Abstract
Enteric viruses like norovirus, rotavirus and astrovirus have long been accepted as spreading in the population through fecal–oral transmission: viruses are shed into feces from one host and enter the oral cavity of another, bypassing salivary glands (SGs) and reaching the intestines to replicate, be shed in feces and repeat the transmission cycle1. Yet there are viruses (for example, rabies) that infect the SGs2,3, making the oral cavity one site of replication and saliva one conduit of transmission. Here we report that enteric viruses productively and persistently infect SGs, reaching titres comparable to those in the intestines. We demonstrate that enteric viruses get released into the saliva, identifying a second route of viral transmission. This is particularly significant for infected infants, whose saliva directly transmits enteric viruses to their mothers’ mammary glands through backflow during suckling. This sidesteps the conventional gut–mammary axis route4 and leads to a rapid surge in maternal milk secretory IgA antibodies5,6. Lastly, we show that SG-derived spheroids7 and cell lines8 can replicate and propagate enteric viruses, generating a scalable and manageable system of production. Collectively, our research uncovers a new transmission route for enteric viruses with implications for therapeutics, diagnostics and importantly sanitation measures to prevent spread through saliva.
Subject terms: Viral transmission, Infection
Enteric viruses replicate in salivary glands, can be propagated in salivary gland-derived spheroids and cell lines, and are released into saliva, which is a new transmission route having implications for therapeutics, diagnostics and sanitation measures.
Main
Saliva forms the first line of defence against many pathogens entering through the oral route and can reflect an individual’s clinical state9,10. Saliva testing is commonly used to diagnose several viruses that infect SGs, such as Epstein–Barr virus, rabies virus, herpes simplex virus and both severe acute respiratory syndrome coronavirus 1 and 2 (refs. 2,3,9–11). Norovirus, rotavirus and astrovirus genomic RNA have been frequently detected in saliva in symptomatic and asymptomatic individuals12–14. These observations were interpreted as gut contaminants because these viruses are thought to mainly transmit through the fecal–oral route by ingestion of contaminated food and water and productively replicate in the intestines1,15–18.
Suckling transmits enteric viruses to mothers
Neonatal mouse pups (less than 10 days old) are excellent models for studying enteric infections because their immature digestive tracts and immune systems make them susceptible to viruses19,20. Indeed, intestinal enteric infections were readily detectable when pups were orally inoculated with murine norovirus 1 (MNV-1) or rotavirus (epizootic diarrhoea of infant mice (EDIM)) (Fig. 1a) and replication measured by either median tissue culture infectious dose (TCID50) (MNV-1) or quantitative PCR (qPCR) (EDIM). Robust intestinal replication of MNV-1 (Fig. 1b) and EDIM (Fig. 1c) was observed in pup intestines with both viruses peaking between 3 and 5 days post-inoculation (dpi) and clearing by 7–10 dpi. Similar findings were also obtained in adult mice (Extended Data Fig. 1a,b).
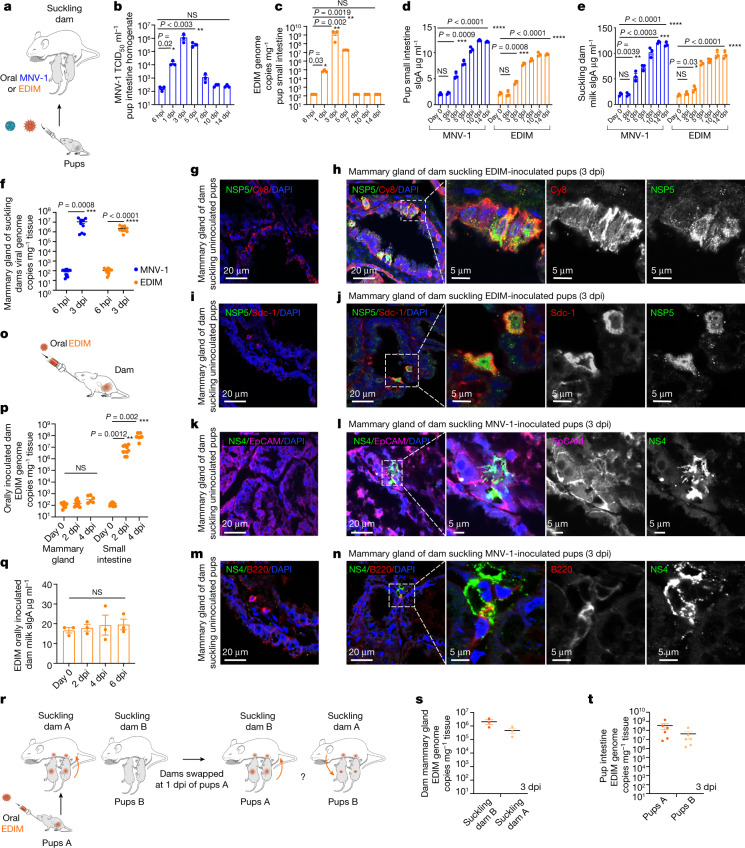
Fig. 1. Suckling infected pups directly transmit enteric viruses to mothers’ mammary glands.
a, Mouse pups orally inoculated with MNV-1or EDIM were left to nurse with mothers (dams). b, MNV-1 titres in pup intestines post-inoculation. c, EDIM replication in pup intestines post-inoculation. d, Intestinal sIgA of MNV-1- and EDIM-infected pups. b–d, n = 3 number of biologically independent experiments, seven animals per experiment; each dot represents one animal. e, Milk sIgA of dams (n = 3, three dams per virus; each dot represents an experiment). f, Viral replication in dam mammary glands suckling MNV-1- or EDIM-inoculated pups (n = 3, 23 animals per virus; each dot represents one animal). g–n, Immunostaining of mammary glands of dams suckling uninoculated or MNV-1- or EDIM-inoculated pups. o, Ten-day post-partum dams were orally inoculated with EDIM. p, EDIM replication in mammary glands and intestines of o (n = 3, eight animals, three each for 6 hpi and 2 dpi and two for 4 dpi per experiment; each dot represents one animal). q, Milk sIgA from dams orally inoculated with EDIM (n = 3, each dot represents an experiment). r, Pup/dam swap experiment scheme. s,t, EDIM levels in the mammary glands of dams (s) and small intestines of pups (t) (n = 3, each experiment included two cages and each cage consisted of one dam and two pups; each dot represents one animal). Data are the mean ± s.e.m. b–d,e,f,p,q, Two-tailed unpaired t-test. Statistical information is found in Supplementary Table 4. The 6-hpi (b,c,f) and day-0 (d,e,p,q) values were taken as the starting input values. Calculations of the limit of detection (LOD) for the TCID50 and qPCR are described in the Methods. In Figs. 1–3, the LOD for TCID50 per millilitre was approximately 102; the LOD for the qPCR data was 78 ± 9 (s.e.m.) genome copies per milligram tissue. All micrograph experiment reproducibility information is found in the Methods.
