Abstract
Deep learning has demonstrated remarkable performance in the medical domain, with accuracy that rivals or even exceeds that of human experts. However, it has a significant problem that these models are “black-box” structures, which means they are opaque, non-intuitive, and difficult for people to understand. This creates a barrier to the application of deep learning models in clinical practice due to lack of interpretability, trust, and transparency. To overcome this problem, several studies on interpretability have been proposed. Therefore, in this paper, we comprehensively review the interpretability of deep learning in medical diagnosis based on the current literature, including some common interpretability methods used in the medical domain, various applications with interpretability for disease diagnosis, prevalent evaluation metrics, and several disease datasets. In addition, the challenges of interpretability and future research directions are also discussed here. To the best of our knowledge, this is the first time that various applications of interpretability methods for disease diagnosis have been summarized.
Keywords: Deep learning, Interpretability methods, Applications, Disease diagnosis
Introduction
In recent years, deep learning has become one of the mainstream approaches widely used in various areas, such as natural language processing [1–3], image processing [4–6], etc. Its performance often far exceeds that of traditional machine learning. In terms of the medical field, a variety of high-performance computer-aided diagnosis systems (CAD) based on deep learning have emerged [7–9], which can not only accurately identify diseases but also enhance diagnostic efficiency. However, due to the fact that the deep learning model is a “black-box” structure, which is opaque, non-intuitive, and difficult for people to understand the decision-making process, this directly leads to these advanced models being somewhat limited in clinical practice. As such, the interpretability of deep learning models has become a potentially hot topic, receiving increasing attention.
Interpretability is a critical property of deep learning, which is able to improve the model transparency and gain user trust. Although a variety of interpretability methods have been proposed and even some projects are related to interpretability [10], there are very few interpretable review papers in the medical field. For example, Singh et al. [11] analyzed the interpretability of deep learning models from the applications of different medical imaging tasks (CT imaging, X-ray imaging, etc.). Tjoa et al. [12] discussed recent research on two aspects of interpretability: perceptive interpretability and mathematical structure interpretability, as well as their corresponding applications in the medical field. Unlike the above works, we provide a comprehensive overview of the current state of research on interpretability in medicine by reviewing several common interpretability methods, summarizing their application in disease diagnosis based on the latest literature, as well as presenting the challenges faced and future directions, thereby better inspiring readers to further explore interpretability in the medical domain and implement CAD systems that are truly applied in clinical practice. The main contributions made in this paper are as follows:
We provide a comprehensive review of the interpretability methods from three categories, i.e., visualization-based interpretability methods, surrogate interpretability methods, and intrinsic interpretability methods. For each category, we provide a detailed description of their principles, implementation steps, as well as their characteristics.
To the best of our knowledge, this is the first time that various applications of interpretability methods for disease diagnosis have been summarized. It is worth noting that we focus on providing different methods for the diagnosis of specific diseases. Therefore, we classify the applications by disease category rather than interpretability methods.
We briefly introduce the challenges of interpretability and potential research directions to better facilitate its further development in the medical field.
The remainder of this paper is organized as follows: Sect. 2 introduces various interpretability methods. Section 3 elaborates on the applications of interpretability methods for disease diagnosis. Section 4 summarizes common datasets and evaluation methods. Section 5 analyzes the challenges of interpretability and its future directions, and Sect. 6 concludes the paper.
Taxonomy of interpretability methods
There has yet to be a standard and clear concept to define interpretability, though there have been a series of studies proposing several interpretability methods [13–15]. But it can be seen as one that generates details or reasons to make the final results easy to understand. In general, interpretability methods can be classified into two main categories, i.e., ante-hoc interpretability and post-hoc interpretability [16, 17]. Besides, post-hoc interpretability can be classified into global interpretability and local interpretability. In this section, we classify some interpretability methods commonly used in the medical field into three categories and provide a detailed description of each category. It is worth noting that classification approaches are not absolute, they can be divided into many overlapping or non-overlapping categories simultaneously, depending on the characteristics of interpretability methods. Figure 1 shows the taxonomy of interpretability methods.
Fig. 1.
Taxonomy of interpretability methods
Visualization-based interpretability methods
Visualization is a promising and intuitive qualitative analysis method for interpretability that greatly helps humans understand the internal mechanisms of deep learning models. It not only highlights the prediction regions directly but also acts as a verification tool to check whether the prediction results are consistent with the true values [18]. Herein, we mainly focus on three visualization-based interpretability methods, namely, back-propagation interpretability methods, CAM-based interpretability methods, and perturbation-based interpretability methods.
Back-propagation interpretability methods
Activation maximization (AM) is one of the most intuitive visualization methods, which makes the output with the highest confidence by finding the preferred input of neurons at a certain layer [19, 20]. It can be represented as:
| 1 |
where x denotes the input image and denotes the fixed parameter of the classifier during activation maximization. is the activation value of the i-th neuron of l-th layer for input x. The basic principle of this method is to find an input image x that is able to maximize the activation of the i-th neuron of l-th layer, making it obtain the optimal . However, it has a serious problem in that as the network grows more complex, the machine can recognize the information by using the AM method, but humans cannot. To find human-interpretable patterns, researchers introduced a regularization term . It can be defined as:
| 2 |
where is a regularization term. In spite of the fact that the AM method is able to reflect the actual behavior of the model more intuitively, as it is an optimization problem in itself, noise and uncertainty during the process of optimization may cause the resulting prototype samples to be difficult to interpret. In addition, since the optimized input can be visualized as abstracted images to reflect the meaning, whereas the text is discrete data and the meaning cannot be abstracted [21]. Thus, the AM method cannot be directly applied to interpret natural language processing models.
Layer-Wise Relevance Propagation (LRP) is a typical backpropagation-based method to predict classification results by calculating the contributions of individual pixels. In general, it starts from the output layer of the network, moves to the opposite direction, and redistributes the relevant score until it reaches the input layer [22, 23]. Simultaneously, it follows the global conservation property, which indicates that whatever a neuron receives must be redistributed in equal amounts to the lower layer. It can be represented as follows:
| 3 |
where f(x) denotes the prediction and denotes a relevance score of each variable. The LRP is defined as:
| 4 |
where i and j are neurons in two consecutive layers. denotes the impact of neuron i on neuron j. Exploiting divides the sum of contributions of all neurons in the lower layer to implement the conservation property. The outer sum over j means that the relevance of neuron i is determined by the sum of its influence on all neurons j of the following layer. In addition to the basic LRP, there are some variants of LRP such as LRP-, LRP-, and more details are covered in [24, 25]. The whole LRP process is illustrated in Fig. 2. Although the LRP is able to provide interpretability by generating heatmaps, experiment studies have revealed that it has a lower sensitivity to the target class [26]. To mitigate this issue, the Contrastive Layer-wise Relevance Propagation (CLRP) and Relevance-weighted Class Activation Map (Relevance-CAM) [27] are proposed.
Fig. 2.

The process of LRP. Each neuron redistributes to the lower layer as much as it has received from the higher layer [25]
Subsequently, Deep Learning Important FeaTures (DeepLIFT) was proposed [28, 29], which is similar to LRP. It attempts to explain the difference in the output from the reference in terms of the difference in the input from the reference. It is worth noting that when the reference activations are set to zero, it becomes an LRP interpretation. Formally, assuming that t denotes some output neurons and denotes some neurons in intermediate layer used to calculate t. Let denote the reference activation of t, then the difference in the output of a neuron between the given input and the reference input can be calculated by . DeepLIFT assigns a contribution score for the input feature. It can be represented as:
| 5 |
where can be considered as a weight, representing the impact of on . According to the above equation, the sum of all the weights is equal to the difference-from-reference output . The contribution scores of DeepLIFT can be calculated via specific rules such as linear rule, rescale rule, etc., which are elaborated in detail in [30, 31]. Moreover, this method has two unique strengths. One is that since the difference-from-reference is continuous, it allows DeepLIFT to propagate critical signals and avoid artifacts caused by discontinuities in the gradient. This makes it superior to most gradient-based methods. The other is that DeepLIFT can show dependencies as it is able to separately considers the effects of positive and negative contributions at nonlinearities. In addition, it also exits several open questions that need to be further explored [28].
CAM-based interpretability methods
Class activation mapping (CAM) [32] is a classic visual technique to provide interpretability for image classification tasks. It is able to generate class activation maps for researchers to highlight the discriminative image regions. To be noticed, the network architecture utilizes a global average pooling layer to replace the fully-connected layer, and then projects the output weights back onto the convolutional feature map of the last convolution layer to localize the important regions. Specifically, for a given image x, the probability of interest class c is easily obtained by feeding it into the network. Then, by projecting the output to the feature maps in the last convolutional layer, the importance of the feature maps with the corresponding weights can be identified. To aggregate features over multiple channels, the CAM technique adopts a linearly weighted combination of activation maps. The class activation map is generated and can be represented as follows:
| 6 |
where denotes the activation of the k-th channel in the last convolutional layer of the model at the spatial location . represents the weight corresponding to class c for the channel. The process of generating class activation maps is illustrated in Fig. 3. Although this method is simple and easy to implement, it is restricted to the specific network structure. If the network without a global average pooling layer, it needs to be modified and retrained. Furthermore, since the output weights are projected back to the last convolutional layer, only this layer can be visualized.
Fig. 3.
The process of generating class activation maps: The predicted score is mapped back to the previous convolutional layer to produce the class activation maps [32]
To tackle these problems, the Gradient-weighted Class Activation Mapping (Grad-CAM) was proposed, which is a generalization of CAM [33]. In contrast to CAM, Grad-CAM is applicable to a broader variety of convolutional neural network models without requiring a specific architecture. It is able to generate a coarse localization map to highlight the important regions. Specifically, for a given image x as an input, the predicted score of the target category c can be obtained by feeding it into the network. Then, the neuron importance weights can be acquired by calculating the gradient of with respect to the feature map activations of the l-th convolutional layer. Ultimately, a coarse localization map can be generated by adopting the global average pooling operation and performing the linear combination of the weights and feature maps. The overview of Grad-CAM is illustrated in Fig. 4. The Grad-CAM can be represented as:
| 7 |
| 8 |
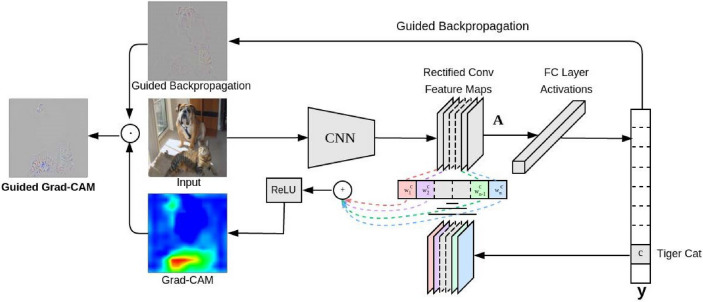
where Z denotes the number of pixels in the feature map. represents the gradient via backprop. denotes the neuron importance weights of the k-th feature map for class c. denotes the k-th feature map of the convolutional layer. Moreover, the Grad-CAM method combined with the Guided Backpropagation method develops a novel interpretability method called Guided Grad-CAM, which is able to generate a fine-grained feature localization map.
