Abstract
Resin acids are tricyclic terpenoids occurring naturally in trees. We investigated the occurrence of resin acid-degrading bacteria on the Arctic tundra near the northern coast of Ellesmere Island (82°N, 62°W). According to most-probable-number assays, resin acid degraders were abundant (103 to 104 propagules/g of soil) in hydrocarbon-contaminated soils, but they were undetectable (<3 propagules/g of soil) in pristine soils from the nearby tundra. Plate counts indicated that the contaminated and the pristine soils had similar populations of heterotrophs (106 to 107 propagules/g of soil). Eleven resin acid-degrading bacteria belonging to four phylogenetically distinct groups were enriched and isolated from the contaminated soils, and representative isolates of each group were further characterized. Strains DhA-91, IpA-92, and IpA-93 are members of the genus Pseudomonas. Strain DhA-95 is a member of the genus Sphingomonas. All four strains are psychrotolerant, with growth temperature ranges of 4°C to 30°C (DhA-91 and DhA-95) or 4°C to 22°C (IpA-92 and IpA-93) and with optimum temperatures of 15 to 22°C. Strains DhA-91 and DhA-95 grew on the abietanes, dehydroabietic and abietic acids, but not on the pimaranes, isopimaric and pimaric acids. Strains IpA-92 and IpA-93 grew on the pimaranes but not the abietanes. All four strains grew on either aliphatic or aromatic hydrocarbons, which is unusual for described resin acid degraders. Eleven mesophilic resin acid degraders did not use hydrocarbons, with the exception of two Mycobacterium sp. strains that used aliphatic hydrocarbons. We conclude that hydrocarbon contamination in Arctic tundra soil indirectly selected for resin acid degraders, selecting for hydrocarbon degraders that coincidentally use resin acids. Psychrotolerant resin acid degraders are likely important in the global carbon cycle and may have applications in biotreatment of pulp and paper mill effluents.
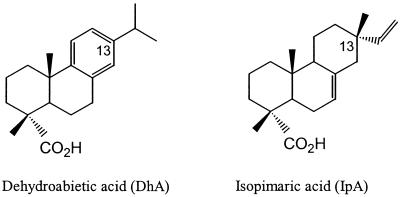
Resin acids are a group of tricyclic diterpenoids synthesized by trees, particularly softwood trees, in which resin acids can constitute up to a few percent of the tree biomass. Resin acids, including abietanes and pimaranes (Fig. 1), appear to protect trees from microbial pathogens and pests (6). Resin acids occur in pulp mill effluents and cause much of the toxicity of these effluents to fish (10, 11, 25, 28, 29). In addition, resin acids are components of pitch, which interferes in papermaking. Consequently, resin acids are considered priority pollutants of the pulp and paper industry.
FIG. 1.
Chemical structures of DhA and IpA. Abietanes like DhA all have the isopropyl side chain at C-13, while pimaranes like IpA have methyl and vinyl substituents at C-13.
Resin acids are biodegradable, and most resin acid-degrading microorganisms so far characterized are mesophilic bacteria which can both mineralize resin acids and use them as sole organic substrates for growth (reviewed in references 13 and 17). Thermophilic resin acid-degrading bacteria also exist (32). Resin acids present in pulp mill effluents usually are biodegraded in aerobic biological treatment systems. In natural environments, most resin acids are presumably biodegraded in forest soils. We hypothesize that resin acid-degrading bacteria are distributed at least as widely as resin acid-producing trees and that these bacteria may be ubiquitous.
We also hypothesize that there exist psychrophilic or psychrotolerant resin acid-degrading bacteria, since resin acids do not appear to accumulate in forest soils of cold regions. Much of the range of softwood trees includes temperate regions where soil temperatures rarely exceed 15°C. In addition, even though pulp mill effluents are generally at elevated temperatures, some resin acids from pulp mills do reach environments where their degradation is governed by the ambient conditions, including low temperature. Psychrotolerant resin acid degraders may be useful for bioaugmentation to enhance resin acid removal in biological treatment systems that suffer from poor resin acid removal during winter (12). Therefore, the isolation and characterization of resin acid degraders and the investigation of resin acid biodegradation at low temperatures are of scientific interest and practical importance.
“Everything is everywhere” (5), “but the environment selects” (3), succinctly states a widely held tenet with much empirical support. However, little is known about the factors and mechanisms which influence the distribution and abundance of catabolic capabilities. We tested the above tenet by looking for psychrotolerant resin acid-degrading bacteria in the extreme northern Arctic, thousands of kilometers from the nearest resin acid-producing trees. We did find such bacteria, and four distinct strains are described here for the first time. A preliminary report of one of the strains, DhA-95, was included in a review article (22). This is the first direct evidence for the mineralization of resin acids at low temperatures. We looked for psychrotolerant resin acid degraders in both pristine and hydrocarbon-polluted Arctic soils, reasoning that the hydrocarbon pollution may have enriched heterotrophic bacteria. Resin acid degraders were associated almost exclusively with the polluted soils, but the pollution did not increase heterotrophic populations. Rather, hydrocarbons appear to have selected bacteria that degrade hydrocarbons and coincidentally also degrade resin acids.
MATERIALS AND METHODS
Soil samples.