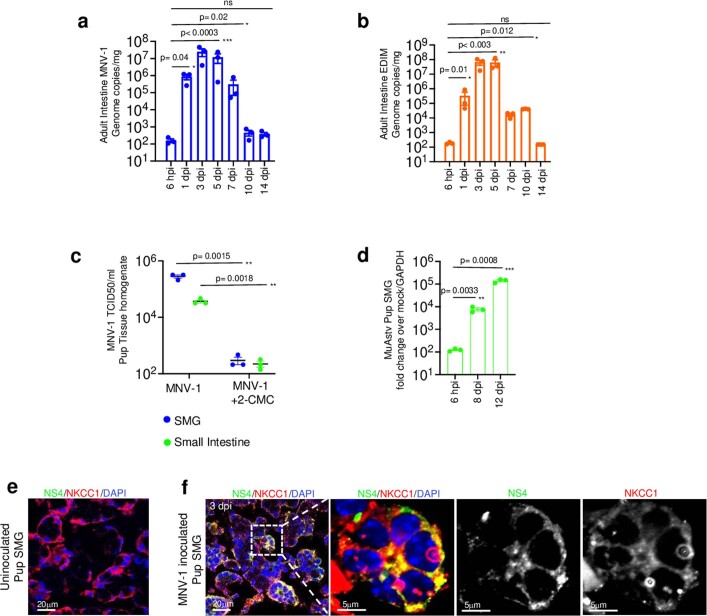
Extended Data Fig. 1. MNV-1, EDIM and Astrovirus replication in SMGs and Intestine.
a, MNV-1, and b, EDIM replication in adult intestines (from 3 independent experiments, each dot represents 1 animal, total 7 animals/experiment). Bar graphs are mean ± Standard Error of Mean (SEM). c, MNV-1 titers in SMGs and small intestine isolated from inoculated pups, +/− 2-CMC treatment (from 3 independent experiments, each dot represents an experiment, 3 animals/experiment). 3 animals/ experiment were only treated with 2-CMC as a negative control. Dot plots are mean ± SEM. d, Murine Astrovirus genome copies in SMGs of inoculated pups (from 3 independent experiments, total 3 animals/experiment). GAPDH was used as control for normalization. Bar graphs are mean ± SEM. e, f Immunostaining of SMGs obtained from uninoculated, and MNV-1 inoculated pups with anti-NS4 and anti-NKCC1. Depicted are representative images obtained from 4 independent experiments (total 4 pups). All statistical tests were performed by two-tailed unpaired t-test. Statistical information is in Supplementary Table 4. All micrograph experiment reproducibility information is in Statistics and Reproducibility in Methods. Input for a, b, d is 6hpi. c, The LOD for TCID50 ml −1 is 102; a, b LOD for qPCR is 78 ± 9 (s.e.m.) genome copies mg −1 tissue.
Like all infant mammals, pups rely on suckling milk to obtain critical immune components such as secretory IgA (sIgA), which is produced by the mammary glands of lactating mothers (called dams in mice)4–6. Milk sIgA protects pups, who are not capable of producing their own sIgA until close to weaning, by helping them combat infections in the gut4–6. We observed a rapid spike in pup small intestine sIgA levels starting at 3 dpi of the pup with either MNV-1 or EDIM (Fig. 1d), which correlated with a rapid spike in the dam’s milk sIgA levels (Fig. 1e). To start with, all dams used in these experiments were EDIM/MNV-1 seronegative (not shown). When we isolated dam mammary glands, we found an approximate 105-fold increase in MNV-1 and EDIM genomic RNA abundances over input, indicating mammary enteric virus replication (Fig. 1f). This was confirmed by immunostaining dam mammary glands with antibodies against EDIM and MNV-1 non-structural replication proteins NSP5 and NS4 respectively, which identified epithelial cells lining the milk ducts (Fig. 1g,h,k,l) and B cells (Fig. 1i,j,m,n) as replication sites for EDIM and MNV-1.
We tested whether replication in mammary glands and the rapid surge in milk sIgA was due to dams being infected by their pups via the conventional fecal–oral route because of possible coprophagia in the shared living space. The entero–mammary pathway during pregnancy and lactation is well known to lead to transfer of immune cells, although not viruses, from the gut to the mammary gland on gut infection or vaccination4. We orally inoculated EDIM seronegative dams (pup-free) with EDIM at 10 days post-partum (Fig. 1o) and measured the levels of EDIM genomic RNA in mammary glands and small intestine (Fig. 1p) and milk sIgA (Fig. 1q). In contrast to dams suckling infected pups(Fig. 1d–f), there was no sIgA surge in the milk of orally inoculateddams (Fig. 1q); their mammary glands did not show any detectable viral RNA (Fig. 1p) whereas their small intestines had a 106-fold increase in viral RNA by 4 dpi (Fig. 1p).
To further interrogate this mode of viral transfer, mouse pups were orally inoculated with EDIM (pups A) and placed back for suckling on their mother (dam A). At 1 dpi, their mother was swapped with a foster dam (dam B) from a cage of uninoculated pups (pups B). Dam A was placed with and suckled pups B whereas pups A were suckled by dam B (Fig. 1r). At 3 dpi, all animals from both cages were killed and virus replication in the dam mammary glands (Fig. 1s) and pup small intestines (Fig. 1t) was measured. We found a 104-fold increase in viral genome levels in the mammary glands of both dams A and B (Fig. 1s) and a 106-fold increase in the genome levels in the small intestines of both pups A and B (Fig. 1t) over input (approximately 102 genome copies per milligram tissue). This indicated that both dams A and B had been infected by suckling the directly inoculated pups A, with dam A infected by 1 dpi with pups A and dam B in the days after. Pups B were probably infected by suckling from dam A’s infected mammary glands or from her feces. Collectively, these findings indicated that enteric viruses backflow from pups to their mothers’ mammary glands through suckling, leading to both an in situ mammary gland infection and rapid triggering of milk sIgA surge that may contribute to clearing the pups’ infection6.
Enteric viruses are transmitted through saliva
We tested whether saliva could be the conduit for enteric viral transmission to mammary glands during suckling. We extracted saliva from adult mice (as it was easier than doing so from pups)21 orally inoculated with EDIM or MNV-1 (Fig. 2a). Immunoblotting with anti-EDIM rotavirus VP6 (Fig. 2b) and anti-MNV-1 VP1 (Fig. 2c) showed each virus to be shed in saliva starting at 2 dpi; MNV-1 TCID50 measurements showed titres at 3 dpi to be approximately 104-fold higher than input (6 h post-inoculation (hpi)) (Fig. 2d). Whereas MNV-1 and EDIM infections are acute and cleared in 7–10 days (ref. 20) (Fig. 1b,c and Extended Data Fig. 1a,b), MNV-3, MNV-4 and the less-well-characterized WU23 murine norovirus strains persistently infect the proximal colon and shed into feces for weeks22–25. We inoculated mice with these strains and found that all were also persistently shed in saliva for at least 3 weeks after inoculation, with titres approximately 103-fold higher than input (6 hpi) (Fig. 2e).
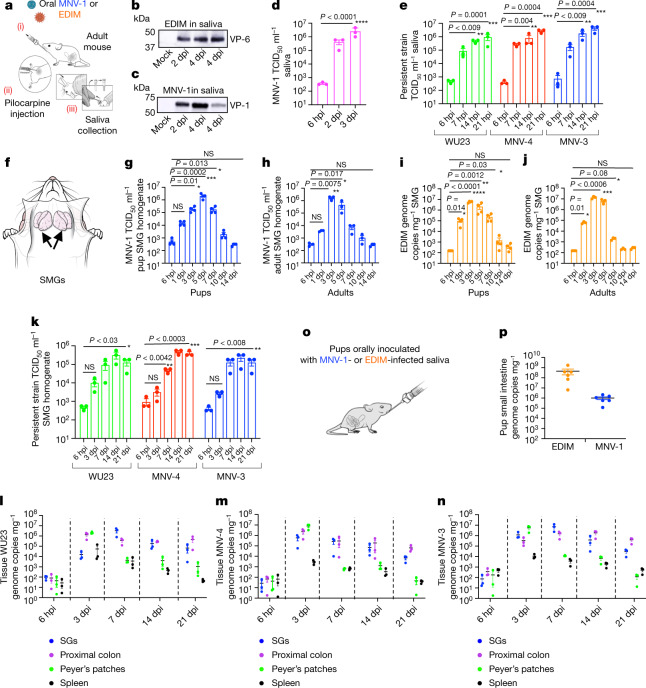
Fig. 2. Enteric viruses replicate acutely and persistently in SGs and are transmitted through saliva.
a, Schematic of saliva collection. b,c, Saliva collected from EDIM-infected (b) or MNV-1-infected (c) mice probed with anti-VP-6 and anti-VP1, respectively (n = 3, five animals per experiment). d, MNV-1 titres in saliva (n = 3, three animals per experiment; each dot represents an experiment). e, WU23, MNV-4 and MNV-3 titres in saliva (n = 3, four animals per experiment; each dot represents an experiment). f, Schematic of mouse SMGs. g,h, MNV-1 titres in pup (n = 4) (g) and adult (h) SMGs (n = 3). i,j, EDIM replication in pup (i) (n = 4) and adult (j) SMGs (n = 3). g–j, Each data point represents one animal, seven animals per experiment. k, WU23, MNV-4 and MNV-3 titres in SMGs (n = 3, five animals per experiment; each dot represents one animal). l–n, Replication in the SGs, proximal colon, Peyer’s patches and spleen of adult mice inoculated with WU23 (l), MNV-4 (m) and MNV-3 (n) (n = 3, five animals per experiment; each dot represents data from one animal). o, Schematic of oral inoculation with infected saliva. p, Viral replication in the small intestines of pups orally inoculated with infected saliva (n = 3, each dot represents one animal, total six animals). Pups were also inoculated with uninfected saliva for baseline reference. Data are the mean ± s.e.m. d,e,g–j,k, Two-tailed unpaired t-test. Statistical information is in Supplementary Table 4. For gel source data, see Supplementary Figure 1. The input for d,e,g–k,l–n, is 6 hpi.
