Summary
The ability to precisely control transgene expression is essential for basic research and clinical applications. Adeno-associated viruses (AAVs) are non-pathogenic and can be used to drive stable expression in virtually any tissue, cell type, or species, but their limited genomic payload results in a trade-off between the transgenes that can be incorporated and the complexity of the regulatory elements controlling their expression. Resolving these competing imperatives in complex experiments inevitably results in compromises. Here, we assemble an optimized viral toolkit (VTK) that addresses these limitations and allows for efficient combinatorial targeting of cell types. Moreover, their modular design explicitly enables further refinements. We achieve this in compact vectors by integrating structural improvements of AAV vectors with innovative molecular tools. We illustrate the potential of this approach through a systematic demonstration of their utility for targeting cell types and querying their biology using a wide array of genetically encoded tools.
Keywords: AAV, vector, neuroscience, circuits
Graphical abstract
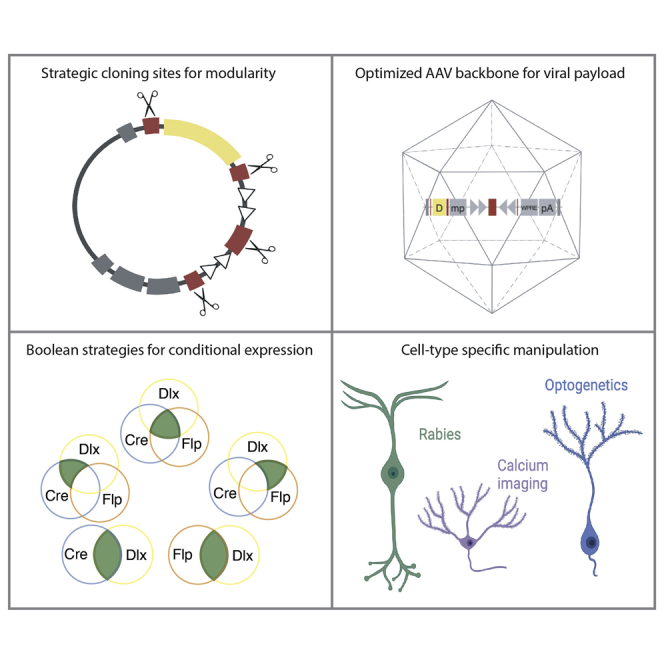
Highlights
-
•
Cell-type-specific targeting is required to study circuits and behaviors
-
•
Factors such as transgene size limit the AAV payload available
-
•
VTK is a set of optimized AAV backbones that overcomes these limitations
Motivation
The diversity of transgenes allowing for precise labeling and manipulation of neurons is ever evolving, but the evolution of viral vectors capable of restricting their expression to specific and functionally defined subtypes lags behind. Here, we present an optimized viral toolkit (VTK) that aims at simplifying the experimental design by increasing the modularity of AAV vectors for the targeting of specific types of neurons.
Pouchelon et al. develop an optimized and modular viral toolkit (VTK) that addresses key AAV limitations and allows for efficient combinatorial targeting of neuronal cell types. This approach allows efficient querying of neuronal biology using a wide array of genetically encoded tools.
Introduction
Exploring biological systems requires the ability to selectively target particular cell types. Nowhere is this more evident than in the nervous system, where specialized cells function through the formation of canonical circuits (Tasic et al., 2018; Fishell and Kepecs, 2019). The use of transgenic animal models for targeted effector and sensor expression has transformed our understanding of the brain (Taniguchi et al., 2011; Madisen et al., 2015; He et al., 2016). However, flexibility in designing experiments that are reliant solely on transgenic animals remains costly, time consuming, and limiting. Given the current explosion in innovative molecular tools, recombinant adeno-associated viruses (AAVs) provide an ideal means for their rapid and efficient implementation (Blankvoort et al., 2018; Deverman et al., 2018; Dimidschstein et al., 2016; Jüttner et al., 2019; Vormstein-Schneider et al., 2020; Hrvatin et al., 2019; Graybuck et al., 2021; Mich et al., 2021; Lambert et al., 2021). Size limitations in AAVs demand careful consideration for attaining an optimal balance between payload and the regulatory elements controlling their expression (Bedbrook et al., 2018; Fenno et al., 2014, 2020). Over the past 10 years, a bewildering array of genetic components has been engineered into AAV backbones. This has resulted in the accumulation of elements included by historic happenstance rather than utility. Here, we create a viral toolkit (VTK) consisting of highly optimized AAV backbones that provides end users with the flexibility to implement complex targeting strategies.
Results
Design of an optimized VTK for direct and combinatorial transgene expression
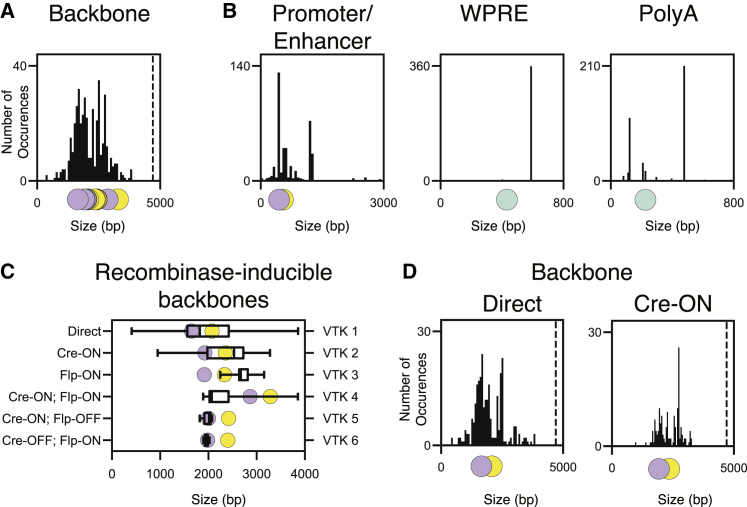
For efficient viral transgene expression, the payload of AAV vectors has to be limited to 4.7 kb (Hirsch et al., 2016), and as such, effective use of AAVs involves finding an optimal compromise between size restriction and cellular specificity. This requires a tradeoff between (1) the requirement for the essential elements needed for AAV expression (i.e., the backbone [Powell et al., 2015]) and (2) components such as enhancers or recombinase sites that provide for selective cell-type expression. Here, we systematically considered all constituent components used in existing AAV constructs. To do so, we began by surveying the most commonly used CNS-related plasmids available at Addgene, a non-profit organization, which has become the largest and most utilized repository for AAV plasmids. For the 428 constructs requested more than 20 times (Table S1; see STAR Methods), we systematized the annotation of all elements included in the AAV payload (i.e., between the internal terminal repeats [ITRs]) and classified them into 5 categories: promoters/enhancers, transgenes, woodchuck hepatitis virus posttranscriptional regulatory elements (woodchuck hepatitis virus posttranscriptional regulatory elements,WPREs), polyA sequences, and recombinase recognition sites (Table S1). For all plasmids, we assessed frequency of use and size of backbones and constituent elements (i.e., all elements between ITRs minus the transgene; Figures 1A, 1B, and S1A). As the addition of recombinase-inducible elements (e.g., complex recombination-site configurations, stop cassette) can easily result in exceeding the packaging capability, we further considered the size of each backbone in terms of their recombinase dependency (Figures 1C, 1D, and S1B). While many of these constructs are Cre dependent (141/428), a small fraction are Flp dependent (10/428) or combine both Flp and Cre (10/428). This analysis revealed that among the commonly used plasmids, there exists a wide variability in construct size, resulting from the historical inclusion of both suboptimal elements and extraneous sequences that provide no functional utility. Hence, this highlighted the lack of standardized, robust, and flexible AAV backbones from which to fully exploit the widest range of possible approaches. This also revealed notable gaps in the strategies devised to achieve cell-type specificity, in particular an underutilization of Boolean subtractive approaches.
Figure 1.
Constitutive elements analysis of Addgene most commonly used AAV vectors
Number of occurrences of the indicated elements among Addgene’s top plasmids sorted by size (base pairs, bp).
(A) Backbones. The dotted line shows the 4.7 kb payload limit.
(B) Promoters. WPRE and polyA sequences.
(C) Distribution of backbones’ sizes for recombinase-inducible plasmids from Addgene. Each row represents specific recombinase dependencies as described on the left y axis, and its corresponding VTK constructs are represented on the right y axis.
(D) Number of occurrences of backbones in direct comparison to Cre-ON conformation.
In (A–D), the colored dots on the x axis indicate the size of the corresponding elements used in VTK vectors. Green dots for common elements to all VTKs (WPRE and polyA), purple dots for VTKS1–6, and yellow dots for VTKD1–6.