Fig. 4.
Grad-CAM overview: Given an image as input, we can obtain the predicted score of the target class by forward propagating the image through the convolutional neural network and task-specific network. Then the gradient of the target class is set to 1, and the others are set to 0. This signal is then backpropagated to the rectified convolutional feature maps of interest, which we combine to compute the coarse Grad-CAM localization, which represents where the model has to look to make the particular decision. Finally, we pointwise multiply the heatmap with guided backpropagation to get Guided Grad-CAM visualizations that are both high-resolution and concept-specific [33]
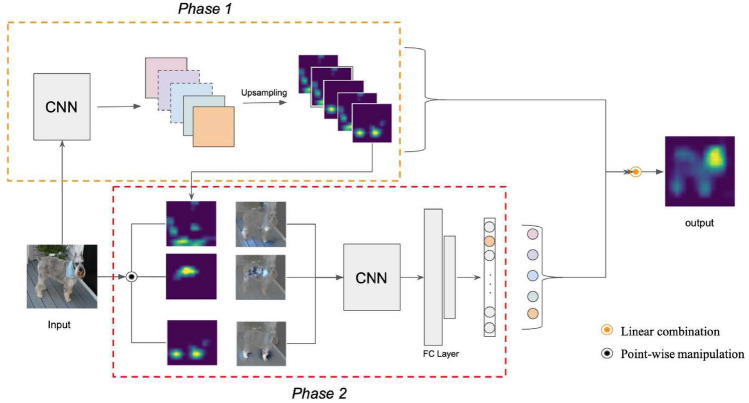
Although the performance of Grad-CAM is superior to that of CAM, it still has several limitations caused by gradients, such as gradient saturation. In response to the gradient problem, several gradient-free interpretability methods have been proposed, such as Score-CAM [34], Ablation-CAM [35], Group-CAM [36], and Relevance-CAM [27], which can not only solve problems caused by gradients but also enhance the interpretability of the model. Taking Score-CAM as an example, it bridges the gap between CAM-based and perturbation-based methods, ultimately providing an understandable localization map through a linear combination of score-based weights and activation maps. The process of the Score-CAM is shown in Fig. 5.
Fig. 5.
The Score-CAM overview: The Score-CAM has two stages. Phase 1 first extracts activation maps, and then each activation works as a mask on the original image, obtaining its forwarding-pass score on the target class. Phase 2 repeats N times. Finally, the result can be generated by a linear combination of score-based weights and activation maps [34]
The whole process of Score-CAM has two stages, where the first stage aims to gain activation maps using the CAM method, whereas the second stage aims to obtain the weight of each activation map via Channel-wise Increase of Confidence (CIC). To be specific, for an input image x, we can get its activation maps by using the CAM method. Then, by upsampling and normalizing these activation maps to the input size and taking them as masks working on the original image, the forward-pass score for the target class can be handily acquired. Ultimately, the importance of the activation map on the feature map can be measured by calculating the CIC, where CIC is defined as:
| 9 |
| 10 |
where denotes the k-th feature map of the l-th convolutional layer. denotes upsampling and normalizing the feature map. denotes the baseline input image. denotes Hadamard Product. Thus, Score-CAM is represented as:
| 11 |
| 12 |
where denotes the CIC score for the activation map. The study showed that the Score-CAM method outperforms other CAM-based methods such as CAM, Grad-CAM and Guided Backpropagation. In addition to the above methods, there are several CAM-based methods with interpretability. We have compiled a general overview in Table 1, including their advantages, disadvantages, and how they work.
Table 1.
Overview of the advantages and disadvantages of common CAM-based methods and how they work
| Method | Advantages | Disadvantages | How they work |
|---|---|---|---|
| CAM [32] |
Identify important areas for image classification |
Need a global average pooling layer Only visualize the output of the last convolution layer |
A linear combination of weights and activation maps |
| Grad-CAM [33] |
Apply to a broader network without requiring a global average pooling layer Visualize any layer of a deep network |
Gradient issue, such as saturation Lack the ability to highlight the fine-grained details Performance drops when localizing multiple occurrences of the same class Cannot capture the entire object in completeness |
Weight the 2D activations by average gradient |
|
Guided Grad-CAM [33] |
Provide a fine-grained localization map Apply to a broader network without requiring a global average pooling layer Visualize any layer of a deep network |
Gradient issue, such as saturation |
Acquire a fine-grained localization map by combination of Grad-CAM and Guided Backpropagation |
| Grad-CAM++ [37] |
More suitable for localizing multiple targets of the same category. |
Using second order derivatives requires more complex computation |
Like Grad-CAM but uses second order gradients |
| Score-CAM [34] |
Gradient-free localization High performance |
– |
Perturbate the image by the scaled activations and measure how the output drops |
| Ablation-CAM [35] |
Gradient-free localization It is able to fuse with pixel-space gradient visualizations to generate high resolution localization maps |
Require more computation time |
Zero out activations and measure how the output drops |
| Group-CAM [36] |
Require less computation. More efficient Can be used as a data augment trick to fine-tune classification models |
- |
Adopt the “split-transform-merge” strategy to generate saliency maps |
Perturbation-based interpretability methods
Sensitivity analysis is a simple yet powerful local interpretability method, used to analyze the impact of the uncertainties of the input variables on the final prediction. Its attributions can be calculated by the following formulation:
| 13 |
where is the target output and is the input variable. And the absolute value of the gradient represents those input features like pixels that can be perturbed the least in order to maximize the alteration in the target output. In addition, the sensitivity analysis method is regarded as a suboptimal model for interpretability as it has been proven to have several inevitable shortcomings, such as gradient shattering [38].
In contrast to the sensitivity analysis method, occlusion sensitivity [30, 39] is considered as another perturbation-based interpretability method, which measures the important features by occluding parts of the input. Specifically, it usually occludes different parts of the input with a gray square to measure the change outcome. If the output of the model changes dramatically, it means that this part has a significant impact on the prediction.
Summary
In this subsection, we mainly review three categories of visual interpretability methods, including back-propagation interpretability methods, CAM-based interpretability methods, and perturbation-based interpretability methods, respectively. Among them, the back-propagation interpretability methods identify the salient features of the input by evaluating the gradient signals of the network from output to input, whereas the perturbation-based interpretability methods measure the input feature relevance by altering or removing parts of the input and observing the altered outcomes. Moreover, we discuss several CAM-based interpretability methods that can be divided into two categories: gradient-based and gradient-free. But most of them adopt a linear combination of weights and activation maps to identify the important regions. Thus, how to obtain the weights of the feature map is crucial for generating the final interpretable localization map. To sum up, all of them have the potential to generate various visual maps, such as heatmaps and CAMs, to provide interpretability.
Surrogate interpretability methods
Surrogate interpretability methods are easy to implement. They seek to train interpretable models (e.g., shallow decision trees, linear models, etc.) to mimic the behavior of “black-box” models. We mainly review two surrogate interpretability methods. Details are described below. The main idea of the surrogate method is shown in Fig. 6.
Fig. 6.
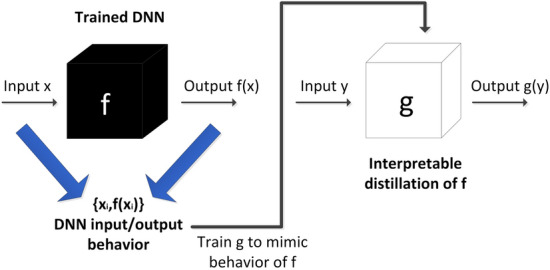
Illustration of surrogate interpretability methods, where g is a white box, which is trained to mimic the behavior of the “black-box”, so that [30]
Local interpretable model-agnostic explanation (LIME) is a popular local interpretability method that is able to approximately explain complex networks by analyzing simple models. Specifically, researchers select a neighborhood of the inputs, then feed them into simple networks to mimic the actual results of the “black-box” as much as possible, and finally provide interpretability for deep learning models by analyzing the interpretable simple networks [40, 41]. The interpretability method of LIME is defined as follows:
| 14 |
where g is an interpretable model (such as linear models, decision trees, etc.) of instance x, whereas G is the collection of possible interpretable models. represents a loss function, which is leveraged to measure how poorly g approximates the original model f in the locality around the data point x. It is worth noting that is a regularization used to measure the complexity of the g, thereby avoiding the model becoming more complex. Although the LIME method is intuitive, flexible, and easy to understand, it is a local approximation of the original model. In addition, for each input instance, this method needs to retrain an interpretable model to fit the decision results of the original model. Thus, the efficiency of this method is relatively low.
In addition, Moraffah et al. [16] mentioned that LIME is a quick approximation of a broader approach called Shapley Additive exPlanations (SHAP) [40]. Note that SHAP is not a surrogate interpretability method, but a classic post-hoc interpretability method inspired by optimal Shapley Value. It seeks to improve interpretability by calculating the importance value of each feature for individual prediction. The core of SHAP is Shapley Value, which is a fair distribution benefits of cooperation by considering the contribution of each participant. This approach considers not only the impact of individual variables, but also the impact of groups of variables and possible synergies between variables. However, in practical problems, there are huge possible coalitions of feature values that need to be computed. Thus, it demands lots of computing time.
Knowledge distillation (KD) [42, 43] is a typical model compression method with interpretability. It is able to solve complex problems by learning a small model from a large model. In contrast to other compression methods, KD not only reduces the size of the network but also saves computing power [44]. The “Student-Teacher” is one of classic KD architectures that can achieve the transfer knowledge from the teacher model to the student model under the supervision of the teacher model. The process is illustrated in Fig. 7.
Fig. 7.
