tubes (Eppendorf, catalog number: 0030120.086)
Cell scraper (Sarstedt, catalog number: 83.3951)
15 mL Polypropylene conical tube (Falcon, catalog number: 352096)
Cell strainer 40 µm (Falcon, catalog number: 352360)
96-well BRAND plate (Life Science Products, catalog number: 781602)
5 mL disposable syringe (BD, catalog number: 309050)
Luna cell counting slides (Biocat, catalog number: L201B1C3GB)
6-cm dish (TTP, catalog number: 93060)
Salmonella enterica serovar Typhimurium SL1344 expressing red fluorescent protein (RFP) ( Birmingham et al. , 2006 ; Wu et al. , 2017 )
Lysogeny broth (LB Broth) Lennox (Roth, catalog number: X964.2)
Glycerol (Sigma, catalog number: G5516-100ML)
Phosphate buffer saline (PBS; Lonza, catalog number: 17-515 F)
Agar-Agar Kobe I (Roth, catalog number: 5210.3)
CASY Cup (OMNI Life Science, catalog number: 5651794)
CASY Ton Buffer (OMNI Life Science, catalog number: 5651808)
Aqua bidest (Fresenius Kabi, catalog number: 16.231)
Gentamicin (Gibco, catalog number: 15750-037)
L-glutamine (Lonza, catalog number: BE17-605E)
Dulbecco′s Modified Eagle′s Medium (DMEM, Pan Biotech TM , catalog number: P04-01500)
Fetal bovine serum (FBS, Pan Biotech TM , catalog number: P30-3031)
BV650 anti-mouse/human CD11b (BioLegend, catalog number: 101239)
APC-R700 rat anti-mouse CD45 (BDHorizon TM , catalog number: 565478)
BV421 rat anti-mouse F4/80 (BD Biosciences; catalog number: 565411)
PerCP-eFluor TM 710 anti-mouse Ly6G (Invitrogen; catalog number: 46-9668-82)
FITC anti-mouse CD3 (BioLegend, catalog number: 100204)
FITC anti-mouse CD19 (ImmunoTools, catalog number: 22220193S)
FITC anti-mouse CD49b (BioLegend, catalog number: 103503)
PE-Cyanine7 anti-mouse iNOS (Invitrogen; catalog number: 25-5920-80)
APC anti-mouse ARG1 (Invitrogen, catalog number: 17-3697-82)
BD Cytofix/Cytoperm TM (BD Biosciences, catalog number: 51-2091K7)
BD PermWash TM (BD Biosciences, catalog number: 51-2090K7)
50 mL Polypropylene conical tube (Falcon, catalog number: 352070)
Ketamine (Livisto, catalog number: 6680219)
Xylazine (Animedica, catalog number: 7630517)
Omnican F syringes (Braun, catalog number: 91615025)
Disposable hypodermic needle 100 Sterican R (Braun, catalog number: 4657519)
Pen-Strep (Lonza, catalog number: DE17-602E)
Erythrocyte lysis buffer (R&D, catalog number: WL2000)
Acridine Orange/propidium iodide stain (Biocat, catalog number: F23001)
LB medium (see Recipes)
LB-medium with 30% Glycerol (see Recipes)
Equipment
Laminar Flow Cabinet; EuroClone Safe Mate Eco 1.2 (Politakis Laborgeräte, catalog number: EN 12 469)
Shaking incubator (VWR, catalog number: GFL 3031)
Heraeus ® HERAcell ® CO 2 Incubator (Thermo Fisher Scientific, catalog number: 3615-45)
Photometer (Eppendorf, BioPhotometer D30, catalog number: 6133000001)
Centrifuge (Hettich Micro 200R and Rotanta 460R, catalog number: Z652113, Z623520)
CASY TT counting system (OMNI Life Science, catalog number: TT-20A-2571)
CytoFLEX S V4-B4-R2-I2 Flow Cytometer (13 detectors, 4 lasers, Beckman Coulter, catalog number: C01161)
LUNA Automated Cell Counter (Biocat, L10001-LG)
Software
-
FlowJo v10.7.0 (BD Biosciences, https://www.flowjo.com/solutions/flowjo )
Note: Any software package for analyzing flow cytometry data can be used with this protocol.