Arctic tundra soil samples were collected at or near Canadian Forces Station Alert. Alert is at the northern end of Ellesmere Island (83°N, 62°W), which is in the Cold-Arctic vegetation zone (14). At the latitude of the sampling sites, there is constant daylight from 8 April to 5 September and constant darkness from 10 October to 1 March. During summer, there are typically 28 frost-free days, during which the average daily high temperature is 6°C. During winter, the temperature drops to −50°C. The region of Alert is a desert with mean annual precipitation of 155 mm.
In June 1998, three Arctic hydrocarbon-contaminated soil samples (Alert-1, Alert-2, and Alert-3) were collected at Alert, and three pristine soil samples (Alert-4, Alert-5, and Alert-6) were collected from the Arctic tundra near Alert. The three pristine soil samples were collected from sites 5 to 10 km away from Alert which showed no evidence of human disturbance. Samples were dug from the top 10 cm with sterile scoops and placed in sterile bottles. These soil samples were kept on ice during transport by air to our laboratory and were stored at 7°C until use. Prior to subsampling, each sample was thoroughly mixed by shaking the sample bottle by hand. The three contaminated soil samples had a strong smell of hydrocarbons and had measured total petroleum hydrocarbons (TPH) ranging from 0.98 to 33.4 mg/g of soil (see Fig. 4). The history of the contaminated sites is unknown. The soils were contaminated with arctic diesel fuel, which contains about 10 to 20% aromatic compounds, primarily naphthalenes. The three pristine samples did not smell of hydrocarbons and did not have detectable TPH (<10 μg/g of soil). Resin acids were not detectable in any of the six soil samples (<0.1 μg/g of soil).
FIG. 4.
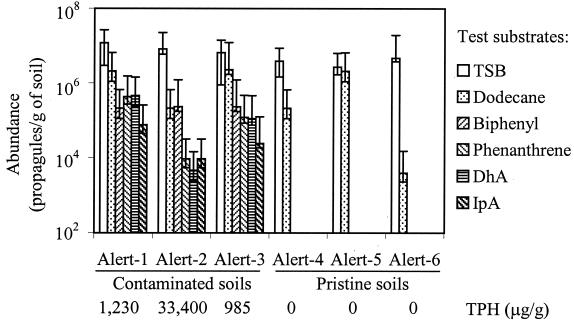
Hydrocarbon (TPH) concentrations and abundance of total heterotrophs, hydrocarbon degraders, and resin acid degraders in the Arctic soils. Error bars indicate two-sided 95% confidence limits.
MPN and plate count assays.
Microorganisms were extracted from soil in precooled 0.1% pyrophosphate solution (1.0 g of soil in 9.5 ml of solution) by shaking by hand for 10 min. Then, serial 10-fold dilutions of the extract were made in precooled PAS medium (4). Total cultureable heterotrophs were counted on tryptic soy agar (TSA) plates (3.0 g/liter; 10% of the normal concentration), which were inoculated by spreading 0.1 ml of the above dilutions. The plates were incubated at 7°C for 4 weeks. Most-probable numbers (MPN) were determined using triplicates of each dilution in PAS medium. Hydrocarbon degraders were assayed in 1.0-ml cultures with 100 mg of the test substrate per liter. Hydrocarbons were added to sterile dry tubes from acetone stock solutions, and the acetone was allowed to volatilize from the tubes for 1 h at room temperature prior to adding the medium and inoculation. Resin acid degraders were assayed in 2.0-ml cultures with 60 mg of the test substrate per liter. All the MPN cultures were incubated at 7°C on a tube roller for 4 weeks, after which substrate concentrations were determined by gas chromatography (see below). A tube was scored positive if its substrate concentration was less than the average of the sterile controls by an amount at least twice the standard deviation of the three sterile controls. MPN were determined using an MPN table (24).
Enrichment and isolation.
Bacterial isolates were obtained by selective enrichment in BR mineral medium (18) containing dehydroabietic acid (DhA) plus isopimaric acid (IpA) and by subsequent isolation on BR plates with either DhA or IpA as the sole organic substrate as described previously (18). The enrichment cultures were initiated by placing 1.0 g of each soil sample into 100 ml of precooled BR medium containing DhA and IpA (400 μM each) of high purity (99%; Helix Biotechnologies, New Westminster, Canada). These and subsequent liquid cultures were incubated at 7°C on a shaker. A sterile control culture was included which was inoculated with an autoclaved soil sample. Subsamples were taken from each enrichment culture for resin acid analysis after 4 weeks incubation, and the enrichment cultures that showed biodegradation of resin acids were transferred (1.0% inoculum) into precooled BR medium containing either DhA or IpA (400 μM). After 26 days, biodegradation of DhA and IpA was confirmed in the secondary cultures and they were streaked on precooled BR plates containing purified agar and either DhA or IpA (200 μM). The plates were incubated at 7°C for 2 weeks, and individual colonies were transferred into BR medium containing DhA or IpA as the sole organic substrate to verify their ability to degrade and grow on each resin acid. Confirmed resin acid degraders were further purified by streaking on TSA plates (3.0 g/liter; 10% of the normal concentration) and were further verified by one passage on liquid BR medium containing either DhA or IpA (400 μM) as the sole organic substrate.
