Abstract
We have applied the soluble pyridine nucleotide transhydrogenase of Pseudomonas fluorescens to a cell-free system for the regeneration of the nicotinamide cofactors NAD and NADP in the biological production of the important semisynthetic opiate drug hydromorphone. The original recombinant whole-cell system suffered from cofactor depletion resulting from the action of an NADP+-dependent morphine dehydrogenase and an NADH-dependent morphinone reductase. By applying a soluble pyridine nucleotide transhydrogenase, which can transfer reducing equivalents between NAD and NADP, we demonstrate with a cell-free system that efficient cofactor cycling in the presence of catalytic amounts of cofactors occurs, resulting in high yields of hydromorphone. The ratio of morphine dehydrogenase, morphinone reductase, and soluble pyridine nucleotide transhydrogenase is critical for diminishing the production of the unwanted by-product dihydromorphine and for optimum hydromorphone yields. Application of the soluble pyridine nucleotide transhydrogenase to the whole-cell system resulted in an improved biocatalyst with an extended lifetime. These results demonstrate the usefulness of the soluble pyridine nucleotide transhydrogenase and its wider application as a tool in metabolic engineering and biocatalysis.
The pyridine nucleotide cofactors NAD and NADP are essential components of the cell, where they act as electron carriers in reduction and oxidation reactions. A large percentage of enzymes are dependent on these coenzymes for their activities (SwissProt database, http://www.expasy.ch/sprot/), and the cofactors are known to be involved in a vast amount of reactions (EcoCyc database). Many of the NAD(P)-dependent oxidoreductases catalyze reactions of commercial interest and have many applications, for instance, in the production of chiral compounds, amino acids, steroids, and other therapeutics for the pharmaceutical industry, in the modification or synthesis of polymers, in the oxidative remediation of pollutants, in the oxyfunctionalization of hydrocarbons, and in the construction of biosensors (14, 18). The high cost of the pyridine nucleotide cofactors which need to be provided in stoichiometric quantities in enzyme reactions is an important commercial issue. Cofactor regeneration is, therefore, an important consideration when processes involving NAD(P)-dependent oxidoreductases are to be applied in a commercial setting.
In cell-free systems the cofactors must be supplied, albeit at a lower-than-stoichiometric concentration (catalytic amounts), when cofactor regeneration is achieved. Alternatively, processes dependent on these cofactors can be carried out in whole cells which are known to have some reserves of the cofactor, but also here cofactor depletion can be a problem. Mostly NAD is regenerated using formate dehydrogenase, while NADP is recycled using glucose dehydrogenase. Recently, Galkin et al. described the synthesis of optically active amino acids from 2-keto acids using recombinant Escherichia coli coexpressing an amino acid dehydrogenase and a formate dehydrogenase for recycling NAD (10), and NADP recycling by recombinant E. coli expressing glucose dehydrogenase has similarly been described (17), although in both cell-free and whole-cell systems the supply of extra substrate, respectively, formate and glucose, is necessary.
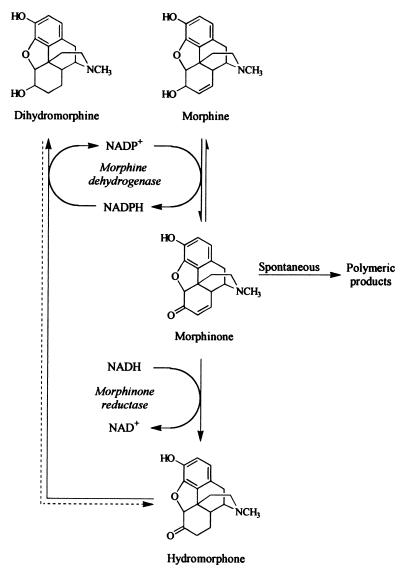
The biological production of the potent analgesic hydromorphone is an example of a biotransformation process that suffers problems of cofactor depletion. The whole-cell recombination process is mediated by two constitutively expressed Pseudomonas enzymes, an NADP+-dependent morphine dehydrogenase (MDH) and an NADH-dependent morphinone reductase (MR) (5, 6, 8, 12) (Fig. 1). Although this biotransformation system was promising in that it was the first to demonstrate the biological production of hydromorphone, some problems were identified (Fig. 1). The unwanted by-product dihydromorphine accumulated; this by-product, together with the depletion of cofactors, the poor solubility of the opiate substrate, and the instability of the intermediate morphinone (22), allowed only a limited amount of material to be transformed to hydromorphone and thereby decreased the lifetime and usefulness of the system. It was envisaged that hydromorphone yields could be improved by increasing the ratio of MR to MDH and by enhancing the transfer of reducing equivalents from NADPH, produced in the MDH reaction, to NADH, needed for the irreversible MR reaction.