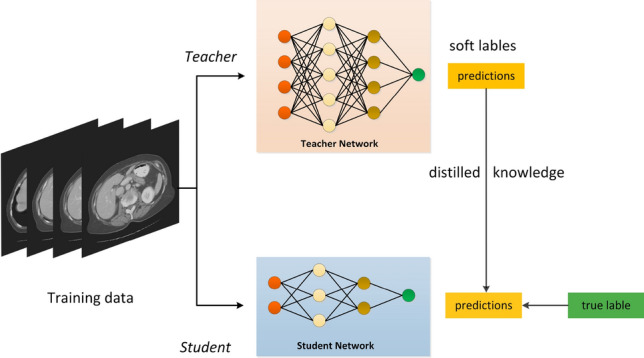
The process of knowledge distillation. A smaller student model is trained to mimic a pretrained larger teacher model [44]
Specifically, the teacher and the student are two neural networks, each taking the same training data as input, in which the student network has fewer layers than the teacher network. For the task of image classification, the logits are used as the carriers of the knowledge from the teacher model. The knowledge distillation is formulated as follows.
| 15 |
where is the logit that is used to as the carrier of the knowledge from the teacher model for the i-th class, and T is temperature parameter. Note that when , it is a standard softmax function. As T increases, the distribution of the class probability becomes softer and provides more information, thereby improving the generalization ability of the student model. Although the KD method is simple enough to understand, it is only a global approximation of the original complex model. Thus, the explanation based on the KD does not reflect the real behavior of the model. In addition, since the process of KD is usually uncontrollable and cannot guarantee that the knowledge learned from the complex model can be effectively transferred to the distillation model, resulting in poor quality of the explanations.
In this subsection, we mainly discuss two kinds of surrogate interpretability methods, namely LIME and KD. Although they have the same goal, that is, using an interpretable model to interpret a complex model and obtain results as similar as possible to the original model, they do have substantial differences in terms of their principles. For the LIME method, it uses a simple or interpretable model to mimic a complex model, whereas for KD, it employs teacher-student networks to solve complex tasks by using a large model to distill a small model, more like model compression. Although both of them can provide interpretability for complex models, their interpretability is not yet ideal as they just provide approximate interpretability to the original model.
Intrinsic interpretability methods
In general, intrinsic interpretability methods convey a concept that the model can be interpreted by itself, which is being non-controversial, simple, and easy to understand [40]. As such, we elaborate on several common intrinsic interpretability methods here.
The attention mechanism [45, 46] is one of the most popular methods, which uses the computer vision system to mimic the human visual system so as to focus on the regions of interest. The purpose of the attention mechanism is to strengthen the regions of interest and suppress irrelevant regions or potential background regions. This method can be divided into two types: soft attention and hard attention. Intuitively, the soft attention model is differentiable. It pays more attention to each part of an image and obtains the average weight of all its parts, whereas hard attention only focuses on some parts of an image. Each part of an image is either discarded or leveraged to gain the context vector. Furthermore, self-attention is also a hot attention mechanism, initially designed to deal with NLP tasks. Gradually, it is used to handle computer vision tasks with excellent performance as well. Recently, there have been some arguments about whether the attention mechanism is interpretable [47–49]. Thus, the interpretability of the attention mechanism may be worth further exploring in the future.
Rule-based extraction [30, 50] is able to provide interpretability by extracting the interpretable rules from the training model. It takes the complex model as an entry point, using comprehensible rule sets to generate interpretable symbol descriptions or extract interpretable models (such as decision trees, linear models, etc.) to make it have similar decision-making capabilities to the original model. However, since it extracts interpretable rules from a large model, the accuracy of the model is often not precise enough, so it can only provide an approximate explanation. Besides, more information about rule-based interpretability algorithms, such as RuleFit, Node Harvest, etc. are discussed in [51].
In addition, the decision tree [16, 50] is also an interpretable model, used to solve the problems of regression and classification. It makes full use of the characteristics of the tree-structured model, which downsizes the dataset into smaller subsets and incrementally develops an associated decision tree. For any decision tree, it includes a root node, internal nodes, and leaf nodes. Internal nodes are used to check whether the features satisfy conditions, and the leaf nodes represent the predictions. The process of making a decision starts from the root node and follows a path until it reaches a leaf node, which follows the “if... then...” rule. Please note that decision trees are only suitable for small networks. Once used in complex models, they are likely to become deep forests due to more features, and then the networks are more difficult to interpret.
In this subsection, we mainly analyze three types of intrinsic interpretability methods: attention mechanisms, rule-based extraction, and decision trees, which can be interpreted by themselves. Among them, the attention mechanism simulates human cognition to focus on regions of interest and suppress irrelevant regions, thereby generating an interpretable attention map. Rule-based extraction is able to extract interpretable rules from a large model to interpret the original model. The decision tree obtains the final prediction result by searching from the root node to leaf nodes. Although all of them are able to provide interpretability and make the model transparent, the effects of the rule-based extraction and decision trees are not accurate enough.
Applications of interpretability methods in disease diagnosis
It is a fact that deep learning models have exceeded the traditional methods in the medical field with high efficiency and accuracy. Several applications are designed to help physicians make more accurate decisions. Thus, the applications of interpretability in disease diagnosis are reviewed here and classified by disease category according to the body part, aiming to introduce the current research state in the medical field. Figure 8 shows a brief overview of the interpretability applications for disease diagnosis.
Fig. 8.

The applications of interpretability methods in disease diagnosis
Applications in eye diseases
Examining the retinal images of the fundus with the naked eye is a long and tedious task, especially for the tiny capillaries in the eye. As such, it is necessary to develop interpretable deep learning models to assist physicians in diagnosing eye diseases. Herein, we summarize several applications in eye diseases.
Diagnosis of diabetic retinopathy
Diabetic retinopathy is a chronic eye disorder caused by diabetes that may lead to blindness. To detect diabetic retinopathy, Kind et al. [52] applied the fast R-CNN ResNet101 architecture to classify the retinal fundus images as healthy or not. To further diagnose the severity of the disease, the researchers designed various interpretable models, exploiting the excellent performance of deep learning. For example, De et al. [53] designed a diabetic retinopathy interpretable classifier whose interpretability is based on the modified LRP. It can not only classify the severity of the disease but also provide interpretability by scoring each significant pixel of the input image and evaluating its contribution. Compared with other back-propagation interpretability methods, this method has a unique way of generating scores. Kumar et al. [54] presented an interpretable system called the CLass-Enhanced Attentive Response Discovery Radiomics (CLEAR-DR) to classify diabetic retinopathy and provide a visual CLEAR-DR map to interpret its decision-making process. Besides, Jiang et al. [55] designed an ensemble model consisting of three models: ResNet152, Inception-V3, and Inception-Resnet-V2. It independently trains these deep models and ensembles their results using the Adaboost algorithm to alleviate the bias of each single model. It can generate class activation maps for each single model to provide an explanation and produce an integrated class activation map for the final result by using the CAM technique, in which the integrated class activation map can effectively alleviate the problems of each single model. Moreover, they proposed a multi-label deep learning model for diabetic retinopathy classification based on the ResNet [56], and utilized the Grad-CAM technique to localize the lesions and provide visual explanations. Afterwards, they combined the Grad-CAM method with Guided Backpropagation to get a fine-grained result. In comparison to the previously proposed ensemble model, the multi-label deep learning model does not require specific structure but performs better on a private dataset with 93.90% sensitivity, 94.40% specificity, 94.2% accuracy, and 0.989 AUC. Furthermore, an EfficientNet architecture was proposed to detect vision-threatening diabetic retinopathy (VTDR) and referable diabetic retinopathy (RDR) [57]. It combines Grad-CAM with fine-grained visualizations to generate a high-resolution class-discriminative visualization. To investigate the validity of the model, the authors detected VTDR and RDR on both EyePACS and APTOS2019 datasets, respectively, and obtained experiment results with faster inference and higher performance.
Diagnosis of glaucoma diseases
Glaucoma is another irreversible eye disease that can lead to vision loss. To diagnose glaucoma accurately, most researchers adopted based-attention methods to localize and identify lesion regions rather than CAM-based methods. This is due to the fact that clinical evidence for glaucoma diagnosis mainly focuses on the optic disc, requiring localization of more tiny lesion regions. For example, Li et al. [58] designed a deep learning model based on attention called AG-CNN for glaucoma detection, which consists of three parts. In particular, an attention prediction subnet is designed first to remove the redundancy of fundus images. Then, a pathological area localization subnet is presented for visualizing the feature maps by the guided back propagation technique. Finally, a classification subnet is designed for classifying positive or negative glaucoma. The experiment results demonstrated that this method is far superior to other advanced methods, not only for glaucoma detection but also for pathological region localization. Similarly, Liao et al. [59] proposed an interpretable model called EAMNet. It is also composed of three parts: ResBlocks as the backbone; the multi-layer average pooling for aggregating different scales of features; and Evidence Activation Mapping (EAM) for highlighting the lesion regions. It is noted that the EAM is a channel-wise attention-based approach that is able to aggregate feature maps of different scales into a single map, containing rich spatial information. Thus, it can comprehensively diagnose glaucoma and identify lesion regions. Moreover, a multi-task architecture was proposed [60], consisting of two parts: segmentation and prediction. The former focuses on localizing the disc and cup regions to provide interpretability, whereas the latter focuses on detecting glaucoma diseases. This is the first work that incorporates interpretability into glaucoma screening with promising outcomes.
Diagnosis of macular diseases
Macular is the central part of the retina that is mainly responsible for central vision. To accurately diagnose macular diseases, Fang et al. [61] presented an attention-based on convolutional neural network (LACNN) model with a lesion-attention module for retinal OCT image classification, in which the lesion-attention module aims to highlight the local lesion features by generating a soft attention map. Experiment results found that by employing the lesion-attention modules, the performance of LACNN is indeed superior to other classical methods. Moreover, Joshi et al. [38] designed a lightweight network to implement the image classification by using weakly supervised segmentation. It utilizes the adapted CAM approach to provide interpretability by generating more accurate heatmaps. In contrast to the normal CAM, the adapted CAM is able to learn features for both segmentation and classification, rather than just for the classification task. Besides, this method has stronger generalization ability for different modalities, not only for diabetic macular edema detection on OCT images but also for breast cancer detection, both with positive results.
Summary
In this subsection, we mainly focus on the application of several interpretability methods in the diagnosis of different eye diseases. Among them, most researchers have used CAM-based and attention-based methods to provide visual interpretability by generating heatmaps. Since the CAM technique only produces a coarse localization, researchers tried to combine it with other methods such as the guided backpropagation method to generate a fine-grained result. To accurately localize and identify smaller lesion areas, attention-based methods were also fully employed to enhance regions of interest and suppress irrelevant regions. In addition, all of the models can not only implement disease diagnosis accurately but also provide interpretability. Part of experimental results are shown in Table 2.
Table 2.