Procedure
-
Preparation of Salmonella Typhimurium ( S. tm) stock
Take an aliquot of Salmonella enterica serovar Typhimurium SL1344 expressing red fluorescent protein (RFP) from -20°C storage.
Thaw the aliquot at room temperature.
Pipette 10 µL of S. tm into 10 mL of LB-medium in a 250-mL Erlenmeyer flask.
Cover the top of the flask using tin foil.
Incubate in a shaking incubator at 200 rpm and 37°C overnight.
The following day, pipette 50 µL of the overnight culture into 10 mL of fresh LB-medium into a 250-mL Erlenmeyer flask.
Discard the overnight culture of S .tm. Wash and sterilize the 250-mL Erlenmeyer flask.
Cover the top of the flask using tin foil.
Incubate the culture in a shaking incubator at 200 rpm and 37°C for 1–2 h.
Calibrate a photometer, using 500 µL of LB-medium in a disposable cuvette as blank.
-
Measure OD 600 , to check if S. tm reached 0.5.
S. tm reaches the optimal logarithmic growth phase when OD 600 is between 0.5–0.7.
Note: If the OD 600 value is below 0.5, continue the incubation of the culture in the 250-mL Erlenmeyer flask as described above, until an OD 600 value of 0.5 is reached. Of note, S.tm density duplicates every 20 min. If the OD 600 value is above 0.7, dilute the culture 1:1 with LB-medium, and incubate it in the 250-mL Erlenmeyer flask, until an OD 600 value of 0.5.
Transfer the culture into a 50-mL conical tube.
Centrifuge the S. tm culture at 4,967 × g at room temperature for 5 min.
Discard the supernatant.
Resuspend the pellet in 1 mL of freshly prepared LB-medium + 30% glycerol.
Prepare aliquots of 50 µL in 0.5-mL Eppendorf tubes, and store them at -20°C.
-
Culture of S. tm to the optimal growth phase
Thaw one aliquot of the S. tm stock.
Pipette 10 µL of this aliquot into 10 mL of LB-medium in a 250-mL Erlenmeyer flask, and cover the top of the flask using tin foil.
Incubate in a shaking incubator at 200 rpm and 37°C overnight.
The following day, pipette 50 µL of this overnight culture into 10 mL of LB-medium in a 250-mL Erlenmeyer flask, and cover the top of the flask using tin foil.
Discard the overnight culture of S .tm. Wash and sterilize the 250 mL Erlenmeyer flask.
Incubate in a shaking incubator at 200 rpm and 37°C for 1–2 h.
Calibrate a photometer, using 500 µL of LB medium in a disposable cuvette as blank.
-
Measure OD 600 to check if S. tm reached 0.5, which is equivalent to the optimal logarithmic growth phase.
Note: If the OD 600 value is below 0.5, continue the incubation of the culture in the 250-mL Erlenmeyer flask as described above, until an OD 600 value of 0.5 is reached. Of note, S.tm density duplicates every 20 min. If the OD 600 value is above 0.7, dilute the culture 1:1 with LB-medium, and incubate it in the 250-mL Erlenmeyer flask, until an OD 600 value of 0.5.
-
Counting viable S. tm using a Casy counting system
Use the 45-µm capillary.
Measure the background by placing a new Casy cup with 10 mL of fresh Casy ton buffer under the measuring unit.
Select the program for background measurement ( Table 1 Background Measurement).
Measure the background. This should be below 30 counts and 1 µm in size. Otherwise, wash the system.
Prepare a new Casy cup with 10 mL of Casy ton buffer, and add 5 µL of S. tm OD 600 0.5.
Shake gently.
Place the sample under the measuring unit.
Select the program for measuring between 1–3 µm ( Table 1 S. tm Measurement).
Measure.
-
Click next, to get the number of viable counts/mL = viable S. tm /mL.
Note: Viable counts from a freshly prepared S.tm culture with an OD 600 of 0.5 should be between 2.5 × 10 8 –3 × 10 8 viable counts/mL.
After the measurement is finished, remove the sample cup, and add a fresh Casy cup with 10 mL of Casy ton buffer.
Perform Casy Clean up to five times.