FIG. 1.
Transformation of morphine by MDH and MR from P. putida M10.
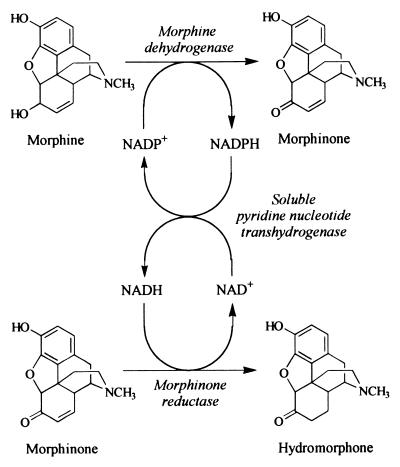
Here we address the problems of cofactor depletion in the biological production of hydromorphone by using the soluble pyridine nucleotide transhydrogenase (STH) of Pseudomonas fluorescens (7), an enzyme which can catalyze the transfer of reducing equivalents according to the following equation: NAD+ + NADPH ⇌ NADH + NADP+. We show that efficient cofactor cycling (Fig. 2) can be achieved using this enzyme and that a reuseable whole-cell biocatalyst is produced. The results demonstrate the usefulness of STH as a tool in biocatalysis and metabolic engineering.
FIG. 2.
Cofactor cycling by STH in the biological production of hydromorphone.
MATERIALS AND METHODS
Chemicals and other materials.
NADH, NADPH, NAD+, NADP+, and thionicotinamide NAD+ were obtained from Sigma (Poole, Dorset, United Kingdom). Restriction enzymes were purchased from New England Biolabs Ltd. (Hitchin, United Kingdom). Morphine alkaloids were kindly donated by Macfarlan Smith Ltd. (Edinburgh, United Kingdom). Other reagents were of analytical or higher grade. MDH, MR, and STH were purified from recombinant E. coli as described previously (5, 7, 8). The specific activities of the enzyme preparations employed were 11 U of MDH per mg, 6 U of MR per mg, and 276 U of STH per mg.
Expression constructs.
The original pBluescript SK+-derived expression construct pMORAB5 containing both morA (which encodes MDH) and morB (which encodes MR) with their respective constitutive Pseudomonas promoters (9) was employed for the whole-cell biotransformations. The P. fluorescens sth gene was introduced into E. coli on a compatible plasmid, ps 1EMBL (21). The sth gene was excised from pSTH1 (7) as a 1.5-kb PstI-SalI fragment and subcloned into the PstI-XhoI sites of the low-copy-number plasmid ps 1EMBL, and the resulting plasmid was denoted pPFSTH4.
A second construct was made where the morA gene was replaced by morA C80S · K244M (25), which encodes a less active and more stable MDH. A 1.2-kb PstI fragment bearing the mutant morA gene complete with its upstream ribosome binding site and promoter sequences was excised from pMDH1.7 (25) and ligated into PstI-digested ps 1EMBL (21). This resulted in the construct pMORA4C80S · K244M, which contained suitable restriction sites for further subcloning. Transformants were selected by growth on Luria-Bertani agar containing kanamycin at 50 μg/ml, 5-bromo-4-chloro-3-indoyl-β-d-galactoside at 32 mg/ml and 0.5 mM isopropyl-β-d-thiogalactopyranoside. Plasmid DNA was prepared from white insert-containing colonies and analyzed by digestion with MslI and PstI to determine the orientation of the insert. A 1.2-kb HindIII/EcoRI fragment carrying the mutant morA gene, ribosome binding site, and promoter region was excised from pMORA4K244M · C80S and ligated into HindIII- and EcoRI-digested pMORB3 (9), which carried a single copy of morB, together with its ribosome binding site and promoter region, creating the construct pMORB3-AC80S · K244M.
Strains and culture conditions.