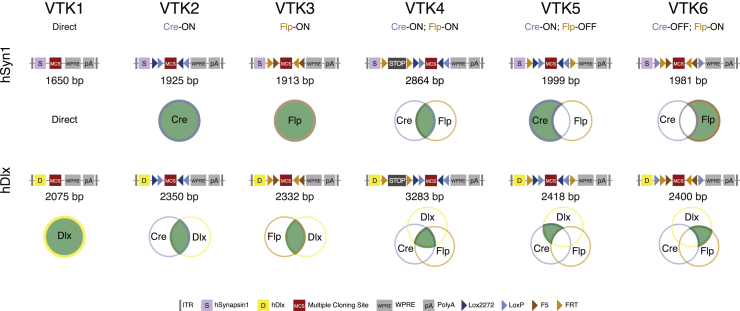
To address these limitations, we designed, assembled, and validated a set of AAV backbones that constitute a generalizable VTK for targeting and manipulating the nervous system. These plasmids vary in their mode of regulation from simple promoter/enhancer-dependent expression to complex combinatorial expression, dependent on both Cre and Flp recombinases (Figure 2; Table S2). To build these vectors, we incorporated a set of structural elements that represent the best compromise between size and efficiency. Specifically, we included a WPRE sequence of 251 bp and a bovine growth hormone (bGH) polyA sequence of 225 bp in each plasmid to increase transgene expression by stabilizing the resulting mRNA, without affecting the titer of the AAV production (Figure S1B; see STAR Methods) (Donello et al., 1998; Kohara et al., 2014; Pfarr et al., 1986). By building these plasmids using direct DNA synthesis (Table S2), we were able to optimize the size of the backbones while further improving their modularity. This allowed us to strategically incorporate restriction-enzyme recognition sites at key locations to facilitate the cloning of constructs as needed (Figure S2). We then used Boolean strategies to create a set of recombinase-dependent backbones for conditional expression: direct expression from the promoter (VTK1), Cre-recombinase-dependent expression (VTK2, Cre-ON), Flp-recombinase-dependent expression (VTK3, Flp-ON), both Cre- and Flp-recombinase-dependent expression (VTK4, Cre-ON; Flp-ON), Cre-recombinase-dependent expression that is turned off by Flp recombinase (VTK5, Cre-ON; Flp-OFF), and Flp-recombinase-dependent expression that is turned off by Cre recombinase (VTK6, Cre-OFF; Flp-ON). For improved recombination efficiency, Lox or FRT inducible sites were placed in a double-inverted orientation (DIO) conformation (Atasoy et al., 2008) for ON systems or facing each other for deletion when in an OFF conformation. Each of these iterations is provided in two variants that include either the human synapsin 1 promoter for neuron-specific expression (hSyn for VTKS1–6) (Nieuwenhuis et al., 2021; Kügler et al., 2003) or the human Dlx5/6 enhancer element to further restrict expression to telencephalic GABAergic interneurons (Dlx,VTKD1–6) (Dimidschstein et al., 2016). All constructs, irrespective of the promoter used, optimized AAV payloads. While VTK1–3 backbones are more compact than most of the commonly used plasmids, VTK4–6 constitute backbones that offer ways of targeting specific populations (see yellow and purple dots in Figures 1 and S1). Together, these constructs provide an overarching architecture for inclusion of complex transgenes while retaining flexibility for the myriad of contexts in which they could be useful.
Figure 2.
Optimization of a viral toolkit (VTK) for direct and combinatorial transgene expression
Diagram representing each VTK plasmid. The size between ITRs and the corresponding Boolean strategy are displayed below the relevant plasmid backbone. Top row: VTKS1–6. Bottom row: VTKD1–6. MCS, multiple cloning site; pA: polyA; LoxP, Lox2272 are Cre-dependent sites; FRT, F5 are Flp-dependent sites.
VTK vectors: Transgene expression and cell-type selectivity
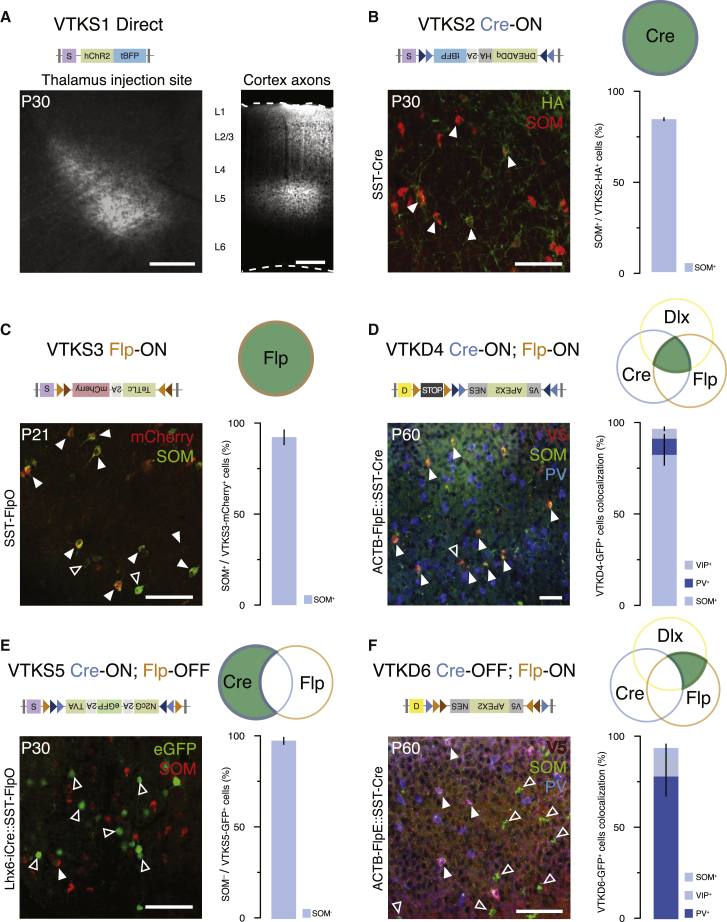
To assess the utility of the VTK backbones, we first examined their ability to direct gene expression and then explored their functional effectiveness for visualizing neuronal function or modulating activity. We first tested the expression and specificity of these vectors by inserting various reporters and effector transgenes in VTK1–6 backbones and injecting the corresponding AAVs in mice at various ages and in various brain regions (Table S3). VTKS1-hChR2-tBFP (direct) was injected in the thalamus of wild-type (WT) mice at postnatal day 10 (P10), which led to local expression of tBFP across all neurons in this structure. Consistent with this, labeled projections of these neurons were observed in the expected cortical areas (Figure 3A). VTKS2-HA-GqDREADD-P2A-tBFP (Cre-ON) was injected in the somatosensory cortex (S1) of SST-Cre transgenic animals, which resulted in strong colocalization between the hemagglutinin (HA) tag associated with the viral transgene and somatostatin (SOM) (Figure 3B; 87.32% ± 1.12%, N = 4). VTKS3-TeTLc-P2A-mCherry (Flp-ON) was injected into S1 of SST-FlpO animals, which showed high levels of colocalization of mCherry expression with SOM+ neurons (Figure 3C; 92.26% ± 4.31%, N = 3). VTKD4-APEX2-V5 (Cre-ON; Flp-ON) was injected into S1 of ACTB-FlpE::SST-Cre animals (i.e., ubiquitous expression of FlpE- and SST-specific expression of Cre). In conjunction with the hDlx enhancer, we observed that the vast majority of cells expressing the viral reporter were SOM+ (Figures 3D and S3B; SOM+: 83.17% ± 5.95%; other GABAergic, with parvalbumin [PV]: 8.97% ± 2.67%; vasoactive intestinal peptide [VIP]: 5.5% ± 1.22; N = 3). VTKS5-Rabies-Helper (Cre-ON; Flp-OFF) was injected in Lhx6-iCre::Sst-FlpO, which led to a strong transgene expression de-targeted from SOM interneurons (Pouchelon et al., 2021) (Figure 3E; SOM-: 97.42% ± 0.19%, N = 3). VTKD6-APEX2-V5 (Cre-OFF; Flp-ON) was injected into the hippocampus (Figure 3F, S1, S3A, and S3B) of ACTB-FlpE::SST-Cre animals. This resulted in effective targeting of GABAergic interneurons with hDlx (PV+: 78.27% ± 10.99%; VIP+: 11.13% ± 2.55%; N = 3) and the expected exclusion of SOM+ cells due to the Cre deletion (SOM-: 95.5% ± 2.5%). Using these strategies as exemplars, we demonstrate the targeting specificity of all constructs and highlight that the VTK4–6 vectors can be used for effective Boolean logic strategies compatible with the use of large transgenes.