The experimental results of diagnosing various eye diseases (such as diabetic retinopathy, glaucoma, etc.) on different datasets
| Task | Author | Network | Dataset | Interpretability | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|---|---|---|---|
|
Diabetic Retinopathy classification |
Jiang et al. [55] |
Integrated model |
Private dataset | CAM | 85.57% | 90.85% | 88.21% | 0.946 |
|
Diabetic Retinopathy Classification |
Jiang et al. [56] | Based on ResNet | Private dataset | Grad-CAM | 93.90% | 94.40% | 94.20% | 0.989 |
|
Detect referable diabetic retinopathy(RDR) |
Chetoui et al. [57] | EfficientNet | EyePACS | Grad-CAM | 91.70% | 98.90% | – | 0.984 |
| APTOS2019 | 91.40% | 97.20% | 0.966 | |||||
|
Detect vision-threatening |
Chetoui et al. [57] | EfficientNet | EyePACS | Grad-CAM | 98.10% | 93.70% | – | 0.99 |
| APTOS2019 | 99.10% | 92.50% | 0.998 | |||||
| Glaucoma Diagnosis | Liao et al. [59] | EAMNet | ORIGA dataset |
Evidence Activation Mapping |
– | – | – | 0.88 |
| Glaucoma Detection | Li et al. [58] | AG-CNN | LAG database | Attention | 95.40% | 95.20% | 95.30% | 0.975 |
| RIM-ONE database | 84.80% | 85.50% | 85.20% | 0.916 | ||||
|
Retinal OCT image classification |
Fang et al. [61] | LACNN | UCSD dataset | Attention | 86.80% | 86.20% | 90.10% | – |
| NEH dataset | 99.33% | 99.39% | – | 0.994 |
Applications in lung diseases
In recent years, the application of deep learning in lung disease diagnosis has received a lot of attention and achieved many excellent outcomes [62–65], not only since lung cancer is the major cancer that causes death, but also due to the current epidemic period of coronavirus disease 2019 (COVID-2019). Therefore, in this subsection, we focus on the application of some interpretable methods in the diagnosis of lung diseases.
Classification of pulmonary nodules
Most of the current lung nodules are benign. Yet there are a few lung nodules that are malignant, which is a sign of lung cancer. To implement the classification of pulmonary nodules, Li et al. [66] designed an interpretable network architecture, including a classification network and an estimation network. Among them, the estimation network aims to generate a visual interpretation by scoring the important pixels of the feature maps, and the classification network is used to diagnose the disease. By comparing it with three popular interpretability methods (CAM, VBP, and LRP), it was found that the proposed method can generate a more accurate and clear visual interpretation map. Similarly, the stacked interpretable sequencing cells (SISC) architecture was introduced [67] to predict lung cancer by generating attentive maps to provide explanations. It can not only achieve remarkable results for lung prediction but also highlight the crucial regions. In addition, Shen et al. [68, 69] proposed an interpretable hierarchical semantic convolutional neural network (HSCNN) to judge whether pulmonary nodules are malignant, which includes three parts: a high-level task module, a feature learning module, and a low-level task module. Among them, the low-level task module is able to predict five semantic diagnostic features for diagnosing malignant lung nodules, and the high-level task module combines information from the feature learning module and the low-level task module to generate a holistic prediction of the malignancy. Experiment results showed that the HSCNN model can greatly improve the performance classification for malignant lung nodule prediction, and the semantic diagnostic feature predictions can improve the interpretability of the model. However, there are still several incorrect situations since the labels in the experiment do not include a higher association with malignancy, leading to the semantic diagnostic feature predictions and malignancy predictions being inconsistent sometimes. Furthermore, Jiang et al. [70] employed the Neural Architecture Search (NAS) technique [71] to build a 3D network for lung nodule classification. The convolutional block attention module (CBAM) was introduced to provide a visual interpretation of the diagnosis process by generating an attention map. This model can implement higher accuracy with less computation. It is worth noting that NAS techniques require a large number of labeled samples, but medical images are generally small samples, so designing NSA techniques based on small samples is promising in the medical field.
Diagnosis of COVID-19
The global pandemic of COVID-19 has severely impacted human life but is accelerating with no sign of ending. As such, early diagnosis and treatment are essential to curb the spread of the disease [72, 73]. Recently, a large number of interpretable deep learning models have been proposed for diagnosing COVID-19. For example, Shi et al. [74] combined channel-wise and spatial-wise attention mechanisms to improve classification interpretability by generating attention maps. The proposed model showed promising results on both the CT and X-ray modality datasets. Wu et al. [75] designed a COVID-19 diagnosis system with a classification and segmentation module, in which the classification module focuses on discriminating against COVID-19 patients, whereas the segmentation module aims to identify the lesion regions. Besides, they applied the activation mapping (AM) method with an image mixing technique in the classification module to provide a visual interpretation, which can identify opacification regions more accurately.
In addition, Brunese et al. [76] proposed an explainable two-stage classification model. The first stage is to judge whether a person has pulmonary disease. The second stage is to detect COVID-19 in patients with pulmonary disease. The Grad-CAM was employed to highlight the predicted areas. Similarly, [77, 78] designed different methods separately for diagnosing normal, pneumonia, and COVID-19. The former adopts the Grad-CAM technique to provide a visual interpretation, whereas the latter employs LRP and Grad-CAM++ techniques to highlight the pathological regions. It has been validated that Grad-CAM++ can produce more precise localization than LRP. Moreover, Alshazly et al. [79] exploited twelve deep networks (i.e., ResNet, DenseNet, etc.) and a transfer learning strategy to achieve COVID-19 detection. They applied t-distributed stochastic neighboring embedding (t-SNE) and Grad-CAM to provide interpretability, in which the t-SNE is used to show the separate clusters of COVID-19 and non-COVID-19, whereas Grad-CAM is used to highlight the predicted regions. Extensive experiments showed that the proposed method has the potential to automatically detect COVID-19. Some of experimental results for diagnosing COVID-19 are illustrated in Table 3.
Table 3.
Some of experimental results for COVID-19 diagnosis
| Task | Author | Network | Dataset | Interpretability method | Accuracy | Precision | Sensitivity/recall | Specificity | F1 score |
|---|---|---|---|---|---|---|---|---|---|
| COVID-19 Detection | Alshazly et al. [79] | ResNet101 | SARS-CoV-2 CT-scan |
t-SNE, Grad-CAM |
99.40% | 99.60% | 99.80% | 99.60% | 99.40% |
| DenseNet201 | COVID19-CT | 92.90% | 91.30% | 93.70% | 92.20% | 92.50% | |||
| COVID-19 Detection | Shi et al. [74] | - |
COVID-19 X-Ray dataset (private) |
Attention | 0.9411 | 0.9673 | 0.978 | – | 0.9726 |
|
COVID-19 CT dataset (private) |
0.8654 | 0.913 | 0.8513 | – | 0.8811 | ||||
| COVID-19 Detection | Brunese et al. [76] | VGG-16 | Private dataset | Grad-CAM | 0.98 | – | 0.87 | 0.94 | 0.89 |
| COVID-19 Detection | Karim et al. [78] | Ensemble model |
Private chest X-ray dataset (balanced /imbalanced dataset) |
LRP, Grad-CAM++ |
– |
0.904 0.877 |
0.905 0.881 |
0.905 0.879 |
– |
| COVID-19 Classification | Wu et al. [75] | Res2Net | Private COVID-CS dataset | Activation mapping | – | – | 95% | 93% | – |
Summary
In this subsection, we mainly discuss several applications with interpretability to cope with lung diseases. It was found that most researchers prefer to exploit the Grad-CAM technique to provide an intuitive interpretation. Also, there are some researchers exploit attention mechanisms to strengthen the regions of interest. In short, both approaches are competent to enhance model transparency and yield satisfactory results.
Applications in brain diseases
The brain is responsible for controlling human thought, behavior, and movement. Once brain disease appears, irreversible damage will occur. In the following, we mainly analyze several interpretable applications for two brain diseases.
Diagnosis of Alzheimer’s disease
Alzheimer’s disease (AD) is an age-related primary degenerative brain disease. Its main clinical manifestations are memory loss and cognitive dysfunction, and patients may gradually lose the ability to work and live independently. To accurately diagnose AD, Tang et al. [80] exploited Guided Grad-CAM and occlusion to generate complementary maps for the Alzheimer’s disease pathologies classification, which intuitively provide plaque predictions motivated by the neural networks. In contrast, Nigri et al. [81] believe that the occlusion method is more suitable for natural image classification instead of medical images. It is due to the fact that natural images can exhibit different backgrounds and color intensities, whereas brain scans are very similar. Also, during the process of training, the occluding patches may interfere with image labels. Thus, they proposed the swap test method to interpret the prediction results. Experiments found that this method is indeed better than the occlusion method. But it is worth noting that when using the swap test method, it is necessary to consider the patch size. Since the model is uncertain, the small patches can enhance the spatial resolution of the heatmap, but they can also increase the noise. Besides, Wang et al. [82] presented three explainable models to automatically detect AD based on language abilities, exploiting the multi-head self-attention mechanism and a 1-dimension convolutional neural network to interpret the decision-making process. Since the model is based on the spoken language, it can be easily applied for AD detection. Moreover, a multilayer multimodal model was proposed [83] to diagnose AD and detect its progression. It uses the SHAP method to provide explanations for each layer of the model. In contrast to other machine learning models, this model achieves remarkable performance. Furthermore, [84, 85] also proposed argumentation-based methods for diagnosing AD, respectively. Both of them have a positive outcome.
Diagnosis of Parkinson’s disease
In addition to AD, Parkinson’s disease (PD) is also a common neurodegenerative brain disease with clinical manifestations only second to AD. To detect early PD, Magesh et al. [86] exploited transfer learning on the VGG16 architecture to classify DaTscan images. Meanwhile, they adopted the LIME technique to provide reasonable interpretability for the prediction results. It can strengthen the region of interest in the image and classify PD and non-PD. Besides, Cavaliere et al. [87] designed an interpretable method based on genetic programming called grammar evolution, which can automatically diagnose PD by analyzing handwriting.
Summary
In this subsection, we focus on analyzing several interpretable applications of brain diseases. Since brain diseases affect the human neurological system, researchers have tried to exploit different approaches to analyze models and provide interpretability. Extensive experiments have found that these methods do achieve promising outcomes. Some experimental results for diagnosing brain diseases are shown in Table 4.
Table 4.
Part of experimental results for diagnosing brain diseases
| Task | Author | Network | Dataset | Interpretability method | Accuracy | Precision | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis of AD | Nigri et al. [81] | AlexNet | private MRI dataset | Swap Test | – | – | – | – | 0.923 |
|
Diagnosis of AD by part-of-speech (Pos) features |
Wang et al. [82] | C-Attention-FT | DementiaBank dataset | Attention | 92.20% | 93.50% | 97.10% | – | 0.971 |
|
Diagnosis of AD by language embedding features |
Wang et al. [82] | C-Attention-Embedding | DementiaBank dataset | Attention | 84.50% | 88.50% | 92% | – | 0.837 |
|
Diagnosis of AD by both Pos and language embedding features |
Wang et al. [82] | C-Attention-FT+Embedding | DementiaBank dataset | Attention | 91.50% | 96.90% | 92.20% | – | 0.977 |
|
Classification of NC (normal control) and AD |
Achilleos et al. [85] | – | ADNI-1 | Rule extraction | 91% | – | 87% | 95% | – |
| Detect PD | Magesh et al. [86] | VGG16 | PPMI database | LIME | 95.20% | – | 97.50% | 90.90% | – |
Note: C-Attention-FT, C-Attention-Embedding and C-Attention-FT+Embedding denote attention model with only part-of-speech (PoS) features, only the language embeddings and a unified architecture (both Pos features and language embeddings), respectively
Applications in other diseases
In addition to the above-mentioned interpretable models, there are also some interpretable models for the diagnosis of common diseases that have been proposed. Herein, we mainly focus on several interpretable applications for three common diseases, namely, heart diseases, skin diseases, and breast diseases.