Select the program for washing ( Table 1 Washing Program).
After the washing is completed, check the background again.
If the background is below 30, the Casy counting system can be turned off. Otherwise, continue the washing.
-
Preparation of bone marrow-derived macrophages (BMDM)
Note: Preparation of BMDM has been described by Zanoni et al. (2012; doi:10.21769/BioProtoc.225.).
A video demonstrating the procedure can be found at: https://www.jove.com/de/v/52347/isolation-intravenous-injection-murine-bone-marrow-derived .
A schematic illustration of the generation of bone marrow-derived macrophages and infection with S.tm is shown in Figure 1 .
-
Anesthetize a wildtype C57Bl/6N mouse, by intraperitoneally injecting 50 µL of 100 mg/kg Ketamine + 10 mg/kg Xylazine.
Note: A video demonstrating the procedure in general can be found at Intraperitoneal Injection in the Mouse: https://researchanimaltraining.com/articles/intraperitoneal-injection-in-the-mouse/ .
Perform euthanization of the deeply anesthetized mouse by cervical dislocation. Therefore, place a large tweezer behind the base of the anesthetized mouse's skull, and pull sharply back on the tail at a 45° angle.
Fixate the animal on a Styrofoam panel, and spray the surface of the animal with 75% alcohol.
Remove skin and muscle tissue from one leg, by cutting upwards from the heel with sterile scissors.
Cut around the femur head.
Cut in the middle of the knee joint. Be careful not to damage the bones.
Cut the ankle joint.
Remove excess muscles with tissue paper.
Pull on the upper leg, to remove the femur head from the hip joint.
Place the bones into PBS containing 1% penicillin and 1% streptomycin on ice.
Move to a laminar flow hood, and perform all steps on ice.
Open the ends of the bones, by cutting with a pair of sterile scissors.
Place a 40-µm cell strainer on a 50-mL Falcon-tube.
-
Flush out the bone marrow:
Use a disposable hypodermic needle and a 5-mL syringe.
Fill the syringe with PBS containing 1% penicillin and 1% streptomycin.
Place a needle on one end of the opened bone.
Flush the bone marrow out onto the 40-µm cell strainer.
Repeat flushing of the bone, until it is completely white.
Wash the cell strainer with 10 mL of PBS containing 1% penicillin and 1% streptomycin.
Using the plunger of the syringe, strain the cells through the cell strainer.
Wash the cell strainer with 10 mL of PBS containing 1% penicillin and 1% streptomycin.
Centrifuge the 50-mL conical tube containing your flushed bone marrow at 300 × g and 4°C for 5 min.
Dilute Erythrocyte Lysis Buffer 1:10 with double distilled water
Discard the supernatant.
Resuspend the pellet in 2 mL of diluted Erythrocyte Lysis Buffer.
Incubate at room temperature for 3 min.
Add 15 mL of PBS containing 1% penicillin and 1% streptomycin on top of the Erythrocyte Lysis Buffer.
Centrifuge the 50-mL conical tube containing your flushed bone marrow at 300 × g and 4°C for 5 min.
Discard the supernatant.
Resuspend the pellet in 15 mL of PBS containing 1% penicillin and 1% streptomycin.
Repeat steps 24–26.
Centrifuge the 50-mL conical tube again, and discard the supernatant.
Resuspend the cell pellet in 45 mL of DMEM media supplemented with 10% FBS, 1% L-glutamine, 1% penicillin, 1% streptomycin, and 50 ng/mL recombinant murine M-CSF.
Pipette 15 mL of the cell suspension into each of three 20-cm Falcon dishes.
Change the medium every second day.
On day 5, cells can be harvested (Procedure E).
-
-
Harvesting and counting of cells
Remove the culture media from the cell culture dishes.
Wash the cells twice with 10 mL of PBS.
Add 8 mL of DMEM medium supplemented with 10% FBS and 1% L-glutamine.
Scrape the cells using a disposable cell scraper.
Wash the dishes with another 2 mL of DMEM medium supplemented with 10% FBS and 1% L-glutamine.
Transfer the cells into a 50-mL conical tube.
Close the tube, and invert the cells 2–3 times.
Place 9 µL of the cell suspension in a 0.5-µL Eppendorf tube.
