Summary
There is no cure for HIV infection, as the virus establishes a latent reservoir, which escapes highly active antiretroviral treatments. One major obstacle is the difficulty identifying cells that harbor latent proviruses. We devised a single-round viral vector that carries a series of versatile reporter molecules that are expressed in an LTR-dependent or LTR-independent manner and make it possible to accurately distinguish productive from latent infection. Using primary human CD4+ T cells, we show that transcriptionally silent proviruses are found in more than 50% of infected cells. The latently infected cells harbor proviruses but lack evidence for multiple spliced transcripts. LTR-silent integrations occurred to variable degrees in all CD4+ T subsets examined, with CD4+ TEM and CD4+ TREG displaying the highest frequency of latent infections. This viral vector permits the interrogation of HIV latency at single-cell resolution, revealing mechanisms of latency establishment and allowing the characterization of effective latency-reversing agents.
Keywords: HIV-1, latency establishment, CD4+ T cell subsets, reporter virus, HIV integration, mass cytometry, HIV persistence, HIV reservoir, CD4+ T memory cells, CD4+ T stem cells
Graphical abstract
Highlights
-
•
HIV pMorpheus-V5 accurately identifies productively and latently infected cells
-
•
More than 50% of all infected primary human CD4+ T cells harbor LTR-silent proviruses
-
•
We identify CD4+ T cell subsets that preferentially support HIV latency
-
•
Naive CD4+ T cells harbor LTR-silent proviruses but lack evidence for productive replication
Motivation
HIV latency is a major obstacle to the eradication of HIV/AIDS. Identification of the infected cells harboring LTR-silent proviruses is technically complicated and labor intensive. Cell culture systems that identify latently infected primary human cells at the single-cell level are necessary to study the mechanisms of latency establishment in a cell-type-specific manner. Moreover, such an experimental system would also allow the discovery of small molecules that either lock or reactivate proviruses. The pMorpheus HIV vector system represents an advancement in the field, as it provides a flexible toolbox for dissecting HIV latency in the context of a single-cycle virus encoding all the accessory genes.
Kim et al. developed a HIV reporter vector (pMorpheus-V5) suitable to identify latently and productively infected primary CD4+ T cells at single-cell resolution. Latent infections were more frequent than productive infections in all seven CD4+ T cell subsets investigated. Expression of markers validated the nature of latent and productive pMorpheus-V5 infections.
Introduction
An HIV/AIDS cure remains elusive because HIV establishes a hard-to-eradicate latent reservoir. Despite the intense efforts over the past forty years, one of the main obstacles to HIV eradication is the difficulty identifying the cells that harbor transcriptionally silent proviruses forming a long-lasting, antiretroviral-resistant, latent reservoir.
CD4+ T cells are the primary targets of HIV infection and contribute to establishing and maintaining the latent reservoir (Churchill et al., 2016). Subsets of these cells differ in their susceptibility to infection and their ability to traffic, exert effector functions, proliferate, and survive (Zerbato et al., 2016; Kulpa et al., 2019; Grau-Exposito et al., 2019). A more detailed and cell type-specific understanding of latency establishment is, therefore, of great relevance.
HIV latency can result from progressive epigenetic silencing of productive infections (Colin and Van Lint, 2009; Siliciano and Greene, 2011; Pearson et al., 2008; Weinberger et al., 2008; Hashemi et al., 2016; Jefferys et al., 2021; Mbonye and Karn, 2014). However, evidence suggest that a substantial portion of HIV integrations occur in an LTR-silent manner (Dahabieh et al., 2013; Chavez et al., 2015; Calvanese et al., 2013). This shift in our understanding of HIV latency establishment was made possible by the use of HIV vector constructs, which encode fluorescent reporters expressed in either an HIV LTR-dependent or LTR-independent manner (Dahabieh et al., 2013; Calvanese et al., 2013).
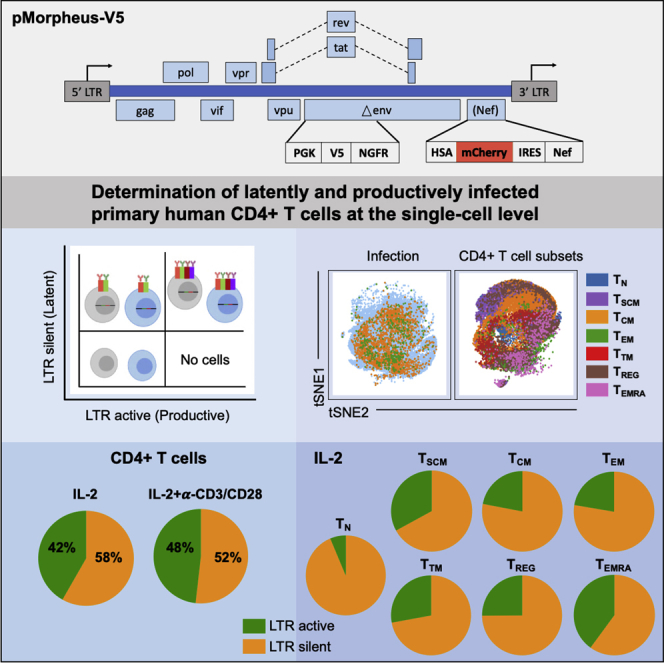
A number of different dual-reporter vectors have been used in the past to investigate latency establishment (Battivelli et al., 2018; Calvanese et al., 2013; Cavrois et al., 2017; Chavez et al., 2015; Dahabieh et al., 2013, 2014; Hashemi et al., 2016; Kim et al., 2019; Matsuda et al., 2015; Cai et al., 2021). They all rely on LTR-dependent or LTR-independent differential reporter expression. We reasoned that positioning the reporters in distinct reading frames undergoing different splicing events would reduce the potential for promoter interference. Moreover, having a full-length functional gag and all accessory proteins, including Nef, is necessary to obtain a physiologically relevant assessment of HIV latency establishment. Last, a constitutively active PGK promoter ensures high expression of the latency protein tags while avoiding rapid silencing by the HUSH complex (Jones et al., 2009; Kim et al., 2019; Cai et al., 2021; Tchasovnikarova et al., 2015). Of note, the resulting HIV molecular clone, pMorpheus-V5, lacks a functional Env, which eases biosafety precautions for downstream applications with live cells and allows pseudotyping with different HIV envelopes.
Detection of infected cells at the single-cell level in a manner that is independent of HIV LTR activity allows probing for potential differences in latency establishment in specific CD4+ T cell subsets. Here we show that primary human CD4+ T cells harboring transcriptionally silent proviruses are more frequent than productively infected cells. This was true for all seven CD4+ T cell subsets investigated in this study, but CD4+ regulatory T cells (CD4+ TREG) and CD4+ effector memory T cells (CD4+ TEM) had the highest frequency of LTR-silent infection. We propose that the pMorpheus-V5 system encoding all the viral accessory proteins as well as different reporter proteins will facilitate obtaining new mechanistic insights into latency establishment and reactivation in different primary human cells, including but not limited to primary human CD4+ T cells.
Results
pMorpheus-V5 construction and identification of productively and latently infected cells
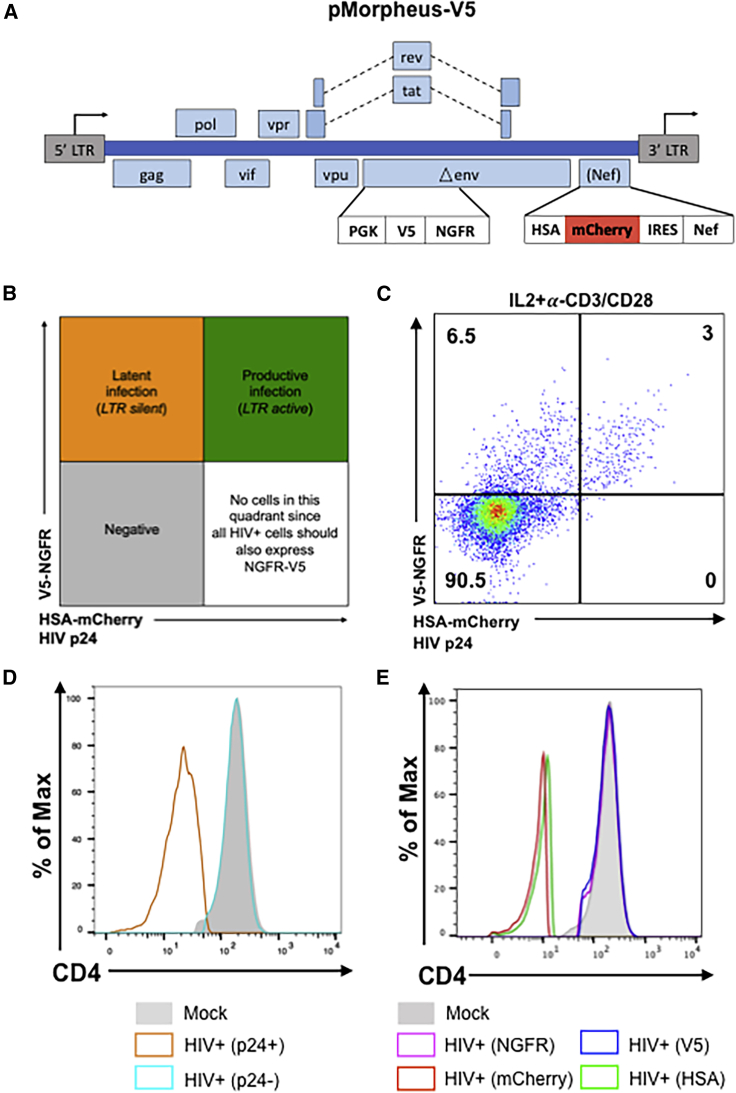
To investigate the cellular processes that govern establishment of HIV latency at the single-cell level in human primary CD4+ T cells, we developed a viral reporter vector (pMorpheus-V5) suitable to detect the LTR-dependent or LTR-independent expression of four reporters, two of which, HSA and V5-NGFR, are membrane bound (Figure 1A). A PGK promoter-driven reporter cassette encoding V5-NGFR was inserted into the Env open reading frame deleting a portion of the HIV envelope. Upon proviral integration, but independently of LTR promoter activation, V5-NGFR localizes to the surface of the infected cell. The LTR-dependent expression cassette includes sequences encoding HSA and mCherry that were cloned upstream of an internal ribosomal site followed by Nef. Thus, LTR-dependent expression of HSA, mCherry and Nef is linked (Young et al., 2018). Cells productively infected with pMorpheus-V5 express four reporters: V5-NGFR driven by PGK as well as HSA and mCherry driven by the HIV-LTR. In contrast, latently infected cells express only V5-NGFR. The combination of fluorescence reporter and membrane bound tags allow easy and specific identification of the different cell population using flow cytometry (Figures 1B and 1C). Cells expressing only HSA and mCherry (lower right quadrant in the cartoon) should not be found, as all productively infected cells harbor proviruses and thus would be positive for V5-NGFR (Figure 1B). To assess the ability to reliably distinguish cells harboring LTR-silent or LTR-active proviruses, we infected primary human CD4+ T cells, stimulated either with IL-2 alone or activated by a combination of IL-2 and α-CD3/CD28 coated beads, with pMorpheus-V5. Dual-tropic envelope pSV III-92 HT593.1 (Gao et al., 1996) was used to pseudotype pMorpheus-V5 in order to broaden the range of CD4+ T cell types susceptible to infection, including naive CD4+ T cells as well as CD4+ memory stem cells. It is important to understand latency establishment in these specific CD4+ T cell types known to express low levels of CCR5 yet critical for the formation of the latent reservoir (Buzon et al., 2014; Mavigner et al., 2018; Zerbato et al., 2019). Of note, pMorpheus-V5 is a single-cycle viral vector, which does not support spreading infection. Primary human CD4+ T cells were infected with pMorpheus-V5 for five days. Day 5 post-infection was chosen because optimization experiments revealed it to be the time point when productive infection peaked. We used flow cytometry or cytometry by time-of-flight (CyTOF) for the analysis (Table S1). The cell viability was comparable between infected and uninfected cells under each experimental condition (Figures S1A, S1B, S2A, and S2B). NGFR and V5 as well as HSA and mCherry provided comparable percentages of latently or productively infected cells as measured using either flow cytometry or CyTOF (Figure 1C, flow cytometry; Figure S2C, CyTOF). The absence of HSA-mCherry alone expressing cells indicates that promoter interference is not a problem with the current vector system.