Physiological tests.
Growth temperatures were tested in BR medium containing either DhA or IpA (200 μM). The relative growth rates were scored by microscopic enumeration (insoluble substrates interfered with optical density measurement). Gram staining, catalase activity, oxidase activity, motility, colony morphology, and cells sizes were analyzed with cultures grown on TSA. Catalase activity was determined on colonies using H202; oxidase activity was determined using the Bacto differentiation disks for oxidase (Difco Laboratories, Detroit, Mich.).
Experiments investigating biodegradation of DhA or IpA and the corresponding cell growth were performed at 15°C in large numbers of replicate 2.0-ml cultures, because the poor solubility of resin acids prevented representative subsampling of cultures. These cultures were in BR medium with DhA or IpA (200 μM). At each sampling time, two replicate culture tubes (one for resin acid analysis, one for protein analysis) were frozen at −20°C for later analysis. Resin acids were extracted directly from the culture tubes as reported previously (18).
Substrate use by each isolate was tested at 15°C in BR medium containing the test compound as the sole organic substrate. Naphthalene, camphor, and citronellol were added to the cultures from acetone stock solutions, as described above. To test toxicity of any residual acetone, cultures were tested for growth on pyruvate plus acetone added as described above. Residual acetone did not inhibit any of the isolates. Sitosterol was from a hexane stock solution, added in the manner of the acetone solutions. Linoleic acid, palmitic acid, n-hexane, benzene, toluene, and Jet A-1 fuel (Shell Chemicals, Ltd.) were added to cultures directly from neat stocks. The Jet A-1 fuel contained less than 20% aromatics, primarily naphthalenes. Betulin was added as a dry powder. Other substrates were added from sterile aqueous stock solutions. Growth was confirmed by microscopy and a second passage on the same medium. The ability to denitrify was tested in tryptic soy broth (3.0 g/liter) with 1.0 g of pyruvate per liter and 1.0 g of nitrate per liter in 8.5-ml tubes with 4.5 ml of medium and 4.0 ml of headspace equipped with inverted Durham tubes to collect N2. These anaerobic cultures were incubated at 7°C for 8 weeks to monitor growth and concomitant N2 production.
Screening hydrocarbon use by previously isolated, mesophilic resin acid degraders.
The following 11 mesophilic resin acid-degrading bacteria were from our culture collection: Burkholderia sp. strain DhA-54, Burkholderia sp. strain IpA-51, Mycobacterium sp. strain DhA-55, Mycobacterium sp. strain IpA-13, Pseudomonas vancouverensis DhA-51, Pseudomonas abietaniphila BKME-9, Pseudomonas multiresinivorans IpA-1, Pseudomonas sp. strain IpA-2, Ralstonia sp. strain BKME-6, Sphingomonas sp. strain DhA-33, and Zoogloea resiniphila DhA-35. These strains were previously isolated from forest soils and biotreatment systems (21). In this study, we tested the ability of these strains to use hydrocarbons as sole organic substrates in BR medium. Octane, dodecane, pristane, cyclohexane, benzene, toluene, and jet fuel were at the same concentrations used for the psychrotolerant isolates (see Table 3). The cultures were incubated for 2 weeks at 30°C on a tube roller and scored for growth by culture turbidity. Viability of the inocula was confirmed in BR medium with 1.0 g of pyruvate per liter. Lack of contamination was confirmed in uninoculated cultures.
TABLE 3.
Substrates used by the psychrotolerant resin acid degraders
| Substratea | Concn (mg/liter) | Substrates used byb:
|
|||
|---|---|---|---|---|---|
| DhA-91 | DhA-95 | IpA-92 | IpA-93 | ||
| Fatty acids | |||||
| Linoleic acid (95) | 1,000 | + | − | − | + |
| Palmitic acid (99) | 1,000 | + | + | + | + |
| Resin acids | |||||
| AbA (96) | 60 | + | + | − | − |
| DhA (99) | 60 | + | + | − | − |
| IpA (99) | 60 | − | − | + | + |
| PiA (88) | 60 | − | − | + | + |
| Other terpenoids | |||||
| Camphorc (96) | 200 | − | − | − | − |
| Citronellolc (95) | 200 | + | − | − | − |
| Betulin (97) | 200 | − | − | − | − |
| Sitosterol (98.7) | 200 | − | − | − | − |
| Aromatic compounds | |||||
| Benzoate (>99) | 240 | + | − | + | + |
| Benzene (99.9) | 200 | − | − | − | + |
| Toluene (99.8) | 200 | − | − | + | + |
| Naphthalenec (99) | 200 | − | − | − | − |
| Alkanes | |||||
| n-Octane (>99) | 500 | + | − | − | − |
| n-Dodecane (99) | 500 | + | + | − | − |
| n-Hexadecane (99.7) | 500 | − | − | − | − |
| Cyclohexane (99.9) | 500 | − | − | − | − |
| Pristane (97) | 500 | − | − | − | − |
| Jet A-1 fuel | 1,000 | + | + | − | − |
| Sugars | |||||
| Arabinose (99) | 1,000 | + | + | + | + |
| Galactose (99) | 1,000 | + | + | + | + |
| Glucose (99) | 1,000 | + | + | + | + |
| Xylose (99) | 1,000 | − | + | − | − |
| Other substrates | |||||
| Acetone (99) | 1,000 | − | − | − | − |
| Pyruvate (99) | 1,000 | + | − | + | + |
Numbers in parentheses indicate the percent purity of the compounds used in this study.