Figure 3.
Validation of VTK vector cell-type specificity
(A) Example of VTKS1-hChR2-tBFP injection in the thalamus and corresponding thalamocortical projections in the cortex. Scale bar: 100 μm (left) and 200 μm (right).
(B) Colocalization of somatostatin (SOM) and Gq-HA tag showing SST-Cre-specific VTKS2-Dreadd-Gq recombination (87.32% ± 1.12%, N = 4). Scale bar: 100 μm.
(C) Colocalization of SOM with mCherry showing the recombination of VTKS3-TeTLC upon SST-FlpO (92.26% ± 4.31%, N = 3). Scale bar: 100 μm.
(D) Left, colocalization of V5 labeling with SOM and parvalbumin (PV). Scale bar: 100 μm. Right, quantification of V5 colocalization with SOM, PV, and vasoactive intestinal peptide (VIP)-expressing interneurons (SOM: 83.17% ± 5.95%; PV: 8.97% ± 2.67%; VIP: 5.5% ± 1.22%, N = 3) showing the recombination of VTKD4-NES-APEX2 in GABAergic interneurons and primarily in SOM+ cells.
(E) Colocalization of EGFP and SOM showing the exclusion of VTKS5-helpers from SOM+ neurons (97.42% ± 0.19%, N = 3). Scale bar: 100 μm.
(F) Left, colocalization of V5 with SOM and PV in the hippocampus. Scale bar, 100 μm. Right, quantification of V5 with SOM, PV, and VIP (PV: 78.27% ± 10.99%; VIP: 11.13% ± 2.55%; SOM: 4.5% ± 2.55%, N = 3) showing the recombination of VTKD6-NES-APEX2 in GABAergic interneurons and a majority of PV+ neurons (full arrows), except for SOM+ cells (empty arrows).
Having demonstrated that these constructs worked independently, we wondered whether AAVs with different enhancers could be used in combination. In testing this, we noticed that the coinjection of multiple AAVs under the control of different enhancers in the same animal can lead to a complete loss of their original specificity. For example, upon coinjecting two AAVs into the cortex—mDlx-EGFP, which only targets inhibitory interneurons (Dimidschstein et al., 2016) and CamKII-tdTomato, which only targets excitatory neurons—we observed an unexpected colocalization of their reporters (Figure S3C). This pattern of interference occurred in most instances where promoters/enhancers with largely distinct expression profiles were used in tandem (data not shown). Therefore, despite the appeal of using different gene-regulatory elements in combination, such experiments should be undertaken with considerable caution.
VTK vectors: Transgene functionality
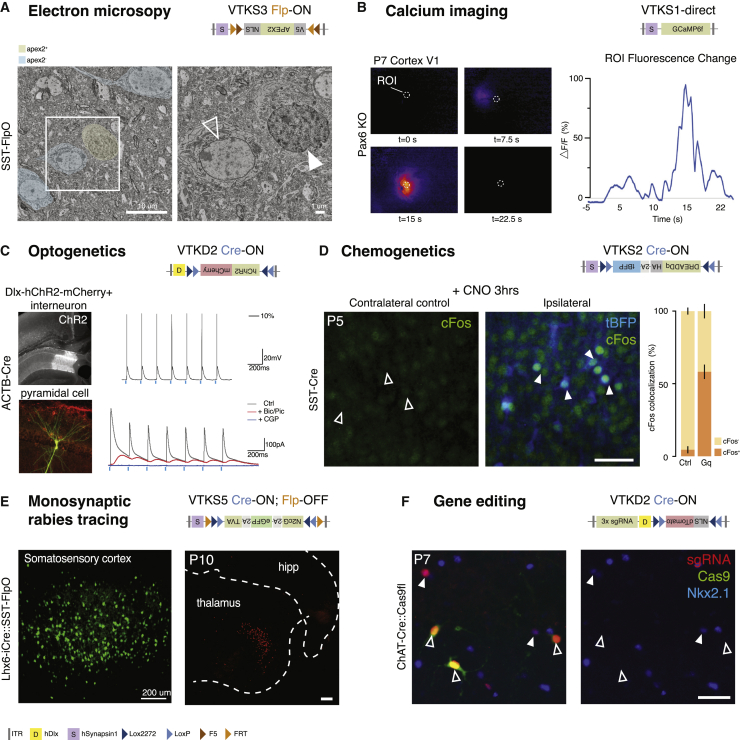
Over the past decade, neurobiologists have developed a diverse armamentarium of genetic tools to examine neuronal ultrastructure, visualization and manipulation of their activity, and connectivity, as well as gene editing. The size of these components varies considerably, and all require different levels of transgene expression. If one wishes to use these tools in the context of complex targeting strategies, this cannot be achieved without integrating them into compact vectors. We thus validated that the VTK backbones can be effectively employed to drive functional expression of genetically encoded tools spanning a wide range of applications: electron microscopy, calcium imaging, optogenetics, chemogenetics, gene editing, and monosynaptic tracing.
Electron microscopy
A nuclear-restricted form of APEX2 (Lam et al., 2015) (NLS-APEX2-V5) was inserted into the VTKS3 backbone and injected in the somatosensory cortex of SST-FlpO transgenic animals. Nuclear APEX labeling allowed for the structural analysis of SST interneurons using electron microscopy (Figure 4A).
Figure 4.
Validation of VTK vectors functionality
(A) VTKS3-NLS-APEX2-V5 injected into the somatosensory cortex of SST-FlpO mice allowed for the labeling of sparse populations of nuclei. SST cells with an APEX2-positive nucleus are labeled in yellow (full arrow) and negative ones in blue (empty arrow). Scale bar: 10 μm (left) and 1 μm (right).
(B) Calcium events from VTKS1-GCaMP6f-infected cells were found in the visual cortex of Pax6 knockout (KO) mice. Waves of spontaneous infected cell activity were detected. Graph represents the difference in fluorescence (ΔF/F%) detected in the region of interest (ROI) over time.
(C) Top, cells infected by VTKD2-hChR2-mCherry in dorsal hippocampus of ACTB-Cre mice show depolarization upon blue-light stimulation; bottom, ChR2-mCherry-negative neighboring pyramidal cells receive light-evoked inhibitory postsynaptic currents (IPSCs). IPSCs are blocked by bicuculline (Bic)/picrotoxin (PIc) and CGP55845 (CGP) and GABAa and GABAb receptor antagonists, respectively.
(D) VTKS2-driven Dreadd-Gq (tBFP) triggers the expression of the immediate-early gene, cFos, after 3 h of CNO activation compared with the control on the contralateral side (Ctrl: 4.47% ± 2.4%, N = 3; after CNO: 58.21% ± 5.19%, N = 6; data are shown as mean ± SEM). Full arrows: cFos + tBFP colocalization upon activation. Empty arrows: absence of cFos in Ctrl. Scale bar: 50 μm.
(E) Left, VTKS5-rabies helpers (N2cG-eGFP-TVA) are injected in the somatosensory cortex of Lhx6-iCre::SST-FlpO animals. Right, example retrograde labeling tracing found at the thalamus level after secondary rabies infection of helper+ cells. Scale bar: 200 μm (left and right).
(F) Three CRISPR guides (sgRNA) targeting Nkx2.1 gene are inserted in VTKD2-NLSdTomato and injected in ChAT-Cre mice::Floxed-Cas9 mice. sgRNA (red) in Cas9-positive (green) cells suppressed Nkx2.1 expression (blue, empty arrows). Cells without colocalization still express Nkx2.1 (full arrows). Scale bar: 50 μm.
Calcium imaging
The calcium sensor GCaMP6f (Chen et al., 2013) integrated in the VTKS1 backbone was used to query changes in neuronal activity in the postnatal visual cortex of a mouse with altered retinal activity (Figure 4B) (Burbridge et al., 2014).
Optogenetics
The light-activated opsin ChR2-mCherry (Boyden et al., 2005) was integrated in VTKD2 and injected into the hippocampus of ACTB-Cre mice to test if ChR2-mCherry expression could be restricted to interneurons despite ubiquitous Cre expression. Blue-light stimulation produced depolarization of GABAergic interneurons (Figures 4C, top, and S4A) and resulted in inhibitory synaptic currents in adjacent pyramidal populations. Postsynaptic currents in pyramidal neurons showed a reverse potential of −60 mV and were completely blocked by GABA receptor antagonists (Figures 4C, bottom, and S4B).