E. coli JM109 was used as the host organism for all cloning transformations and whole-cell biotransformations. Plasmids were introduced into E. coli JM109 using the method of Hanahan (13). In cases where cells were to contain pPFSTH4 in addition to other plasmids, E. coli JM109 was transformed sequentially with the individual plasmids. Transformants were maintained on Luria-Bertani agar plates (23). Cells were routinely grown in SOB medium (23) at 37°C on a rotating shaker with the addition of ampicillin or carbenicillin at 100 μg/ml and/or kanamycin at 50 μg/ml to select for the presence of plasmids when appropriate. When cells were to be used for biotransformation purposes, a small culture, grown for approximately 16 h as described above, was used as a 5 to 10% (vol/vol) inoculum in 500 ml of fresh medium with antibiotics, which was then grown to stationary phase at 37°C over approximately 16 h.
Cell-free biotransformations.
Cell-free transformations were performed at 30°C in a solution containing 50 mM Tris-HCl (pH 8.0), 1 mM dithiothreitol, 20 mM morphine, and purified preparations of MDH, MR, and STH in a total volume of 200 μl. Transformations were performed in the presence of catalytic amounts of cofactors, either with NADPH and NAD+, both at a concentration of 0.2 mM, or with the four cofactors at the following concentrations: 2 mM NAD+, 0.15 mM NADH, 0.25 mM NADP+, and 0.2 mM NADPH. Samples (30 μl) were taken at regular intervals, and proteins were removed by precipitation with glacial acetic acid and centrifugation prior to high-performance liquid chromatography (HPLC) analysis.
Whole-cell incubations.
Cells grown as indicated above were harvested by centrifugation at 17,310 × g for 15 min at 4°C. Cells were then washed with 50 mM Tris-HCl (pH 8.0) and recentrifuged. The supernatant was removed, and the pelleted cells were used immediately or stored on ice for a few hours prior to use in biotransformations. Small-scale whole-cell biotransformations (3-ml total volume) were carried out using recombinant E. coli JM109 at a final cell density of 0.17 g (wet weight) per ml with 20 mM morphine in 50 mM Tris-HCl (pH 8.0). Biotransformations were carried out at 30°C on a rotary shaker, and samples (100 to 200 μl) were taken at regular intervals. Samples were clarified by centrifugation and analyzed for opiate content by HPLC.
Chromatography of alkaloids.
Samples were analyzed for opiate content by reverse-phase HPLC using a 5-μm (inside diameter) C18 Spherisorb column on a Waters Corporation (Milford, Mass.) model 2690 separation module with a Waters model 996 photodiode array detector. The mobile phase consisted of 20% (vol/vol) acetonitrile and 80% (vol/vol) 15 mM phosphate buffer (pH 3.5) (prepared by adjusting the pH of 15 mM KH2PO4 to 3.5 with H3PO4). A flow rate of 1 ml/min was used, and alkaloids were detected by absorbance at 230 nm. The method was essentially as described by French et al. (9) but without prior extraction, and analysis was performed using Millennium software (Waters Corporation).
MDH and MR assays.
Cell extracts of recombinant E. coli JM109 were prepared by French pressing cell suspensions with a density of 0.5 g (wet weight) per ml in 50 mM Tris-HCl (pH 8.0) at 2,500 lb/in2, followed by centrifugation at 31,180 × g 4°C for 1 h. Extracts were analyzed for MDH and MR activities using previously described assays (5, 8). One unit of MDH activity is the amount of enzyme necessary to reduce 1 μmol of NADP+ in 1 min at 30°C using morphine as the substrate, and 1 U of MR is the amount of enzyme required to oxidize 1 μmol of NADH in 1 min at 30°C using codeinone as the substrate. STH activity was measured as described by French et al. (7), who defined 1 U of activity as the amount of activity reducing 1 μmol of thionicotinamide NAD+ per min under the conditions stated.
RESULTS
Cell-free biotransformations.
The feasibility of cofactor cycling by the STH was initially tested in a cell-free system using purified enzyme preparations. The transformation of 20 mM morphine in the presence of 0.2 mM concentrations of the cofactors NADPH and NAD+ was evaluated in the presence and absence of 1.25 U of STH per ml. The transforming enzymes MDH and MR were used at three different ratios of activity by using equal amounts of MDH and MR (1.25 U/ml each), excess MDH (6.25 U of MDH per ml and 1.25 U of MR per ml), and excess MR (6.25 U of MR per ml and 1.25 U of MDH per ml). The biotransformations were performed at pH 8.0 as a compromise between the pH optima of the three enzymes (pH 9.6 for MDH [4], pH 7 to 8 for MR [8], and pH 7 to 8 for STH [not shown]). The enzymes were stable over the course of the biotransformation, since no loss of activity was observed when they were incubated in 50 mM Tris-HCl (pH 8.0) at 30°C for 9 h.