Symbols: +, supports growth; −, does not support growth.
Substrate added from solution in acetone.
Chemical analyses.
Protein was quantified as reported previously (19). The concentrations of resin acids were analyzed by gas chromatography as previously described (18). TPH were also quantified by gas chromatography as previously described (20).
Phylogenetic analyses.
The 16S ribosomal DNA (rDNA) of the isolates was amplified by PCR from individual colonies of pure cultures grown on TSA plates, as reported by Zon et al. (35), using primers Eub-27f and Eub-1525r (8). After confirmation of the product size, the nearly complete 16S rDNA sequences were cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.). The cloned 16S rDNA was sequenced as reported previously (32). The determined 16S rDNA sequences were initially analyzed by using BLAST (National Center for Biotechnology Information) (1) and SIMILARITY_RANK of the Ribosomal Database Project (15) to determine the most similar sequences in the databases. Sequences for comparison were then retrieved, in the aligned form, from the Ribosomal Database Project. The 16S rDNA sequences of DhA-91, DhA-95, IpA-92, IpA-93, and other resin acid-degrading bacteria obtained from previous studies (18, 21, 31, 32) were manually aligned against the above-mentioned aligned sequences using GeneDoc (23). Ambiguous positions were not included in the manual alignment. Evolutionary distances (9) were calculated, and a neighbor-joining tree (26) was inferred by using the Phylogeny Inference Package (version 3.5c) (7). Genomic DNA was extracted using the method of Yu and Mohn (33), and a degenerate PCR assay was performed to detect possible ditA1 homologues, as previously described (34).
Nucleotide sequence accession numbers.
The 16S rDNA sequences determined in this study were deposited in the GenBank under the accession numbers as follows: DhA-91, AF177916; DhA-95, AF177917; IpA-92, AF177918; IpA-93, AF177919.
RESULTS
Enrichment and isolation.
After 4 weeks of incubation at 7°C, the enrichment cultures inoculated with the three contaminated soils, Alert-1, Alert-2, and Alert-3, removed from their medium all of the 400 μM IpA and most of the 400 μM DhA (93, 99, and 100%, respectively). However, the enrichment cultures inoculated with the pristine soils did not show significant IpA or DhA removal. The only exception was the culture inoculated with the pristine soil sample, Alert-6, which removed 70% of the added DhA from the medium. The transfer of the enrichment culture inoculated with Alert-6 failed to grow on DhA, while all the transfers of enrichment cultures inoculated with the contaminated soil samples grew on DhA and IpA. Seven strains capable of growth on DhA and four strains capable of growth on IpA were isolated from the contaminated soils, but no resin acid degraders were obtained from the pristine soils. Four isolates were from the Alert-1 enrichment culture: DhA-91, DhA-92, IpA-91, and IpA-92; five isolates were from the Alert-2 enrichment culture: DhA-93, DhA-94, DhA-95, DhA-96, and IpA-93; and two isolates were from the Alert-3 enrichment culture: DhA-97 and IpA-94. The isolates were in four groups based on comparison of their partial (ca. 500-bp) 16S rDNA sequences. The four groups are represented by strains DhA-91 (six isolates), DhA-95 (two isolates), IpA-92 (one isolate), and IpA-93 (two isolates). These four representative strains were further characterized.
Phylogeny.
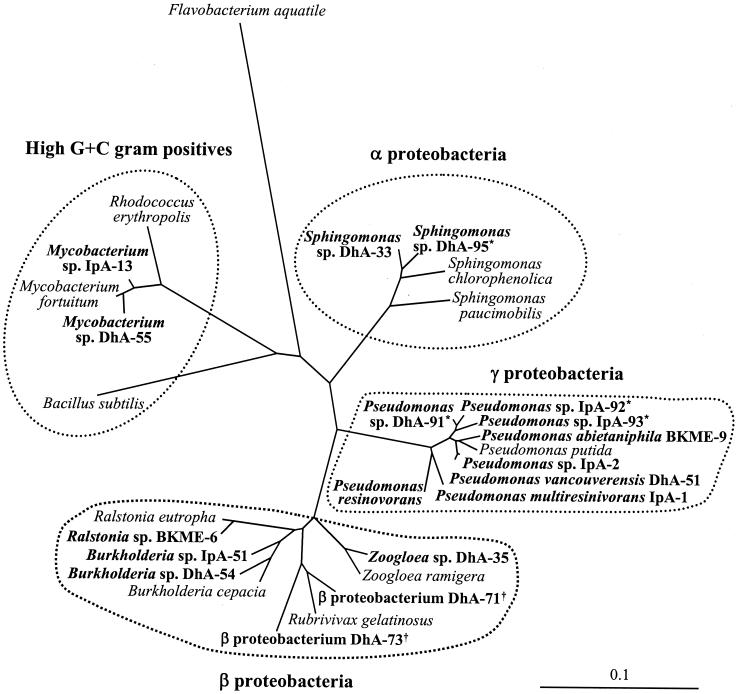
The nearly complete sequences of the 16S rDNA, from position 27 to 1525 (Escherichia coli numbering), of strains DhA-1, DhA-95, IpA-92, and IpA-93 were determined. Cluster analysis of these sequences indicates that DhA-91, IpA-92, and DhA-93 group with the genus Pseudomonas, while DhA-95 groups with Sphingomonas (Fig. 2). The sequence similarity among all three Pseudomonas isolates is less than 98.7%. The sequence of each isolate was compared to the most-similar 16S rDNA sequences available in GenBank. The 16S rDNA sequence of DhA-91 is 98.9% similar to that of Pseudomonas veronii, that of IpA-92 is 98.9% similar to that of Pseudomonas rhodesiae, and that of IpA-93 is 99.2% similar to that of Pseudomonas mandelii. These similarities tentatively suggest that DhA-91 and IpA-92 may represent new species, while IpA-93 may belong to P. mandelii. The sequence of DhA-95 is most similar, 99.5%, to the 16S rDNA sequence of Sphingomonas sp. strain UN1F1. Thus, these two Sphingomonas strains may belong to the same species.