Mix 1 µL of Acridine Orange/ Propidium Iodide stain solution with the cell aliquot.
Place 10 µL of the mixture into a Luna cell counting slide.
-
Count the cells using the LUNA-FL fluorescent and bright field automated cell counter.
Note: Approximately 4.5 × 10 7 cells are obtained from one mouse, after isolating and culturing the bone marrow of both hind legs.
Seed the cells in 6-cm dishes, at a density of 1.5 × 10 6 cells/mL in DMEM medium supplemented with 10% FBS and 1% L-glutamine.
Seed four additional dishes for fluorescent minus one (FMO) control (see step G3).
Let the cells settle in a cell incubator overnight.
-
In vitro infection of bone marrow-derived macrophages with S. tm
-
Infect BMDM with S. tm at a multiplicity of infection 10 (MOI 10). Therefore, add 10 times more S. tm than cells.
Note: Unused S.tm culture with OD 600 of 0.5 can be used for preparing new S.tm aliquots (Procedure A), or be discarded.
Incubate the cells in a cell incubator (5% CO 2 , 37°C) for 1 h.
Remove the medium containing non-phagocytosed S. tm.
Wash the cells twice with 1 mL of PBS + 25 µg/mL gentamicin.
Add 1 mL of DMEM medium supplemented with 10% FBS, 1% L-glutamine, and 25 µg/mL gentamicin.
Incubate the cells in a cell incubator for 4 h.
-
-
Flow Cytometry staining of cultured BMDM
-
Extracellular staining
Harvest the BMDM by scraping in culture medium, using a disposable cell scraper.
Transfer the cells to a 15-mL Falcon tube, and pellet by centrifugation at 300 × g and 4°C for 5 min.
Discard the supernatant.
Resuspend the pellet in 1 mL of PBS, and transfer it into an 1.5-µL Eppendorf tube.
Pellet the cells by short spinning at 10,860 × g and 4°C for 30 s.
Discard the supernatant.
Resuspend the pellet in 50 µL of surface antibody mix (all antibodies 1:200 in PBS; Table 2 ).
Incubate in the dark at 4°C for 10 min.
Wash with 500 µL of PBS.
Pellet the cells by short spinning (10,860 × g , 4°C, 30 s).
Discard the supernatant.
Resuspend the pellet in 100 µL of Cytofix/CytoPerm TM Buffer, to permeabilize and fix the cells.
Incubate in the dark at 4°C for 20 min.
Dilute the PermWash TM Buffer 1:10 with Aqua bidest.
Add 500 µL of the diluted PermWash TM Buffer on top of the 100 µL of Cytofix/CytoPerm TM Buffer.
Pellet the cells by short spinning (10,860 × g , 4°C, 30 s).
Discard the supernatant.
-
Intracellular stain
Prepare the intracellular antibody mix in diluted PermWash TM Buffer (iNOS 1:100, and ARG1 1:100; Table 3 ).
Resuspend the samples in 50 µL of intracellular antibody mix.
Incubate the samples protected from light at room temperature for 45 min.
Wash the cells once with 500 µL of diluted PermWash TM . Pellet the cells by short spinning (10,860 × g , 4°C, 30 s).
Discard the supernatant, and resuspend the pellets in 200 µL of PBS.
Transfer the samples into a flat bottom 96-well plate, via a 40-µm strainer.
Analyze directly in a flow cytometry device.
-
Fluorescent minus one (FMO) control
Perform extracellular staining of BMDM, as described in step G1, in the additionally seeded uninfected and infected BMDM samples (see step E13).
Discard the supernatant after washing with diluted PermWash TM Buffer on top of the 100 µL of Cytofix/CytoPerm TM Buffer.
Resuspend one uninfected and one infected BMDM cell pellets in the intracellular antibody mix with only the antibody against ARG1. Resuspend the other uninfected and infected BMDM cell pellets in the intracellular antibody mix with only the antibody against iNOS.
Incubate the samples protected from light at room temperature for 45 min.
Wash the cells once with 500 µL of diluted PermWash TM .
Pellet the cells by short spinning (10860 × g , 4°C, 30 s).
Discard the supernatant, and resuspend the pellets in 200 µL of PBS.