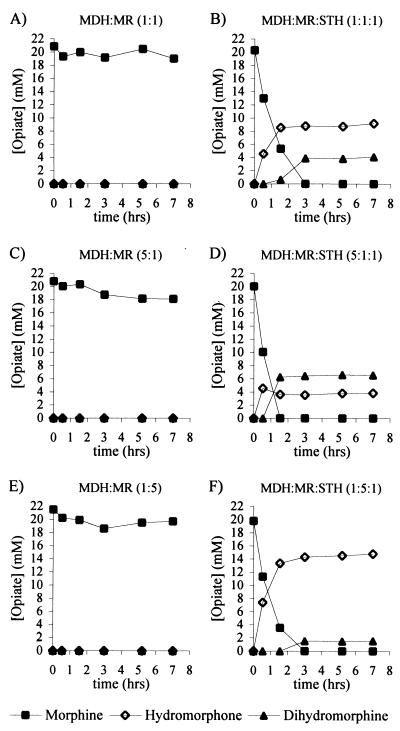
Samples were taken from the cell-free biotransformation at regular intervals and analyzed for opiate content (Fig. 3). In incubations with only MDH and MR, there was no conversion of morphine (Fig. 3A, C, and E); however, when STH was included in the incubation mixture, morphine was found to be converted to hydromorphone and dihydromorphine (Fig. 3B, D, and F).
FIG. 3.
Cell-free biotransformation of morphine at various ratios of MDH to MR in the presence and absence of STH. The transformations were performed without (A, C, and E) or with (B, D, and F) 1.25 U of STH per ml. MDH and MR were used as the ratios 1:1 (1.25 U of each per ml) (A and B), 5:1 (6.25 U of MDH per ml and 1.25 U of MR per ml (C and D), 1:5 (1.25 U of MDH per ml and 6.25 U of MR per ml (E and F). The cofactors NADPH and NAD+ were supplied at a concentration of 0.2 mM.
Various amounts of hydromorphone and the unwanted side-product dihydromorphine were observed depending on the ratio of MDH and MR. With equal amounts of MDH and MR, 46% hydromorphone and 20% dihydromorphine were produced (Fig. 3B). An excess of MDH resulted in the low yields of hydromorphone (21%) and high levels of dihydromorphine (34%) (Fig. 3D). An excess of MR reduced the amount of the unwanted side-product dihydromorphine (8.4%) and increased hydromorphone yields (78%) (Fig. 3F).
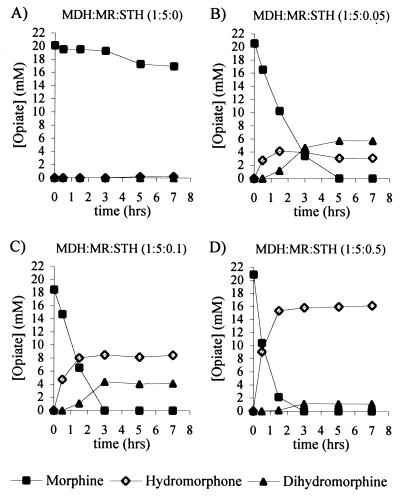
Using an excess amount of MR (6.25 U/ml) compared to that of MDH (1.25 U/ml), the effect of various amounts of STH on the biotransformation was investigated (Fig. 4). This analysis was performed using catalytic amounts of cofactor at ratios likely to be found in the cell (2, 16, 26). The cofactor concentrations used (2 mM NAD+, 0.15 mM NADH, 0.25 mM NADP+, and 0.2 mM NADPH) accommodated the production of only 0.15 mM (0.75% yield) hydromorphone without cofactor cycling. With as little as 0.0625 U of STH per ml compared to 6.25 U of MR per ml, conversion of morphine was observed, but only low levels of product accumulated (17% hydromorphone and 28% dihydromorphine) (Fig. 4B). By increasing the amount of STH, yields of hydromorphone were elevated while dihydromorphine levels decreased until an optimum was reached, after which no further improvement was observed (Fig. 4D). Hydromorphone yields of up to 84% were obtained, while as little as 4.5% dihydromorphine could be seen to accumulate.
FIG. 4.