Figure 1.
Description and characterization of the pMorpheus-V5 viral reporter vector
(A) Representation of the HIV reporter viral vector pMorpheus-V5 (pLAI2-V5-NGFR HSA-mCherry-IRES-Nef).
(B) Gating strategy to identify cells latently or productively infected by pMorpheus-V5 using flow cytometry or CyTOF is depicted.
(C) Latently (V5+, NGFR+) or productively (HSAPOS, mCherryPOS, V5POS, NGFRPOS, p24POS) infected, IL-2 + α-CD3/CD28-stimulated CD4+ T cells were identified using flow cytometry after infection with pMorpheus-V5. The data show one representative example of four independent experiments using cells from different healthy donors. The percentage of CD4+ T cells in each quadrant is indicated.
(D) Nef down-regulates CD4 levels during pMorpheus-V5 infection. CD4+ T cells from two different donors were stimulated with IL-2 + α-CD3/CD28 and infected with pMorpheus-V5. CyTOF was performed 5 days post-infection (dpi). Histogram plots show the CD4 expression for uninfected cells (shaded light gray), p24POS cells (orange), and p24NEG cells (light blue). The data were analyzed using FlowJo.
(E) Cells were stimulated with IL-2 + α-CD3/CD28 prior to infection with p-Morpheus-V5. Histogram analysis of CD4 expression levels of NGFR (magenta), V5 (blue), mCherry (red), or HSA (green) positive cells. The data were analyzed using FlowJo. Data from two independent donors.
As HIV-1 Nef is known to down-regulate the CD4 receptor (Garcia and Miller, 1991; Guy et al., 1987), we next compared the expression of CD4 in latently and productively infected CD4+ T cells with that of mock-infected cells. As expected, HIV Gag p24-positive CD4+ T cells expressed substantially less CD4 compared with p24-negative cells or mock-infected cells (Figures 1D, 1E, S1C, S1D, S2D, and S2E). Down-regulation of CD4 was also observed in HSAPOS or mCherryPOS cells but not in NGFRPOS or V5POS-only cells. Thus, CD4 expression in CD4+ T cells harboring only LTR-silent proviruses (as identified by NGFRPOS/V5POS but HSANEG/mCherryNEG expression) remained comparable with that of mock-infected cells (Figures 1D and 1E; Figures S1C and S1D [flow cytometry] and S2D and S2E [CyTOF]).
Taken together, infection with pMorpheus-V5 permits to distinguish between latently infected and productively infected primary CD4+ T cells using either flow cytometry or CyTOF.
Primary human CD4+ T cells harboring LTR-silent proviruses are as frequent as those harboring LTR-active proviruses
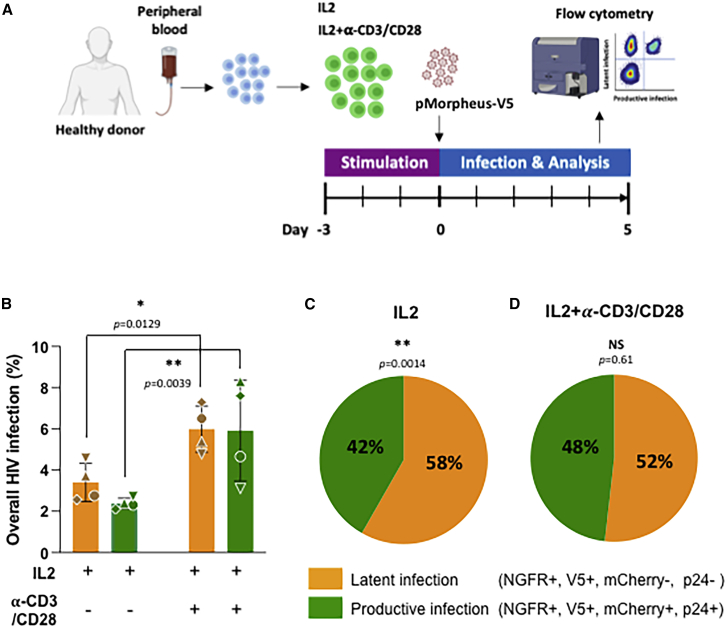
We next measured the proportion of CD4+ T cells latently and productively infected with pMorpheus-V5. We specifically compared the expression of the different viral reporters and/or proteins including mCherry and/or intracellular p24, NGFR, and V5 in CD4+ T cells obtained from four independent healthy donors (Figures 2A and 2B; Table S1). Overall, we observed 3.4% ± 1.2% of latently infected cells and 2.4% ± 0.2% of productively infected cells in the presence of IL-2 alone (total percentage of infected cells 5.8%; Figure 2B; Table S1). In the presence of IL-2 ± α-CD3/CD28, these proportions rose to 6% ± 1.3% of latently infected cells and 6% ± 2.5% of productively infected cells (total percentage of infected cells 11.9%; Figure 2B; Table S1).
Figure 2.
Identification of latently and productively infected CD4+ T cells
(A) Experimental design to analyze CD4+ T cells infected with pMorpheus-V5. CD4+ T cells from four healthy donors were stimulated with IL-2 or a combination of IL-2 with α-CD3/CD28 antibody-coated beads for 3 days prior to infection with pMorpheus-V5. The frequency of latently and productively infected cells was determined 5 dpi using flow cytometry.
(B) Frequency of latently (V5POS, NGFRPOS; yellow) and productively (mCherryPOS, V5POS, NGFRPOS, p24POS; green) infected CD4+ T cells stimulated with IL-2 alone or IL-2/α-CD3/CD28. Cells were analyzed using flow cytometry 5 dpi. Each of the four healthy donors is identified by symbol. Error bars represent the average ± SD, and statistical significance was established using unpaired t test.
(C) Relative proportion of pMorpheus-V5 latently (yellow) or productively (green) infected CD4+ T cells stimulated with IL-2 is shown. The data represent the average of the four healthy donors. Average LTR-silent 58% ± 4% versus average LTR-active 42% ± 4% (p = 0.0014, unpaired t test).
(D) Relative proportion of pMorpheus-V5 latently (yellow) or productively (green) infected CD4+ T cells activated with α-CD3/CD28. The data represent the average of the four individual healthy donors. Average LTR-silent 52% ± 8% versus average LTR-active 48% ± 8% (p = 0.61, t test).
The fraction of cells harboring LTR-silent proviruses was significantly higher than the number of productively infected in IL-2-only stimulated primary human CD4+ T cells (Figure 2C; average LTR-silent 58% ± 4% versus average LTR-active 42% ± 4%, p = 0.0014). This difference was reduced when CD4+ T cells were activated with IL-2 + α-CD3/CD28 (Figure 2D; average LTR-silent 52% ± 8% versus average LTR-active 48% ± 8%, p = 0.61).
Thus, under both conditions (i.e., IL-2 only and IL-2 + α-CD3/CD28), more than half of the infected primary human CD4+ T cells harbor LTR-silent proviruses (Figures 2C and 2D). These results imply that experimental settings relying solely on viral production-based readouts (e.g., p24 production, LTR-driven reporter gene expression) under-estimate the frequency of overall HIV infection by at least 2-fold (e.g., 5.8% latently plus productively infected cells versus 2.4% productively infected CD4+ T cells; Figure 2B).
Detection of HIV proviruses and transcripts in latently infected primary human CD4+ T cells
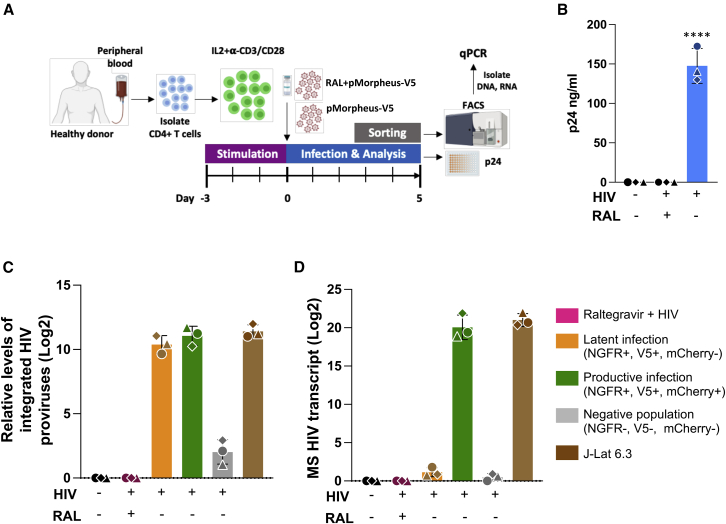
We next measured the level of integration and multiply spliced HIV transcripts in LTR-silent and LTR-active infected CD4+ T cells (Figure 3A). Briefly, we infected IL-2 + α-CD3/CD28-stimulated primary human CD4+ T cells from three different healthy donors with pMorpheus-V5 in the presence and absence of the FDA-approved integrase inhibitor raltegravir. Five days later, culture supernatants were collected for p24 analysis, and the live CD4+ T cells were sorted using fluorescence-activated cell sorting (FACS) (latently infected: V5-NGFRPOS/HSA-mCherryNEG; productively infected: V5-NGFRPOS/HSA-mCherryPOS; negative: V5-NGFRNEG/HSA-mCherryNEG). As anticipated, raltegravir prevented both latent and productive infections (Figure 3B).
Figure 3.
Quantification of proviruses and multiply spliced HIV transcripts in FACS-sorted CD4+ T cells
(A) Representation of the experimental design to analyze provirus integration or multiply spliced transcripts in productive or latently infected cells from three different healthy donors. CD4+ T cells were activated with IL-2 and α-CD3/CD28 antibody-coated beads for 3 days prior to infection with pMorpheus-V5. CD4+ T cells were infected with pMorpheus-V5 in the presence and absence of raltegravir. Latently (orange), productively (green), and the reporter negative cell populations were sorted using FACS 5 dpi with pMorpheus-V5. RNA and DNA were extracted from the same samples.