Chemogenetics
The activating GqDREADD-tBFP (Alexander et al., 2009; Armbruster et al., 2007) was incorporated into VTKS2 and injected into the somatosensory cortex of SST-Cre animals. Three hours following N-oxide (CNO) administration, we observed a strong and specific upregulation of the immediate-early gene cFos in cells expressing the viral reporter (Figure 4D; control [Ctrl]: 4.47% ± 2.4%, N = 3; after CNO: 58.21% ± 5.19%, N = 6).
Monosynaptic rabies tracing
The rabies helper transgenes required for monosynaptic retrograde labeling include the coding sequence of a reporter, the cellular receptor for subgroup A avian leukosis viruses (TVA), and rabies glycoprotein, amounting to a 3 kb construct. These transgenes were integrated into VTKS5 and injected into the somatosensory cortex of Lhx6-iCre::SSTFlpO mice (Pouchelon et al., 2021). Following injection of rabies viruses, we observed monosynaptic retrograde labeling from the AAV-targeted cells in the expected thalamic nucleus (Figure 4E).
Gene editing
Nkx2.1 CRISPR guide RNA (single guide RNA [sgRNA]) (Sanjana et al., 2014) was inserted upstream of the promoter, which allowed for the insertion of multiple promoter-sgRNA-polyA (3xU6-sgRNA-pA), permitting the incorporation of the NLS-dTomato reporter into a VTKD2 backbone. This was injected in the cholinergic basal forebrain (Magno et al., 2011) of ChAT-Cre::Floxed:stop:Cas9 mice. In cells coexpressing Cas9 with sgRNA and dTomato, Nkx2.1 expression was absent (Figure 4F).
To expand the utility of the set of backbones presented in this study, we produced additional plasmids containing dTomato, GFP, ChR2, or GCaMP6f in either VTKD4–6 or VTKS4–6 backbones. Altogether, this represents a collection of 12 ready-to-use backbones (see Table S2) and 41 effector-containing plasmids (see Table S3) available for AAV production for the neuroscience community via Addgene.
Discussion
By surveying the breadth of existing genetic elements utilized in AAV production and selecting streamlined components, we have assembled an optimized set of backbones that can be employed in both direct and combinatorial strategies. This set of tools provides the means for targeting and examining the biological functions of virtually any desired neural population. While our initial aim was to simplify and provide more consistency in AAV-based neuron targeting with the use of VTK, our toolkit also allows for adapting to complex strategies, including the use of the largest effectors commonly employed in neuroscience (i.e., rabies helper transgenes and single-vector multiple CRISPR-gRNA with reporter). While interference at present precludes effective coinjection of multiple AAVs, we expect that further innovation will ultimately overcome this limitation and greatly increase the ability to target different neuronal populations in tandem. We view VTK not as a static toolset but as a platform upon which further innovation will allow for ever greater flexibility in querying and manipulating neural circuits. We expect this will include both in vitro and in vivo applications, notably including free animal behavior. Moreover, while this approach has been optimized for use in rodents, we envision that it will ultimately prove to be efficacious across vertebrate models including primates.
Limitations of the study
Leaky expression
While VTK simplifies the use of AAVs for complex genetics and effector expression, their efficiency and specificity are dependent upon the experimental context. For instance, while driver lines are extremely efficient for targeted deletion or expression of a transgene, mouse lines driving recombinase are not equally effective; this is especially true when recombinases are under an activity-dependent promoter. For instance, VTKS5 did not provide selective deletion from NPY-FlpO animals (data not shown).
Experimental parameters
Additional considerations for experimental design are the use of specific serotypes (Bedbrook et al., 2018), the mode of administration (i.e., intraparenchymal, systemic) (Challis et al., 2019), titer, mode of production (i.e., HEK293 triple-transfection versus Baculovirus [Carinhas et al., 2009; Nasimuzzaman et al., 2018]) and species (i.e., rodents, primates), all of which can considerably influence the level of infection and specificity. In this study, we used AAV serotype 1 for local injection to assess the functionality of the VTK backbone in mice. As such, we recommend that users validate the specificity and functionality of the VTK vectors using the specific parameters imposed by their experimental design. These various parameters have been extensively tested, and the corresponding studies constitute a valuable resource to guide the experimental design (Dimidschstein et al., 2016; Haery et al., 2019; Royo et al., 2008).
AAV quality
Moreover, an essential consideration is the quality of the AAV production; non-specific expression commonly results from the production of AAV particles containing transgenes where recombination occurred during production (Dimidschstein et al., 2016; Haery et al., 2019; Royo et al., 2008). This is typically caused by the presence of latent recombinase activity during the production of AAVs, which especially affects smaller transgenes. To avoid such contamination, the bacterial growth of AAV plasmids for viral production must be tightly controlled (see STAR Methods for details).
Recombinases as transgenes
Due to their high catalytic activity, incorporating recombinases into AAVs can be problematic. This is particularly complicated when the expression of Cre recombinase is placed under the control of enhancer elements (e.g., the mDlx or E2 enhancers; Dimidschstein et al., 2016; Vormstein-Schneider et al., 2020), which can lead to a complete loss of selective targeting. While, in principle, using attenuated versions of Cre recombinase (i.e., Cre, iCre, dCre) or different modes of injection (i.e., systemic, local) might improve the observed specificity, our efforts to do so proved unsuccessful (data not shown). Nonetheless, several groups have, in particular contexts, achieved specificity of expression in select circumstances. For example, the Feng laboratory used the hDlx-Cre construct in Shank3-floxed animals and found that 80% of cells knocked down for Shank3 were interneurons (Fischer et al., 2019). Together, these data suggest that the high level of catalytic activity of these proteins can trigger non-specific recombination even at low levels of expression. This effect can, in principle, be mitigated by using less-active recombinases or in contexts where recombinase-dependent sites are less accessible (e.g., the chromatin of the R26 locus of RCE and Sun1-GFP mouse lines is likely more accessible than the Shank3 locus). As such, the effective use of recombinases in AAVs requires their use to be validated on a case-by-case basis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-somatostatin | Peninsula Laboratories International | T-4103.0050, RRID:AB_518614 |
| guinea pig anti-cFos | Synaptic Systems | #226004, RRID:AB_2619946 |
| anti-tagFP | Synaptic Systems | FluoTag-X2 N0502-At542 RRID:AB_2744624 |
| anti-tagFP | Synaptic Systems | FluoTag-X2 At647 RRID:AB_2744626 |
| rat anti-HA | Sigma | #11867423001 RRID:AB_390918 |
| chicken anti-GFP | Aves Labs | #1020 RRID:AB_10000240 |
| rat anti-RFP | Chromotek | #5f8 RRID:AB_2336064 |
| mouse anti-V5 | Thermofisher | #R960-25 RRID:AB_2556564 |
| guinea-pig anti-PV | Swant | GP-72 RRID:AB_2665495 |
| rabbit anti-SST | Millipore | MAB354 RRID:AB_2255365 |
| rabbit anti-Nkx2.1 | Abcam | #ab76013 RRID:AB_1310784 |
| rabbit anti-VIP | Joan Vaughan at the Saghatelian lab | N/A |
| Alexa Fluor 405-, 488-, 546-, 594-, or 647-conjugated secondary antibodies | Thermo Fisher Science or Jackson ImmunoResearch | N/A |
| Bacterial and virus strains | ||
| Tables S2 and S3 | Addgene and this paper | N/A |
| Deposited data | ||
| Tables S2 and S3 | In this paper as indicated in the tables | N/A |
| Experimental models: Organisms/strains | ||
| C57Bl/6 | Jackson Laboratories | RRID:IMSR_JAX:000664 |
| SST-Cre | Jackson Laboratories | RRID:IMSR_JAX:013044 |
| SST-FlpO | Jackson Laboratories | RRID:IMSR_JAX:031629 |
| Lhx6-iCre | Jackson Laboratories | RRID:IMSR_JAX:026555 |
| ACTB-FlpE | Jackson Laboratories | RRID:IMSR_JAX:005703 |
| ACTB-Cre | Jackson Laboratories | RRID:IMSR_JAX:003376 |
| ChAT-Cre | Jackson Laboratories | RRID:IMSR_JAX:031661 |
| Sun1-GFP | Jackson Laboratories | RRID:IMSR_JAX:021039 |
| RCE | Jackson Laboratories | RRID:IMSR_JAX:32037 |
| Cas9eGFP | Jackson Laboratories | RRID:IMSR_JAX:026179 |
| Pax6ɑ-Cre | (Marquardt et al., 2001) | N/A |
| ϐ2-nAChRs floxed | (Burbridge et al., 2014) | N/A |
| Recombinant DNA | ||
| Tables S2, S3 and S4 | Addgene and In this paper as indicated in the table | N/A |
| Software and algorithms | ||
| EMBOSS needle 6.5.0 | (Madeira et al., 2019) | N/A |
| SnapGene Server version 1.1.58 | http://www.snapgene.com | RRID:SCR_015052 |
| Addgene data analysis code | This paper | N/A |
| Fiji | https://imagej.nih.gov/ij/ | RRID:SCR_002285 |
| Cell Counter | https://github.com/Jessdlin/CellCounter/ | N/A |
| pClamp | http://www.moleculardevices.com/products/software/pclamp.html | RRID:SCR_011323 |
| Atlas Zeiss software | https://www.zeiss.fr/microscopie/produits/microscope-software/atlas5.html | RRID:SCR_020928 |
| MATLAB | MathWorks | RRID:SCR_001622 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gabrielle Pouchelon (pouchel@cshl.edu) and Jordane Dimidschstein (jordane.dimidschstein@gmail.com).