Transfer the samples into a flat bottom 96-well plate, via a 40-µm strainer.
Analyze directly in a flow cytometry device.
-
Table 1. Programs CASY TT Counting system.
|
Background Measurement |
Measurement of S. Typhimurium |
Washing Program | |
|---|---|---|---|
| Capillary | 45 µm X-Axis: 5 µm | 45 µm X-Axis: 3 µm | 45 µm X-Axis: 5 µm |
| Sample Volume | 200 µL Cycles: 1 | 200 µL Cycles: 3 | 200 µL Cycles: 10 |
| Dilution | 1.001e+00 | 2.001e+03 | 1.001e+00 |
| Y-Axis | Auto | Auto | Auto |
| Eval.Cursor | 1.00–4.89 µm | 0.75–2.93 µm | 0.00–5.00 µm |
| Norm. Cursor | 0.5–4.89 µm | 0.3–2.93 µm | 0.00–5.00 µm |
| %Calculation | %Via Debris: On | %Via Debris: On | %Via Debris: On |
| Aggr. Correct: | Auto | Auto | Auto P |
| Interface | Par P.Feed: On | Par P.Feed: On | Par P.Feed: On |
| Print Mode | Manual Graphic: On | Manual Graphic: On | Manual Graphic: On |
Figure 1. Schematic representation of the experimental setup, showing the generation of bone marrow-derived macrophages and infection with S .tm.
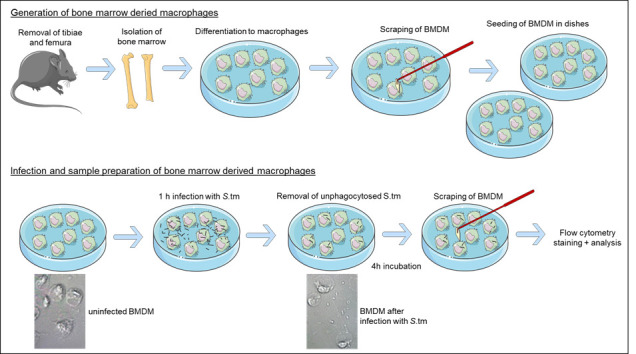
The different morphological shapes of uninfected and infected BMDM are visualized in a ScanR Imaging Platform (pictures kindly provided by Demetz E.).
Table 2. Antibodies for flow cytometry – extracellular stain.
| Antibody | Clone | Company | Catalog number | expressed on |
|---|---|---|---|---|
| CD3 FITC | 17.A2 | BioLegend | 100204 | T cells |
| CD19 FITC | PeCa1 | ImmunoTools | 22220193S | B cells |
| CD49b FITC | HMa2 | BioLegend | 103503 | NK cells |
| CD11b BV650 | M170 | BioLegend | 101239 | neutrophils, monocytes |
| CD45 APC-R700 | 30-F11 | BD Horizon | 565478 | leukocytes |
| F4/80 BV421 | T45-2342 | BD Horizon | 565411 | macrophages |
| Ly6G PerCP-eFlour 710 | 1AB-Ly6g | Invitrogen | 46-9668-82 | neutrophils |
Note: To be sure that the bone marrow-derived macrophages are not contaminated by T cells, B cells, or NK cells, antibodies against CD3, CD19, and CD49b are added to the flow cytometry panel. All these antibodies are labeled with FITC; therefore, all FITC + cells can be excluded in the gating strategy ( Figure 2 ). The typical cell distribution of a BMDM culture (uninfected and infected with S.tm) is shown in Table 4 .
Table 3. Antibodies for flow cytometry – intracellular stain.
| Antibody | Clone | Company | Catalog number | expressed on |
|---|---|---|---|---|
| iNOS PE-Cyanine7 | CXNFT | Invitrogen | 25-5920-80 | pro-inflammatory macrophages |
| ARG1 APC | A1exF5 | Invitrogen | 17-3697-82 | anti-inflammatory macrophages |
Data analysis
The FlowJo software was used to analyze the data. The gating strategy is described in Figure 2 .
Figure 2. Gating strategy for the infected bone marrow-derived macrophages – upper panel, and the uninfected bone marrow-derived macrophages – lower panel.