(B) Gag p24 concentration of culture supernatants was measured using ELISA at 5 dpi. Bar graph shows the production of p24 in pMorpheus-V5 or mock-infected cells in the presence and absence of raltegravir (RAL). Each donor is identified by a specific symbol. The average of three donors is shown (±SD) (∗∗∗∗p < 0.0001, unpaired t test).
(C) HIV integration was measured using a two-step quantitative Alu-PCR assay using total DNA from each sorted cell population (latent infection, yellow; productive infection, green; reporter expression-negative cells, gray). The bar graph depicts relative amounts of integrated proviruses. J-Lat 6.3 cells were used as a positive control. The mean ± SD of three independent experiments/donors are shown. Sorted cells from the same donor (N = 3) are identified by the same symbol.
(D) Multiply spliced (MS) HIV transcripts were quantified using semi-nested real-time qPCR. RNA was extracted from the same samples as in (C). RNA extracted from stimulated J-Lat 6.3 cells was used as a positive control. The bar graph depicts the mean ± SD from three independent experiments/donors. Sorted cells from the same healthy donor (N = 3) are identified by the same symbol.
Genomic DNA was extracted from sorted cells to measure integration using Alu-1 PCR (Butler et al., 2001), while RNA was extracted from the same samples to detect multiply spliced HIV tat/rev transcripts using qRT-PCR assay (Pasternak et al., 2008; Zerbato et al., 2021) (Figure 3A). As a positive control, we used the T cell line J-Lat, which harbors a single provirus, stimulated with PMA (16 nM) and ionomycin (0.5 μM) for three days. We found that the levels of integrated provirus were comparable between latently and productively infected cells (Figure 3C). Raltegravir treatment effectively blocked HIV integration. A small portion of the cells lacking expression of all four reporters (“negative cell population”) did contain integrated proviruses. Such positive qPCR signal could derive from integrated defective viruses, wherein the reporters have been deleted, or cellular pathways silenced expression of both reporter cassettes.
Multiply spliced HIV transcripts were detected only in the productively infected CD4+ T cells (V5-NGFRPos/HSA and mCherryPos) and not in the LTR-silent (V5-NGFRPos) or reporter-negative CD4+ T cells (Figure 3D). Similarly, the raltegravir-treated cells lacked expression of multiply spliced HIV transcripts, while such expression in stimulated J-Lat cells was comparable with that of productively infected primary human CD4+ T cells.
Collectively, these data reinforce the notion that the difference between LTR-active and LTR-silent cells is not due to differences in the level of integrated proviruses but rather to low HIV gene expression rate, as revealed by the assessment of HIV early gene Tat/Rev RNA expression.
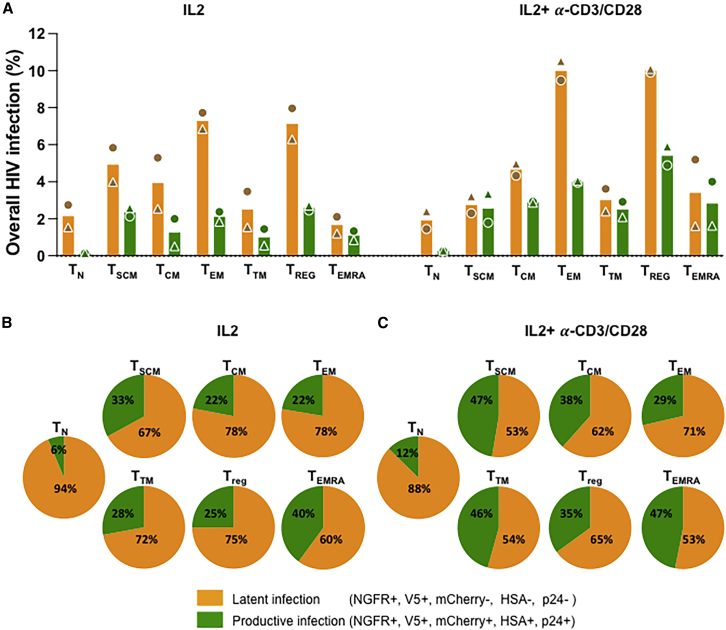
Identification of CD4+ T cell subpopulations that preferentially support HIV latency
We next used CyTOF (Kotecha et al., 2010) to determine whether CD4+ T cell subsets differ in their propensity to support latency establishment. Primary human CD4+ T cells stimulated with IL-2 alone or IL-2-α-CD3/CD28 were infected with pMorpheus-V5 and stained for CyTOF. We first barcoded the individual infections with CD45 antibodies and then stained with a custom antibody cocktail recognizing the HIV-encoded reporters as well as 29 other cellular markers (Figure S3A).
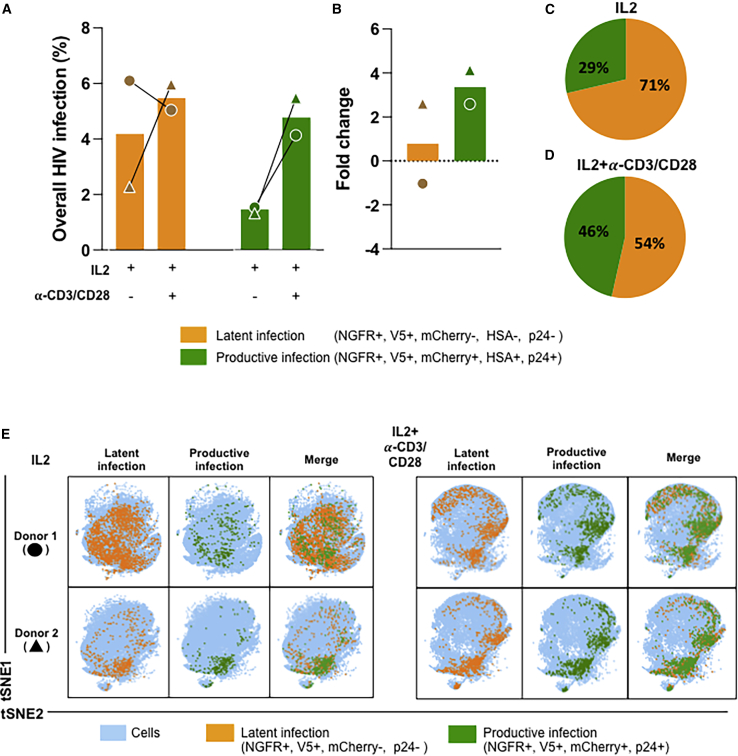
Multi-dimensional analysis identified the protein tags of latent infection (V5-NGFRPOS only) in more than 50% of the cells analyzed (Figures 4A, 4C, 4D, 5A, and 5B). The overall frequency of infection (both latent and productive) was 6.05% (donor 1) and 2.31% (donor 2) in IL-2-treated cells. Stimulation of CD4+ T cells with IL-2-α-CD3/CD28 increased the percentages of productively infected cells to 5.02% and 5.92%. The percentages of latently infected cells (V5-NGFRPOS) were 2.3% and 6% (IL-2 alone) and 5% and 5.9% when stimulated with IL-2-α-CD3/CD28. The productively (V5-NGFGPOS, mCherryPOS, HSAPOS, p24POS) infected cells instead amounted to 1.6% and 1.3% with IL-2 alone and 2.1% and 5.5% with IL-2-α-CD3/CD28 stimulation (Figure 4A). In line with our previous data generated using flow cytometry, the relative percentage of productively infected cells increased overall with activation (compare IL-2-α-CD3/CD28 with IL-2 alone in Figure 4B), while the proportion of latently infected cells was higher in the cells treated only with IL-2 (Figures 4C and 4D).
Figure 4.
Characterization of latently and productively infected CD4+ T cell populations using CyTOF
(A) pMorpheus-V5-infected primary human CD4+ T cells stimulated with IL-2 or activated with IL-2/α-CD3/CD28 beads were analyzed using CyTOF at 5 dpi. Latently (V5POS, NGFRPOS; yellow) or productively (HSAPOS, mCherryPOS, V5POS, NGFRPOS, p24POS; green) infected cells were identified. Data for each donor (±SD) are indicated by a specific symbol.
(B) The bar graph shows the fold change of latently or productively infected cells depending on cellular activation. The average of two individual donors is shown.
(C) The proportion of latently or productively infected CD4+ T cells stimulated with IL-2 for three days. The average of two individual donors is shown with the percentage and the total number of cells.
(D) The proportion of latently or productively infected CD4+ T cells stimulated with IL-2 and α-CD3/CD28. The average of two individual donors is shown with the percentage and the total number of cells.
(E) viSNE analysis of pMorpheus-V5-infected CD4+ T cells stimulated with IL-2 alone or with IL-2 and α-CD3/CD28. The tSNE1 and tSNE2 axes are based on the relevant markers of the subsets defined by the Cytobank program. The results are based on 66,000 cells for each condition from each donor identified by the circle and triangle symbols in (A).
Figure 5.
Determination of latent and productive infection in different CD4+ T cell subsets
(A) pMorpheus-V5-infected primary human CD4+ T cells from healthy donors stimulated with IL-2 or with IL-2 and α-CD3/CD28 were analyzed using CyTOF to determine levels of NGFR, V5, mCherry, HSA, and p24 in different CD4+ T cell subsets. The bar graphs represent percentage of latent or productive infection of each T cell subsets. The average of two individual donors is shown. Each donor is indicated by a unique symbol and each stimulated group is color coded.
(B) The proportion of latent (yellow) and productive (green) infection of CD4+ T cells upon pMorpheus-V5 infection. CD4+ T cells from 2 donors were stimulated with IL-2 for 3 days. The average of two individual donors is shown.
(C) CD4+ T cells were stimulated with IL2 and α-CD3/CD28 beads for 3 days and infected with pMorpheus-V5. The proportion of latently (yellow) or productively (green) infected cells is depicted. The average of two individual donors is shown.
We visualized the latently and productively infected cells at the single-cell level using viSNE, which uses t-stochastic neighbor embedding (t-SNE) to generate a two-dimensional map of the latency protein tag expression in different CD4+ T cell populations (Figure 4E).
The CyTOF antibody panel used allowed the identification and visualization of the distribution of seven different CD4+ T cell subsets using viSNE (Figure S4E). Of note, activation with IL-2 + α-CD3/CD28 modified the distribution of the CD4+ T cell subsets in the absence of pMorpheus-V5 infection (Figures S4A–S4C).