Diagnosis of heart diseases
The heart is one of the most crucial organs, carrying blood to all the body tissues. Once heart disease occurs, it will seriously affect human life. At present, there are several research studies detecting heart disease by analyzing the beat changes of the electrocardiogram (ECG) [88, 89]. Besides, Clough et al. [90] proposed a deep learning architecture using variational autoencoders (VAE) to detect the presence of coronary artery disease, which encodes image segmentations into a low-dimensional latent space with a VAE and then uses latent vectors for classification. This model not only performs classification well but also provides global interpretability by using concept activation vectors (CAV). Similarly, Puyol-Ant’on et al. [91] designed a framework for image-based classification using a VAE. It encodes the existing clinical knowledge into the model with a secondary classifier, which can interpret the output of the model and improve the interpretability of classification results. Although the experiment indicated that the performance of this classifier is similar to the baseline VAE, it can interpret its decision-making process for clinicians, which is the key novelty. Moreover, Aghamohammadi et al. [92] presented an effective classification technique to predict heart attacks that combines the advantages of the genetic algorithm, neural network, and fuzzy logic, using explainable graphs to provide interpretability for the final prediction. Experiment results demonstrated that the performance of the proposed algorithm is quite satisfactory. Furthermore, a novel architecture for cardiac MRI image segmentation was proposed called SAU-Net [5], which mainly focuses on the interpretability and robustness of the model. In addition, it can achieve multi-level interpretability by using the dual-attention decoder module. Compared with other post-hoc interpretable methods, it is able to provide different levels and multi-resolution saliency maps during the forward pass without additional computation.
Diagnosis of skin diseases
Skin cancer has the highest incidence and is steadily increasing year by year [93]. As such, several interpretable applications have been proposed to help people diagnose skin diseases effectively. Barata et al. [94] developed a CAD system to diagnose skin lesions that combines multi-task convolutional neural networks with attention modules to improve the interpretability of the model. This system can mimic dermatologists to make hierarchical decisions. Likewise, an attention U-Net architecture [95] was presented to automatically detect skin lesions, which adopts multi-scale images as input. Each type of lesion attribute is trained as a separate network to solve the negative effects of the class imbalance. It can achieve the best results in detecting skin lesions by using fewer parameters and computations. In addition, a light-weight attention mechanism-based deep learning architecture was designed [96] to distinguish 11 types of skin diseases on histopathological images, which uses the CAM technique to provide visual explanations for deep models. This model can not only output the disease name but also highlight the lesion regions. Moreover, Gu et al. [97] proposed a CA-Net architecture that utilizes a comprehensive attention-based neural network to deal with the task of skin lesion segmentation. In contrast to the other interpretable methods that only consider spatial information, this method integrates spatial, channel, and scale attention to provide a comprehensive interpretation for the prediction results. At the same time, this method can directly obtain the attention coefficients without requiring additional computations. Furthermore, Stieler et al. [98] designed a deep skin classifier that integrates the LIME and ABCD-rule (a diagnostic approach of dermatologists) to distinguish melanocytes and non-melanocytes. Although this method allows us to draw the conclusion of which features are more important, the information needs to be converted into a user-friendly explanation.
Diagnosis of breast diseases
Due to the fact that breast cancer is one of the high-risk diseases for women, a variety of interpretable applications for diagnosing breast diseases are presented to aid clinicians in making more-informed decisions. For example, Velden et al. [99] proposed a regression convolutional neural network method to estimate the density of the breast on MRI, which utilizes the Deep Shapley Additive exPlanations values to provide interpretability. Experiments found that this method is effective with promising results. Shen et al. [100] presented the globally-aware multiple instance classifier (GMIC) to predict benign and malignant lesions in screening mammograms. Specifically, it first uses a global module on the whole image to generate sailency maps that provide coarse localization, and then it uses a local module to extract fine-grained features. Finally, it applies a fusion module to integrate the information from both to predict the final result. Because of the model’s generic design, it can be widely applied to high-resolution image classification tasks. In addition, an explainable semantic classifier was developed for detecting breast cancer [101]. Although the performance of this approach is not ideal, it can provide semantic interpretation. Moreover, Beykikhoshk et al. [102] designed the DeepTRIAGE model to classify the breast cancer sub-types. It utilizes attention mechanisms to generate personalized biomarker scores, highlighting important features. This model can not only accurately classify cancer subtypes but also assign interpretable and personalized biomarker scores to each patient. Furthermore, Gu et al. [103] presented an extreme gradient boosting method (XGBoost) based on the decision tree model to predict breast cancer recurrence and exploited the case-based reasoning (CBR) method to provide explanations for the results. Since this method ensembles several classifiers, it can achieve higher accuracy and better generalization ability.
Summary
In this subsection, we mainly discuss some interpretable applications for diagnosing three common high-incidence diseases. For heart diseases, VAE was introduced to build interpretable models, which is an effective way to observe the beat changes of EGG to diagnose the disease. For skin diseases, most researchers are willing to use attention-based models to diagnose the diseases, which can strengthen the interest regions to enhance the interpretability. For breast diseases, we reviewed several interpretable applications, from estimating breast density to predicting cancer and then predicting breast cancer recurrence, all of them with promising performance. In addition to the above-mentioned, there are various interpretable applications with remarkable performance [104–108]. All of them can improve diagnostic efficiency and facilitate the development of deep learning in the medical field.
Evaluation metrics and Datasets
In this section, we focus on some common evaluation metrics and datasets in the medical field, which are compiled from applications of interpretable methods in the references of Sect. 3. These evaluation metrics and datasets are useful for evaluating the performance of disease diagnosis. In contrast, there are no standard evaluation metrics for interpretability, and most of the performance is judged by humans. Therefore, these evaluation metrics are important for evaluating the interpretability performance of deep learning models in the medical domain, aiming at providing references for readers.
Evaluation metrics
Evaluation metrics is an essential way to measure the performance of the models, in which the confusion matrix can calculate several metrics (i.e. accuracy, precision, etc.) by calculating the number of true positive (TP), false positive(FP), true negative (TN) and false negative (FN). They can be denoted as:
| 16 |
| 17 |
| 18 |
| 19 |
| 20 |
Besides, we introduce two popular metrics, Dice coefficient, Jaccard index (IoU), based on the overlap measures to evaluate the performance of the model. We assume A and B are two given sets. Then they can be represented as:
| 21 |
| 22 |
Disease Datasets
We list some common disease datasets, including anatomy, modality, dataset size, and so on. More detailed contents are shown in Table 5.
Table 5.
The common disease datasets
| Anatomy | Medical diagnosis | Dataset | Modality | Dataset size | Url |
|---|---|---|---|---|---|
| Eye | Diabetic Retinopathy | Messidor-2 | Retinal funds images | 1748 images | https://www.adcis.net/en/third-party/messidor2/ |
|
Retinopathy grade Risk of macular edema |
Messidor | Fundus color numerical images | 1200 images | https://www.adcis.net/en/third-party/messidor/ | |
|
Diabetic retinopathy grade Diabetic macular edema |
IDRiD | Color fundus images | 516 images | https://idrid.grand-challenge.org/ | |
| Diabetic Retinopathy | e-ophtha-EX | Color fundus images | 82 images | https://www.adcis.net/en/third-party/e-ophtha/ | |
| e-ophtha-MA | Color fundus images | 381 images | |||
| Diabetic retinopathy (DR) | DiaRetDB1 V2.1 | Digital images of eye fundus | 89 images | https://www.it.lut.fi/project/imageret/diaretdb1_v2_1/ | |
| Processing a retinal image | STARE | Retinal funds images | 400 images | http://cecas.clemson.edu/~ahoover/stare/ | |
| Diabetic retinopathy grade | APTOS 2019 | Fundus photography |
3662 train_images 1928 test_images |
https://www.kaggle.com/c/aptos2019-blindness-detection/overview/aptos-2019 | |
| Lung | Lung nodules | LIDC-IDRI | CT scans | 1018 images | http://ncia.nci.nih.gov/ |
| Lung cancer | NIH Chest X-ray | X-ray images | Over 100,000 images | https://nihcc.app.box.com/v/ChestXray-NIHCC | |
| Lung nodules | LUNA 16 | CT scans | 888 images | https://luna16.grand-challenge.org/Data/ | |
|
14 Common Thorax Disease Categories |
ChestX-ray14 | X-ray images | 112,120 images | https://nihcc.app.box.com/v/ChestXray-NIHCC/file 220660789610 | |
| Brain | Alzheimer’s disease | ANDI1 |
Multi-site MRI and PET data |
Participant: 200 Normal Controls, 400 mild cognitive impairment(MCI), 200Mild AD | http://adni.loni.usc.edu/study-design/#collaborative-studies-container |
| ADNI GO | Multi-site MRI and PET data | Participant: 200 the mildest symptomatic phase of AD (EMCI), 500 Normal Controls and MCI | |||
| ADNI2 | Multi-site MRI and PET data | Participant: 150 Normal Controls, 450–500 CN and MCI, 350 EMCI, 150 LMCI, 200 mild AD | |||
| ADNI3 | Multi-site MRI and PET data |
Participant: 430-830Normal Controls, 425-835 MCI, 215-335 Mild Alzheimer’s, disease dementia |
|||
| Parkinson’s disease | PPMI | DaTscan SPECT images | 642 images | https://www.ppmi-info.org/ | |
| https://challenge2018.isic-archive.com/ | |||||
| Skin | Skin lesions | ISIC 2018 challenge Task1 | Dermoscopic lesion images | 2594 images and 2594 corresponding ground truth response masks | |
| ISIC 2018 challenge Task2 | Dermoscopic lesion images |
2594 images and 12,970 corresponding ground truth response masks (5 for each image) |
|||
| ISIC 2018 challenge Task3 | Dermoscopic lesion images | 10,015 images and 1 ground truth response CSV file |
Challenges and future directions
Medical diagnosis with interpretability is making convincing progress [109, 110], but there is still some way to go before it can be truly applied in clinical practice. In this section, we point out several challenges of interpretability that need to be faced, as well as its future directions for further research in the medical field.