Enteric virus replication in SGs
The major SG complex (Fig. 2f) consists of parotid, sublingual and submandibular glands (SMG)26,27. SMGs were chosen as a representative SG because they are the largest and provide basal salivary secretion. After oral inoculation of pups and adults with the MNV-1, MNV-3, MNV-4 or WU23 strains, an approximate 104-fold increase in titres over each virus respective input level (6 hpi) was measured in the SMGs (Fig. 2g,h,k). Replication was also sensitive to 2′-C-methylcytidine (2-CMC) norovirus polymerase inhibitor28 (Extended Data Fig. 1c). Similarly, in the SMGs of EDIM- and murine astrovirus-inoculated29 mice, approximately 105- and 103-fold increases in viral genome copies were measured, respectively (Fig. 2i,j and Extended Data Fig. 1d). Notably, for MNV-3, MNV-4 and WU23, the level and duration of SMG replication was comparable to that of the proximal colon (Fig. 2l–n). By contrast, viral RNA levels were significantly lower and ultimately cleared from Peyer’s patches and spleen (Fig. 2l–n). We also tested the CR6 murine norovirus, which shares with MNV-3 and MNV-4 critical sequence motifs in its VP1 and NS1/2 proteins needed for systemic infection and persistence, respectively25,30. CR6 did not replicate in SMGs (Extended Data Fig. 2), pointing to differences among persistent murine norovirus strains.
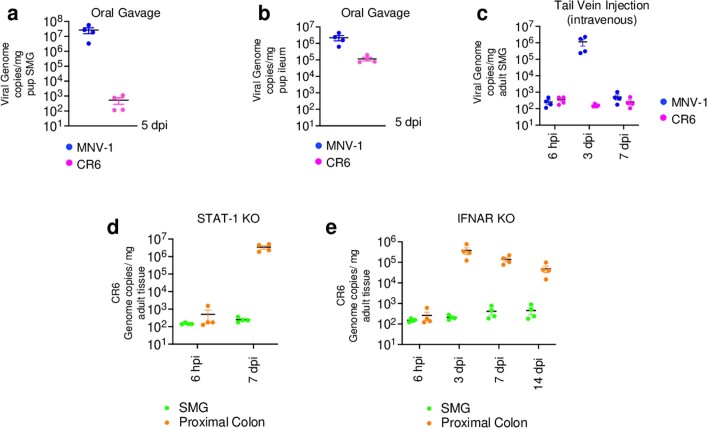
Extended Data Fig. 2. Investigating SMG infection of CR6.
MNV-1 and CR6 viral genome copies were measured in a, SMG and b, ileum of orally inoculated mice (n = 2, with 2 animals/experiment, each data point represents data from 1 animal. Data are mean ± SEM). Similarly in c, viral genome copies for MNV-1 and CR6 were measured in SMGs after tail vein injection of each virus (n = 4, with 3 animals/experiment each dot represents data from 1 animal. Data are mean ± SEM). CR6 genome copies were measured in d, STAT-1, and e, IFNAR knockout mice. In both experiments animals were orally gavaged and SMG and proximal colon tissues monitored for replication. For d, n = 2 with 2 animals/ experiment and e, n = 4 with 4 animals/ experiment, each dot represents data from 1 animal. Data are mean ± SEM. Input for c, d, e is 6 hpi. a–e, The LOD for qPCR data is 78 ± 9 (s.e.m) genome copies mg−1 tissue.
In addition to suckling, we tested whether saliva could transmit infection to others through the conventional oral route. We orally inoculated pups with saliva obtained from MNV-1- or EDIM-infected adults (Fig. 2o) and measured intestinal viral genome levels at 3 dpi. This showed significant EDIM and MNV-1 replication (Fig. 2p), as in pups orally inoculated with fecal EDIM or MNV-1 (Fig. 1b,c).
Replication in immune and epithelial cells
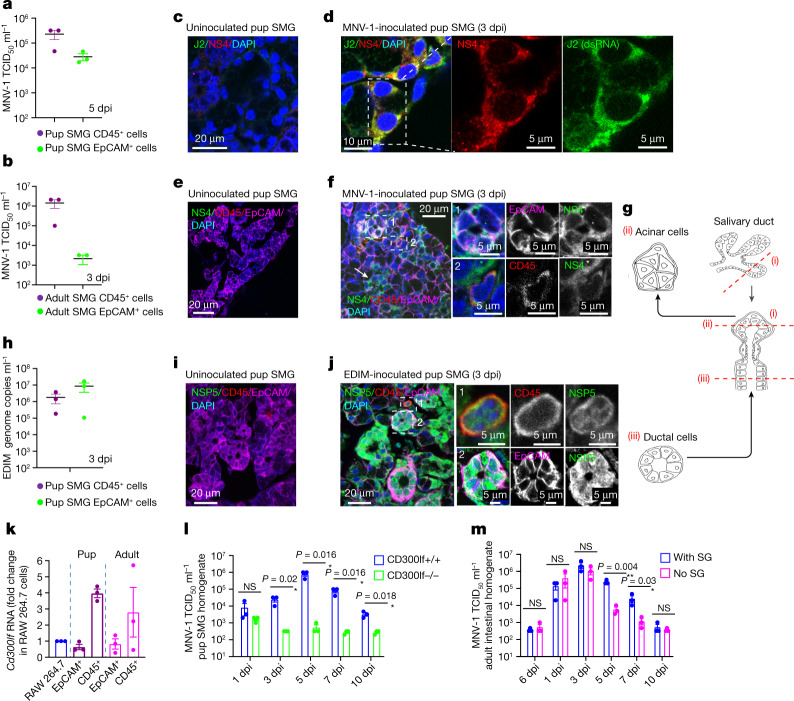
TCID50 measurements showed significant MNV-1 titres in the CD45+ immune and EpCAM+ (epithelial cell adhesion molecule) epithelial SMG cell populations (Fig. 3a,b and Extended Data Fig. 3). This was confirmed by coimmunostaining SMGs with antibodies against MNV-1 replication reporters NS4 and double-stranded RNA (dsRNA) (J2) and epithelial (EpCAM) and immune cell (CD45) markers (Fig. 3c–f). NS4 staining was localized to acinar epithelial cells (Fig. 3f,g), marked with NKCC1 (Extended Data Fig. 1e,f). EDIM also replicated in both CD45+ and EpCAM+ SMG cell populations (Fig. 3h), but replication in the latter was mainly in ductal epithelial cells (Fig. 3g,i,j).