Across the CD4+ T cell subsets, the proportion of cells harboring LTR-silent proviruses (V5-NGFRpos only) was higher than that of productively infected cells (Figures 5A and 5B). We noted that LTR-silent infections were more common in cells stimulated with IL-2, while cell activation with IL-2 + α-CD3/CD28 reduced the relative proportion of latently infected cells (Figures 5B and 5C). viSNE maps further visualize LTR-silent and LTR-active proportions in each distinct CD4+ T cell populations (Figures S5A and S5B). Of note, the vast majority of infected naive T cells harbored latent proviruses. We observed that CD4+ TEM and CD4+ TREG have the highest total number of latently infected cells, and IL-2/α-CD3/CD28 stimulation increases the percentage of productive infection in all subsets with the exception of TEM. Taken together, our data are in agreement with previous reports (Buzon et al., 2014) suggesting that latent infections occur across all seven CD4+ T cell subsets analyzed with CD4+ TEM and CD4+ TREG contributing heavily to the latent reservoir formation. Naive T cells are regarded as being refractory to infection, but several studies have shown that in vivo naive T cells bear integrated provirus, suggesting that they can be infected and are part of the latent reservoir (Venanzi Rullo et al., 2020; Zerbato et al., 2019). Indeed, we observe that a portion of naive cells support infection up to the level of integration (latent infection in naive T cells 1.53%–2.745% [IL-2] and 1.44%–2.395% [IL-2 + α-CD3/CD28]). Further studies will determine the extent to which naive T cells play a role in the establishment of HIV-1 latent reservoir.
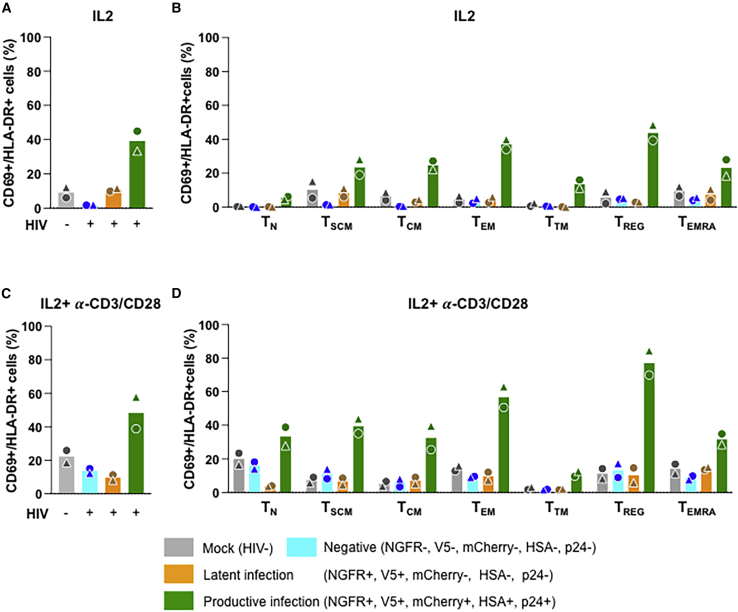
Expression of activation and exhaustion markers confirms the latent and productive nature of infections with pMorpheus-V5
Expression of the activation markers CD69 and HLA-DR associated with productive infection (Marini et al., 2008; Sahu et al., 2006) but not with latent infection (Moso et al., 2019). We therefore analyzed the expression of CD69 and HLA-DR in latently and productively infected CD4+ T cells upon pMorpheus-V5 infection. We observed that both markers were expressed at higher levels in productively infected CD4+ T cells compared with latently infected, the negative cell populations, or mock-infected cells (Figures 6A and 6C). In the absence of overt T cell activation, all the memory T cell populations showed higher expression of these two activation markers when productively infected (Figure 6B). The same was true following stimulation with IL-2 + α-CD3/CD28, with the exception of CD4+ T transitional memory (TTM) cells. This T cell subset displayed only a marginally higher expression of CD69/HLA-DR in productively infected cells compared with latently infected, the negative populations, or mock-infected cells (Figure 6D). Importantly, latently infected CD4+ T cells showed CD69/HLA-DR expression profiles that were comparable with the negative cells or mock-infected cells, providing validation for the fact that latently infected cells identified by pMorpheus-V5 infection are fundamentally distinct from the productively infected cells.
Figure 6.
Validation of latent and productive infection by measuring early activation markers CD69 and HLA-DR in different CD4+ T cell subsets
(A) CD69/HLA-DR expression on latently and productively pMorpheus-V5-infected primary human CD4+ T cells stimulated with IL-2 was determined using CyTOF. Bar graphs represent levels of CD69 and HLA-DR in total CD4+ T cells. Each donor is indicated by a unique symbol. The infection status of each CD4+ T cell subset is color coded in the figure. The average of two individual donors is shown.
(B) CD69/HLA-DR expression on latently and productively pMorpheus-V5-infected CD4+ T subsets stimulated with IL-2 was determined. The levels of CD69+/HLA-DR+ in mock-infected, reporter-negative cells, latently infected, and productively infected CD4+ T cell subsets stimulated were measured using CyTOF. The average of two individual donors is shown.
(C) CD69/HLA-DR expression on latently and productively pMorpheus-V5-infected primary human CD4+ T cells stimulated with IL-2 and α-CD3/CD28 was determined using CyTOF. Bar graphs represent levels of CD69 and HLA-DR in total CD4+ T cells. Each donor is indicated by a unique symbol. Results are from two independent donors.
(D) CD69/HLA-DR expression on latently and productively pMorpheus-V5-infected CD4+ T subsets stimulated with IL-2 and α-CD3/CD28. The levels of CD69/HLA-DR in mock-infected, reporter-negative cells, latently infected, and productively infected CD4+ T cell subsets were measured using CyTOF. The average of two individual donors is shown.
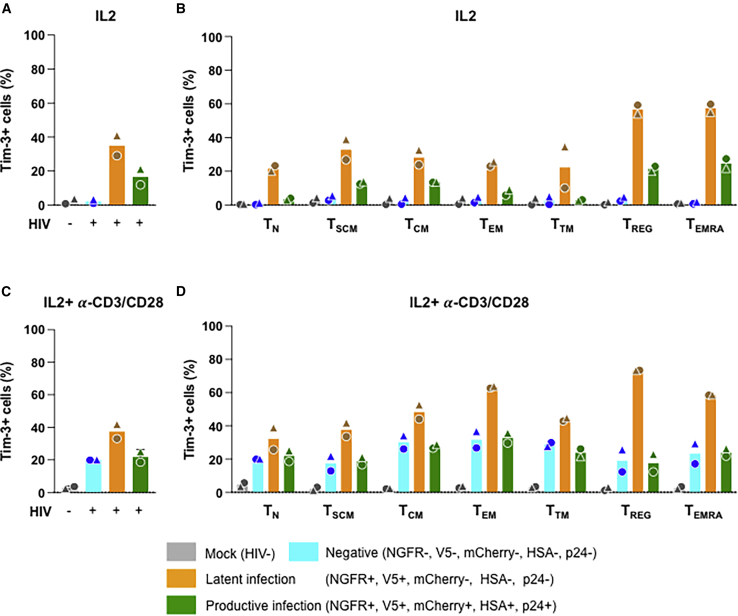
Recent studies have indicated that, among other markers, Tim-3 is enriched in CD4+ T cells harboring latent proviruses (Evans et al., 2018). We therefore analyzed the expression of this exhaustion marker on CD4+ T cells latently and productively infected with pMorpheus-V5. We found that Tim-3 was enriched in latently infected CD4+ T cells compared with productively infected cells as well as negative and mock-infected cells. The Tim-3 enrichment of latently infected cells was more pronounced in cells treated with IL-2 alone than with IL-2 + α-CD3/CD28 (Figures 7A and 7C). When analyzing the different CD4+ T cell populations, we found that the most pronounced differences were observed in the IL-2-only treated CD4+-naive T cells and in the CD4+ TTM, with an average 6.3-fold difference (range 5.9–6.7) and 8.2-fold (range 3.3–13), respectively (Figure 7B). Overall, Tim-3 expression increased in all populations with IL-2 + α-CD3/CD28 treatment except for the mock-infected cells (Figure 7D).
Figure 7.
Investigation of immune exhaustion maker Tim-3 in latently and productively infected cells in different CD4+ T cell subsets
(A) Tim-3 expression on latently and productively pMorpheus-V5-infected primary human CD4+ T cells stimulated with IL-2 was determined using CyTOF. Bar graphs represent levels of Tim-3 in total CD4+ T cells. Each donor is indicated by a unique symbol. The infection status of each CD4+ T cell subset is color coded in the figure. Results are from two independent donors.
(B) IL-2 stimulated CD4+ T cells were infected with pMorpheus-V5. The levels of Tim-3POS in mock-infected, reporter-negative cells, latently infected and productively infected CD4+ T cell subsets stimulated were measured using CyTOF. The average of two individual donors is shown.
(C) Tim-3 expression on latently and productively pMorpheus-V5-infected primary human CD4+ T cells stimulated with IL-2 and α-CD3/CD28 beads was determined using CyTOF. Bar graphs represent levels of Tim-3 in total CD4+ T cells. Each donor is indicated by a unique symbol. Results are from two independent donors.
(D) Tim-3 expression on latently and productively pMorpheus-V5-infected CD4+ T subsets stimulated with IL-2 and α-CD3/CD28 beads was determined. The levels of Tim-3POS in mock-infected, reporter-negative cells, latently infected, and productively infected CD4+ T cell subsets were measured using CyTOF. The average of two individual donors is shown.
Taken together, our CyTOF experiments detail the CD69/HLA-DR and Tim-3 expression profiles of the CD4+ T cell subsets undergoing productive and latent infection. Furthermore, the stark differences in expression of these T cell activation and exhaustion markers represent an important independent confirmation of the ability of pMorpheus-V5 to distinguish productive from latent infections.
Discussion
A more comprehensive understanding of the mechanisms supporting HIV latency is needed to guide HIV cure strategies. The ability to detect and enrich for latently infected primary human cells is essential to dissect mechanisms controlling HIV latency establishment, maintenance, and provirus reactivation. Dual-color HIV reporters are powerful tools to distinguish latently and productively infected cells from uninfected cells (Calvanese et al., 2013; Dahabieh et al., 2014; Battivelli et al., 2018). However, many of these dual-reporter HIV vectors are suboptimal with respect to detecting latently infected cells because of promoter interference between the LTR and the LTR-independent promoter controlling expression of the phenotypic markers for productive and latent infection (Battivelli et al., 2018; Calvanese et al., 2013; Chavez et al., 2015; Dahabieh et al., 2013, 2014; Hashemi et al., 2016; Kim et al., 2019; Matsuda et al., 2015).
In this study, we generated a quadruple-reporter HIV vector (pMorpheus-V5) that efficiently identifies productive or latently infected primary CD4+ T cells (Figures 1A and 1B) with negligible promoter interference (Figures 1C and S2C). In addition, pMorpheus encodes all the accessory proteins, including Nef. Indeed, the down-regulation of the CD4+ receptor exclusively on the surface of cells harboring LTR-active proviruses indicates that pMorpheus-V5 expresses a functional Nef alby Vpu also does contribute somewhat to CD4 down-regulation (Wildum et al., 2006).