FIG. 2.
An unrooted phylogenetic tree inferred from 16S rDNA sequences. Resin acid degraders are shown in boldface type. The distance scale corresponds to 0.1 mutation per nucleotide position. Symbols: ∗, psychrotolerant strains reported in this study; †, thermophilic strains.
The PCR assay for homologues of ditA1 did not produce any product of the expected size (ca. 930 bp) when DNA from strains DhA-91, DhA-95, IpA-92, and IpA-93 was used as the template. The ditA1 gene encodes the large subunit of a DhA-hydroxylating dioxygenase found in P. abietaniphila BKME-9 (16). Previously, we found putative ditA1 homologues in some other resin acid-degrading isolates (34). These new psychrotolerant isolates do not appear to have a closely related homologue of this dioxygenase.
Physiological characterization of the isolates.
The four characterized isolates are psychrotolerant. Strains DhA-91 and DhA-95 grew at temperatures from 4 to 30°C, and IpA-92 and IpA-93 grew at temperatures from 4 to 22°C (Table 1). The optimal growth temperature on resin acids was 15 to 22°C for DhA-91, DhA-95, and IpA-92 and 22°C for IpA-93. All the isolates are gram-negative, motile rods with similar physiological characteristics and colony morphologies (Table 2).
TABLE 1.
Growth of isolates on resin acids at different temperaturesa
| Temp (°C) | Time of growth
|
|||
|---|---|---|---|---|
| DhA-91 (on DhA) | DhA-95 (on DhA) | IpA-92 (on IpA) | IpA-93 (on IpA) | |
| 4 | 3 wk | 3 wk | 3 wk | 3 wk |
| 7 | 3 days | 3 days | 3 days | 3 days |
| 15 | 1 day | 2 days | 2 days | 2 days |
| 22 | 1 day | 2 days | 2 days | 1 day |
| 30 | 2 days | 1 wk | — | — |
| 37 | — | — | — | — |
The relative growth rates were indicated as the time required for each culture to reach a cell density of approximately 20 cells/field of view under a microscope (magnification × 1,000). —, no growth.
TABLE 2.
Characteristics of psychrotolerant resin acid-degrading bacteriaa
| Characteristic | DhA-91 | DhA-95 | IpA-92 | IpA-93 |
|---|---|---|---|---|
| Gram stain | − | − | − | − |
| Catalase | + | + | + | + |
| Oxidase | + | + | + | + |
| Denitrification | − | − | + | + |
| Motility | + | + | + | + |
| Size (μm) | 1.0–2.0 by 0.9 | 1.0–1.5 by 1.0 | 1.5–2.0 by 0.75 | 2.0 by 1.0 |
| Colony morphology | Pale yellow, circular, raised, smooth, convex, translucent | Yellowish, circular, raised, smooth, convex, translucent | Yellowish, circular, raised, smooth, convex, translucent | Yellowish, circular, raised, smooth, convex, translucent |
+, positive; −, negative.
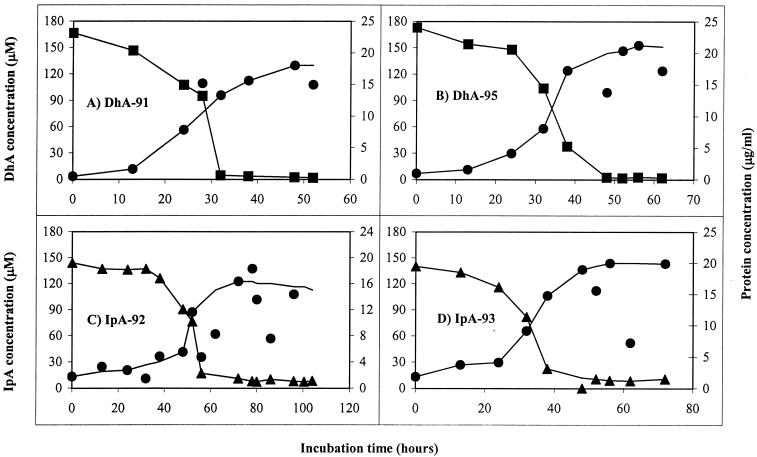
The four isolates completely removed from their medium the resin acid on which each was isolated (Fig. 3). Concomitantly with resin acid removal, growth occurred, as measured by protein concentration. No degradation intermediates were found accumulating in the culture medium, suggesting mineralization of the resin acids by these isolates. The lag periods before growth and the growth rates varied substantially among the isolates. The DhA isolates, DhA-91 and DhA-95, grew faster than the IpA isolates, IpA-93 and IpA-92. At 15°C, the approximate doubling times for DhA-91, DhA-95, IpA-92, and IpA-93 were 4.9, 7.2, 13, and 9.3 h, respectively.