Cell-free biotransformation of morphine in the presence of increasing amounts of STH. The transformations were performed with 1.25 U of MDH per ml and 6.25 U of MR per ml and various amounts of STH as follows: none (A), 0.0625 U/ml (B), 0.125 U/ml (C), and 0.625 U/ml (D). With 1.25 and 6.25 U of STH per ml, the result was similar to that shown in panel D. The cofactors used were 2 mM NAD+, 0.15 mM NADH, 0.25 mM NADP+, and 0.2 mM NADPH.
Construction of recombinant E. coli coexpressing MDH, MR, and STH.
In the original biotransformation system, the genes morA and morB, which encode MDH and MR, respectively, were coexpressed from the same pBluescript SK+-derived plasmid, pMORAB5 (9). We cloned the P. fluorescens sth gene on a compatible low-copy-number plasmid, ps 1EMBL (21), which resulted in the plasmid pPFSTH4. The two plasmids pMORAB5 and pPFSTH4 are compatible in E. coli due to their different origins of replication.
The original construct pMORAB5 expressed higher levels of MDH than MR, and as indicated in the cell-free biotransformations, this was unfavorable. Furthermore, MDH became inactivated by morphinone through the formation of a covalent adduct with Cys80, and it was found that changing this residue to a serine greatly enhanced the stability of MDH (25). A mutation, K244M, leading to an approximately 10-fold-less-active MDH was combined with the C80S mutation, to give morA C80S · K244M (25). The mutant morA was subcloned into the pBluescript SK+-based construct pMORB3, which expressed morB (9), resulting in the plasmid pMORB3-AC80S · K244M. The relative activities of MDH and MR could in this way be altered without the need of modifying gene expression, for instance, by the use of promoters of different strengths or induction methods.
Recombinant E. coli JM109 strains with and without pPFSTH4 were generated. The enzyme activities obtained from the different recombinant E. coli JM109 strains are shown in Table 1.
TABLE 1.
Enzyme activities obtained from expression constructs in E. coli JM109a
| Construct in E. coli JM109 | Sp act (U/mg) of:
|
||
|---|---|---|---|
| MDH | MR | STH | |
| pMORAB5 | 5.76 ± 1.02 | 1.92 ± 0.33 | — |
| pMORAB5/pPFSTH4 | 4.19 ± 0.07 | 1.60 ± 0.08 | 0.58 ± 0.01 |
| pMORB3-AC80S · K244M | 0.030 ± 0.001 | 0.84 ± 0.05 | — |
| pMORB3-C80S · K244M/pPFSTH4 | 0.040 ± 0.004 | 1.03 ± 0.07 | 0.45 ± 0.05 |
Figures shown are the averages of triplicate measurements ± the standard deviations. Crude cell extracts were prepared as described in the text. Units of enzyme activities are as defined in the text. —, no detectable activity.
Whole-cell biotransformations.
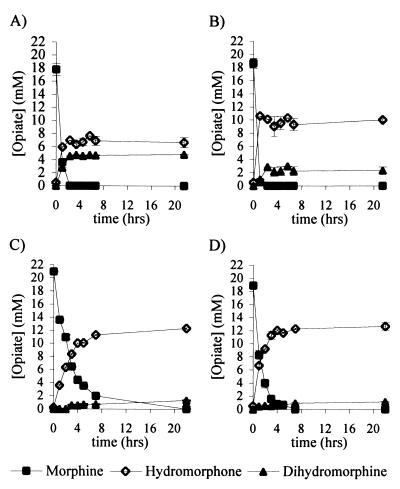
Whole-cell incubations with 20 mM morphine were performed with the recombinant strains (Fig. 5). Biotransformations with the original construct E. coli JM109/pMORAB5 demonstrated the rapid conversion of morphine and the rapid but low-level accumulation of hydromorphone to a final yield of 37.1% ± 2.4% (Fig. 5A). High levels (25.6% ± 0.5%) of the unwanted by-product dihydromorphine were also found to accumulate. When STH was present in the engineered strain E. coli JM109/pMORAB5/pPFSTH4, yields of hydromorphone were found to be improved (51.7% ± 1.0%) and less dihydromorphine accumulated (12.3% ± 2.7%) (Fig. 5B).
FIG. 5.
Whole-cell biotransformation of morphine with E. coli JM109/pMORAB5 (A), E. coli JM109/pMORAB5/pPFSTH4 (B), E. coli JM109/pMORB3-AC80S · K244M (C), and E. coli JM109/pMORB3-AC80S · K244M/pPFSTH4 (D). Figures shown are the averages of duplicate measurements, and error bars represent standard deviations.