Evaluation methods of interpretability
Although the deep learning model with interpretability has yielded some positive outcomes in the medical field, there is still no standard concept or evaluation metrics for interpretability. In general, the effect of interpretability is evaluated by intuitively observing the visual images, but in the medical domain, such qualitative evaluation may potentially have an impact on the final diagnosis by judging with the naked eye. To make the interpretability of the model more reliable and convincing, it is necessary to focus on the quantitative analysis of the interpretability methods and pay more attention to the effectiveness of applications in actual clinical practice. In addition, the method of grade assessment by experts could be designed to judge the performance of model interpretability. Only with well-defined and interpretable evaluation methods can the applications of deep learning models in the medical field gain the trust of physicians and be applied to clinical practice as early as possible.
Trade-off between interpretability and accuracy
Deep learning is widely used in various fields with remarkable performance, attributed to the ability of deep networks to learn richer information. However, as the network structure gets deeper and the accuracy becomes higher, the model becomes more difficult to interpret. To address this problem, we can consider breaking the black box of neural networks into modules with specific functions. For the medical field, we consider classifying diseases according to their locations and diagnostic methods, encapsulating them into separate modules, and eventually explaining the entire decision-making process of the model by explaining each simple module. This will greatly improve the accuracy and interpretability of the model. In this way, the deep learning model will not be limited to an “end-to-end” structure but will be like building blocks that directly compose middle layer functional modules to achieve specific functions with interpretability.
Multiple and complete interpretability
Most researchers adopt a single method or modality to diagnose diseases and provide interpretability [111]. However, adopting multiple modalities, such as patients’ records and medical images, in the decision-making process is one of the future trends. Thus, we can consider attributing the model decision-making process to each modality, thereby simulating the physician’s diagnostic process. For example, to detect whether a patient has pulmonary nodules, medical images need to be considered along with whether the patient has a genetic history, smokes, etc. Such simultaneous consideration of the patient’s medical images and physical parameters to make the final decision can not only potentially enhance the generalization and accuracy of the model, but also interpret these phenomena more holistically.
Design specific methods for medical diagnosis
Currently, a large number of interpretability methods have emerged, but most of them are extended based on the existing methods. For example, the Grad-CAM method is an extension of the CAM method that enables us to visualize any target layer of the network without structural changes. The Score-CAM method solves the gradient problem of the Grad-CAM method. Most of them are designed as general frameworks and not specifically for sensitive fields like the medical domain. In order to make interpretable techniques more suitable for the medical field while providing a more holistic interpretation, it is necessary to involve experts in the medical field in the model design process, not only to provide some expertise but also to understand the decision-making process of the model. This greatly enhances the trust of physicians and patients in the model, which in turn promotes the application of computer-aided diagnostic systems in clinical medical practice.
Conclusion
Interpretability has received considerable attention, especially in the medical domain. It is an effective way to break the “black-box” structure, provide interpretability of deep learning models, and ultimately gain user trust. In this paper, we have comprehensively reviewed some interpretability methods that have been widely applied to the medical domain, sorted out various applications with interpretability by categorizing the disease locations, and analyzed the challenges that need to be faced as well as their potential research directions. We hope this paper will help readers better understand the current state of interpretability in the medical field, so as to promote its applications to clinical practice as early as possible.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (61976106, 61772242); China Postdoctoral Science Foundation (2017M611737); Six talent peaks project in Jiangsu Province, China (DZXX-122); Key special projects of health and family planning science and technology in Zhenjiang City, China (SHW2017019); Innovation capacity building Foundation of Jilin Provincial Development and Reform Commission (2021C038-7).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Otter DW, Medina JR, Kalita JK. A survey of the usages of deep learning for natural language processing. IEEE Trans. Neural Netw. Learn. Syst. 2020;32(2):604–624. doi: 10.1109/TNNLS.2020.2979670. [DOI] [PubMed] [Google Scholar]
- 2.Minaee S, Kalchbrenner N, Cambria E, Nikzad N, Chenaghlu M, Gao J. Deep learning-based text classification: a comprehensive review. ACM Comput. Surv. 2021;54(3):1–40. doi: 10.1145/3439726. [DOI] [Google Scholar]
- 3.Chrysostomou, G., Aletras, N.: Improving the faithfulness of attention-based explanations with task-specific information for text classification (2021). at preprint arxiv:2105.02657
- 4.Schwartz E, Giryes R, Bronstein AM. Deepisp: Toward learning an end-to-end image processing pipeline. IEEE Trans. Image Process. 2018;28(2):912–923. doi: 10.1109/TIP.2018.2872858. [DOI] [PubMed] [Google Scholar]
- 5.Sun, J., Darbehani, F., Zaidi, M., Wang, B.: Saunet: Shape attentive u-net for interpretable medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 797–806 (2020) 10.1007/978-3-030-59719-1_77.Springer
- 6.Tian C, Xu Y, Zuo W. Image denoising using deep CNN with batch renormalization. Neural Netw. 2020;121:461–473. doi: 10.1016/j.neunet.2019.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Murcia FJ, Ortiz A, Ramírez J, Górriz JM, Cruz R. Deep residual transfer learning for automatic diagnosis and grading of diabetic retinopathy. Neurocomputing. 2021;452:424–434. doi: 10.1016/j.neucom.2020.04.148. [DOI] [Google Scholar]
- 8.Sun, M., Huang, Z., Guo, C.: Automatic diagnosis of alzheimer’s disease and mild cognitive impairment based on cnn+ svm networks with end-to-end training. In: 2021 13th International Conference on Advanced Computational Intelligence (ICACI), pp. 279–285 (2021) 10.1109/ICACI52617.2021.9435894. IEEE [DOI] [PMC free article] [PubMed]
- 9.Goel T, Murugan R, Mirjalili S, Chakrabartty DK. Optconet: an optimized convolutional neural network for an automatic diagnosis of covid-19. Appl. Intell. 2021;51(3):1351–1366. doi: 10.1007/s10489-020-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunning D, Aha D. Darpa’s explainable artificial intelligence (xai) program. AI Magazine. 2019;40(2):44–58. doi: 10.1609/aimag.v40i2.2850. [DOI] [Google Scholar]
- 11.Singh A, Sengupta S, Lakshminarayanan V. Explainable deep learning models in medical image analysis. J. Imaging. 2020;6(6):52. doi: 10.3390/jimaging6060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tjoa E, Guan C. A survey on explainable artificial intelligence (xai): toward medical xai. IEEE Trans. Neural Netw. Learn. Syst. 2020;32(11):4793–4813. doi: 10.1109/TNNLS.2020.3027314. [DOI] [PubMed] [Google Scholar]
- 13.Messalas, A., Kanellopoulos, Y., Makris, C.: Model-agnostic interpretability with shapley values. In: 2019 10th International Conference on Information, Intelligence, Systems and Applications (IISA), pp. 1–7 (2019). 10.1109/IISA.2019.8900669. IEEE
- 14.Da Cruz HF, Pfahringer B, Martensen T, Schneider F, Meyer A, Böttinger E, Schapranow M-P. Using interpretability approaches to update black-box clinical prediction models: an external validation study in nephrology. Artif. Intell. Med. 2021;111:101982. doi: 10.1016/j.artmed.2020.101982. [DOI] [PubMed] [Google Scholar]
- 15.Pedapati T, Balakrishnan A, Shanmugam K, Dhurandhar A. Learning global transparent models consistent with local contrastive explanations. Adv. Neural. Inf. Process. Syst. 2020;33:3592–3602. [Google Scholar]
- 16.Moraffah R, Karami M, Guo R, Raglin A, Liu H. Causal interpretability for machine learning-problems, methods and evaluation. ACM SIGKDD Explor. Newslett. 2020;22(1):18–33. doi: 10.1145/3400051.3400058. [DOI] [Google Scholar]
- 17.Murdoch WJ, Singh C, Kumbier K, Abbasi-Asl R, Yu B. Definitions, methods, and applications in interpretable machine learning. Proc. Natl. Acad. Sci. 2019;116(44):22071–22080. doi: 10.1073/pnas.1900654116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes M, Meier R, Pereira S, Silva CA, Dahlweid F-M, Tengg-Kobligk HV, Summers RM, Wiest R. On the interpretability of artificial intelligence in radiology: challenges and opportunities. Radiology. 2020;2(3):190043. doi: 10.1148/ryai.2020190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen A, Yosinski J, Clune J. Explainable AI: Interpreting, Explaining and Visualizing Deep Learning. Cham: Springer; 2019. Understanding neural networks via feature visualization: A survey; pp. 55–76. [Google Scholar]
- 20.Qin, Z., Yu, F., Liu, C., Chen, X.: How convolutional neural network see the world-a survey of convolutional neural network visualization methods (2018). at print
- 21.Yuan, H., Chen, Y., Hu, X., Ji, S.: Interpreting deep models for text analysis via optimization and regularization methods. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, pp. 5717–5724 (2019) 10.1609/aaai.v33i01.33015717
- 22.Xu, F., Uszkoreit, H., Du, Y., Fan, W., Zhao, D., Zhu, J.: Explainable ai: A brief survey on history, research areas, approaches and challenges. In: CCF International Conference on Natural Language Processing and Chinese Computing, pp. 563–574 (2019) 10.1007/978-3-030-32236-6_51. Springer