Materials availability
Plasmids generated in this study have been deposited to Addgene and are listed in Tables S2 and S3.
Experimental model and subject details
Mice
All experiments were approved by and in accordance with Harvard Medical School IACUC protocol number IS00001269, Broad Institute IACUC protocol number 0156-03-17-1 and the National Institutes of Health. C57Bl/6 mice were used for breeding with transgenic mice. Transgenic mice, SST-Cre (stock number: 013044), SST-FlpO (stock number: 031629), Lhx6-iCre (stock number: 026555), ACTB-FlpE (stock number: 005703), ACTB-Cre (stock number: 003376), ChAT-Cre (stock number: 031661), Sun1-GFP (stock number: 021039), RCE (stock number: 32037), Cas9eGFP (stock number: 026179) are available at Jackson Laboratories. Both female and male animals were used for all experiments. PAX cKO mice were created by crossing Pax6ɑ-Cre (Marquardt et al., 2001) to ϐ2-nAChRs floxed mice (Burbridge et al., 2014). Both males and females were included in all experiments.
Method details
Addgene plasmids selection and analysis
A list of commonly used neuroscience-related AAV plasmids was generated from Addgene’s internal database. We defined "commonly used" as having been distributed more than 20 times since the plasmid was made available on Addgene’s website. The list was further refined by manually removing plasmids containing promoters specific to tissues other than the brain, such as the liver-specific TBG promoter. Plasmid features were identified using SnapGene Server version 1.1.58 with the common features that were available by April 2021. For each Addgene plasmid sequence, a list of plasmid features was obtained from SnapGene that included the feature name, type, direction, and location within the sequence (start and end nucleotide). The size of each feature was calculated based on the feature location information. The nucleotide sequence for each feature was extracted from the full plasmid sequence using the start and end nucleotides (see Table S1). For each of the 445 most-used plasmids obtained from Addgene we used SnapGene’s 1.1.58 annotation to identify the promoter, WPRE, polyA, and inducible elements. Since not every feature was recognized by SnapGene, we improved the annotation in between bacterial origins of replication (oris): (1) 17 plasmids were removed since they did not contain both an ori and an f1 ori; (2) if no ITR was detected in proximity to each ori or f1 ori, the region 62–191 bp from the ori or 85–214 bp from the f1 ori was classified as an ITR, which is based on the mode ofITR location in other plasmids. Thus, 41 ITR annotations have been added; (3) since SnapGene had only annotated WPREs of size 589 bp, with the exception of one WPRE of length 588 bp (whereas the WPRE in VTK is only 432 bp), the EMBOSS needle tool was used to annotate additional WPREs. The tool identified 18 regions that were not yet annotated as WPRE, and after manual inspection, we added annotations for 11 of these; (4) we carefully examined each type of annotation and reclassified elements when needed. For example, IRES was classified as a miscellaneous feature instead of a Coding Sequence (CDS), while FRT and Lox features were reclassified as inducible sites; (5) the five largest unannotated regions per plasmid after removing CDS elements were inspected and classified manually; (6) if a Kozak sequence together with a T2A, E2A, F2A or P2A element was in between two CDS elements, the entire region between the two CDS elements was also labeled as CDS. The code that was used to perform these steps has been added to the manuscript (Data S1).
Elements listed below were reported in Figure 1:
Backbone
-
-
ITR-delimited payload from which all identified “Transgenes” were subtracted
Promoters/enhancers/introns
-
-
As labeled by features in SnapGene 1.1.58
-
-
Largest unlabeled space manually annotated as promoters or introns
Inducible sites
-
-
As labeled by SnapGene 1.1.58
Transgenes
-
-
As labeled by SnapGene 1.1.58
-
-
Largest unlabeled space manually annotated
WPRE/PolyA
-
-
As labeled by SnapGene 1.1.58
-
-
Added CW3SL using EMBOSS needle
Stereotactic and systemic AAV injections and Rabies injection
Mouse local
Stereotactic injections were performed at postnatal time P0-1 (VTKS2, VTKS3, VTKS5, VTKD2-sgRNA, VTKS1-GCaMP6f) or P10 (VTKS1, VTKS3-APEX2-NLS). P0-1 stereotaxic injections were achieved using a neonate adapter (Harvard apparatus). Mouse pups were anesthetized by hypothermia and stereotactically micro-injected using NanojectIII at 1nl/second with the rAAVs either in somatosensory cortex at P0-1 (from lambda AP+1.4, ML-1.8, DV-0.2) or in the thalamus at P10 (from lambda AP+1.8, ML-1.3, DV-3.1).
Local injections in two-month-old adult mice (VTKD4, VTKD6, VTKD2-ChR2-mCherry, VTKD2-APEX-V5) were performed by stereotactically-guided injections into the S1 cortex (from bregma AP+1, ML ± 2.9, DV-0.45/-0.7) with 75 nL of virus and the hippocampal area for VTKD6 (from bregma AP+1, ML ± 2.9, DV-1.5). VTKD2-ChR2-mCherry was injected in the dorsal hippocampus (from bregma AP+2; ML ± 1.5; DV -1.1) with 350-600 nL of virus at 100 nL/min. Mice recovered for 2 weeks before being used for recording or IHC.
Rabies retrograde tracing
After VTKS5-helpers injection, EnvA-N2dGcRV-H2B-tdTomato (rabies) was injected after SST-FlpO deletion occurred, 5 days after AAV infection in the somatosensory cortex (from lambda AP+2, ML-2, DV-025) (Fischer et al., 2019). After VTKS2-3xsgRNA-helpers injection, EnvA-N2cRV was injected at P30 (from bregma AP-1, ML-3, DV-0.87) and animals were perfused 7 days later.
VTK constructs cloning
VTKS/D constructs containing the promoter and inducible sites were synthesized (see Table S2) and cloned using ApaI/AscI into a backbone containing ITRs, WPRE and bGHp polyA (from Addgene #52473, a generous gift from Ian Wickersham) (Kohara et al., 2014). For VTKS1-hChR2-tBFP, we cloned the hChR2 coding sequence from the Addgene plasmid #26976 (a generous gift from Karl Deisseroth) and tBFP from the Addgene plasmid #80086 (a generous gift from Connie Cepko); for VTKS2-hM3Dq-P2A-tBFP, we amplified the hM3Dq-P2A coding sequence from Addgene plasmid #83897 (Chen et al., 2020) and tBFP from the Addgene plasmid #80086; for VTKS3-TeTLc-P2A-mCherry, the coding sequence of TeTLc-P2A-mCherry was a generous gift from Rosa Cossart; for VTKD4-V5-APEX2-NES and VTKD6-V5-APEX2-NES, we amplified the V5-APEX2-NES coding sequence from the plasmid Addgene #72480 (a generous gift from Alice Ting); for VTKS5-N2cG-P2A-eGFP-P2A-TVA, we amplified the eGFP-P2A-TVA coding sequence from Addgene plasmid #52473 (a generous gift from Tom Jessell) and we cloned N2cG from pAM-CAGGS-FLEX-H2B-GFP-P2A-N2c(G) (Pouchelon et al., 2021).