In the system which possessed lower levels of MDH than of MR, E. coli JM109/pMORB3-AC80S · K244M, the yield of hydromorphone increased to 58.7% ± 0.6% while the level of dihydromorphine was reduced to 6.3% ± 0.1% (Fig. 5C). In comparison, the E. coli strain containing STH, JM109/pMORB3-AC80S · K244M/pPFSTH4, appeared to convert morphine more rapidly (Fig. 5D), and final yields were slightly improved (67.1% ± 2.8% hydromorphone and 6.0% ± 0.6% dihydromorphine).
Reuse of biocatalyst.
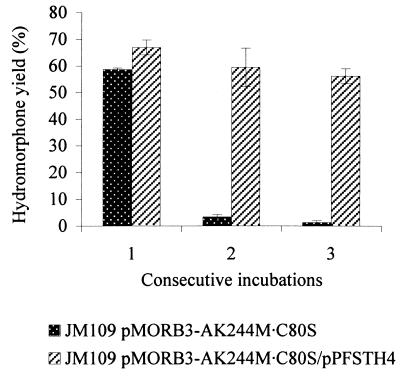
The ability to reuse the same batch of cells was investigated using E. coli JM109/pMORB3-AC80S · K244M and E. coli JM109/pMORB3-AC80S · K244M/pPFSTH4. A series of biotransformations was carried out using the same batch of cells which were harvested and washed between incubations. Enzyme assays of cells prior to each biotransformation indicated that the levels of MDH, MR, and STH did not differ significantly from the initial values determined at the point of initial cell harvesting (data not shown). E. coli JM109/pMORB3-AC80S · K244M cells were able to transform morphine efficiently only once. The cells were then exhausted, resulting in a very limited amount of hydromorphone being produced in a second biotransformation (Fig. 6). In contrast, biotransformations with E. coli JM109/pMORB3-AC80S · K244M/pPFSTH4, which contained STH, showed that these cells were able to transform morphine efficiently over three cycles of incubations, with only a small reduction in efficiency (Fig. 6).
FIG. 6.
Reuse of cells. The graph shows hydromorphone yields obtained from consecutive incubations of recombinant E. coli JM109 with 20 mM morphine. Biotransformations were carried out as described in the text. Cells were harvested once the biotransformation had reached completion (12 to 22 h) and washed with 50 mM Tris-HCl (pH 8.0) prior to the start of the next biotransformation. Figures shown are the averages of duplicate measurements, and error bars represent the standard deviations.
An exhaustive attempt was made to measure the levels of NAD and NADP in the cells, both before and after the biotransformations, using a variety of methods (11, 20, 24, 26); however, it proved impossible to obtain reproducible data, possibly as the critical extraction procedure is difficult to perform on resting cells or on cells that have undergone several manipulations (such as centrifugation, washing, resuspension, etc.).
DISCUSSION
These experiments demonstrate that STH from P. fluorescens is able to regenerate the cofactors needed for the production of hydromorphone in both cell-free and whole-cell systems (Fig. 2). The cell-free biotransformation studies show that STH is able to cycle the cofactors, thereby allowing the efficient transformation of morphine in the presence of catalytic amounts of cofactors to take place. The ratio of the three enzymes is a critical parameter for optimum hydromorphone yields. By keeping MR and STH activities high such that MDH activity becomes rate limiting buildup of the unwanted side-product dihydromorphine can be kept to a minimum and opiate loss due to the spontaneous oxidation of the intermediate morphinone can also be minimized since high levels of MR rapidly convert morphinone to the more stable hydromorphone.
The inclusion of STH in the whole-cell biocatalyst clearly improved the biotransformation of morphine and resulted in a reuseable biocatalyst, implying that STH is able to recycle the cofactors and maintain useful intracellular cofactor balances. An apparent opiate loss (of up to 35%) can still be observed in the biotransformations. Poor solubility of morphine combined with the chemical instability of the intermediate morphinone, which breaks down and can form polymeric products, are likely causes of this apparent loss. The extended lifetime of the biocatalyst, however, raises the possibility of overcoming the problem of poor substrate solubility through the use of a continuous substrate-cycling system.