Fig. 3. Murine norovirus and rotavirus replicate in the epithelial and immune cells of SMGs.
a,b, MNV-1 titres in epithelial (EpCAM+) and immune (CD45+) cells sorted from the SMGs of inoculated pups (a) and adults (b) (n = 3, each dot represents data from two animals, six animals in total). c–f, Immunostaining of SMGs from uninoculated and MNV-1-inoculated pups with anti-NS4, anti-J2 (dsRNA), anti-EpCAM and anti-CD45. MNV-1 replication in acinar (arrows and box 1) and immune cells (box 2). g, Schematic of salivary duct structure. h, EDIM replication in EpCAM+ and CD45+ cells sorted from the SMGs of inoculated pups (n = 3, each dot represents two animals, six animals in total). i,j, Immunostaining of SMGs from uninoculated (i) and EDIM-inoculated (j) pups with anti-NSP5, anti-CD45 and anti-EpCAM. EDIM replication in immune (box 1) and ductal cells (arrows and box 2). k, Cd300lf expression in SMGs (n = 3, three adults and three pups per experiment). RAW246.7 and HeLa cells were used as positive and negative controls, respectively. l, MNV-1 titres in the SMGs of inoculated Cd300lf−/− and Cd300lf+/+ mice (n = 3, each dot represents one animal, five animals per experiment for each group). m, MNV-1 titres in the intestines of inoculated SG-removed or SG-intact adult mice (n = 3, each dot represents one animal, six animals per experiment for each group). a,b,h,k, Gating strategies are depicted in Extended Data Fig. 3. Data are the mean ± s.e.m. l,m, Two-tailed unpaired t-test between two groups for each time point. Statistical information is given in Supplementary Table 4. m, Input is 6 hpi. All micrograph experiment reproducibility information is found in the Methods.
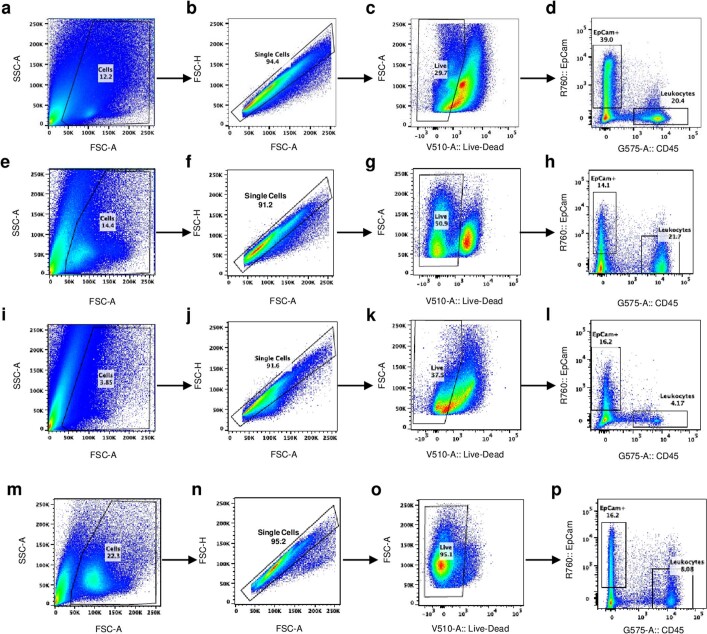
Extended Data Fig. 3. Gating strategies for cell sorting.
Panels a–d, e–h, i–l and m–p represents gating strategies for Fig. 3a, b, h and k. Briefly, a panel represents the gating for cells, eliminating the cell debris followed by single cell selection in b, eliminating doublets. Panel c represents gating strategy for live/dead cells. Live cells selected from c, was subjected to cell sorting based on EpCam+ and CD45+ cells in d. Sorted cells obtained from d was analyzed in Fig. 3a. The same chronology is maintained for panels: e–h, i–l and m–p.
Cd300lf, encoding the functional intestinal receptor for all known murine norovirus strains23,31, was also highly expressed in SMG EpCAM+ and CD45+ cell populations (Fig. 3k). Cd300lf−/− mouse pups inoculated with MNV-1 did not show any productive infection of their SGs (Fig. 3l) indicating that CD300lf receptors are essential for SMG infection by murine noroviruses. Notably, partial extraction of the SGs from adult mice before oral MNV-1 inoculation led to faster clearing of the intestinal infection (Fig. 3m). These data, together with MNV-3/MNV-4/WU23 persistence (Fig. 2e,k–n) and slower clearance of acute viruses from SMGs (Fig. 2g–j) relative to intestines (Fig. 1b,c), indicates that SGs may act as enteric virus reservoirs.
Ex vivo salivary replication models
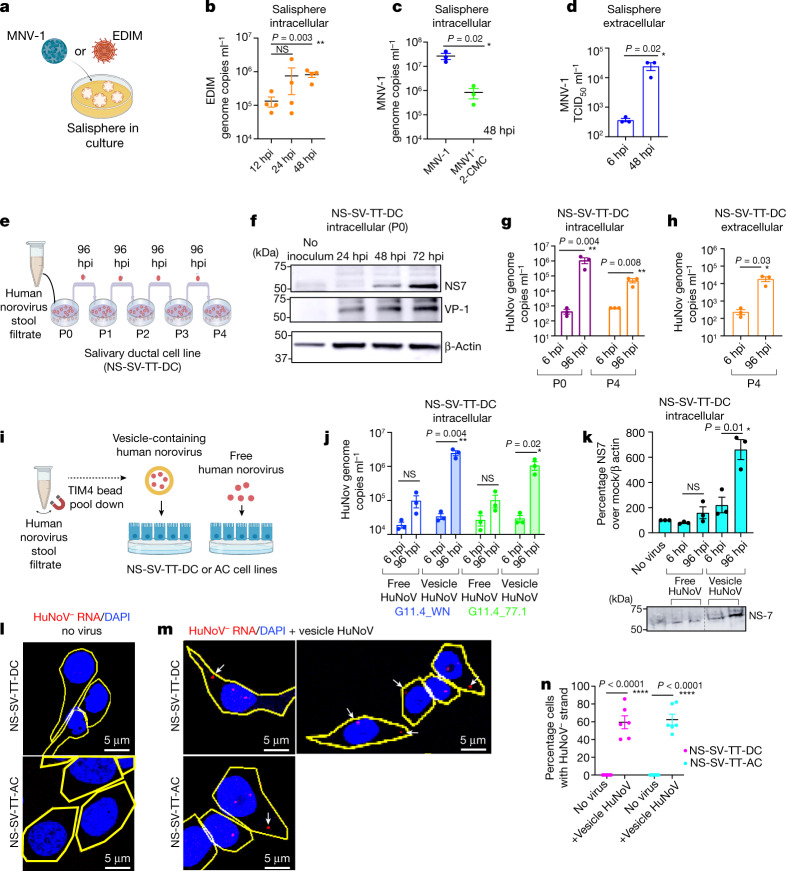
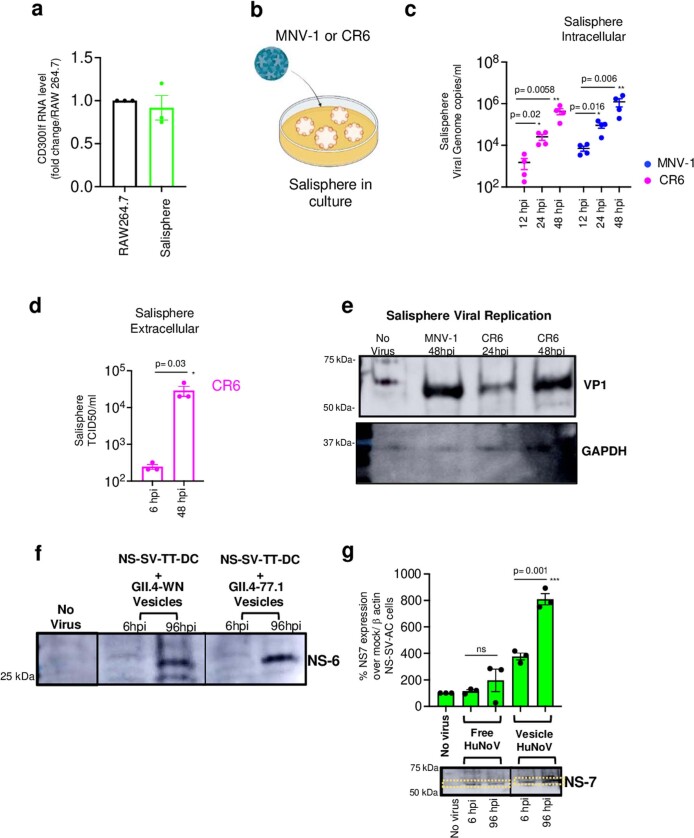
In vitro models of virus replication are important tools for studying viral life cycles, therapeutics and vaccines. We investigated whether spheroids assembled from murine SG cells (salispheres)7 could be used as ex vivo culture systems for murine enteric viruses. In EDIM-inoculated salispheres, we measured a tenfold increase in EDIM genomic RNA copies between 12 hpi and 48 hpi (Fig. 4a,b). As salispheres endogenously express CD300lf (Extended Data Fig. 4a), we inoculated them with MNV-1. This resulted in 2-CMC-sensitive replication (Fig. 4c) and MNV-1 egress into culture supernatants at 48 hpi (Fig. 4d). CR6, which had not replicated in SMGs in vivo (Extended Data Fig. 2), robustly replicated in salispheres (Extended Data Fig. 4b–d).