FIG. 3.
Growth on and removal of DhA or IpA at 15°C by strains DhA-91 (A), DhA-95 (B), IpA-92 (C), and IpA-93 (D). Uninoculated control cultures had no resin acid biodegradation. Symbols: squares, DhA; triangles, IpA; circles, protein.
Two patterns of resin acid specificity were found. The DhA isolates used another abietane, abietic acid (AbA), but did not use pimaranes (Table 3). The IpA isolates used another pimarane, pimaric acid (PiA), but did not use abietanes. The DhA isolates used jet fuel and n-alkanes. Strain DhA-91 also used citronellol and benzoate. The IpA isolates used aromatic compounds and were able to denitrify. Neither of the IpA isolates grew on alkanes or Jet A-1 fuel; although Jet A-1 fuel contains an aromatic fraction, this fraction consists of naphthalenes which the IpA isolates do not use. All the isolates could use a number of sugars.
Growth of previously isolated resin acid degraders on hydrocarbons.
In general, the 11 previously isolated, mesophilic resin acid degraders tested did not use hydrocarbons as growth substrates. Mycobacterium sp. strain DhA-55 grew on dodecane and pristane, and Mycobacterium sp. strain IpA-13 grew on octane and pristane. The other mesophilic resin acid degraders, including all the gram-negative strains tested, did not use any of the hydrocarbons tested.
Soil populations of catabolic groups.
The pristine and contaminated soil samples did not significantly differ in populations of heterotrophs or alkane degraders (Fig. 4). The mean populations of total cultureable heterotrophs in the contaminated and the pristine soils were 6.7 × 106 (standard deviation [SD] = 5.3 × 106) and 3.8 × 106 (SD = 1.0 × 106) propagules/g of soil, respectively. The mean populations of dodecane degraders in the contaminated and the pristine soils were 1.5 × 106 (SD = 1.2 × 106) and 7.7 × 105 (SD = 1.2 × 106) propagules/g of soil, respectively. The proportional abundance of dodecane degraders in the pristine soil samples was highly variable, from 0.1 to 78%. Biphenyl degraders, phenanthrene degraders, DhA degraders, and IpA degraders were only detected in the contaminated soil samples, suggesting a positive correlation between hydrocarbon contamination and the abundance of bacteria degrading these compounds. Alert-2, the soil sample with the highest TPH concentration, had the lowest densities of most catabolic groups assayed (Fig. 4). This suggests that the TPH concentration, which was greater than 30 mg/g of this soil, may have been inhibitory to some microorganisms.
DISCUSSION
The psychrotolerant resin acid-degrading bacteria that we isolated from the Arctic soils have characteristics generally consistent with those of previously reported mesophilic and thermophilic resin acid degraders (17, 21). The Arctic isolates are members of the genera Pseudomonas and Sphingomonas, which include most of the previously described resin acid degraders (Fig. 2). The Arctic isolates also have typical specificities for resin acids, using either abietanes or pimaranes, but not both classes of resin acids (Table 3). The Arctic isolates resemble previously described mesophilic isolates in that they use resin acids as sole organic substrates and they appear to mineralize the resin acids (Fig. 3). However, the Arctic isolates are the first reported psychrotolerant resin acid degraders. Also, these strains are unusual in being able to grow on both resin acids and hydrocarbons.
This study is the first demonstration of resin acid biodegradation at low temperatures. The lowest temperature for resin acid biodegradation previously reported was 20°C (31). The existence of psychrotolerant resin acid degraders supports the inference that resin acids must be biodegraded and their carbon must be biologically cycled in cold environments. This cycling is necessary and likely substantial, as resin acids are very abundant in the biosphere. We estimate that the resin acid content of the annually harvested softwood trees, a small fraction of the standing biomass, is on the order of 5 Tg. The existence of psychrotolerant resin acid degraders also indicates the potential for resin acid removal from pulp and paper mill effluents in biological treatment systems in cold regions, which have low winter operating temperatures. However, studies showed significant decreases in resin acid removal in such treatment systems (12). Bioaugmentation with psychrotolerant resin acid degraders is a possible strategy to assist these treatment systems to efficiently remove resin acids during winter.