Recombination sensitive cloning and viruses production
AAV production
The enhancers, reporters, and effectors were cloned using the Gibson Cloning Assembly Kit (New England BioLabs, catalog no. NEB-E5510S). After cloning and sequencing, the growth time of the transformed DH10B Competent Cells was kept below 12 hours on plates and 10 hours in flask at 37°C. DNA from several clones was recovered with an endotoxin free midi-kit (Zymo D4202). This allows for consistently obtaining low recombination rates detected by PCR. Primers were designed for amplification of plasmids at the junction of each possible recombined form and compared to recombination-free controls. Only clones with a recombined/not-recombined ratio of 1.0E+05 or greater were considered for further AAV production. The rAAVs were produced using standard production methods. Polyethylenimine was used for transfection and OptiPrep gradient (Sigma) was used for viral particle purification. Serotype 1 was used to produce the AAVs for local injections in mice. Serotype PHPeB was used for systemic and local injections in mice. Titer was estimated by quantitative PCR with primers for the WPRE sequence that is common to all constructs. All batches produced were in the range of 5.0E+11 to 5.0E+13 viral genomes per milliliter (see Table S3). VTKS1-hChR2-BFP (local injection in thalamus, injection to perfusion: P10–P30); VTKS2-hM3Dq-P2A-BFP (injection in primary somatosensory cortex, injection to perfusion: P0–P30 (cell type specificity) or P0–P5 (function)); VTKS3-TeTLc-P2A-mCherry (injection in primary somatosensory cortex, injection to perfusion: P1–P15); VTKD4-V5-APEX2-NES (local injection in primary somatosensory cortex, injection to perfusion: P60); VTKS5-N2cG-P2A-eGFP-P2A-TVA (local injection in primary somatosensory cortex, injection to perfusion: P0–P30); VTKD6-V5-APEX2-NES (injection in primary somatosensory cortex and hippocampus, injection to perfusion: P60); VTKS3-NLS-APEX2-V5 (injection in primary somatosensory cortex, injection to perfusion: P10-P30); VTKD2-ChR2-mCherry (injection in dorsal hippocampus, injection to perfusion: adult, 2 weeks); VTKD2-x3 U6sgRNApA-NLStdTomato (injection in basal forebrain; injection to perfusion: P0/P1-P7); VTKD2-APEX-V5 (injection in primary somatosensory cortex; injection to perfusion: P60-P70), VTKS1-GCaMP6f (injection in primary visual cortex, injection to perfusion: P1 - P7); CamKII-ndTom (rAAV1, titer = 6.5E+12 vg/mL, injection in primary somatosensory cortex, injection to perfusion: P70–P77). AAV-mDlx-GFP (rAAV1, titer = 1.6E+12 vg/mL, injection in primary somatosensory cortex, injection to perfusion: P70–P77).
Rabies production
EnvA-pseudotyped CVS-N2c(DeltaG)-H2B-tdTomato (titer = 1.4E+07 IU/mL) was generously produced by Kimberly Ritola.
Histology
Tissue processing
Mice at P28–P32 and P60 for the adult time point, at P5 and P15 for the developmental time point (N = 3 to 6 for each condition) were transcardially perfused with 4% paraformaldehyde (PFA) and brains were fixed overnight or 2hrs in 4% PFA at 4°C. Brains were sectioned at 40 μm or 50 μm using a Leica VTS1000 vibrosector. Floating sections containing AAV-infected cells or their projections were processed for immunohistochemistry. Brain sections were incubated 1–2hrs at room temperature in a blocking solution containing 3% Normal Donkey serum and 0.3% Triton X-100 in PBS, then incubated overnight or 48hrs at 4°C with primary antibodies: rabbit anti-somatostatin (1:3,000; Peninsula Laboratories International, catalog no. T-4103.0050), guinea pig anti-cFos (1:300, Synaptic Systems, catalog no. #226004), anti-tagFP (1:500; Synaptic Systems, catalog no. FluoTag-X2 N0502, At542 or At647), rat anti-HA (1:1,000; Sigma, catalog no. #11867423001), chicken anti-GFP (1:1,000; Aves Labs, catalog no. #1020), rat anti-RFP (1:1,000; Chromotek, catalog no. #5f8), mouse anti-V5 (1:500; ThermoFisher, catalog no. #R960-25), guinea-pig anti-PV (1:2,000; Swant, catalog no. GP-72), rabbit anti-SST (1:500, Millipore, catalog no. MAB354), rabbit anti-Nkx2.1 (1:2000, Abcam, catalog no. ab76013). The rabbit anti-VIP antibody was a gift from the Salk Institute; it was generated by Joan Vaughan at the Saghatelian lab, and validated by Paul Sawchenko and Josselyn Vergara. Sections were rinsed four times in PBS and incubated for 60–90 min at room temperature or overnight at 4 °C with the Alexa Fluor 405-, 488-, 546-, 594-, or 647-conjugated secondary antibodies (1:1000; Thermo Fisher Science or Jackson ImmunoResearch).
Imaging
Each brain section containing labeled cells was acquired as a tiled image on a motorized Zeiss Axio Imager A1 or a Zeiss confocal LSM800 using the 10x objective. For quantification of co-localization, cells expressing the indicated reporter were counted using Fiji (ImageJ) using only the corresponding color channel and then, among these cells, the number of cells co-expressing the marker of interest was manually counted or using automated quantification (https://github.com/jessdlin/cellcounter). A cell was considered to be positive for a given marker if the corresponding signal was above background fluorescence. The ratio of cells co-expressing both markers over the total number of cells expressing only the reporter was then calculated and reported as mean ± s.e.m. Quantifications were performed using a minimum of two independent biological replicates. Several sections from the same animal were used when indicated. Data collection and analysis were not performed blind to the conditions of the experiments, but experimenters from different research groups performed the quantification.
Slice electrophysiology
Mice were anesthetized with isoflurane and then decapitated. The brain was dissected out in ice-cold sucrose substituted artificial cerebrospinal fluid (SSaCSF) containing the following (in mM): 90 sucrose, 80 NaCl, 3.5 KCl, 24 NaHCO3, 1.25 NaH2PO4, 4.5 MgCl, 0.5 CaCl2, and 10 glucose, saturated with 95% O2 and 5% CO2. Transverse hippocampal slices (300-350 μm, in coronal or horizontal sections respectively) were cut using a VT-1200S vibratome (Leica Microsystems) and incubated submerged in the above solution at 32-34°C for 30–40 min and then maintained at room temperature until use in the SSaCSF. For patch-clamp recordings following recovery slices were transferred to an upright microscope (Zeiss Axioskop), perfused with aCSF consisting of the following (in mM): 130 NaCl, 3.5 KCl, 24 NaHCO3, 1.25 NaH2PO4, 1.5 MgCl2, 2.5 CaCl2, and 10 glucose, saturated with 95% O2 and 5% CO2 at 2-3 mL/min at a temperature of 32°C-34°C. For pharmacological dissection of GABA transmission aCSF was supplemented as indicated with (in μM): 50 picrotoxin, 10 bicuculline, 2 CGP 55845 all from Tocris (UK). Individual cells were visualized using a 40x objective using fluorescence and IR-DIC video microscopy. Electrodes were pulled from borosilicate glass (World Precision Instruments) to a resistance of 3–5 mΩ using a vertical pipette puller (Narishige, PP-830). Whole-cell patch-clamp recordings were made using a Multiclamp 700A amplifier (Molecular Devices), and signals were digitized at 20 kHz (Digidata 1322A, filtered at 3 kHz) for collection on a PC computer equipped with pClamp 9.2 or 10.4 software (Molecular Devices). Intracellular recording solution was composed of (in mM): 130 K-gluconate, 5 KCl, 10 HEPES, 3 MgCl2, 2 Na2ATP, 0.3 NaGTP, 0.6 EGTA, and 0.2% biocytin (ECl- ∼ −65 mV). Uncompensated series resistance ranged from 10 to 20 mΩ and was monitored continuously throughout recordings with −5 mV voltage steps. For current-clamp recordings of RFP+ cells membrane potential was biased between −55 and −60mV. Optogenetic events were evoked with brief blue light (470 nm, 2.5−5 ms) stimulation from an LED source (CoolLED PE4000, Andover, UK) through the 40x water-immersion objective. After biocytin filling during whole-cell recordings, slices were fixed with 4% paraformaldehyde and stored at 4°C then permeabilized with 0.3% Triton X-100 and incubated with Alexa Fluor 488-conjugated streptavidin. In addition, fixed sections were processed for mCherry staining with rabbit anti-RFP at 1:1,000 (Antibodies Online, catalog no. ABIN129578) primary and Alexa-555-conjugated secondary at 1:500 (Invitrogen). Resectioned slices were mounted on gelatin-coated slides using Mowiol mounting medium.