Upon infection with pMorpheus-V5, cells harboring latent proviruses are distinguished from productively infected cells using flow cytometry or CyTOF (Figures 2B–2D, 4A, 4C, and 4D). On average, 64% of the non-activated infected cells and 51% of the activated infected cells harbored an LTR-silent provirus (total: six different healthy donors; Figures 2C, 2D, 4C, and 4D; Table S1). These data indicate that the level of latency establishment in primary CD4+ T cells might have been previously under-estimated (Battivelli et al., 2018). This could be because the two reporter expression cassettes in pMorpheus-V5 (LTR-dependent and LTR-independent) are distantly positioned in a manner that minimizes promoter interference. Indeed, some reporter viral vectors (Battivelli et al., 2018) carry both reporters in the Nef ORF, possibly mediating promoter interference between the HIV-1 5′ LTR and the LTR-independent EF1-α promoter (Greger et al., 1998). Furthermore, the strength of the PGK promoter present in pMorpheus-V5, which might overcome the action of epigenetic factors that cause HIV latency through LTR silencing, could contribute to these observations. For instance, data reported about the impact the HUSH complex, that mediates trimethylation of H3K9 on both endogenous and exogenous retro-elements (Tchasovnikarova et al., 2015; Robbez-Masson et al., 2018), and thought to play a major role in latency (Chougui et al., 2018; Yurkovetskiy et al., 2018), show that the PGK promoter, unlike retroviral LTRs and other cellular promoters, is only minimally affected such activity (Tchasovnikarova et al., 2015).
Our data indicate that HIV latency was established upon integration given that “early transcription” of multiply spliced RNA of regulatory proteins, Tat and Rev, is absent in latently infected cells (Figure 3D). Additionally, in both latent and productive infections, we show similar levels of integration (Figure 3C). It is worth noting that a small portion of the cells lacking expression of all four reporters (“negative cell population”) harbor integrated proviruses but lack evidence of HIV multiple splice transcripts. This might be due to the presence of integrated defective proviruses or epigenetic silencing of both promoters. Indeed, defective proviruses are common, especially in patients undergoing antiretroviral treatment (Bruner et al., 2016). A broad range of deletions have been observed in the genome of these proviruses, some of which could still provide antigenic activity (Bruner et al., 2016; Pollack et al., 2017). It will be of interest to determine whether these “reporter-negative CD4+ T cells” display a distinct integration landscape or are enriched for defective proviruses.
We identified the CD4+ T cells subsets most prone to latency establishment using CyTOF (Figures 5A and 5B). By measuring expression of activation and exhaustion markers, CD69/HLA-DR as well as Tim-3, we also present independent validation that V5-NGFR-only positive cells are bona fide latently infected (Figures 6A–6D and 7A–7D). In the future, it will be of interest to study the kinetics of latency establishment and confirm CD4+ T cells subset specific latency by assess to what extent other exhaustion markers, such as PD-1, TIGIT, and LAG-3, shown to be of relevance to the latent reservoir (Fromentin et al., 2016), are enriched in latently infected cells detected by pMorpheus-V5. We found that CD4+ TEM and TREG are the subsets where HIV latency establishment occurred with higher frequency (Figures 5A and 5B). Previous reports show that CD4+ TEM cells contain the largest reservoir in which HIV reactivation has been observed (Kulpa et al., 2019; Buzon et al., 2014). CD4+ TEM cells also harbor more intact proviruses (Hiener et al., 2017), while maintaining a higher proliferation rate and a short half-life (Vukmanovic-Stejic et al., 2006; Farber et al., 2014). TREGs also represent an important HIV reservoir because of their potential to expand in chronically HIV-infected patients (Moreno-Fernandez et al., 2012). Moreover, TREGs contribute more to the HIV cellular reservoir than non-TREGs in HIV-infected patients on antiretroviral therapy (ART) (Tran et al., 2008). Their impact on the rapid establishment of the HIV reservoir could be due to their ability to reverse the immune activation status of CD4+ T cells (Moreno-Fernandez et al., 2012). Although with much lower frequency than for the T central memory (TCM) and TTM compartments, proviruses are almost always detected in naive cells of people with HIV (Chomont et al., 2009; Bacchus et al., 2013; Douek et al., 2002; Lambotte et al., 2002; Zerbato et al., 2016). Similarly, we observed establishment of latency in the CD4+-naive T cells subset following either stimulation or activation (Figures 5A–5C). Indeed, CD4+-naive T cells were the more prone subset to harbor LTR-silent proviruses. These results suggest that CD4+-naive T cells may serve as an important reservoir of LTR-silent proviruses.
The molecular mechanisms supporting these differences require further targeted investigations, which now become feasible using pMorpheus-V5. For example, cell-to-cell transmission, through the formation of virologic synapses is a critical mode of HIV infection (Groot et al., 2008; Hubner et al., 2009; Jolly et al., 2004), as it is exponentially more efficient than cell-free transmission and far more antibody neutralization-resistant (Chen et al., 2007). Recently it has also been reported that cell-to-cell transmission from an activated CD4+ T cell to a resting one preferentially results in latent infection (Agosto et al., 2018). Although the current version of pMorpheus-V5 generates single round viruses, one can envision studies that will involve a replication competent version of our reporter virus that will enable us to gain new insight in latency establishment in the context of HIV cell-to-cell transmission.
Last not least, pMorpheus-V5 will be an excellent tool for screening for drugs targeting cellular signaling networks and epigenetic complexes (Agosto et al., 2015; Gagne et al., 2019) that selectively prevent or reverse latency establishment in primary human CD4+ T cell subpopulations.
Limitations of the study
The pMorpheus system was built with two reporter cassettes inserted in different HIV regions in order to decrease the chance of promoter interference. The PGK promoter was chosen as an LTR-independent promoter because of the constitutive nature of its activity in combination with its low susceptibility to silencing by epigenetic mechanisms such as the HUSH complex (Tchasovnikarova et al., 2015). Nonetheless, there are some characteristics of the system that deserve further clarification in the future. For example, the insertion of the NGFR cassette deleted a cryptic exon, contained in the Env ORF and typically present in HIV IIIB. This cryptic exon forms a fusion protein called Tnv of unknown properties. Its deletion has been reported to be without major consequences (Gottlinger et al., 1992), but this deletion could modify the pMorpheus phenotype. Likewise, it might be of interest to investigate whether cryptic splicing is responsible for the absence of PGK driven Nef expression, as Nef is preceded by an IRES that, theoretically, could lead to its translation. The CyTOF experiments allowed us to begin investigating latency establishment in CD4+ T subset-dependent manner. The results obtained provided us with insights about the potential importance of naive CD4+ T cells in the establishment of the latent reservoir and, moreover, provided independent validation of the nature of pMorpheus-V5 infections. Nonetheless, the CyTOF experiments presented here were performed using cells from only two independent healthy donors, resulting in some expected donor variability (Table S1), and future experiments with additional donors are needed to confirm and extend our observations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD45 (HI30)-Y89 | Fluidigm | Cat#: 3089003B, RRID: AB_2661851 |
| Anti-mCherry 142 Nd, rabbit pAb, coupled in house | Abcam | Cat#: ab167453, RRID: AB_2571870 |
| Anti-human CD45RA (HI100)143Nd | Fluidigm | Cat#: 3143006B, RRID: AB_2651156 |
| Anti-human CD69 (FN50) 144Nd | Fluidigm | Cat#:314401, RRID: AB_2687849 |
| Anti-human CD4 (REA623) 145Nd | Miltenyi Biotec | Cat#: 130-122-283, RRID: AB_2801864 |
| Anti-human CD127 (A019D5) 149Sm | Fluidigm | Cat#: 3149011B, RRID: AB_2661792 |
| Anti-mouse CD24 (M1/69) 150Nd | Fluidigm | Cat#: SKU 3150009B, RRID: AB_2916042 |
| Anti-V5 tag (R960-25) 152Sm, coupled in house | ThermoFisher | Cat#: R960-25, RRID: AB_2556564 |
| Anti-human TIM-3 (F38-2E2) 154Sm | Fluidigm | Cat#: 3154010B, RRID: AB_2893002 |
| Anti-human CD27 (REA499) 155Gd, coupled in house | Miltenyi Biotec | Cat#: 130122295, RRID: AB_2801876 |
| Anti-human Foxp3 (PCH101) 162Dy | Fluidigm | Cat#: 3162011A, RRID: 2687650 |
| Anti-human CD197 (CCR7) (REA546) 167Er, coupled in house | Miltenyi Biotec | Cat#: 130-122-300, RRID: AB_2801881 |
| Anti-human CD3 (REA613) 168Er, coupled in house | Miltenyi Biotec | Cat#: 130-113-138, RRID: AB_2725966 |
| Anti-human CD95 (DX2) 171Yb, coupled in house | Biolegned | Cat#: 305602, RRID: AB_314540 |
| Anti-human HLA-DR 174 Yb, clone REA805 | Miltenyi Biotec | Cat#: 130-122-299, RRID: AB_2801880 |
| Anti-human CD45RO (UCHL1) 176Yb, coupled in house | Biolegned | Cat#: 304202, RRID: AB_314418 |
| COULTER CLONE KC57-RD1 165Ho, coupled in house | Beckman Coulter | Cat#: 6604667, RRID: AB_1575989 |
| Anti-human Alexa Fluor 647 CD271 (C40-1457) | BD Pharmingen | Cat#: 560326, RRID: AB_1645403 |
| Anti-V5 tag monoclonal antibody (2F11F7) Alexa Fluor 488 | ThermoFisher | Cat#: 37-7500-A488, RRID: AB_2610630 |
| Bacterial and virus strains | ||
| pMorpheus-V5 HIV viral vector | This paper | n/a |
| Biological samples | ||
| Human peripheral mononuclear cells, PBMC (Leukopak) | New York Blood Bank | n/a |
| Chemicals, peptides, and recombinant proteins | ||
| Dextran | Millipore Sigma | Cat#: 9004-54-0 |
| Bovine Serum Albumin (BSA) | ThermoFisher | Cat#: A1113803 |
| Human recombinant IL-2 | NIH AIDS Reagent | Cat#: 136 |
| Dynabeads human T- Activator CD3/CD28 | Thermo Fisher Scientific | Cat#: 11161D, AB_2916088 |
| Cell-ID Intercalator-103 Rh | Fluidigm | Cat#: 201103A |
| Cell-ID IdU-127I | Fluidigm | Cat#: 201127 |
| Cell-ID Intercalator- 191 Ir | Fluidigm | Cat#: 201192A |
| Cell-ID Intercalator- 193 Ir | Fluidigm | Cat#: 201192A |
| Critical commercial assays | ||
| Human CD4+ T cell isolation Kit | Miltenyi Biotec | Cat#: 130-096-533, RRID: AB_2916089 |
| LIVE/DEAD Fixable dead cell stain kits | Invitrogen | Cat#: L10119 |
| Experimental models: Cell lines | ||
| Human: HEK293T | ATCC | ATCC#: CRL-3216, RRID: CVCL_0063 |
| Oligonucleotides | ||
| Oligonucleotides, primers and probes used in this study | This paper | See Table S2 |
| Recombinant DNA | ||
| pMorpheus-V5 | This paper | n/a |
| Software and algorithms | ||
| FlowJo | FlowJo, LLC | http://www.flowjo.com, RRID: SCR_008520 |
| PRISM 9.0 | Graphpad | http://graphpad.com, RRID: SCR_005375 |
| Cytobank | (Kotecha et al., 2010) | http://www.cytobank.org, RRID: SCR_014043 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Viviana Simon (Viviana.simon@mssm.edu).