Fig. 4. Enteric viruses replicate in salispheres and SG cell lines.
a, Schematic of MNV-1 or EDIM inoculation in salispheres. b, EDIM genome copies in EDIM-inoculated salisphere lysates (n = 4). c, MNV-1 replication in salispheres with 2-CMC (n = 3). d, MNV-1 titres in MNV-1-inoculated salisphere supernatants (n = 3). e, NS-SV-TT-DC inoculation scheme with HuNoV. f, Immunoblot of P0 cell lysates probed with anti-NS7 and anti-VP1. β-Actin was used as the loading control (n = 3). g,h, HuNoV genomic RNA levels in P0 and P4 cell lysates (g) and P4 supernatants (h) (n = 3). i, Scheme of vesicle pulldown with TIM-4 beads from stool filtrate11. j, HuNoV genomic RNA levels in cell lysates of vesicle-cloaked or free virus-inoculated NS-SV-TT-DC cultures (n = 3). k, Cellular lysates of vesicle-cloaked or free virus-inoculated NS-SV-TT-DC cultures were analysed by immunoblotting with anti-NS7 (n = 3) and anti-β-actin. l,m, FISH with probes against the (−) HuNoV strand was performed on uninoculated (l) and vesicle-cloaked HuNoV (GII.4-WN)-inoculated (m) NS-SV-TT-DC and NS-SV-TT-AC cell cultures. Cell outlines are shown in yellow; the arrows point to (−) HuNoV RNA. n, Percentage of NS-SV-TT-DC and NS-SV-TT-AC cells with (−) HuNoV strand FISH staining (n = 6). Data are the mean ± s.e.m. b–d,g,h,j,k,n, Each dot represents a biologically independent experiment; two-tailed unpaired t-test between two groups. Statistical information is shown in Supplementary Table 4. d,g,h,j,k, The input was 6 hpi. b,c,g,h,j, The LOD for qPCR was 102 genome copies ml−1. d, The LOD for TCID50 per millilitre was 2 × 102. All micrograph experiment reproducibility information is found in the Methods. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 4. Ex-vivo replication of MNV-1, CR6 and human norovirus.
a, CD300lf expression in uninoculated salispheres (from 3 independent). RAW246.7 cells were used as positive control for comparison. Bar graph is mean ± SEM. b, Schematic of MNV-1/CR6 inoculation into salisphere culture. c, CR6 and MNV-1 replication in salispheres (from 4 independent experiments represented by a dot). Genome copies/ml was calculated based on standard curve plotted on log scale. Dot plots are mean ± SEM. d, Supernatants collected from CR6 inoculated salispheres (3 independent experiments, each dot represents an experiment) and analyzed by TCID50 and plotted on log scale. Bar graphs are mean ± SEM. e, Immunoblots of lysates from MNV-1 (48hpi) and CR6 (24 hpi & 48 hpi) inoculated salisphere cultures probed against VP-1 and GAPDH (blot representative of 2 independent experiments). f, Lysates from NS-SV-TT-DC cultures inoculated with vesicle-cloaked human noroviruses from two separate isolates (GII.4-WN and GII. 4-77.1) for 6 hpi and 96 hpi (but washed at 6 hpi) were probed with anti-NS6 antibody (from 3 independent experiments). g, Lysates from NS-SV-TT-AC cultures inoculated with vesicle-cloaked human noroviruses from isolate (GII. 4-77.1) for 6 hpi and 96 hpi (but washed at 6 hpi) were probed with anti-NS7 and anti-β actin (from 3 independent experiments represented by each dot) and analyzed by densitometry (dashed line box) and plotted as a bar graph; mean ± SEM. Samples were derived from the same experiment and blots were processed in parallel. Statistical test was performed using two-tailed unpaired t-test between 6 hpi and 96 hpi for each inoculum groups. Statistical information is in Supplementary Table 4. For gel source data, see Supplementary Figure 1. Input for d, g is 6 hpi and c is 12 hpi. c, The LOD for qPCR data from salisphere was 102 genome copies ml−1. d, The LOD for TCID50 ml−1 was approximately 2 x 102.
Salivary cell lines replicate human norovirus
Human enteroid and B cell culture models currently used to replicate human noroviruses (HuNoVs) require supplements (bile acids and microbiota) and are costly32,33. Therefore, we tested whether HuNoV could replicate in human SG cell lines. We chose SV40-transformed adherent salivary cell lines, NS-SV-TT-DC (ductal) and NS-SV-TT-AC (acinar), that routinely grow as a monolayer8 and adapted them to serum-free media. The ductal NS-SV-TT-DC cells were inoculated with a fixed volume of HuNoV stool filtrate (GII.4 Sydney) (Fig. 4e), which was washed off at 6 hpi. The P0 lysate collected at intervals post-wash and immunoblotted with antibodies against capsid VP1 and polymerase NS7 showed time-dependent increases in both proteins over input (6 hpi), indicating replication (Fig. 4f). Next, HuNoV was sequentially passaged four times through NS-SV-TT-DC cells, each time inoculating the supernatant from the previous passage in a new culture, washing off inoculum at 6 hpi and collecting the cell lysates or supernatants at 96 hpi (Fig. 4e) to measure HuNoV genomic copies (Fig. 4g,h). In P0, we measured an approximate 1,000-fold increase in intracellular HuNoV (genome copies per millilitre) between 6 and 96 hpi. Although by P4 replication had attenuated, with intracellular and extracellular virus levels approximately 100-fold (Fig. 4g) and tenfold higher, respectively, than the inputs for each (6 hpi) (Fig. 4h), the replication measured approximated those reported in enteroid cultures33.
Rotaviruses and noroviruses egress from cells non-lytically as viral clusters inside vesicles18. Compared to free fecal viruses (for example, due to vesicle lysis), vesicle-cloaked viruses are protected from fecal proteases and nucleases, enabling high multiplicities of infection (MOIs) and leading to enhanced infectivity18,34,35. Indeed, only vesicle-cloaked, HuNoV-enriched inocula replicated efficiently in the ductal and acinar salivary cell lines as evidenced by qPCR, immunoblots for viral non-structural proteins (NS7, NS6) and single-molecule fluorescence in situ hybridization (FISH)34 against the (−) replicative HuNoV RNA strands (Fig.4i–n and Extended Data Fig. 4f,g). This finding may have implications for enhancing infectivity in other HuNoV culture models32,33.
Annually, norovirus, rotavirus and astrovirus combined infect approximately one billion people across developed and developing countries, leading to significant morbidity and mortality. The diverse sanitation practices do not support the fecal–oral route as the sole route of their transmission. We have shown that SGs are significant replication sites for these viruses on par with the intestines; saliva transmits infection to others including nursing mothers. Furthermore, SGs may act as reservoirs, thus continuing to spread enteric viruses by saliva in the absence of diarrhoea. Our results bring focus on the enteric viral infection of SGs and saliva as a potentially more significant transmission route through talking, coughing, sneezing and kissing compared to the accepted transmission mode, fecal contamination. Hence our findings indicate that sanitation measures in addition to ones preventing fecal spread may be needed to prevent transmission of enteric viruses in the population.