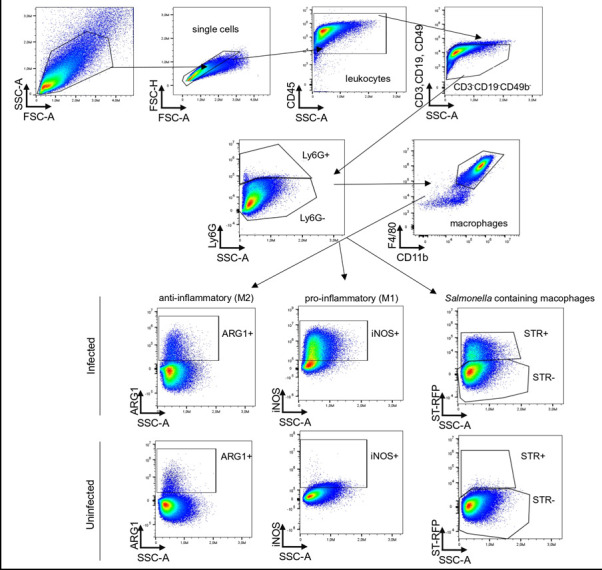
After exclusion of doublets, the leukocytes are described as CD45 + . T-cells, B-cells, and NK cells are excluded (CD3–CD19–CD49b–), and macrophages are gated as Ly6G–CD11b + F4/80 + . Depending on the research question, there is the possibility to characterize pro-inflammatory macrophages as iNOS-expressing cells, and anti-inflammatory macrophages as ARG1–expressing cells. Furthermore, macrophages containing S. tm are characterized by expression of the red fluorescent protein (RFP) in the PE channel.
In BMDM generated from C57BL/6N mice that were either left uninfected or were infected with S. tm, and afterwards further incubated for 4h, typical values for analyzed cell subsets are depicted in Table 4 .
Table 4. Typical cell distribution of a BMDM culture.
| Cell type | Gating | Mean ± SEM Uninfected | Mean ± SEM Infected |
|---|---|---|---|
| Cells | FSC-A to SSC-A | 94.7 ± 2.1 | 95.2 ± 2.1 |
| Single Cells | FSC-A to FSC-H | 90.0 ± 2.7 | 91.7 ± 2.2 |
| Leukocytes | CD45 to FSC-A | 98.6 ± 1.3 | 99.5 ± 0.3 |
| FITC- negative cells | FITC (CD3, CD19, CD49b) to FSC-A | 98.6 ± 0.7 | 97.7 ± 0.6 |
| Monocytes (Ly6G-) | Ly6G to FSC-A | 98.8 ± 0.7 | 98.4 ± 0.5 |
| Macrophages | F4/80 to CD11b | 98.0 ± 0.9 | 98.8 ± 1.1 |
| Anti-inflammatory macrophages | ARG1 to FSC-A | 1.8 ± 0.5 | 1.4 ± 0.3 |
| Pro-inflammatory macrophages | iNOS to FSC-A | 4.2 ± 2.3 | 77.7 ± 3.8 |
| Salmonella containing macrophages | STR to FSC-A | 0 | 24.7 ± 1.78 |
Troubleshooting
-
Cultivation of S .tm (see Procedures A and B)
Salmonella grow best at 37°C, and need oxygen for optimal growth. Therefore, they should be cultivated in an Erlenmeyer flask which is covered with tin foil. The temperature of 37°C must be strictly ensured.
-
Preparation of BMDM (see Procedure D)
For the generation of bone marrow-derived macrophages, it is important to work in a sterile manner. Cells should only be handled in a laminar flow cabinet. It is important to isolate the entire tibiae and femura, and to cut them open only under sterile conditions, using autoclaved scissors and tweezers.
-
Harvesting, counting, and seeding of BMDM (see Procedure E)
Harvesting of BMDM by scraping needs to be performed rather gently, to obtain high cell viability. We recommend scraping away from one’s body, exerting not much force on the disposable cell scraper.
Cells are seeded for infection in antibiotic free media. Therefore, the day before seeding, it is recommended to incubate 5 mL of the antibiotic free medium in a cell culture dish placed in a cell incubator overnight, to be sure that the medium is not contaminated by bacteria.