The P. fluorescens STH is encoded by a single gene, enabling straightforward regulated expression in recombinant systems. As the reaction is not energy linked and is freely reversible, it shows no predisposition towards a particular cofactor pool and is likely to be of benefit to recombinant systems with different cofactor requirements. While commonly used enzymatic methods for NAD and NADP regeneration are dependent on additional supplies of substrate (10, 14, 17), the simultaneous regeneration of both cofactors, one reduced and the other oxidized, can be achieved by STH without the addition of any other substrates.
E. coli has recently been found to possess a gene encoding an STH; however, the activity was not detected in crude extracts (3). As hydromorphone is found to accumulate at concentrations up to 12 mM in whole-cell incubations without the recombinant STH present, a cell obviously contains significant reserves of the cofactors or is to some extent able to regenerate the cofactors due to other indigenous enzymes in the cell. Such indigenous cofactor regeneration may possibly be envisaged, for instance, for the two E. coli transhydrogenases. However, the membrane-bound transhydrogenase couples the transfer of reducing equivalents from NADH to NADP+ with proton import and only in extreme conditions operates in the reverse reaction (15), and the STH activity of E. coli is not detected. These activities in the cell are, in any case, apparently not sufficient for maintaining useful cofactor concentrations for the efficient production of hydromorphone, and the introduction of P. fluorescens STH facilitates cofactor cycling and thereby improves the biotransformation.
Recently, Anderlund et al. (1) reported an attempt to use a recombinant membrane-bound pyridine nucleotide transhydrogenase from E. coli to alter cofactor levels during anaerobic fermentation of Saccharomyces cerevisiae. However, this enzyme was found to remain localized in the endoplasmic reticulum, to have essentially no effect on NAD+ regeneration, and to be unable to decrease glycerol formation. A cytoplasmic transhydrogenase from Azotobacter vinelandii also did not positively affect the amount of glycerol produced by S. cerevisiae, as the NAD+ pool became limiting (19). To the best of our knowledge, this paper offers the first successful demonstration of the use of a recombinant STH for cofactor regeneration. Although problems with plasmid stability have not been encountered, it can be envisaged that integrating the P. fluorescens sth gene into the E. coli chromosome would further enhance the suitability of the biocatalyst for commercial applications and also lead to a more general applicable cofactor regeneration strain.
The results demonstrate the usefulness of STH as a tool in biocatalysis and metabolic engineering programs where different cofactor requirements limit product formation.
ACKNOWLEDGMENTS
We extend special thanks to M. McPherson of Macfarlan Smith Ltd., Edinburgh, Scotland, for support and helpful discussions throughout this work.
The work was supported by Macfarlan Smith Ltd., Edinburgh, Scotland, through a DTI LINK award. B.B. acknowledges the Norwegian Research Council for funding.
REFERENCES
- 1.Anderlund M, Nissen T L, Nielsen J, Villadsen J, Rydström J, HahnHagerdal B, KiellandBrandt M C. Expression of the Escherichia coli pntA and pntB genes, encoding nicotinamide nucleotide transhydrogenase, in Saccharomyces cerevisiae and its effect on product formation during anaerobic glucose fermentation. Appl Environ Microbiol. 1999;65:2333–2340. doi: 10.1128/aem.65.6.2333-2340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen K B, von Meyenburg K. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J Biol Chem. 1977;252:4151–4156. [PubMed] [Google Scholar]
- 3.Boonstra B, French C E, Wainwright I, Bruce N C. The udhA gene of Escherichia coli encodes a soluble pyridine nucleotide transhydrogenase. J Bacteriol. 1999;181:1030–1034. doi: 10.1128/jb.181.3.1030-1034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruce N C, Willey D L, Coulson A F W, Jeffery J. Bacterial morphine dehydrogenase further defines a distinct superfamily of oxidoreductases with diverse functional activities. Biochem J. 1994;299:805–811. doi: 10.1042/bj2990805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce N C, Wilmot C J, Jordan K N, Stephens L D G, Lowe C R. Microbial degradation of the morphine alkaloids: purification and characterisation of morphine dehydrogenase from Pseudomonas putida M10. Biochem J. 1991;274:875–880. doi: 10.1042/bj2740875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce N C, Wilmot C J, Jordan K N, Trebilcock A E, Stephens L D G, Lowe C R. Microbial degradation of the morphine alkaloids: identification of morphinone as an intermediate in the metabolism of morphine by Pseudomonas putida M10. Arch Microbiol. 1990;154:465–470. doi: 10.1007/BF00245229. [DOI] [PubMed] [Google Scholar]