Methods
Experimental models
Cell culture
For murine norovirus propagation and performing TCID50, RAW264.7 cells (ATCC TIB-71, research resource identifier: CVCL_0493) were grown in 10% FCS (catalogue no. EF-0500-A; Atlas Biologicals) supplemented with DMEM (catalogue no. 11995-065; Thermo Fisher Scientific). HeLa cells were used for checking the baseline level of CD300lf expression. HeLa cells (ATCC CCL-2) were maintained in DMEM supplemented with 10% FCS and 2 mM l-glutamine. All cell lines were maintained in incubators at 37 °C and 5% CO2. NS-SV-DC and NS-SV-AC cells have been described previously8. Mycoplasma-free cells were established and grown in serum-free medium of KGM-2 Keratinocyte Growth Medium-2 BulletKit (Lonza) consisting of KBM-2 Basal Medium and KGM-2 SingleQuots Supplements (Lonza). Identity was established by short tandem repeat analysis and found to be distinct from other published profiles at the D16S539 locus, and the lines are referred to as NS-SV-TT-DC and NS-SV-TT-AC. SG organoid culture or salispheres were processed and propagated as per the established protocol26.
Mouse model
Specific-pathogen-free 6-week-old BALB/c adult female mice and litters of 10-day-old BALB/c pups including males and females were used for the experiments mentioned; 10-day-old C57BL/6J (stock no. 000664) and B6.129S(Cg)-Stat1tm1Dlv/J (stock no. 012606) mice were purchased from The Jackson Laboratory. C57BL/6J-CD300lfem1Cbwi/J breeding pairs were a gift from C. B. Wilen (Yale School of Medicine). The Cd300lf−/− allele was created by H. W. Virgin (Washington University at St Louis) using CRISPR–Cas9 endonuclease-mediated genome editing in C57BL/6J mouse zygotes. The B6 Ifnar1−/− mouse breeding pairs were a gift from D. Verthelyi (U.S. Food and Drug Administration (FDA)). The animals were maintained at the National Heart, Lung, and Blood Institute (NHLBI) animal care facility and 10-day-old mouse pups were used for the experiments. Genotyping for all breeding pairs and mouse pups was done by Transnetyx using tail cuts to confirm the knockout gene. All animals were tested to be seronegative for EDIM, MNV-1 and murine astroviruses. All BALB/c adults and pups were either purchased from The Jackson Laboratory (strain no. 000651) or housed and bred in-house (animals aged more than 6 weeks) in accordance with the procedure outlined in the guide for the Care and Use of Laboratory Animals under an animal study proposal approved by the NHLBI Animal Care and Use Committee.
In brief, the animal facility temperature was maintained at 22 °C with 40–50% humidity. Animals were housed in ventilated racks and the cages were supplied with hardwood bedding and nest packs. Animals were provided the NIH 31 feed and autoclaved water. A 6:00–18:00 light cycle was followed. Animals were infected randomly without any bias as to sex, and all animal experiments were validated double blinded. The age of the animals selected for each experiment is given in the figure legends. All animal experiments were performed in an American Association for Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility. Animals were killed as per the AAALAC guidelines: adult animals were killed by CO2 exposure (USP Grade A) at 3 l min−1 and mouse pups were killed by cervical dislocation. It took approximately 2–3 min to anaesthetize the animals with a 10–30% filled CO2 chamber, at which time lack of respiration and faded eye colour were observed. CO2 flow was maintained for a minimum of 1 min after respiration ceased.
Virus stock
Murine rotavirus
The EDIM strain, a gift from H. Greenberg (Stanford University), was propagated in BALB/c mice. The DD50 (the virus dose determined empirically to cause diarrhoea in 50% of mouse pups) of the stock in 10-day-old mouse pups was 105 plaque-forming units. Five-day-old mouse pups were orally inoculated with 100-fold-diluted stock virus in PBS; at 2 dpi, when the animals started to have diarrhoea, they were killed by cervical dislocation. Intestines were surgically extracted and tissues were homogenized in M199 medium and centrifuged at 5,000 r.p.m. for 10 min at 4 °C to isolate the virus from tissue. The stock virus was aliquoted and stored at −80 °C until ready to use.
Murine norovirus
MNV-1 (ref. 36) was a gift from K. Green (NIAID) and CR6 was a gift from M. Baldridge (Washington University at St Louis). Strains were propagated in RAW264.7 cells according to a previously published protocol36. In brief, 1 × 107 cells were seeded in 150-mm cell culture dishes and incubated for 48 h. Cells were infected with MNV-1 or CR6 at an MOI of 0.1 and incubated for 1 h at 37 °C and 5% CO2. Finally, cells were rinsed and further incubated with pre-warmed serum-free medium until the cytopathic effect (CPE) was visible. Cell cultures were freeze-thawed three times, medium was collected, and cell debris was removed by centrifugation (1,000g for 15 min). The TCID50 per millilitre was calculated to be 8 × 107. Cleared stock virus was aliquoted and stored at −80 °C until ready to use.
WU23 (8.75 × 109 TCID50 ml−1) and MNV-3 (5.79 × 1010 TCID50 ml−1) were gifts from S. Karst, Department of Molecular Genetics and Microbiology, University of Florida College of Medicine; MNV-4 (1 × 107 TCID50 ml−1) was a gift from C. Wobus, Department of Microbiology and Immunology, University of Michigan Medical School. Both were similarly propagated in RAW cells.
Murine astrovirus
The strain, a natural isolate denoted as murine astrovirus-Y, was a gift from S. Compton (Yale School of Medicine). The virus was propagated as per the published protocol37. In brief, 20 μl of murine Astrovirus-strain Y was orally inoculated into 6-week-old BALB/c mice. At 5 dpi, mice were killed and the colon was extracted and processed as a 10% homogenate in DMEM. Pooled colon homogenates were clarified by centrifugation and sterilized by passing through a 0.22-μm filter; stocks were stored at −80 °C.
HuNoV
HuNoV GII.4-77.1 (NIHIC 77.1) was a gift from K. Green (NIAID) and GII.4-WN (Sydney Strain) was a gift from W. A. Henderson (University of Connecticut). The GII.4-77.1 stool sample was first filtered through a 70-µm sieve to eliminate fats and other debris, followed by a finer filtration through a 20-µm sieve and lastly a 0.3-µm sieve to eliminate bacteria. The filtrate was then concentrated and diluted in 500 µl of PBS after ultracentrifugation at 100,000g for 1.5 h. Thereafter the stock was inoculated into NS-SV-TT-DC cells for 6 h, which were then washed with PBS twice and then incubated in fresh culture medium for 96 h. This generation of cells receiving the stock virus is denoted as P0. Virus was collected from P0 after 96 h and centrifuged at 3,000g for 15 min to eliminate cell debris. The supernatant obtained was used to infect a fresh cell culture (P1) that was incubated for 96 h; the virus was passaged for three more generations, termed P2–P4.
Vesicle isolation from human stool samples
All stools were diluted to 50% vol/vol with Dulbecco’s PBS (DPBS). Then 1-ml aliquots of diluted stools were subjected to differential centrifugation to clear the stool of debris at 4 °C as follows: 500g for 10 min; 1,000g for 10 min; 2,000g for 10 min; 4,000g for 10 min; 7,000g for 10 min three times. Pellets of stool debris were discarded and supernatant was transferred to a new microcentrifuge tube. Vesicles were isolated from cleared stool solutions using the MagCapture Exosome Isolation Kit PS (catalogue no. 293-77601; Fujifilm Wako) according to the manufacturer specifications with the following modifications: cleared stool solutions were subjected to three sequential pulldowns using 120 µl of beads for each pulldown at 4 °C in a rotator at 15 r.p.m. The first pulldown was performed overnight and the rest for 2–3 h. Vesicles were eluted twice in 2 × 50 µl of elution buffer at 37 °C for 10 min. The isolated vesicles from all pulldowns were combined and stored at 4 °C until needed for the infectivity assays. The rest of the fraction was labelled as free virus and stored similarly.