-
Infection of BMDM (see Procedure F)
After infection, it is necessary to thoroughly wash away the unphagocytosed S .tm. It is important to remove the medium completely, and wash the cells at least twice with PBS + gentamicin. A microscope can be used to see whether S. tm are still present in the culture. If yes, washing steps should be repeated.
It is important to add gentamicin to the PBS for washing, and to the DMEM for further incubation. Gentamicin inhibits the proliferation of S .tm, by blocking protein biosynthesis.
Recipes
-
LB medium
a.d. with
2% LB-Broth
Autoclave (at 121°C for 20 min, and at 50°C for 10 min)
-
LB medium with 30% Glycerol
Add 300 µL of Glycerol to 700 µL of LB medium
Acknowledgments
G.W. is supported by grants from the Christian Doppler Society and an ERA-NET grant by the FWF (EPICROSS, I-3321), and N.B. was supported by the FWF doctoral college project W1253 HOROS.
Salmonella Typhimurium expressing red fluorescent protein were a kind gift from Prof. Dr. Dirk Bumann (University of Basel, Switzerland). This protocol was adapted and modified after Fritsche et al. (2008).
Competing interests
The authors declare no conflicts of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Bhutta Z. A. , Gaffey M. F. , Crump J. A. , Steele D. , Breiman R. F. , Mintz E. D. , Black R. E. , Luby S. P. and Levine M. M. ( 2018. ). Typhoid Fever: Way Forward . Am J Trop Med Hyg 99(3_Suppl): 89-96 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Birmingham C. L. , Smith A. C. , Bakowski M. A. , Yoshimori T. and Brumell J. H. ( 2006. ). Autophagy controls Salmonella infection in response to damage to the Salmonella -containing vacuole . J Biol Chem 281 ( 16 ): 11374 - 11383 . [DOI] [PubMed] [Google Scholar]
- 3. Bogdan C. ( 2015. ). Nitric oxide synthase in innate and adaptive immunity: an update . Trends Immunol 36 ( 3 ): 161 - 178 . [DOI] [PubMed] [Google Scholar]
- 4. Brigo N. , Pfeifhofer-Obermair C. , Tymoszuk P. , Demetz E. , Engl S. , Barros-Pinkelnig M. , Dichtl S. , Fischer C. , L. Valente De Souza , Petzer V. , et al. .( 2021. ). Cytokine-Mediated Regulation of ARG1 in Macrophages and Its Impact on the Control of Salmonella enterica Serovar Typhimurium Infection . Cells 10 ( 7 ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchmeier N. A. and Heffron F. ( 1991. ). Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium . Infect Immun 59 ( 7 ): 2232 - 2238 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Muylder G. , Daulouede S. , Lecordier L. , Uzureau P. , Morias Y. , J. Van Den Abbeele , Caljon G. , Herin M. , Holzmuller P. , Semballa S. , et al. .( 2013. ). A Trypanosoma brucei kinesin heavy chain promotes parasite growth by triggering host arginase activity . PLoS Pathog 9 ( 10 ): e1003731 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El Kasmi K. C. , Qualls J. E. , Pesce J. T. , Smith A. M. , Thompson R. W. , Henao-Tamayo M. , Basaraba R. J. , Konig T. , Schleicher U. , Koo M. S. , et al. .( 2008. ). Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens . Nat Immunol 9 ( 12 ): 1399 - 1406 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fritsche G. , Nairz M. , Werner E. R. , Barton H. C. and Weiss G. ( 2008. ). Nramp1-functionality increases iNOS expression via repression of IL-10 formation . Eur J Immunol 38 ( 11 ): 3060 - 3067 . [DOI] [PubMed] [Google Scholar]