The results of this study provide only qualified support for the premise that “everything is everywhere.” Resin acid degraders were found in tundra soils of the northern Arctic. These soils were collected thousands of kilometers from the nearest source of resin acids and contained no detectable resin acids (<0.1 μg/g of soil). However, resin acid degraders were not detectable (<3 propagules/g of soil) in the pristine soil samples. The removal of DhA from only one of the enrichment cultures inoculated with pristine Arctic soil samples suggests that resin acid degraders are indigenous to, but rare in, pristine Arctic soils. Since no isolates were obtainable from this active enrichment culture, it is possible that any such indigenous resin acid degraders are distinct from the bacteria isolated from the contaminated soils.
Surprisingly, dodecane degraders were, on average, as abundant in soils from pristine sites as in soils from hydrocarbon-contaminated sites, although their populations were highly variable in the latter soils (Fig. 4). This is not consistent with previous findings that hydrocarbon degraders constitute about 0.1% of the total heterotrophs in pristine soils (reviewed in reference 2). It is not clear why the pristine Arctic soil samples—especially Alert-5, which had no sign of human disturbance—harbor abundant dodecane degraders but not other hydrocarbon degraders assayed.
It is possible that the resin acid degraders found at the polluted sites were transported there by human activities. Consistent with this possibility, the Arctic isolates are members of Pseudomonas spp. and Sphingomonas sp. which lack dispersal and survival stages that would help them to be cosmopolitan (27). It is not clear whether such resin acid degraders can colonize the soils in the study region without human intervention or how long this process would take.
Conditions at the polluted sites clearly selected for relatively large populations of resin acid degraders, as well as for biphenyl and phenanthrene degraders (Fig. 4). The enrichment of resin acid degraders in polluted soils was not simply a consequence of relatively large populations of total heterotrophs in those soils, since the pristine and the polluted sites had similar populations of total psychrotolerant, cultureable heterotrophs. Our values for populations of psychrotolerant heterotrophs in the polluted soils from Alert are similar to those determined by Whyte et al. in a previous study (30). In contrast to our findings, Whyte et al. reported that a single pristine soil sample had 10- to 100-fold-lower heterotrophic populations than in three polluted soil samples. One might expect higher microbial populations in polluted soils than in comparable pristine soils due to the substantial organic matter added by the hydrocarbon pollutants. The similar heterotrophic population sizes that we found suggest that heterotrophs in the polluted soils were limited by something other than organic substrates, such as available nitrogen. This hypothesis is consistent with the observations by Whyte et al. (30) and ourselves (unpublished) that the addition of nitrogen and phosphorus to microcosms of polluted soil from Alert stimulated hydrocarbon biodegradation.
The evidence suggests that resin acid degraders were enriched at the polluted sites by virtue of their abilities to use hydrocarbon substrates and that their abilities to use resin acids are purely coincidental. All of the resin acid degraders characterized in this study can use hydrocarbons (Table 3), whereas most previously described resin acid degraders cannot do so. Further, it appears unlikely that the catabolism of resin acids is directly related to the catabolism of hydrocarbons. None of the characterized isolates used alkanes with structural features similar to those of resin acids, such as alicyclic rings or branching. Also, the isolates were specific in the use of resin acids and did not use other structurally related terpenoids, with the exception of DhA-91 which used citronellol. While it appears unlikely that catabolism of resin acids and that of hydrocarbons are directly related, the large populations (4.6 × 103 to 4.6 × 104 propagules/g of soil) of resin acid degraders at the polluted sites (Fig. 4) suggest that a substantial proportion of hydrocarbon degraders can also use resin acids. It is possible that bacteria that use both resin acids and hydrocarbons have features which are generally adaptive for growth on small hydrophobic molecules (i.e., there may be similarities in the niches of resin acid degraders and of hydrocarbon degraders). Thus, it appears that the environment at this northern Arctic site selected for resin acid-degrading bacteria by a mechanism which was not previously predictable.
ACKNOWLEDGMENTS
This research was supported by a grant from the Sustainable Forest Management Network and by the Canadian Department of National Defence.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Atlas R M. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol Rev. 1981;45:180–209. doi: 10.1128/mr.45.1.180-209.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baas-Becking L G M. Geobiologie of inleiding tot de milieukunde. The Hague, The Netherlands: Van Stockum and Zoon; 1934. [Google Scholar]
- 4.Bedard D L, Unterman R, Bopp L H, Brennan M J, Haberl M L, Johnson C. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol. 1986;51:761–768. doi: 10.1128/aem.51.4.761-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beijerinck M W. Jaarboek van de Koninklijke Akademie voor Wetenschappen. Amsterdam, The Netherlands: Müller; 1913. De infusies en de ontdekking der backteriën. [Google Scholar]