Materials availability
Availability of the plasmid encoding the pMorpheus-V5 reporter construct is subject to Material Transfer Agreements.
Experimental model and subject details
Primary CD4+ T cell isolation and cell culture
Leukopaks were obtained from anonymous healthy blood donors from the New York Blood Center. Peripheral blood mononuclear cells were purified by Ficoll paque (GE Healthcare) density centrifugation and primary CD4+ T cells were purified by negative selection of CD4+ cells using CD4+ T cell isolation kit (Miltenyi Biotec) according to the manufacturer’s instructions. CD4+ T cells were cultured in RPMI 1640 supplemented with 10% FBS (Gibco/Thermo Fisher), 100 IU/mL penicillin, 100 μg/mL streptomycin, 0.1 M HEPES, 2 mM L-glutamine in the presence of recombinant human IL-2 (20 U/mL) (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH) at 37°C in a 5% CO2 humidified incubator.
Method details
Construction of pMorpheus-V5 and virus stock production
The backbone of the pLAI2-V5-NGFR-HSA-mCherry-IRES-Nef plasmid (renamed in the text to pMorpheus-V5) is based on the pLAI2 HSA-mCherry-IRES-Nef full length viral construct (Young et al., 2018), which encodes the full-length HIV-1 strain LAI (Peden et al., 1991) as well as the HSA-mCherry-IRES-Nef cassette in the Nef ORF. The cassette encoding the PGK promoter followed by V5-NGFR (Wroblewska et al., 2018) was inserted in the envelope region encompassing the SalI and SmaI restriction sites. The construct was verified by sequencing. The virus stocks were produced by transfecting HEK 293T cells with 20 μg of pLAI2-V5-NGFR-HSA-mCherry-IRES-Nef and 5 μg of expression plasmid encoding the Env of pSV III-92 HT 593.1 subtype B virus R5 aid X4 (dual-tropic). After approximately 20 h, the transfection medium was changed, and the viral supernatants were collected at 48 and 72 hrs after transfection. The viral supernatants were first filtered (0.45 μm) and then concentrated using the Lenti-X concentrator (Takara) following manufacturer’s instruction.
The p24 Gag concentration were determined by p24 ELISA (XpressBio) according to the manufacturers’ instructions. The optical density was read at 450 nm using a microtitration plate reader (VICTOR3TM, Perkin Elmer Precisely). Viral stocks were tested in triplicate. Aliquots of the pMorpheus-V5 pseudotyped with the HIV Env were stored at −80°C until use.
Infection of primary human CD4+ T cells with pMorpheus-V5
Primary human CD4+ T cells were incubated with 20 U/mL interleukin-2 (IL-2) alone or in combination with α-CD3/CD28 antibody-coated beads (100 μL/30 × 106 cells) (Gibco/Thermo Fisher) for 72 hours. Prior to infection, α-CD3/CD28 antibody-coated beads were removed as directed by the product manufacturer, and cells were infected with 100 ng of p24 equivalent of pseudotyped pMorpheus-V5. For each donor, approximately ∼1 × 106 cells of non-activated or activated cells were infected by spinoculation at 1,200 rpm for 2 hrs with equal amount of pMorpheus-V5 in presence of diethylaminoethyl (DEAE)-dextran (0.2 μg/mL). After the spinoculation, the cells were washed and cultured in the presence of IL-2 (20 U/mL) for five days. The number of infected cells were determined by quantifying NGFR, V5, mCherry, p24-expressing cells by Flow cytometry or by CyTOF.
As a negative control, we included cells treated with 2 μM Raltegravir (NIH HIV Reagent Program) in some experiments.
Flow cytometry analysis
After infection with pMorpheus-V5, cells were stained with the LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Thermo Fisher Scientific) for 30 min at 4°C. Cells were washed and stained with antibodies against cell surface molecules in PBS containing 4% of human serum (Valley Biomedical). NGFR was detected with a 20 μL per million cells of Alexa Flour 647 conjugated α-NGFR (BD Bioscience), and V5 detected with a 1:100 dilution of fluorescein isothiocyanate (FITC)-conjugated α-mV5 (Invitrogen) in FACS buffer. Prior to flow cytometry analysis, cells were fixed in 1% (vol/vol) formaldehyde for 30 min at 4°C and analyzed on an LSRFortessa flow cytometer (BD Biosciences). For intracellular p24 staining, cells were permeabilized for 30 min at 4°C with Perm/Wash buffer (BD Biosciences) and stained with α-p24 KC57 antibody (Beckman Coulter) for an additional 45 min at room temperature. Cells were washed and resuspended in PBS for subsequent flow cytometry analysis. CD4 downregulation was measured using PE-Cy5 antibody (Biolegend) with a 5 μL per million cells in 100 μL FACS buffer by flow cytometry. The viability of cells was first determined by the LIVE/DEAD Fixable Dead Cell stain (Thermo Fisher Scientific) with forward vs side scatter (FSC versus SSC). Data were gated on single CD3+ CD8- cells, and CD4 downregulation was assessed on cells expressing level of NGFR, V5, mCherry or p24. Gates were set by comparison with uninfected cells stained as above. An average of 1 × 105 cells were acquired per sample, and data were analyzed using FlowJo software v10.7 (FlowJo, LLC).
Measurements of HIV integration and transcription in FACS sorted CD4+ T cells
Viral stocks were pretreated with Dnase I (10 U/mL, New England Biolabs Inc, MA, USA) and Benzonase nuclease (50 U/mL, Millipore sigma) at room temperature for 1 hour before infection to remove background signal from plasmid DNA. Primary human CD4+ T cells stimulated with IL-2 alone or with α-CD3/CD28 antibody-coated beads (Gibco/Thermo Fisher) were infected for 5 days. Infected cells treated with 2 μM of Raltegravir were used as negative control. Activated J-Lat 6.3 T cells were used as a positive control (Jordan et al., 2003). The J-Lat 6.3 cells were activated with phorbol 12-myristate 13-acetate (PMA, 16 nM, Sigma-Aldrich) and Ionomycin (0.5 μM, Sigma) (Spina et al., 2013).
pMorpheus-V5 infected cells were stained with LIVE/DEAD Fixable Near-IR Dead Cell Stain (Thermo Fisher Scientific) or V5-FITC Abs (Invitrogen) with a 1:100 dilution. Latently infected cells (V5pos population), productively infected cells (mCherrypos/V5pos population), and negative cells (V5neg/mCherryneg population) were sorted with a BD FACS Aria II Cell Sorter (BD Biosciences). After sorting the specific cell populations, cell pellets were lysed to isolate DNA and RNA using AllPrep DNA/RNA Kits (QIAGEN, Valencia, CA, USA) following the manufacturer’s protocol.
We measured HIV integration by Alu-1 PCR (Butler et al., 2001; Tan et al., 2006). Briefly, each PCR reaction contained 100 ng of DNA from each donor sample and 300 nM LM667 and 100 nM Alu-1 primers, and CloneAMP master mix (Takara). The PCR cycles initial denaturation at 95°C for 5 mins, then 20 cycles of 95°C for 10 secs, 55°C for 10 secs, and 72°C for 170 secs, and final extension of 72°C for 5 mins. We then diluted 1:5 the amplicons from the first round of PCR. Each nested real-time PCR reaction included 2 μL of the diluted amplicons, and 300 nM each of primers LR1 and LR2, and 100 nM of ZXF-P probe containing Taqman universal master mix (PE-Applied Biosystems) and amplified on a LightCycler (LightCycler 480 II, Roche Life Science, Roche Diagnostics Corporation, IN, USA). After incubation at 50°C for 2 min followed by 95°C for 10 min, 40 cycles of amplification were carried that included 15 sec at 95°C followed by 90 sec at 60°C. The 1 μL RNAseP qPCR Probe reaction (Integrated DNA technologies), which included the 1 μL DNA from each sample and Taqman universal master mix, was included as an internal control. The relative abundance of the integration events was calculated by ΔCt method (RnaseP-sample) and shown on a log2 scale.
Multiply spliced HIV transcripts products were detected by semi-nested real-time quantitative-PCR as previously described (Pasternak et al., 2008; Zerbato et al., 2021). Briefly, we synthesized first-strand cDNA from 100 ng of total RNA isolated from the sorted samples using iScript™ cDNA synthesis kit (BioRad), following manufacturer’s instructions, 25°C for 5 mins, 46°C for 20 mins, 95°C for 1 min. Two rounds of PCR were performed with 2 μL of cDNA, 10 μL of CloneAMP (Takara), and 300 nM each of primers ks1 and mf83. The PCR conditions were: 15 cycles of 94°C for 3 mins, 55°C for 30 secs, 72°C for 1 min, and followed by final extension of 72°C for 5 mins. Subsequently, the semi-nested real-time qPCR reactions (LightCycler 480 II) were performed with 1:5-diluted 1st PCR product, and 300 nM of each primer mf83 and mf84, and 100 nM of the probe ks2-tq. Each reaction contained Taqman universal master mix (PE-Applied Biosystems). After incubation at 50°C for 2 min followed by 95°C for 10 min, 45 cycles of amplification were carried out including 15 sec at 95°C followed by 30 sec at 60°C. The β-actin reaction, which included the 2 μL cDNA of each sample, 400 nM BGF, BGR primers, 300 nM BGX-P and Taqman universal master mix, was used as an internal control. Table S1 summarizes the primers for the first PCR assay and subsequent nested qPCR assay. The quantity of multiply spliced HIV transcripts was determined by delta-Ct method (β-actin-sample) and shown on a log2 scale.