- 23.Lapuschkin S, Wäldchen S, Binder A, Montavon G, Samek W, Müller K-R. Unmasking clever hans predictors and assessing what machines really learn. Nat. Commun. 2019;10(1):1–8. doi: 10.1038/s41467-019-08987-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohlbrenner, M., Bauer, A., Nakajima, S., Binder, A., Samek, W., Lapuschkin, S.: Towards best practice in explaining neural network decisions with lrp. In: 2020 International Joint Conference on Neural Networks (IJCNN), pp. 1–7 (2020). 10.1109/IJCNN48605.2020.9206975. IEEE
- 25.Montavon, G., Binder, A., Lapuschkin, S., Samek, W., Müller, K.-R.: Layer-wise relevance propagation: an overview. Explainable AI: interpreting, explaining and visualizing deep learning, 193–209 (2019) 10.1007/978-3-030-28954-6_10
- 26.Gu, J., Yang, Y., Tresp, V.: Understanding individual decisions of cnns via contrastive backpropagation. In: Asian Conference on Computer Vision, pp. 119–134 (2018) 10.1007/978-3-030-20893-6_8. Springer
- 27.Lee, J.R., Kim, S., Park, I., Eo, T., Hwang, D.: Relevance-cam: Your model already knows where to look. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 14944–14953 (2021)
- 28.Shrikumar, A., Greenside, P., Kundaje, A.: Learning important features through propagating activation differences. In: International Conference on Machine Learning, pp. 3145–3153 (2017). PMLR
- 29.Shrikumar, A., Greenside, P., Shcherbina, A., Kundaje, A.: Not just a black box: Learning important features through propagating activation differences. arXiv preprint arXiv:1605.01713 (2016). at reprint
- 30.Ras G, Xie N, van Gerven M, Doran D. Explainable deep learning: a field guide for the uninitiated. J. Artif. Intell. Res. 2022;73:329–397. doi: 10.1613/jair.1.13200. [DOI] [Google Scholar]
- 31.Ancona M, Ceolini E, Öztireli C, Gross M. Explainable AI: Interpreting, Explaining and Visualizing Deep Learning. Cham: Springer; 2019. Gradient-based attribution methods; pp. 169–191. [Google Scholar]
- 32.Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2921–2929 (2016)
- 33.Selvaraju, R.R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., Batra, D.: Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 618–626 (2017)
- 34.Wang, H., Wang, Z., Du, M., Yang, F., Zhang, Z., Ding, S., Mardziel, P., Hu, X.: Score-cam: Score-weighted visual explanations for convolutional neural networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 24–25 (2020)
- 35.Ramaswamy, H.G.: Ablation-cam: Visual explanations for deep convolutional network via gradient-free localization. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 983–991 (2020)
- 36.Zhang, Q., Rao, L., Yang, Y.: Group-cam: Group score-weighted visual explanations for deep convolutional networks. arXiv preprint arXiv:2103.13859 (2021). at reprint
- 37.Chattopadhay, A., Sarkar, A., Howlader, P., Balasubramanian, V.N.: Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), pp. 839–847 (2018). IEEE
- 38.Joshi, A., Mishra, G., Sivaswamy, J.: Explainable disease classification via weakly-supervised segmentation. In: Interpretable and Annotation-Efficient Learning for Medical Image Computing, pp. 54–62 (2020). 10.1007/978-3-030-61166-8_6
- 39.Samek W, Montavon G, Lapuschkin S, Anders CJ, Müller K-R. Explaining deep neural networks and beyond: a review of methods and applications. Proc. IEEE. 2021;109(3):247–278. doi: 10.1109/JPROC.2021.3060483. [DOI] [Google Scholar]
- 40.Pintelas E, Livieris IE, Pintelas P. A grey-box ensemble model exploiting black-box accuracy and white-box intrinsic interpretability. Algorithms. 2020;13(1):17. doi: 10.3390/a13010017. [DOI] [Google Scholar]
- 41.Mi J-X, Li A-D, Zhou L-F. Review study of interpretation methods for future interpretable machine learning. IEEE Access. 2020;8:191969–191985. doi: 10.1109/ACCESS.2020.3032756. [DOI] [Google Scholar]
- 42.Wang J, Gou L, Zhang W, Yang H, Shen H-W. Deepvid: Deep visual interpretation and diagnosis for image classifiers via knowledge distillation. IEEE Trans. Vis. Comput. Graph. 2019;25(6):2168–2180. doi: 10.1109/TVCG.2019.2903943. [DOI] [PubMed] [Google Scholar]
- 43.Gou J, Yu B, Maybank SJ, Tao D. Knowledge distillation: a survey. Int. J. Comput. Vis. 2021;129(6):1789–1819. doi: 10.1007/s11263-021-01453-z. [DOI] [Google Scholar]
- 44.Wang L, Yoon K-J. Knowledge distillation and student-teacher learning for visual intelligence: a review and new outlooks. IEEE Trans. Pattern Anal. Mach. Intell. 2021 doi: 10.1109/TPAMI.2021.3055564. [DOI] [PubMed] [Google Scholar]
- 45.Du M, Liu N, Hu X. Techniques for interpretable machine learning. Commun. ACM. 2019;63(1):68–77. doi: 10.1145/3359786. [DOI] [Google Scholar]
- 46.Mohankumar, A.K., Nema, P., Narasimhan, S., Khapra, M.M., Srinivasan, B.V., Ravindran, B.: Towards transparent and explainable attention models. arXiv preprint arXiv:2004.14243 (2020). at reprint
- 47.Serrano, S., Smith, N.A.: Is attention interpretable? arXiv preprint arXiv:1906.03731 (2019). at reprint
- 48.Jain, S., Wallace, B.C.: Attention is not explanation. arXiv preprint arXiv:1902.10186 (2019). at reprint
- 49.Wiegreffe, S., Pinter, Y.: Attention is not not explanation. arXiv preprint arXiv:1908.04626 (2019) at reprint
- 50.Arrieta AB, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, García S, Gil-López S, Molina D, Benjamins R. Explainable artificial intelligence (xai): Concepts, taxonomies, opportunities and challenges toward responsible ai. Inf. Fus. 2020;58:82–115. doi: 10.1016/j.inffus.2019.12.012. [DOI] [Google Scholar]
- 51.Margot, V., Luta, G.: A new method to compare the interpretability of rule-based algorithms. AI 2(4), 621–635 (2021). 10.3390/ai2040037
- 52.Kind, A., Azzopardi, G.: An explainable ai-based computer aided detection system for diabetic retinopathy using retinal fundus images. In: International Conference on Computer Analysis of Images and Patterns, pp. 457–468 (2019) 10.1007/978-3-030-29888-3_37. Springer
- 53.de La Torre J, Valls A, Puig D. A deep learning interpretable classifier for diabetic retinopathy disease grading. Neurocomputing. 2020;396:465–476. doi: 10.1016/j.neucom.2018.07.102. [DOI] [Google Scholar]
- 54.Kumar D, Taylor GW, Wong A. Discovery radiomics with clear-dr: interpretable computer aided diagnosis of diabetic retinopathy. IEEE Access. 2019;7:25891–25896. doi: 10.1109/ACCESS.2019.2893635. [DOI] [Google Scholar]
- 55.Jiang, H., Yang, K., Gao, M., Zhang, D., Ma, H., Qian, W.: An interpretable ensemble deep learning model for diabetic retinopathy disease classification. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 2045–2048 (2019) 10.1109/EMBC.2019.8857160. IEEE [DOI] [PubMed]
- 56.Jiang, H., Xu, J., Shi, R., Yang, K., Zhang, D., Gao, M., Ma, H., Qian, W.: A multi-label deep learning model with interpretable grad-cam for diabetic retinopathy classification. In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pp. 1560–1563 (2020) 10.1109/EMBC44109.2020.9175884. IEEE [DOI] [PubMed]
- 57.Chetoui, M., Akhloufi, M.A.: Explainable diabetic retinopathy using efficientnet. In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pp. 1966–1969 (2020). 10.1109/EMBC44109.2020.9175664. IEEE [DOI] [PubMed]
- 58.Li, L., Xu, M., Wang, X., Jiang, L., Liu, H.: Attention based glaucoma detection: a large-scale database and cnn model. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 10571–10580 (2019)
- 59.Liao W, Zou B, Zhao R, Chen Y, He Z, Zhou M. Clinical interpretable deep learning model for glaucoma diagnosis. IEEE J. Biomed. Health Inform. 2019;24(5):1405–1412. doi: 10.1109/JBHI.2019.2949075. [DOI] [PubMed] [Google Scholar]
- 60.Mojab, N., Noroozi, V., Philip, S.Y., Hallak, J.A.: Deep multi-task learning for interpretable glaucoma detection. In: 2019 IEEE 20th International Conference on Information Reuse and Integration for Data Science (IRI), pp. 167–174 (2019). 10.1109/IRI.2019.00037. IEEE
- 61.Fang L, Wang C, Li S, Rabbani H, Chen X, Liu Z. Attention to lesion: Lesion-aware convolutional neural network for retinal optical coherence tomography image classification. IEEE Trans. Med. Imaging. 2019;38(8):1959–1970. doi: 10.1109/TMI.2019.2898414. [DOI] [PubMed] [Google Scholar]
- 62.Liu, J., Zhao, G., Fei, Y., Zhang, M., Wang, Y., Yu, Y.: Align, attend and locate: Chest x-ray diagnosis via contrast induced attention network with limited supervision. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10632–10641 (2019)
- 63.Guan Q, Huang Y. Multi-label chest x-ray image classification via category-wise residual attention learning. Pattern Recogn. Lett. 2020;130:259–266. doi: 10.1016/j.patrec.2018.10.027. [DOI] [Google Scholar]
- 64.Huang, Z., Fu, D.: Diagnose chest pathology in x-ray images by learning multi-attention convolutional neural network. In: 2019 IEEE 8th Joint International Information Technology and Artificial Intelligence Conference (ITAIC), pp. 294–299 (2019). 10.1109/ITAIC.2019.8785431. IEEE
- 65.Zhang, X., Chen, T.: Attention u-net for interpretable classification on chest x-ray image. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 901–908 (2020) 10.1109/BIBM49941.2020.9313354 IEEE
- 66.Li, Y., Gu, D., Wen, Z., Jiang, F., Liu, S.: Classify and explain: An interpretable convolutional neural network for lung cancer diagnosis. In: ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1065–1069 (2020) 10.1109/ICASSP40776.2020.9054605 IEEE
- 67.Kumar D, Sankar V, Clausi D, Taylor GW, Wong A. Sisc: End-to-end interpretable discovery radiomics-driven lung cancer prediction via stacked interpretable sequencing cells. IEEE Access. 2019;7:145444–145454. doi: 10.1109/ACCESS.2019.2945524. [DOI] [Google Scholar]
- 68.Shen S, Han SX, Aberle DR, Bui AA, Hsu W. An interpretable deep hierarchical semantic convolutional neural network for lung nodule malignancy classification. Expert Syst. Appl. 2019;128:84–95. doi: 10.1016/j.eswa.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen, S., Han, S.X., Aberle, D.R., Bui, A.A., Hsu, W.: Explainable hierarchical semantic convolutional neural network for lung cancer diagnosis. In: CVPR Workshops, pp. 63–66 (2019) [DOI] [PMC free article] [PubMed]
- 70.Jiang H, Shen F, Gao F, Han W. Learning efficient, explainable and discriminative representations for pulmonary nodules classification. Pattern Recogn. 2021;113:107825. doi: 10.1016/j.patcog.2021.107825. [DOI] [Google Scholar]
- 71.Elsken T, Metzen JH, Hutter F. Neural architecture search: a survey. J. Mach. Learn. Res. 2019;20(1):1997–2017. [Google Scholar]