- 9. Gordon S. and Martinez-Pomares L. ( 2017. ). Physiological roles of macrophages . Pflugers Arch 469 ( 3-4 ): 365 - 374 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hannemann N. , Cao S. , Eriksson D. , Schnelzer A. , Jordan J. , Eberhardt M. , Schleicher U. , Rech J. , Ramming A. , Uebe S. , et al. .( 2019. ). Transcription factor Fra-1 targets arginase-1 to enhance macrophage-mediated inflammation in arthritis . J Clin Invest 129 ( 7 ): 2669 - 2684 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iniesta V. , Carcelen J. , Molano I. , Peixoto P. M. , Redondo E. , Parra P. , Mangas M. , Monroy I. , Campo M. L. , Nieto C. G. , et al. .( 2005. ). Arginase I induction during Leishmania major infection mediates the development of disease . Infect Immun 73 ( 9 ): 6085 - 6090 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lahiri A. , Lahiri A. , Iyer N. , Das P. and Chakravortty D. ( 2010. ). Visiting the cell biology of Salmonella infection . Microbes Infect 12 ( 11 ): 809 - 818 . [DOI] [PubMed] [Google Scholar]
- 13. Mastroeni P. and Grant A. J. ( 2011. ). Spread of Salmonella enterica in the body during systemic infection: unravelling host and pathogen determinants . Expert Rev Mol Med 13 : e12 . [DOI] [PubMed] [Google Scholar]
- 14. Mills C. D. ( 2012. ). M1 and M2 Macrophages: Oracles of Health and Disease . Crit Rev Immunol 32 ( 6 ): 463 - 488 . [DOI] [PubMed] [Google Scholar]
- 15. Mosser D. M. and Edwards J. P. ( 2008. ). Exploring the full spectrum of macrophage activation . Nat Rev Immunol 8 ( 12 ): 958 - 969 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murray P. J. ( 2017. ). Macrophage Polarization . Annu Rev Physiol 79 : 541 - 566 . [DOI] [PubMed] [Google Scholar]
- 17. Nairz M. , Schleicher U. , Schroll A. , Sonnweber T. , Theurl I. , Ludwiczek S. , Talasz H. , Brandacher G. , Moser P. L. , Muckenthaler M. U. , et al. .( 2013. ). Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection . J Exp Med 210 ( 5 ): 855 - 873 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Navarre W. W. , Zou S. B. , Roy H. , Xie J. L. , Savchenko A. , Singer A. , Edvokimova E. , Prost L. R. , Kumar R. , Ibba M. , et al. .( 2010. ). PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica . Mol Cell 39 ( 2 ): 209 - 221 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ostuni R. and Natoli G. ( 2011. ). Transcriptional control of macrophage diversity and specialization . Eur J Immunol 41 ( 9 ): 2486 - 2490 . [DOI] [PubMed] [Google Scholar]
- 20. Paduch K. , Debus A. , Rai B. , Schleicher U. and Bogdan C. ( 2019. ). Resolution of Cutaneous Leishmaniasis and Persistence of Leishmania major in the Absence of Arginase 1. J Immunol 202 ( 5 ): 1453 - 1464 . [DOI] [PubMed] [Google Scholar]
- 21. Piccolo V. , Curina A. , Genua M. , Ghisletti S. , Simonatto M. , Sabo A. , Amati B. , Ostuni R. and Natoli G. ( 2017. ). Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk . Nat Immunol 18 ( 5 ): 530 - 540 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schleicher U. , Paduch K. , Debus A. , Obermeyer S. , Konig T. , Kling J. C. , Ribechini E. , Dudziak D. , Mougiakakos D. , Murray P. J. , et al. .( 2016. ). TNF-Mediated Restriction of Arginase 1 Expression in Myeloid Cells Triggers Type 2 NO Synthase Activity at the Site of Infection . Cell Rep 15 ( 5 ): 1062 - 1075 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weiss G. and Schaible U. E. ( 2015. ). Macrophage defense mechanisms against intracellular bacteria . Immunol Rev 264 ( 1 ): 182 - 203 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu A. , Tymoszuk P. , Haschka D. , Heeke S. , Dichtl S. , Petzer V. , Seifert M. , Hilbe R. , Sopper S. , Talasz H. , et al. .( 2017. ). Salmonella Utilizes Zinc To Subvert Antimicrobial Host Defense of Macrophages via Modulation of NF-kappaB Signaling . Infect Immun 85 ( 12 ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zanoni I. , Ostuni R. and Granucci F. ( 2012. ). Generation of Mouse Bone Marrow-Derived Macrophages(BM-MFs) . Bio-protocol 2 ( 12 ): e225 . [Google Scholar]