- 6.Feliciano A S, Gordaliza M, Salinero M A, del Corral J M M. Abietane acids: sources, biological activities, and therapeutic uses. Planta Med. 1993;59:485–490. doi: 10.1055/s-2006-959744. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 8.Giovannoni S. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 177–204. [Google Scholar]
- 9.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 10.Kovacs T G, Voss R H. Biological and chemical characterization of newsprint/specialty mill effluents. Water Res. 1992;26:771–780. [Google Scholar]
- 11.Leach J M, Thakore A N. Identification of toxic constituents of kraft mill effluents that are toxic to juvenile coho salmon (Oncorhynchus kisutch) J Fish Res Board Can. 1973;30:470–484. [Google Scholar]
- 12.Liss S N, Allen D G. Microbiological study of a bleached kraft pulp mill aerated lagoon. J Pulp Paper Sci. 1992;18:J216–J221. [Google Scholar]
- 13.Liss S N, Bicho P A, Saddler J N. Microbiology and biodegradation of resin acids in pulp mill effluents: a minireview. Can J Microbiol. 1997;75:599–611. doi: 10.1139/m97-086. [DOI] [PubMed] [Google Scholar]
- 14.Longton R E. The role of bryophytes and lichens in polar ecosystems. In: Woodin S J, Marquiss M, editors. Ecology of Arctic environments. Oxford, United Kingdom: Blackwell Science Ltd.; 1997. pp. 69–96. [Google Scholar]
- 15.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The ribosomal database project. Nucleic Acid Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin V J, Mohn W W. A novel three-component dioxygenase from the diterpenoid-degrading bacterium, Pseudomonas abietaniphila BKME-9. J Bacteriol. 1999;181:2675–2682. doi: 10.1128/jb.181.9.2675-2682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin V J, Yu Z, Mohn W W. Recent advances in understanding resin acid biodegradation: microbial diversity and metabolisms. Arch Microbiol. 1999;172:131–138. doi: 10.1007/s002030050752. [DOI] [PubMed] [Google Scholar]
- 18.Mohn W W. Bacteria obtained from a sequencing batch reactor that are capable of growth on dehydroabietic acid. Appl Environ Microbiol. 1995;61:2145–2150. doi: 10.1128/aem.61.6.2145-2150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohn W W, Stewart G R. Bacterial metabolism of chlorinated dehydroabietic acids occurring in pulp and paper mill effluents. Appl Environ Microbiol. 1997;63:3014–3020. doi: 10.1128/aem.63.8.3014-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohn W W, Stewart G R. Limiting factors for hydrocarbon biodegradation at low temperature in Arctic soils. Soil Biol Biochem. 2000;32:1161–1172. [Google Scholar]
- 21.Mohn W W, Wilson A E, Bicho P A, Moore E R B. Physiological and phylogenetic diversity of bacteria growing on resin acids. Syst Appl Microbiol. 1999;22:68–78. doi: 10.1016/S0723-2020(99)80029-0. [DOI] [PubMed] [Google Scholar]
- 22.Mohn W W, Yu Z, Moore E R B, Muttray A F. Lessons learned from Sphingomonas species that degrade abietane triterpenoids. J Indust Microbiol Biotechnol. 1999;23:374–379. doi: 10.1038/sj.jim.2900731. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas K B, Nicholas H B, Jr, Deerfield D W., II GeneDoc: analysis and visualization of genetic variation. EMBNEW News. 1997;4:14. [Google Scholar]
- 24.Oblinger J L, Koburger J A. The most probable number technique. In: Speck M L, editor. Compendium of methods for the microbiological examination of foods. Washington, D.C.: American Public Health Association; 1984. p. 108. [Google Scholar]
- 25.Priha M H, Talka E T. Biological activity of bleached kraft mill effluent (BKME) fractions and process streams. Pulp Paper Can. 1986;87:143–147. [Google Scholar]
- 26.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Staley J T, Gosink J J. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu Rev Microbiol. 1999;53:189–215. doi: 10.1146/annurev.micro.53.1.189. [DOI] [PubMed] [Google Scholar]
- 28.Taylor B, Yeager K, Abernethy S, Westlake G. Scientific criteria document for development of provincial water quality objectives: resin acids. Report 0-7729-4347-8. Toronto, Ontario, Canada: Water Resources Branch, Ontario Ministry of the Environment; 1988. [Google Scholar]
- 29.Walden C C, Howard T E. Toxicity of pulp and paper mill effluent: a review. Pulp Paper Can. 1981;83:T143–148. [Google Scholar]
- 30.Whyte L G, Bourbonnière L, Bellerose C, Greer C W. Bioremediation assessment of hydrocarbon-contaminated soils from the high Arctic. Bioremediation. 1999;3:69–79. [Google Scholar]
- 31.Wilson A E, Moore E R B, Mohn W W. Isolation and characterization of isopimaric acid-degrading bacteria from a sequencing batch reactor. Appl Environ Microbiol. 1996;62:3164–3151. doi: 10.1128/aem.62.9.3146-3151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Z, Mohn W W. Isolation and characterization of thermophilic bacteria capable of degrading dehydroabietic acid. Can J Microbiol. 1999;45:513–519. doi: 10.1139/w99-028. [DOI] [PubMed] [Google Scholar]
- 33.Yu Z, Mohn W W. Killing two birds with one stone: simultaneous extraction of DNA and RNA from activated sludge biomass. Can J Microbiol. 1999;45:269–272. [Google Scholar]
- 34.Yu Z, Martin V J J, Mohn W W. Occurrence of two resin acid-degrading bacteria and a gene encoding resin acid biodegradation in pulp and paper mill effluent biotreatment systems assayed by PCR. Microb Ecol. 1999;38:114–125. doi: 10.1007/s002489900163. [DOI] [PubMed] [Google Scholar]
- 35.Zon L I, Dorfman D M, Orkin S H. The polymerase chain reaction colony miniprep. BioTechniques. 1989;7:696–698. [PubMed] [Google Scholar]