- 72.Ramchandani A, Fan C, Mostafavi A. Deepcovidnet: An interpretable deep learning model for predictive surveillance of covid-19 using heterogeneous features and their interactions. IEEE Access. 2020;8:159915–159930. doi: 10.1109/ACCESS.2020.3019989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casiraghi E, Malchiodi D, Trucco G, Frasca M, Cappelletti L, Fontana T, Esposito AA, Avola E, Jachetti A, Reese J. Explainable machine learning for early assessment of covid-19 risk prediction in emergency departments. IEEE Access. 2020;8:196299–196325. doi: 10.1109/ACCESS.2020.3034032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi, W., Tong, L., Zhuang, Y., Zhu, Y., Wang, M.D.: Exam: an explainable attention-based model for covid-19 automatic diagnosis. In: Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, pp. 1–6 (2020)
- 75.Wu Y-H, Gao S-H, Mei J, Xu J, Fan D-P, Zhang R-G, Cheng M-M. Jcs: an explainable COVID-19 diagnosis system by joint classification and segmentation. IEEE Trans. Image Process. 2021;30:3113–3126. doi: 10.1109/TIP.2021.3058783. [DOI] [PubMed] [Google Scholar]
- 76.Brunese L, Mercaldo F, Reginelli A, Santone A. Explainable deep learning for pulmonary disease and coronavirus covid-19 detection from x-rays. Comput. Methods Programs Biomed. 2020;196:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh RK, Pandey R, Babu RN. Covidscreen: explainable deep learning framework for differential diagnosis of covid-19 using chest x-rays. Neural Comput. Appl. 2021;33(14):8871–8892. doi: 10.1007/s00521-020-05636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karim, M.R., Döhmen, T., Cochez, M., Beyan, O., Rebholz-Schuhmann, D., Decker, S.: Deepcovidexplainer: explainable covid-19 diagnosis from chest x-ray images. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 1034–1037 (2020) 10.1109/BIBM49941.2020.9313304.IEEE
- 79.Alshazly H, Linse C, Barth E, Martinetz T. Explainable covid-19 detection using chest CT scans and deep learning. Sensors. 2021;21(2):455. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang Z, Chuang KV, DeCarli C, Jin L-W, Beckett L, Keiser MJ, Dugger BN. Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline. Nat. Commun. 2019;10(1):1–14. doi: 10.1038/s41467-019-10212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nigri, E., Ziviani, N., Cappabianco, F., Antunes, A., Veloso, A.: Explainable deep cnns for mri-based diagnosis of alzheimer’s disease. In: 2020 International Joint Conference on Neural Networks (IJCNN), pp. 1–8 (2020) IEEE
- 82.Wang, N., Chen, M., Subbalakshmi, K.P.: Explainable cnn-attention networks (c-attention network) for automated detection of alzheimer’s disease. arXiv preprint arXiv:2006.14135 (2020) at reprint
- 83.El-Sappagh S, Alonso JM, Islam S, Sultan AM, Kwak KS. A multilayer multimodal detection and prediction model based on explainable artificial intelligence for Alzheimer’s disease. Sci. Rep. 2021;11(1):1–26. doi: 10.1038/s41598-021-82098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeng, Z., Shen, Z., Tan, B.T.H., Chin, J.J., Leung, C., Wang, Y., Chi, Y., Miao, C.: Explainable and argumentation-based decision making with qualitative preferences for diagnostics and prognostics of alzheimer’s disease. In: Proceedings of the International Conference on Principles of Knowledge Representation and Reasoning, vol. 17, pp. 816–826 (2020)
- 85.Achilleos, K.G., Leandrou, S., Prentzas, N., Kyriacou, P.A., Kakas, A.C., Pattichis, C.S.: Extracting explainable assessments of alzheimer’s disease via machine learning on brain mri imaging data. In: 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), pp. 1036–1041 (2020) 10.1109/BIBE50027.2020.00175. IEEE
- 86.Magesh PR, Myloth RD, Tom RJ. An explainable machine learning model for early detection of parkinson’s disease using lime on datscan imagery. Comput. Biol. Med. 2020;126:104041. doi: 10.1016/j.compbiomed.2020.104041. [DOI] [PubMed] [Google Scholar]
- 87.Cavaliere, F., Della Cioppa, A., Marcelli, A., Parziale, A., Senatore, R.: Parkinson’s disease diagnosis: towards grammar-based explainable artificial intelligence. In: 2020 IEEE Symposium on Computers and Communications (ISCC), pp. 1–6 (2020) 10.1109/ISCC50000.2020.9219616. IEEE
- 88.Van Steenkiste, T., Deschrijver, D., Dhaene, T.: Interpretable ecg beat embedding using disentangled variational auto-encoders. In: 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), pp. 373–378 (2019) 10.1109/CBMS.2019.00081. IEEE
- 89.Mousavi S, Afghah F, Acharya UR. Han-ecg: an interpretable atrial fibrillation detection model using hierarchical attention networks. Comput. Biol. Med. 2020;127:104057. doi: 10.1016/j.compbiomed.2020.104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clough, J.R., Oksuz, I., Puyol-Antón, E., Ruijsink, B., King, A.P., Schnabel, J.A.: Global and local interpretability for cardiac MRI classification. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 656–664 (2019) 10.1007/978-3-030-32251-9_72.Springer
- 91.Puyol-Antón, E., Chen, C., Clough, J.R., Ruijsink, B., Sidhu, B.S., Gould, J., Porter, B., Elliott, M., Mehta, V., Rueckert, D.: Interpretable deep models for cardiac resynchronisation therapy response prediction. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 284–293 (2020) 10.1007/978-3-030-59710-8_28.Springer [DOI] [PMC free article] [PubMed]
- 92.Aghamohammadi, M., Madan, M., Hong, J.K., Watson, I.: Predicting heart attack through explainable artificial intelligence. In: International Conference on Computational Science, pp. 633–645 (2019) 10.1007/978-3-030-22741-8_45. Springer
- 93.Jones OT, Ranmuthu CK, Hall PN, Funston G, Walter FM. Recognising skin cancer in primary care. Adv. Ther. 2020;37(1):603–616. doi: 10.1007/s12325-019-01130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barata C, Celebi ME, Marques JS. Explainable skin lesion diagnosis using taxonomies. Pattern Recogn. 2021;110:107413. doi: 10.1016/j.patcog.2020.107413. [DOI] [Google Scholar]
- 95.Nguyen, D.M.H., Ezema, A., Nunnari, F., Sonntag, D.: A visually explainable learning system for skin lesion detection using multiscale input with attention u-net. In: German Conference on Artificial Intelligence (Künstliche Intelligenz), pp. 313–319 (2020) 10.1007/978-3-030-58285-2_28 Springer
- 96.Jiang S, Li H, Jin Z. A visually interpretable deep learning framework for histopathological image-based skin cancer diagnosis. IEEE J. Biomed. Health Inform. 2021;25(5):1483–1494. doi: 10.1109/JBHI.2021.3052044. [DOI] [PubMed] [Google Scholar]
- 97.Gu R, Wang G, Song T, Huang R, Aertsen M, Deprest J, Ourselin S, Vercauteren T, Zhang S. Ca-net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Trans. Med. Imaging. 2020;40(2):699–711. doi: 10.1109/TMI.2020.3035253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stieler, F., Rabe, F., Bauer, B.: Towards domain-specific explainable ai: model interpretation of a skin image classifier using a human approach. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 1802–1809 (2021)
- 99.van der Velden, B.H., Ragusi, M.A., Janse, M.H., Loo, C.E., Gilhuijs, K.G.: Interpretable deep learning regression for breast density estimation on MRI. In: Medical Imaging 2020: Computer-Aided Diagnosis, vol. 11314, p. 1131412 (2020) International Society for Optics and Photonics. 10.1117/12.2549003
- 100.Shen Y, Wu N, Phang J, Park J, Liu K, Tyagi S, Heacock L, Kim SG, Moy L, Cho K. An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization. Med. Image Anal. 2021;68:101908. doi: 10.1016/j.media.2020.101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sabol, P., Sinčák, P., Ogawa, K., Hartono, P.: explainable classifier supporting decision-making for breast cancer diagnosis from histopathological images. In: 2019 International Joint Conference on Neural Networks (IJCNN), pp. 1–8 (2019) 10.1109/IJCNN.2019.8852070.IEEE
- 102.Beykikhoshk A, Quinn TP, Lee SC, Tran T, Venkatesh S. Deeptriage: interpretable and individualised biomarker scores using attention mechanism for the classification of breast cancer sub-types. BMC Med. Genomics. 2020;13(3):1–10. doi: 10.1186/s12920-020-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gu D, Su K, Zhao H. A case-based ensemble learning system for explainable breast cancer recurrence prediction. Artif. Intell. Med. 2020;107:101858. doi: 10.1016/j.artmed.2020.101858. [DOI] [PubMed] [Google Scholar]
- 104.Thomas SM, Lefevre JG, Baxter G, Hamilton NA. Interpretable deep learning systems for multi-class segmentation and classification of non-melanoma skin cancer. Med. Image Anal. 2021;68:101915. doi: 10.1016/j.media.2020.101915. [DOI] [PubMed] [Google Scholar]
- 105.Li, H., Zhou, J., Zhou, Y., Chen, J., Gao, F., Xu, Y., Gao, X.: Automatic and interpretable model for periodontitis diagnosis in panoramic radiographs. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 454–463 (2020) 10.1007/978-3-030-59713-9_44.Springer
- 106.Vasquez-Morales GR, Martinez-Monterrubio SM, Moreno-Ger P, Recio-Garcia JA. Explainable prediction of chronic renal disease in the Colombian population using neural networks and case-based reasoning. IEEE Access. 2019;7:152900–152910. doi: 10.1109/ACCESS.2019.2948430. [DOI] [Google Scholar]
- 107.Penafiel S, Baloian N, Sanson H, Pino JA. Predicting stroke risk with an interpretable classifier. IEEE Access. 2020;9:1154–1166. doi: 10.1109/ACCESS.2020.3047195. [DOI] [Google Scholar]
- 108.Chary M, Boyer EW, Burns MM. Diagnosis of acute poisoning using explainable artificial intelligence. Comput. Biol. Med. 2021;134:104469. doi: 10.1016/j.compbiomed.2021.104469. [DOI] [PubMed] [Google Scholar]
- 109.Liu, F., Wu, X., Ge, S., Fan, W., Zou, Y.: Exploring and distilling posterior and prior knowledge for radiology report generation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 13753–13762 (2021)
- 110.Kwon BC, Choi M-J, Kim JT, Choi E, Kim YB, Kwon S, Sun J, Choo J. Retainvis: Visual analytics with interpretable and interactive recurrent neural networks on electronic medical records. IEEE Trans. Visual Comput. Graphics. 2018;25(1):299–309. doi: 10.1109/TVCG.2018.2865027. [DOI] [PubMed] [Google Scholar]
- 111.Lucieri, A., Bajwa, M.N., Dengel, A., Ahmed, S.: Achievements and challenges in explaining deep learning based computer-aided diagnosis systems. arXiv preprint arXiv:2011.13169 (2020) at reprint