Electron microscopy
Brains were prepared in the same manner and as previously described (Hua et al., 2015). Briefly, an anesthetized animal was first transcardially perfused with 10mL 0.1 M Sodium Cacodylate (cacodylate) buffer, pH 7.4 (Electron microscopy sciences (EMS)) followed by 20 mL of fixative containing 2% paraformaldehyde (EMS), 2.5% glutaraldehyde (EMS) in 0.1 M Sodium Cacodylate (cacodylate) buffer, pH 7.4 (EMS). The brain was removed and placed in fixative for at least 24 hours at 4C. The brain was then mounted to the vibratome (Leica VT1200) on the anterior side, and carefully sectioned down through the cerebellum until it just touched the posterior cortex. The brain was then completely sectioned into a series of 300 μm vibratome sections, placed in order into a 24 well dish, and incubated into fixative for another 24 hours at 4°C. Apex2 precipitation and polymerization was performed by washing slices extensively in the cacodylate buffer at room temperature, followed by incubation in 50 mg/mL 3,3′diaminobenzidine (DAB) in the cacodylate buffer for 1 hour at room temperature. 0.003% (v/v) H202 until a visible precipitate forms (15-20 minutes). Slices were washed extensively in cacodylate buffer then further reduced in 0.8%(w/v) Sodium Hydrosulfite in 60%(v/v) 0.1 M Sodium Bicarbonate 40%(v/v) 0.1 M Sodium Carbonate for 20 minutes, and washed in cacodylate buffer, 3 exchanges, 10 minutes each. A small region (∼1 × 1 mm) around the DAB precipitate was excised from the vibratome sections and prepared for EM by staining sequentially with 2% osmium tetroxide (EMS) in cacodylate buffer, 2.5% potassium ferrocyanide (Sigma-Aldrich), thiocarbohydrazide, unbuffered 2% osmium tetroxide, 1% uranyl acetate, and 0.66% Aspartic acid buffered Lead (II) Nitrate with extensive rinses between each step with the exception of potassium ferrocyanide. The sections were then dehydrated in ethanol and propylene oxide and infiltrated with 812 Epon resin (EMS, Mixture: 49% Embed 812, 28% DDSA, 21% NMA, and 2.0% DMP 30). The resin-infiltrated tissue was cured at 60C for 3 days. Using a commercial ultramicrotome (Powertome, RMC), the cured block was trimmed to a ∼1.0mm × .8 mm rectangle and 40nm thick sections were collected from the tissue block on polyamide tape (Kapton) using an automated tape collecting device (ATUM, RMC) and assembled on silicon wafers as previously described (Kasthuri et al., 2015). The sections were imaged using backscattered electron detection with a Gemini 300 scanning electron microscope (Carl Zeiss), equipped with ATLAS software for wafer imaging.
Wide-field calcium imaging
Mice were given analgesic (Meloxicam) one hour before surgery and then anesthetized with isoflurane (3% briefly for induction and surgery, then 0.75%-1% for headbar preparation) before preparing for imaging by removing the skin above primary visual cortex and affixing a clear glue-cap which was attached to head bars at the anterior and posterior of the imaging region. Skull thickness is very thin at early postnatal ages (P7, in this study) and calcium sensor signal is bright enough at this age to image directly through a hardened glue cap and the skull into V1. After recovery from anesthesia for at least 30 minutes spontaneous activity was recorded in 0.2s increments for five minute increments using LED stimulation, a 4x objective, and a camera. After two or three movies the animal was again anesthetized and sacrificed. Images were generated with Fiji (https://imagej.nih.gov/ij/) and movies analyzed with MATLAB as previously described (Ackman et al., 2012).
Quantification and statistical analysis
For each experiment, animals with fewer than 10 infected cells were removed from the study.
For cell specificity quantification, Fiji software (https://imagej.nih.gov/ij/) was used both for manual or automated quantifications. Data collection and analysis were not performed blind to the conditions of the experiments, but experimenters from different research groups performed the quantification. Two to five independent biological replicates (N), animals were used for the analysis. Several sections from the same animal were averaged when indicated. The number of animals (N) was determined based on previously published work (Pouchelon et al., 2021; Vormstein-Schneider et al., 2020). Data are represented as mean ± s.e.m.
Electrophysiological data were acquired on pClamp 9.2 or 10.4 software (Molecular Devices) on one animal from 4 pyramidal cells in different slices. All of them showed the response illustrated in the figure. Electron microscopy was replicated on four animals, with staining detected in all. Two animals were imaged and one illustrated in the figure. Calcium imaging experiment was replicated on 10 animals.
Acknowledgments
We thank S. Gerard, P. Delvenne, C. Parotte, D. Joseph, O. Lis, P. Copine, Ma. Matou, P. Noir, Mi. Mi, and Co. Co. for their useful comments on the manuscript. We also thank Kimberly Ritola for sharing EnvA-N2c-dG rabies and her advice on recombination-free AAV production and Bernardo Rudy, Robert Machold, and Benjamin Schuman for discussing their VTKS5 experiment using NPY-FlpO animals. J.D. is supported by NIH grants R01-MH111529 and UG3MH120096 in addition to support from Simons Foundation Award 566615. G.P. was supported by an EMBO Long-Term fellowship, early and advanced Swiss Foundation postdoctoral fellowships, and a Hearst foundation grant, and G.F. was supported by grants from the NIH (MH071679, NS08297, NS074972, and MH095147) as well as support from the Simons Foundation (SFARI).
Author contributions
G.P. and J.D. conceived the project. G.P., J.D., and G.F. wrote the manuscript. G.P., J.V., and J.M. performed injections and the neuron-specificity experiments. G.P. performed the tracing and c-Fos experiments. J.D.L. and D.V.-S. performed the interference experiments. T.J.B. performed calcium imaging. G.A.W. performed electron microscopy. K.C.A., M.F.-O., and M.S. performed the CRISPR-based experiment. K.P., R.C., J.C.W., and C.J.M. injected mice and performed optogenetic recordings. G.P., J.V., J.M., and B.L.G. analyzed the Addgene data and prepared the figures. M.F., J.L.N., and J.S.N. provided the sequence data for all Addgene plasmids used in this study.
Declaration of interests
J.D. is founder, shareholder, and employee of Regel Therapeutics. G.F. is founder and shareholder of Regel Therapeutics. The work presented in this manuscript is unrelated to any aspect of Regel Therapeutics’ focus, work, or interest.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Published: May 26, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100225.
Contributor Information
Gabrielle Pouchelon, Email: pouchel@cshl.edu.
Jordane Dimidschstein, Email: jordane.dimidschstein@gmail.com.