Mass cytometry time-of-flight (CyTOF)
Ten million uninfected and infected human primary CD4+ T cells were stained with the Iridium (Ir) DNA intercalator, IdU_Ir193Di, to analyze DNA content (Behbehani et al., 2012) and with the Rhodium (Rh) cationic nucleic acid intercalator, Rh103Di, to measure cellular viability. The cells were then washed in Cell Staining Media (CSM) buffer (0.5% BSA+0.02% NaN3 in PBS), stained with 2X CD298+ ß2M antibodies for barcoding (CD45_Cd111Di, CD45_Cd112Di, CD45_Cd114Di, CD45_Cd116Di, CD45_Pt195Di, CD45_Pt196Di), and pooled. The cells were washed with CSM buffer and resuspended in the membrane antibody mixture containing CD45RA_Nd143Di, CD45RO_Yb176Di, CD3_Er168DI, CD4_Nd145Di, CD24_Nd150Di, CD45_Y89Di, CD69_Nd144Di, CD127_Sm149Di, CD27_Gd155Di, CCR7_Gd156Di, CD25_Tm169Di, CD95_Yb171Di, Tim-3_Sm149Di, HLA-DR_Yb174Di. After 30 min, the cells were washed in CSM. For fixation and permeabilization, the cells were fixed with paraformaldehyde in PBS for 10 min, followed by dropwise addition of cold methanol and storage at −80°C. The cells were washed in CSM containing heparin. The cells were washed in CSM and stained with intracellular antibody mixture containing FoxP3_Dy162Di, NGFR_Er170Di, V5_Sm151Di, mCherry_Nd140Di, HSA_Nd150Di, p24_KC57-RD_Ho165Di (BECKMAN COULTER) for 30 min on ice. The cells were washed and resuspended in Fix solution containing iridium intercalator. Between 200,000 and 550,000 CD4+ T cells were acquired per individual sample on the CyTOF2 or Helios mass cytometer (both Fluidigm). CyTOF settings were parameterized following the quality control of the instrument. After acquisition, the data were normalized using bead-based normalizing using CyTOF software (should list). The data was gated to exclude residual normalization beads, debris, and dead cells for clustering and high dimensional analysis. On acquisition, gating was done manually using the dot plot display on FlowJo or Cytobank. We gated only live cells for further analysis based on cellular viability (Rh103) and DNA content (Ir193). Seven different CD4+ T cell subpopulations were identified: naïve T cells, stem cell memory (TSCM), central memory (TCM), effector memory (TEM), transitional memory (TTM), T regulatory (TREG), and effector memory T cells re-expressing CD45RA (TEMRA) cells (Figure S3E). Figures S3B–S3D depicts the gating strategy for the CD45RO+ memory compartment. Briefly we first gated on the CD45RO cells followed by gating on CD27 and CCR7 to discriminate between TCM, TEM, and TTM (Figure S3B) (Flynn et al., 2014). We distinguished naïve T cells from TSCM using the CD95 gate on CD45RA+, CCR7+, and CD27+ (Figure S3C) (Tabler et al., 2014; Klatt et al., 2014). TREG were identified as being CD127-, Foxp3+ (Ndure et al., 2017), and TEMRA (Tian et al., 2017) were identified by CCR7-, and CD45RA+ (Figure S3D) as previously described. The markers used to distinguish for each CD4+ T cell subset is in summarized Figure S3E.
viSNE analysis
CyTOF data were debarcoded using Single Cell Debarcoder (Zunder et al., 2015) using post-assignment debarcode stringency filter and outlier trimming. We applied Cytobank viSNE (visualized t-stochastic neighbor embedding) (Amir et al., 2013) and the Barnes-Hut implementation of the t-SNE algorithm to generate all viSNE plots of uninfected and pMorpheus-V5 infected cells from differently stimulated CD4+ T cells. The viSNE plots show the distribution of productively or latently infected cells present in each CD4+ T cell subpopulation based on an equal sampling of 66,000 cells from each file for cells stimulated with IL-2 alone or IL-2 and α-CD3/CD28 antibody-coated beads. In the viSNE plots, the position of each dot represents an individual cell. Dot-plot visualizations were performed using Cytobank software (Mountain View, CA).
Quantification and statistical analysis
Statistical analyses were performed using Prism 9 software (GraphPad). All results are presented as mean +/- standard deviation. We used either t-tests or ANOVA for the comparison of the flow cytometry data. p values are provided in the text as well as in the figures.
Acknowledgments
We thank Dr. Adeeb Rahman and his team at the Human Immune Monitoring Core of Icahn School of Medicine at Mount Sinai for expertise and assistance with the CyTOF experiments. The following reagents were obtained through the National Institutes of Health (NIH) HIV Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID): J-Lat Full Length Cells (6.3), ARP-9846, contributed by Dr. Eric Verdin, and raltegravir (Isentress; MK-0518), ARP-11680, contributed by Merck & Company, Inc. The graphical abstract and Figures 2A and 3A were created using BioRender.com. This research was supported in part by NIH/NIAID grant R01AI136916 (V.S.) and NIH grant AI150355 (L.C.F.M).
Author contributions
E.H.K. and V.S. designed the research. E.H.K. performed the experiments. L.C.F.M. designed the proviral vectors. E.H.K. performed and analyzed data. E.H.K. generated figures with the input of L.C.F.M., L.M., and V.S. E.H.K., L.C.F.M., and V.S. interpreted the data. B.D.B. provided critical reagents. M.S. and L.M. provided critical insight for flow data analysis and interpretation. E.H.K. drafted the manuscript. E.H.K., L.C.F.M., L.M., and V.S. revised the final manuscript. All authors approved the final version.
Declaration of interests
The authors declare no competing interests.
Published: June 13, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100238.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this manuscript is available from the lead contact upon request.
References
- Agosto L.M., Gagne M., Henderson A.J. Impact of chromatin on HIV replication. Genes. 2015;6:957–976. doi: 10.3390/genes6040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto L.M., Herring M.B., Mothes W., Henderson A.J. HIV-1-Infected CD4+ T cells facilitate latent infection of resting CD4+ T cells through cell-cell contact. Cell Rep. 2018;24:2088–2100. doi: 10.1016/j.celrep.2018.07.079. [DOI] [PubMed] [Google Scholar]
- Amir E.a.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., Pe'er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchus C., Cheret A., Avettand-Fenoel V., Nembot G., Melard A., Blanc C., Lascoux-Combe C., Slama L., Allegre T., Allavena C., et al. A single HIV-1 cluster and a skewed immune homeostasis drive the early spread of HIV among resting CD4+ cell subsets within one month post-infection. PLoS One. 2013;8:e64219. doi: 10.1371/journal.pone.0064219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battivelli E., Dahabieh M.S., Abdel-Mohsen M., Svensson J.P., Tojal Da Silva I., Cohn L.B., Gramatica A., Deeks S., Greene W.C., Pillai S.K., Verdin E. Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4(+) T cells. Elife. 2018;7 doi: 10.7554/elife.34655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbehani G.K., Bendall S.C., Clutter M.R., Fantl W.J., Nolan G.P. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner K.M., Murray A.J., Pollack R.A., Soliman M.G., Laskey S.B., Capoferri A.A., Lai J., Strain M.C., Lada S.M., Hoh R., et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016;22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler S.L., Hansen M.S., Bushman F.D. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Buzon M.J., Sun H., Li C., Shaw A., Seiss K., Ouyang Z., Martin-Gayo E., Leng J., Henrich T.J., Li J.Z., et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat. Med. 2014;20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Gao H., Zhao J., Hu S., Liang X., Yang Y., Dai Z., Hong Z., Deng K. Infection with a newly designed dual fluorescent reporter HIV-1 effectively identifies latently infected CD4(+) T cells. Elife. 2021;10:e63810. doi: 10.7554/elife.63810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V., Chavez L., Laurent T., Ding S., Verdin E. Dual-color HIV reporters trace a population of latently infected cells and enable their purification. Virology. 2013;446:283–292. doi: 10.1016/j.virol.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrois M., Banerjee T., Mukherjee G., Raman N., Hussien R., Rodriguez B.A., Vasquez J., Spitzer M.H., Lazarus N.H., Jones J.J., et al. Mass cytometric analysis of HIV entry, replication, and remodeling in tissue CD4+ T cells. Cell Rep. 2017;20:984–998. doi: 10.1016/j.celrep.2017.06.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez L., Calvanese V., Verdin E. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Hubner W., Spinelli M.A., Chen B.K. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 2007;81:12582–12595. doi: 10.1128/jvi.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N., El-Far M., Ancuta P., Trautmann L., Procopio F.A., Yassine-Diab B., Boucher G., Boulassel M.R., Ghattas G., Brenchley J.M., et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chougui G., Munir-Matloob S., Matkovic R., Martin M.M., Morel M., Lahouassa H., Leduc M., Ramirez B.C., Etienne L., Margottin-Goguet F. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat. Microbiol. 2018;3:891–897. doi: 10.1038/s41564-018-0179-6. [DOI] [PubMed] [Google Scholar]
- Churchill M.J., Deeks S.G., Margolis D.M., Siliciano R.F., Swanstrom R. HIV reservoirs: what, where and how to target them. Nat. Rev. Microbiol. 2016;14:55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- Colin L., Van Lint C. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology. 2009;6:111. doi: 10.1186/1742-4690-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahabieh M.S., Ooms M., Brumme C., Taylor J., Harrigan P.R., Simon V., Sadowski I. Direct non-productive HIV-1 infection in a T-cell line is driven by cellular activation state and NFkappaB. Retrovirology. 2014;11:17. doi: 10.1186/1742-4690-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahabieh M.S., Ooms M., Simon V., Sadowski I. A doubly fluorescent HIV-1 reporter shows that the majority of integrated HIV-1 is latent shortly after infection. J. Virol. 2013;87:4716–4727. doi: 10.1128/jvi.03478-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek D.C., Brenchley J.M., Betts M.R., Ambrozak D.R., Hill B.J., Okamoto Y., Casazza J.P., Kuruppu J., Kunstman K., Wolinsky S., et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- Evans V.A., Van Der Sluis R.M., Solomon A., Dantanarayana A., Mcneil C., Garsia R., Palmer S., Fromentin R., Chomont N., Sekaly R.P., et al. Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS. 2018;32:1491–1497. doi: 10.1097/qad.0000000000001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber D.L., Yudanin N.A., Restifo N.P. Human memory T cells: generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.K., Paukovics G., Cashin K., Borm K., Ellett A., Roche M., Jakobsen M.R., Churchill M.J., Gorry P.R. Quantifying susceptibility of CD4+ stem memory T-cells to infection by laboratory adapted and clinical HIV-1 strains. Viruses. 2014;6:709–726. doi: 10.3390/v6020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromentin R., Bakeman W., Lawani M.B., Khoury G., Hartogensis W., Dafonseca S., Killian M., Epling L., Hoh R., Sinclair E., et al. CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne M., Michaels D., Schiralli Lester G.M., Gummuluru S., Wong W.W., Henderson A.J. Strength of T cell signaling regulates HIV-1 replication and establishment of latency. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Morrison S.G., Robertson D.L., Thornton C.L., Craig S., Karlsson G., Sodroski J., Morgado M., Galvao-Castro B., Von Briesen H., et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID networks for HIV isolation and characterization. J. Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J.V., Miller A.D. Serine phosphorylation-independent downregulation of cell-surface CD4 by Nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- Gottlinger H.G., Dorfman T., Cohen E.A., Haseltine W.A. The role of the tnv protein and tnv RNA splicing signals in replication of HIV-1 IIIB isolates. Virology. 1992;189:618–628. doi: 10.1016/0042-6822(92)90585-d. [DOI] [PubMed] [Google Scholar]