Virus infection
Cell culture
Viruses to infect the salispheres of MNV-1 and CR6 were inoculated at an MOI = 1; for EDIM, 20 μl of stock was added in a 24-well format and incubated for 2 h before cells were washed with PBS three times (each wash involving centrifugation at 1,000 r.p.m. for 5 min and resuspension of pellets with PBS) and replaced with fresh medium. Thereafter, cells were incubated for various time points, collected for RNA isolation and processed for qPCR analysis. Vesicles and free virus fractions obtained from human stool samples were tested for infectivity in the NS-SV-TT-DC and NS-SV-TT-AC cell lines. Cells were inoculated with 10 μl of each fraction and incubated for 6 h before washing and replacing with fresh medium. Thereafter, cells were incubated for 96 h and further processed for RNA and protein extraction for qPCR and immunoblotting; for staining and the FISH assay, cells were fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature and washed three times with PBS.
Mouse adult and pup infections
For all adult infections with EDIM, MNV-1, CR6, WU23, MNV-3 and MNV-4, 6-week-old mice (for BALB/c, Stat1−/− and Ifnar1−/−) were inoculated pre-orally with a tenfold dilution of the stock virus (EDIM) or 6 × 106 TCID50 units of stock murine noroviruses (MNV-1, CR6, WU23, MNV-3 and MNV-4) in a volume of 100 μl. For all experiments involving pups, pups were infected at 10 days of age with EDIM (100-fold dilution of stock) or MNV-1 (105 TCID50 units) or murine astrovirus (tenfold dilution of stock) with units of each virus strain in a volume of 100 μl delivered by oral gavage using a 22-gauge plastic feeding tube. Mock-infected mice or pups were inoculated with an equal volume of DMEM or PBS. After infection, one set of animals was always killed to collect tissue samples at 6 hpi to measure viral input to tissue; the rest were killed at the indicated time points as per the experiments. Tissues were stored at −20 °C for RNA isolation or kept in 4% PFA at 4 °C overnight for cryosectioning and immunostaining. Similarly, 10-day-old C57BL/6J Cd300lf−/− and wild-type C57BL/6J mouse pups were inoculated with MNV-1 (dosage as mentioned earlier) to measure viral replication in SMGs and small intestines.
Milk extraction from dams
Milking of dams was started a day before inoculation of the pups with MNV-1/EDIM. In brief, dams were separated from pups 2 h before milking. First, dams were injected with 2 IU kg−1 of oxytocin (catalogue no. O3251; Sigma-Aldrich). After 5 min, with gentle squeezing of the nipples, milk was extracted and collected by suction using a 200-μl pipetter as described in ref. 38. Dams were placed back in the cages with the pups after the procedure.
Foster mother experiment
Ten-day-old pups were orally inoculated with EDIM virus; 24 hpi the natural dam was replaced with a foster dam introduced from an age-matched uninfected cage. At the same time, the natural mother from the cage of the infected pups was introduced to feed the uninfected pups from the foster dam cage. After swapping dams between these cages, animals were monitored for 2 days; at 3 dpi, pups and dams from both cages were killed for tissue isolation (pups: small intestine; dams: mammary gland) to quantify viral replication by qPCR.
2-CMC treatment
For MNV-1 replication validation in SMGs, 2-CMC, a nucleoside polymerase inhibitor of MNV-1 replication, was used to monitor viral replication in 10-day-old mouse pups in vivo and in salisphere culture in vitro. The dosage in vivo and in vitro was adapted from an established protocol with a few modifications28. In brief, for in vivo treatment, administration was initiated by an intraperitoneal injection of 50 μg kg–1 day−1 dissolved in PBS, divided into two daily treatments 1 day before oral inoculation of MNV-1 until 5 dpi (time determined for maximum replication in SMGs). To compare the effects of the dosage, a mock group was only administered 2-CMC; another group inoculated with MNV-1 received a similar volume of PBS intraperitoneally twice a day. At 5 dpi, pups were killed and tissues (SMGs and small intestine) were extracted for viral quantification. For the in vitro analysis, salisphere cultures received 12.5 μg ml−1 of 2-CMC dissolved in dimethylsulfoxide (DMSO) 1 h before MNV-1 inoculation. After 2 h of inoculation, the inoculum was removed and the cultures were washed as mentioned earlier and plated with fresh medium along with 2-CMC and monitored for 48 h. Spheres were later collected for RNA extraction. A mock group (without MNV-1 inoculation) was treated with DMSO.
Salivectomy or SG removal from BALB/c adult mice
The surgical area was sanitized with povidone-iodine or chlorhexidine scrub and prepped by alternating three times with alcohol and povidone-iodine or chlorhexidine solution. Then the 6-week-old mice were anaesthetized with isoflurane (1–3%) delivered through a gas anaesthesia machine vaporizer with an appropriate anaesthetic gas scavenging system. Animals were intubated with a nose cone for gas delivery after induction in an inhalation anaesthetic chamber. Ketamine and xylazine (80–120 mg kg−1 ketamine and 5–25 mg kg−1 xylazine) were injected intraperitoneally to anaesthetize the animals. Once they were anaesthetized, hair was removed from the ventral cervical area and ophthalmic ointment was applied to the eyes. The animal was then placed on a heated water blanket on the operating table. The animal was placed in dorsal recumbency and the surgical site was disinfected using chlorhexidine scrub followed by 70% alcohol. This sequence was repeated three times, alternating between the scrub solution and alcohol. The surgical site was then aseptically draped. A ventral midline cervical incision was made in the skin, which was dissected free of the underlying tissue and retracted laterally. Using a blunt dissection instrument, submandibular and sublingual SGs were separated from the connective and surrounding tissue holding them in place. Once the glands were free, the ducts were cut or cauterized to complete the removal. Both pairs of glands were removed as a single unit. The left and right parotid glands were then individually removed in an analogous manner. The skin incision was closed with a simple interrupted or subcutaneous suture pattern. Bupivacaine was infiltrated along the incision line for pain relief. The animal was placed in a warmed cage for recovery and returned to the regular housing cage when mobile. Warmed fluids (2–3 ml) were given subcutaneously to ensure adequate hydration. Analgesics (bupivacaine, up to 2 mg kg−1 given intradermally or topically on the incision site) were administered to alleviate pain during the post-operative period (3 days). MNV-1 was inoculated on the fifth day after the procedure and the animal monitored for viral replication for 10 days; animals were killed at the intervals mentioned earlier. Age-matched control groups without salivectomy were used to compare the effect of the surgical removal of SGs on MNV-1 infection.
Saliva collection and infection
Six-week-old female BALB/c mice were inoculated with EDIM or MNV-1 (five mice per experiment, three independent experiments) and the persistent strains WU23, MNV-3 and MNV-4 (five mice per experiment, three independent experiments). Two hours before saliva collection animals were deprived of food and water to avoid contamination in saliva during further processing. Isoflurane (1–4%) or sevoflurane (2–6%) delivered through a gas anaesthesia machine vaporizer with an appropriate anaesthetic gas scavenging system was used to anaesthetize the animals before saliva collection; 2% pilocarpine (national drug code: 61314-206-15) was injected intraperitoneally to stimulate salivation, followed by a 2-min wait. A cotton swab was inserted into the oral cavity over a 10 min period and then the wet swab was placed in a small hole punctured at the bottom of a 0.6-ml uncapped microfuge tube, which was placed further into a 2-ml tube. Then saliva was recovered by centrifugation for 2 min at 7,500g at 4 °C. As soon as mice recovered, they were returned to cages; saliva was similarly collected at 3 and 4 dpi. For immunoblotting, 40 µl saliva was aliquoted for all the time points from each individual mouse; the rest of the saliva was pulled into a single tube to infect the pups. To test the infectivity of saliva collected from EDIM- or MNV-1-infected adult mice, 10-day-old pups were infected with a volume of 100 µl saliva followed by collection of the small intestines at different time points. RNA was isolated from these tissues to quantify virus replication.