- 7.French C E, Boonstra B, Bufton K A J, Bruce N C. Cloning, sequence, and properties of the soluble pyridine nucleotide transhydrogenase of Pseudomonas fluorescens. J Bacteriol. 1997;179:2761–2765. doi: 10.1128/jb.179.8.2761-2765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French C E, Bruce N C. Purification and characterisation of morphinone reductase from Pseudomonas putida M10. Biochem J. 1994;301:97–103. doi: 10.1042/bj3010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French C E, Hailes A M, Rathbone D A, Long M T, Willey D L, Bruce N C. Biological production of semisynthetic opiates using genetically engineered bacteria. Bio/Technology. 1995;13:674–676. doi: 10.1038/nbt0795-674. [DOI] [PubMed] [Google Scholar]
- 10.Galkin A, Kulakova L, Yoshimura T, Soda K, Esaki N. Synthesis of optically active amino acids from alpha-keto acids with Escherichia coli cells expressing heterologous genes. Appl Environ Microbiol. 1997;63:4651–4656. doi: 10.1128/aem.63.12.4651-4656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrigues C, Loubiere P, Lindley N D, CocaignBousquet M. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD(+) ratio. J Bacteriol. 1997;179:5282–5287. doi: 10.1128/jb.179.17.5282-5287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hailes A M, Bruce N C. Biological synthesis of the analgesic hydromorphone, an intermediate in the metabolism of morphine, by Pseudomonas putida M10. Appl Environ Microbiol. 1993;59:2166–2170. doi: 10.1128/aem.59.7.2166-2170.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Hummel W. Large-scale applications of NAD(P)-dependent oxidoreductases: recent developments. Trends Biotechnol. 1999;17:487–492. doi: 10.1016/s0167-7799(98)01207-4. [DOI] [PubMed] [Google Scholar]
- 15.Jackson J B. The proton-translocation nicotinamide adenine dinucleotide transhydrogenase. J Bioenerg Biomembr. 1991;23:715–741. doi: 10.1007/BF00785998. [DOI] [PubMed] [Google Scholar]
- 16.Karl D M. Cellular nucleotide measurements and applications in microbial ecology. Microbiol Rev. 1980;44:739–796. doi: 10.1128/mr.44.4.739-796.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kataoka M, Rohani L P S, Wada M, Kita K, Yanase H, Urabe I. Escherichia coli transformant expressing the glucose dehydrogenase gene from Bacillus megaterium as a cofactor regenerator in a chiral alcohol production system. Biosci Biotechnol Biochem. 1998;62:167–169. doi: 10.1271/bbb.62.167. [DOI] [PubMed] [Google Scholar]
- 18.May S W. Applications of oxidoreductases. Curr Opin Biotechnol. 1999;10:370–375. doi: 10.1016/S0958-1669(99)80067-6. [DOI] [PubMed] [Google Scholar]
- 19.Nissen T L, Hamann C W, KiellandBrandt M C, Nielsen J, Villadsen J. Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast. 2000;16:463–474. doi: 10.1002/(SICI)1097-0061(20000330)16:5<463::AID-YEA535>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Pinder S, Clark J B, Greenbaum A L. The assay of intermediates and enzymes involved in the synthesis of the nicotinamide nucleotides in mammalian tissues. Methods Enzymol. 1971;18:20–31. [Google Scholar]
- 21.Poustka A, Rackwitz H R, Frischauf A M, Hohn B, Lehrach H. Selective isolation of cosmid clones by homologous recombination in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:4129–4133. doi: 10.1073/pnas.81.13.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapoport H, Baker D R, Reist H N. Morphinone. J Org Chem. 1957;15:1489–1492. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Theobald U, Mailinger W, Baltes M, Rizzi M, Reuss M. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae. 1. Experimental observations. Biotechnol Bioeng. 1997;55:305–316. doi: 10.1002/(SICI)1097-0290(19970720)55:2<305::AID-BIT8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Walker E H, French C E, Rathbone D A, Bruce N C. Mechanistic studies of morphine dehydrogenase and stabilization against covalent inactivation. Biochem J. 2000;345:687–692. [PMC free article] [PubMed] [Google Scholar]
- 26.Wimpenny J W T, Firth A. Levels of nicotinamide adenine dinucleotide and reduced nicotinamide adenine dinucleotide in facultative bacteria and the effect of oxygen. J Bacteriol. 1972;111:24–32. doi: 10.1128/jb.111.1.24-32.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]