Supplemental information
Presence of inducible sites is indicated by a check mark. The recombinase-dependency is specified by the corresponding VTK element.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
All original code is available in this paper’s supplemental information.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ackman J.B., Burbridge T.J., Crair M.C. Retinal waves coordinate patterned activity throughout the developing visual system. Nature. 2012;490:219–225. doi: 10.1038/nature11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander G.M., Rogan S.C., Abbas A.I., Armbruster B.N., Pei Y., Allen J.A., Nonneman R.J., Hartmann J., Moy S.S., Nicolelis M.A., et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster B.N., Li X., Pausch M.H., Herlitze S., Roth B.L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. U S A. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D., Aponte Y., Su H.H., Sternson S.M. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 2008;28:7025–7030. doi: 10.1523/jneurosci.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook C.N., Deverman B.E., Gradinaru V. Viral strategies for targeting the central and peripheral nervous systems. Annu. Rev. Neurosci. 2018;41:323–348. doi: 10.1146/annurev-neuro-080317-062048. [DOI] [PubMed] [Google Scholar]
- Blankvoort S., Witter M.P., Noonan J., Cotney J., Kentros C. Marked diversity of unique cortical enhancers enables neuron-specific tools by enhancer-driven gene expression. Curr. Biol. 2018;28:2103–2114.e5. doi: 10.1016/j.cub.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E.S., Zhang F., Bamberg E., Nagel G., Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Burbridge T.J., Xu H.P., Xu H., Ackman J.B., Ge X., Zhang Y., Ye M.-J., Zhou Z.J., Xu J., Contractor A., Crair M.C. Visual circuit development requires patterned activity mediated by retinal acetylcholine receptors. Neuron. 2014;84:1049–1064. doi: 10.1016/j.neuron.2014.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carinhas N., Bernal V., Yokomizo A.Y., Carrondo M.J.T., Oliveira R., Alves P.M. Baculovirus production for gene therapy: the role of cell density, multiplicity of infection and medium exchange. Appl. Microbiol. Biotechnol. 2009;81:1041–1049. doi: 10.1007/s00253-008-1727-4. [DOI] [PubMed] [Google Scholar]
- Challis R.C., Ravindra Kumar S., Chan K.Y., Challis C., Beadle K., Jang M.J., Kim H.M., Rajendran P.S., Tompkins J.D., Shivkumar K., et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 2019;14:379–414. doi: 10.1038/s41596-018-0097-3. [DOI] [PubMed] [Google Scholar]
- Chen Q., Deister C.A., Gao X., Guo B., Lynn-Jones T., Chen N., Wells M.F., Liu R., Goard M.J., Dimidschstein J., et al. Dysfunction of cortical GABAergic neurons leads to sensory hyper-reactivity in a Shank3 mouse model of ASD. Nat. Neurosci. 2020;23:520–532. doi: 10.1038/s41593-020-0598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.-W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman B.E., Ravina B.M., Bankiewicz K.S., Paul S.M., Sah D.W.Y. Gene therapy for neurological disorders: progress and prospects. Nat. Rev. Drug Discov. 2018;17:641–659. doi: 10.1038/nrd.2018.110. [DOI] [PubMed] [Google Scholar]
- Dimidschstein J., Chen Q., Tremblay R., Rogers S.L., Saldi G.-A., Guo L., Xu Q., Liu R., Lu C., Chu J., et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donello J.E., Loeb J.E., Hope T.J. Woodchuck Hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L.E., Mattis J., Ramakrishnan C., Hyun M., Lee S.Y., He M., Tucciarone J., Selimbeyoglu A., Berndt A., Grosenick L., et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Meth. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L.E., Ramakrishnan C., Kim Y.S., Evans K.E., Lo M., Vesuna S., Inoue M., Cheung K.Y.M., Yuen E., Pichamoorthy N., et al. Comprehensive dual- and triple-feature intersectional single-vector delivery of diverse functional payloads to cells of behaving mammals. Neuron. 2020;107:836–853.e11. doi: 10.1016/j.neuron.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K.B., Collins H.K., Callaway E.M. Sources of off-target expression from recombinase-dependent AAV vectors and mitigation with cross-over insensitive ATG-out vectors. Proc. Natl. Acad. Sci. 2019;116:27001–27010. doi: 10.1073/pnas.1915974116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G., Kepecs A. Interneuron types as attractors and controllers. Annu. Rev. Neurosci. 2020;43:1–30. doi: 10.1146/annurev-neuro-070918-050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybuck L.T., Daigle T.L., Sedeño-Cortés A.E., Walker M., Kalmbach B., Lenz G.H., Morin E., Nguyen T.N., Garren E., Bendrick J.L., et al. Enhancer viruses for combinatorial cell-subclass-specific labeling. Neuron. 2021;109:1449–1464.e13. doi: 10.1016/j.neuron.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haery L., Deverman B.E., Matho K.S., Cetin A., Woodard K., Cepko C., Guerin K.I., Rego M.A., Ersing I., Bachle S.M., et al. Adeno-associated virus technologies and methods for targeted neuronal manipulation. Front. Neuroanat. 2019;13:93. doi: 10.3389/fnana.2019.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Tucciarone J., Lee S., Nigro M.J., Kim Y., Levine J.M., Kelly S.M., Krugikov I., Wu P., Chen Y., et al. Strategies and tools for combinatorial targeting of GABAergic neurons in mouse cerebral cortex. Neuron. 2016;91:1228–1243. doi: 10.1016/j.neuron.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch M.L., Wolf S.J., Samulski R.J. Delivering transgenic DNA exceeding the carrying capacity of AAV vectors. Methods Mol. Biol. 2016;1382:21–39. doi: 10.1007/978-1-4939-3271-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S., Tzeng C.P., Nagy M.A., Stroud H., Koutsioumpa C., Wilcox O.F., Assad E.G., Green J., Harvey C.D., Griffith E.C., et al. A scalable platform for the development of cell-type-specific viral drivers. Elife. 2019;8:e48089. doi: 10.7554/elife.48089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Laserstein P., Helmstaedter M. Large-volume en-bloc staining for electron microscopy-based connectomics. Nat. Commun. 2015;6:7923. doi: 10.1038/ncomms8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüttner J., Szabo A., Gross-Scherf B., Morikawa R.K., Rompani S.B., Hantz P., Szikra T., Esposti F., Cowan C.S., Bharioke A., et al. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci. 2019;22:1345–1356. doi: 10.1038/s41593-019-0431-2. [DOI] [PubMed] [Google Scholar]
- Kasthuri N., Hayworth K.J., Berger D.R., Schalek R.L., Conchello J.A., Knowles-Barley S., Lee D., Vázquez-Reina A., Kaynig V., Jones T.R., et al. Saturated Reconstruction of a Volume of Neocortex. Cell. 2015;162:648–661. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- Kohara K., Pignatelli M., Rivest A.J., Jung H.-Y., Kitamura T., Suh J., Frank D., Kajikawa K., Mise N., Obata Y., et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat. Neurosci. 2014;17:269–279. doi: 10.1038/nn.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kügler S., Kilic E., Bähr M. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 2003;10:337–347. doi: 10.1038/sj.gt.3301905. [DOI] [PubMed] [Google Scholar]
- Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., Ting A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.T., Su-Feher L., Cichewicz K., Warren T.L., Zdilar I., Wang Y., Lim K.J., Haigh J.L., Morse S.J., Canales C.P., et al. Parallel functional testing identifies enhancers active in early postnatal mouse brain. Elife. 2021;10:e69479. doi: 10.7554/elife.69479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y. mi, Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Garner A.R., Shimaoka D., Chuong A.S., Klapoetke N.C., Li L., van der Bourg A., Bourg A. van der, Niino Y., Egolf L., et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magno L., Kretz O., Bert B., Ersözlü S., Vogt J., Fink H., Kimura S., Vogt A., Monyer H., Nitsch R., et al. The integrity of cholinergic basal forebrain neurons depends on expression of Nkx2 .1. Eur. J. Neurosci. 2011;34:1767–1782. doi: 10.1111/j.1460-9568.2011.07890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T., Ashery-Padan R., Andrejewski N., Scardigli R., Guillemot F., Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Mich J.K., Graybuck L.T., Hess E.E., Mahoney J.T., Kojima Y., Ding Y., Somasundaram S., Miller J.A., Kalmbach B.E., Radaelli C., et al. Functional enhancer elements drive subclass-selective expression from mouse to primate neocortex. Cell Rep. 2021;34:108754. doi: 10.1016/j.celrep.2021.108754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasimuzzaman M., van der Loo J.C., Malik P. Production and purification of Baculovirus for gene therapy application. J. Vis. Exp. 2018:57019. doi: 10.3791/57019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis B., Haenzi B., Hilton S., Carnicer-Lombarte A., Hobo B., Verhaagen J., Fawcett J.W. Optimization of adeno-associated viral vector-mediated transduction of the corticospinal tract: comparison of four promoters. Gene Ther. 2021;28:56–74. doi: 10.1038/s41434-020-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr D.S., Rieser L.A., Woychik R.P., Rottman F.M., Rosenberg M., Reff M.E. Differential effects of polyadenylation regions on gene expression in mammalian cells. Dna. 1986;5:115–122. doi: 10.1089/dna.1986.5.115. [DOI] [PubMed] [Google Scholar]
- Pouchelon G., Dwivedi D., Bollmann Y., Agba C.K., Xu Q., Mirow A.M.C., Kim S., Qiu Y., Sevier E., Ritola K.D., et al. The organization and development of cortical interneuron presynaptic circuits are area specific. Cell Rep. 2021;37:109993. doi: 10.1016/j.celrep.2021.109993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S.K., Rivera-Soto R., Gray S.J. Viral expression cassette elements to enhance transgene target specificity and expression in gene therapy. Discov. Med. 2015;19:49–57. [PMC free article] [PubMed] [Google Scholar]
- Royo N.C., Vandenberghe L.H., Ma J.-Y., Hauspurg A., Yu L., Maronski M., Johnston J., Dichter M.A., Wilson J.M., Watson D.J. Specific AAV serotypes stably transduce primary hippocampal and cortical cultures with high efficiency and low toxicity. Brain Res. 2008;1190:15–22. doi: 10.1016/j.brainres.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Meth. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., He M., Wu P., Kim S., Paik R., Sugino K., Kvitsiani D., Kvitsani D., Fu Y., Lin Y., et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B., Yao Z., Graybuck L.T., Smith K.A., Nguyen T.-N., Bertagnolli D., Goldy J., Garren E., Economo M.N., Viswanathan S., et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563:72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vormstein-Schneider D., Lin J.D., Pelkey K.A., Chittajallu R., Guo B., Arias-Garcia M.A., Allaway K., Sakopoulos S., Schneider G., Stevenson O., et al. Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat. Neurosci. 2020;23:1629–1636. doi: 10.1038/s41593-020-0692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Presence of inducible sites is indicated by a check mark. The recombinase-dependency is specified by the corresponding VTK element.
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
All original code is available in this paper’s supplemental information.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.