- Grau-Exposito J., Luque-Ballesteros L., Navarro J., Curran A., Burgos J., Ribera E., Torrella A., Planas B., Badia R., Martin-Castillo M., et al. Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger I.H., Demarchi F., Giacca M., Proudfoot N.J. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 1998;26:1294–1301. doi: 10.1093/nar/26.5.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot F., Welsch S., Sattentau Q.J. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood. 2008;111:4660–4663. doi: 10.1182/blood-2007-12-130070. [DOI] [PubMed] [Google Scholar]
- Guy B., Kieny M.P., Riviere Y., Peuch C.L., Dott K., Girard M., Montagnier L., Lecocq J.P. HIV F/3' orf encodes a phosphorylated GTP-binding protein resembling an oncogene product. Nature. 1987;330:266–269. doi: 10.1038/330266a0. [DOI] [PubMed] [Google Scholar]
- Hashemi F.B., Barreto K., Bernhard W., Hashemi P., Lomness A., Sadowski I. HIV provirus stably reproduces parental latent and induced transcription phenotypes regardless of the chromosomal integration site. J. Virol. 2016;90:5302–5314. doi: 10.1128/jvi.02842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiener B., Horsburgh B.A., Eden J.S., Barton K., Schlub T.E., Lee E., Von Stockenstrom S., Odevall L., Milush J.M., Liegler T., et al. Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep. 2017;21:813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner W., Mcnerney G.P., Chen P., Dale B.M., Gordon R.E., Chuang F.Y.S., Li X.D., Asmuth D.M., Huser T., Chen B.K. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferys S.R., Burgos S.D., Peterson J.J., Selitsky S.R., Turner A.M.W., James L.I., Tsai Y.H., Coffey A.R., Margolis D.M., Parker J., Browne E.P. Epigenomic characterization of latent HIV infection identifies latency regulating transcription factors. PLoS Pathog. 2021;17:e1009346. doi: 10.1371/journal.ppat.1009346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Kashefi K., Hollinshead M., Sattentau Q.J. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 2004;199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Peng P.D., Yang S., Hsu C., Cohen C.J., Zhao Y., Abad J., Zheng Z., Rosenberg S.A., Morgan R.A. Lentiviral vector design for optimal T cell receptor gene expression in the transduction of peripheral blood lymphocytes and tumor-infiltrating lymphocytes. Hum. Gene Ther. 2009;20:630–640. doi: 10.1089/hum.2008.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A., Bisgrove D., Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Cameron P.U., Lewin S.R., Anderson J.L. Limitations of dual-fluorescent HIV reporter viruses in a model of pre-activation latency. J. Int. AIDS Soc. 2019;22:e25425. doi: 10.1002/jia2.25425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt N.R., Bosinger S.E., Peck M., Richert-Spuhler L.E., Heigele A., Gile J.P., Patel N., Taaffe J., Julg B., Camerini D., et al. Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N., Krutzik P.O., Irish J.M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 2010 doi: 10.1002/0471142956.cy1017s53. Chapter 10, Unit10 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa D.A., Talla A., Brehm J.H., Ribeiro S.P., Yuan S., Bebin-Blackwell A.G., Miller M., Barnard R., Deeks S.G., Hazuda D., et al. Differentiation into an effector memory phenotype potentiates HIV-1 latency reversal in CD4(+) T cells. J. Virol. 2019;93 doi: 10.1128/jvi.00969-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambotte O., Demoustier A., De Goer M.G., Wallon C., Gasnault J., Goujard C., Delfraissy J.F., Taoufik Y. Persistence of replication-competent HIV in both memory and naive CD4 T cell subsets in patients on prolonged and effective HAART. AIDS. 2002;16:2151–2157. doi: 10.1097/00002030-200211080-00007. [DOI] [PubMed] [Google Scholar]
- Marini A., Harper J.M., Romerio F. An in vitro system to model the establishment and reactivation of HIV-1 latency. J. Immunol. 2008;181:7713–7720. doi: 10.4049/jimmunol.181.11.7713. [DOI] [PubMed] [Google Scholar]
- Matsuda Y., Kobayashi-Ishihara M., Fujikawa D., Ishida T., Watanabe T., Yamagishi M. Epigenetic heterogeneity in HIV-1 latency establishment. Sci. Rep. 2015;5:7701. doi: 10.1038/srep07701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavigner M., Habib J., Deleage C., Rosen E., Mattingly C., Bricker K., Kashuba A., Amblard F., Schinazi R.F., Lawson B., et al. Simian immunodeficiency virus persistence in cellular and anatomic reservoirs in antiretroviral therapy-suppressed infant rhesus macaques. J. Virol. 2018;92 doi: 10.1128/jvi.00562-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye U., Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454–455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Fernandez M.E., Presicce P., Chougnet C.A. Homeostasis and function of regulatory T cells in HIV/SIV infection. J. Virol. 2012;86:10262–10269. doi: 10.1128/jvi.00993-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moso M.A., Anderson J.L., Adikari S., Gray L.R., Khoury G., Chang J.J., Jacobson J.C., Ellett A.M., Cheng W.J., Saleh S., et al. HIV latency can be established in proliferating and nonproliferating resting CD4+ T cells in vitro: implications for latency reversal. AIDS. 2019;33:199–209. doi: 10.1097/qad.0000000000002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndure J., Noho-Konteh F., Adetifa J.U., Cox M., Barker F., Le M.T., Sanyang L.C., Drammeh A., Whittle H.C., Clarke E., et al. Negative correlation between circulating CD4(+)FOXP3(+)CD127(-) regulatory T cells and subsequent antibody responses to infant measles vaccine but not diphtheria-tetanus-pertussis vaccine implies a regulatory role. Front. Immunol. 2017;8:921. doi: 10.3389/fimmu.2017.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Adema K.W., Bakker M., Jurriaans S., Berkhout B., Cornelissen M., Lukashov V.V. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J. Clin. Microbiol. 2008;46:2206–2211. doi: 10.1128/jcm.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R., Kim Y.K., Hokello J., Lassen K., Friedman J., Tyagi M., Karn J. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 2008;82:12291–12303. doi: 10.1128/jvi.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K., Emerman M., Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- Pollack R.A., Jones R.B., Pertea M., Bruner K.M., Martin A.R., Thomas A.S., Capoferri A.A., Beg S.A., Huang S.H., Karandish S., et al. Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe. 2017;21:494–506.e4. doi: 10.1016/j.chom.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbez-Masson L., Tie C.H.C., Conde L., Tunbak H., Husovsky C., Tchasovnikarova I.A., Timms R.T., Herrero J., Lehner P.J., Rowe H.M. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018;28:836–845. doi: 10.1101/gr.228171.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu G.K., Lee K., Ji J., Braciale V., Baron S., Cloyd M.W. A novel in vitro system to generate and study latently HIV-infected long-lived normal CD4+ T-lymphocytes. Virology. 2006;355:127–137. doi: 10.1016/j.virol.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Siliciano R.F., Greene W.C. HIV latency. Cold Spring Harb. Perspect. Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina C.A., Anderson J., Archin N.M., Bosque A., Chan J., Famiglietti M., Greene W.C., Kashuba A., Lewin S.R., Margolis D.M., et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler C.O., Lucera M.B., Haqqani A.A., Mcdonald D.J., Migueles S.A., Connors M., Tilton J.C. CD4+ memory stem cells are infected by HIV-1 in a manner regulated in part by SAMHD1 expression. J. Virol. 2014;88:4976–4986. doi: 10.1128/jvi.00324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Dong Z., Wilkinson T.A., Barbas C.F., 3rd, Chow S.A. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 2006;80:1939–1948. doi: 10.1128/jvi.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchasovnikarova I.A., Timms R.T., Matheson N.J., Wals K., Antrobus R., Gottgens B., Dougan G., Dawson M.A., Lehner P.J. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–1485. doi: 10.1126/science.aaa7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Babor M., Lane J., Schulten V., Patil V.S., Seumois G., Rosales S.L., Fu Z., Picarda G., Burel J., et al. Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat. Commun. 2017;8:1473. doi: 10.1038/s41467-017-01728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.A., De Goer De Herve M.G., Hendel-Chavez H., Dembele B., Le Nevot E., Abbed K., Pallier C., Goujard C., Gasnault J., Delfraissy J.F., et al. Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PLoS One. 2008;3:e3305. doi: 10.1371/journal.pone.0003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venanzi Rullo E., Pinzone M.R., Cannon L., Weissman S., Ceccarelli M., Zurakowski R., Nunnari G., O'doherty U. Persistence of an intact HIV reservoir in phenotypically naive T cells. JCI Insight. 2020;5 doi: 10.1172/jci.insight.133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukmanovic-Stejic M., Zhang Y., Cook J.E., Fletcher J.M., Mcquaid A., Masters J.E., Rustin M.H., Taams L.S., Beverley P.C., Macallan D.C., Akbar A.N. Human CD4+ CD25hi Foxp3+ regulatory T cells are derived by rapid turnover of memory populations in vivo. J. Clin. Invest. 2006;116:2423–2433. doi: 10.1172/jci28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L.S., Dar R.D., Simpson M.L. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat. Genet. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- Wildum S., Schindler M., Munch J., Kirchhoff F. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J. Virol. 2006;80:8047–8059. doi: 10.1128/jvi.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewska A., Dhainaut M., Ben-Zvi B., Rose S.A., Park E.S., Amir E.A.D., Bektesevic A., Baccarini A., Merad M., Rahman A.H., Brown B.D. Protein barcodes enable high-dimensional single-cell CRISPR screens. Cell. 2018;175:1141–1155.e16. doi: 10.1016/j.cell.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G.R., Terry S.N., Manganaro L., Cuesta-Dominguez A., Deikus G., Bernal-Rubio D., Campisi L., Fernandez-Sesma A., Sebra R., Simon V., Mulder L.C.F. HIV-1 infection of primary CD4(+) T cells regulates the expression of specific human endogenous retrovirus HERV-K (HML-2) elements. J. Virol. 2018;92 doi: 10.1128/jvi.01507-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Guney M.H., Kim K., Goh S.L., Mccauley S., Dauphin A., Diehl W.E., Luban J. Primate immunodeficiency virus proteins Vpx and Vpr counteract transcriptional repression of proviruses by the HUSH complex. Nat. Microbiol. 2018;3:1354–1361. doi: 10.1038/s41564-018-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbato J.M., Khoury G., Zhao W., Gartner M.J., Pascoe R.D., Rhodes A., Dantanarayana A., Gooey M., Anderson J., Bacchetti P., et al. Multiply spliced HIV RNA is a predictive measure of virus production ex vivo and in vivo following reversal of HIV latency. EBioMedicine. 2021;65:103241. doi: 10.1016/j.ebiom.2021.103241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbato J.M., Mcmahon D.K., Sobolewski M.D., Mellors J.W., Sluis-Cremer N. Naive CD4+ T cells harbor a large inducible reservoir of latent, replication-competent human immunodeficiency virus type 1. Clin. Infect. Dis. 2019;69:1919–1925. doi: 10.1093/cid/ciz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbato J.M., Serrao E., Lenzi G., Kim B., Ambrose Z., Watkins S.C., Engelman A.N., Sluis-Cremer N. Establishment and reversal of HIV-1 latency in naive and central memory CD4+ T cells in vitro. J. Virol. 2016;90:8059–8073. doi: 10.1128/jvi.00553-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder E.R., Finck R., Behbehani G.K., Amir E.a.D., Krishnaswamy S., Gonzalez V.D., Lorang C.G., Bjornson Z., Spitzer M.H., Bodenmiller B., et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this manuscript is available from the lead contact upon request.