RNA isolation
The SGs and small intestines from 6-week-old adult mice and 10-day-old pups and mammary glands from dams extracted for the various experiments used in the study were first homogenized in DNA/RNA shield buffer provided with the Quick-RNA Miniprep Plus Kit (catalogue no. R2001; Zymo Research) supplemented with proteinase. Homogenized tissues were further digested in the buffer for 5 h at 55 °C and centrifuged at 5,000 r.p.m. for 5 min; RNA lysis buffer was added to the pellet and the pellet processed according to the manufacturer’s instructions. RNA was eluted in RNase/DNase-free water provided in the kit. For the salispheres, after the incubation time points mentioned earlier, cells were spun down at 1,000 r.p.m. for 5 min, resuspended in PBS for washing and spun down at 1,000 r.p.m. for 5 min. The pellet was resuspended in 200 μl of RNA lysis buffer provided in Quick-RNA Microprep Plus Kit (catalogue no. R1050; Zymo Research) and processed for RNA isolation according to the manufacturer’s instructions. Similarly, after incubation, NS-SV-TT-DC and NS-SV-TT-AC cells were collected and resuspended in 200 μl of RNA lysis buffer from the Quick-RNA Microprep Plus Kit and processed to isolate RNA.
qPCR
qPCR by SYBR Green
For all animal tissue samples (Figs. 1–3 and Extended Data Figs. 1a,b,d and 2) RNA was isolated as mentioned earlier and cDNA was prepared using the Maxima First Strand cDNA Synthesis Kit for qPCR (catalogue no. K1672; Thermo Fisher Scientific). The primers used for EDIM, MNV-1, murine astrovirus, WU23, MNV-3, MNV-4 and CR6 are listed in Supplementary Table 2. cDNA obtained from the samples was run along with the corresponding primers and with SYBR Green (catalogue no. 1725124; Bio-Rad Laboratories) in the Roche LightCycler 96 System (catalogue no. 05815916001). The thermal cycling conditions included a pre-incubation step 95 °C for 90 s followed by 45 cycles at 95 °C for 10 s, 54 °C for 10 s and 72 °C for 110 s.Each sample were run in duplicate for each experiment. For EDIM, MNV-1, WU23, MNV-3, MNV-4 and CR6, standards were run along with each experiment to measure virus genome copies per milligram of tissue. A detailed description of the standard curve preparation for each virus is given in the section ‘Standard curve preparation for EDIM, MNV-1, CR6, WU23, MNV-3, MNV-4’.
For murine astrovirus (Extended Data Fig. 1d), samples were analysed with the Gapdh housekeeping gene (Supplementary Table 2) along with a mock-infected sample. The fold change obtained was measured over the mock-normalized by cycle threshold (Ct) values obtained for the housekeeping gene.
Standard curve preparation for EDIM, MNV-1, CR6, WU23, MNV-3 and MNV-4
Amplicons for each primer set were synthesized by Integrated DNA Technologies; on the basis of the amount and length of amplicons, the number of copies (molecules) was calculated for each primer set:
where X is the amount of amplicons in nanograms, N is the length of the dsDNA amplicon, 660 g mol−1 is the average mass of 1 bp dsDNA and 6.0221 × 1023 is Avogadro’s number.
Amplicons were resuspended in 1 ml of DNase/RNase-free water to obtain the copy number per millilitre for each primer set. Thereafter, each amplicon copy number per millilitre was serially diluted from 10 to 1010 in tenfold increments and subjected to qPCR to obtain the standard curves for each individual set of primers. The initial calculation of genome copies per millilitre indicated the number of viral genomes per millilitre based on standard curves that quantified viral genomes per millilitre of 10 μl of reaction volume used to run the qPCR. This viral genomes per millilitre unit is a standard unit of qPCR calculation usedto quantify the total number of identified RNA copy numbers per millilitre of reaction volume in PCR. For tissues, we additionally included the weight of the tissues used to extract the RNA to make it genome copies per milligram of tissue, which signifies the estimate of viral genome copies in 1 mg of tissue. The Ct values obtained were plotted against the log of copy numbers per millilitre of each to derive the standard curve, which corresponds to the linear equation and R2 value. On the basis of the standard curve Ct values obtained from unknown samples, the copy number per millilitre was obtained for EDIM, MNV-1, CR6, WU23, MNV-3 and MNV-4:
Please refer to Supplementary Table 3 for the amplicons and standards.
LOD for qPCR
qPCR was performed on tissues (salivary, mammary, intestine) of both mock-inoculated (with PBS) and virus-inoculated (EDIM, MNV-1, MNV-3, MNV-4, WU23) animals, the latter at 6 hpi (input time point). The Ct values obtained were converted to genome copies per milligram of tissue using the standard curves for the corresponding primers. The copies per milligram of tissue for mock-infected animals ranged from 124 (upper limit) to 22 (lower limit) (mean = 78, s.e.m. = 9). This probably corresponds to false positive signals arising from non-specific binding of primers. The copies per milligram of tissue obtained from virus-inoculated animals (6 hpi) also had a similar range. Thus, the mean 78 ± 9 (s.e.m) was taken as the LOD, above which qPCR-based viral RNA detection by a particular primer was deemed to be positive for the corresponding virus.
For HuNoV, qPCR was performed following two methods: (1) with SYBR Green and several GII primers probing for various regions of the whole genome (Fig. 4g,h) and (2) by using GII.4 primers with TaqMan probe (Supplementary Table 2). (1) RNA fromAQ15 HuNoV-infected NS-SV-TT-DC cells was isolated as mentioned in the ‘RNA isolation’ section and cDNA was prepared using the Maxima First Strand cDNA Synthesis Kit for qPCR. cDNA derived from the samples was run along with the GII primers and SYBR Green in the PCR system. The GII whole genome (Supplementary Table 3) was run along with the samples as a standard because the primers recognize several regions of the whole genome. A standard curve was prepared in a similar way with the GII whole genome as outlined for the murine rotavirus and norovirus. GII copy number per millilitre was (−3.62 × Ct) + 46.206, R2 = 0.89. The Ct values obtained from running the samples were used to measure HuNoV genome copies per millileter in cell lysate or supernatant (Fig. 4g,h) using the standard curve obtained. (2) For the TaqMan probe, RNA isolation from cells inoculated with GII.4-77.1 (Fig. 4j) was performed similarly using the Quick-RNA Microprep Plus Kit. RNA was subjected to one-step qPCR by kit (TaqMan Fast Virus 1-Step Master Mix, catalogue no. 4444432; Thermo Fisher Scientific) using a TaqMan probe. The sequences of primers and probes are given in the Supplementary Tables 2 and 3. qPCR was performed by reverse transcription at 52 °C for 10 min, RT inactivation at 95 °C for 20 s and initiation of denaturation followed by 45 cycles of amplification at 95 °C for 15 s, 60 °C for 60 s. (3) The standard for the GII Quant Primer amplicon was run alongside each experiment as stated in the previous section and the HuNoV copy number per millilitre was calculated based on the plotted standard curve. Information about primers, probes and amplicons is found in Supplementary Tables 2 and 3. The GII.4 copy number per millilitre was (−0.2673 × Ct) + 13.453, R2 = 0.97.
For the Cd300lf analysis by qPCR (Fig. 3k and Extended Data Fig. 4a), cDNA was prepared similarly to the RNA isolated by the Quick-RNA Microprep Plus Kit from CD45+, EpCAM+ RAW264.7 and HeLa cells (for the Cd300lf experiment) and processed for qPCR using the primers listed in Supplementary Table 2. The Ct value was obtained for each cell type and calculated over RAW264.7 as positive control by the dCt method. HeLa cells were used as the negative control because Cd300lf was undetectable in the analysis with the Cd300lf primer.
Murine norovirus and rotavirus replication in salispheres were also studied by qPCR (Fig. 4b and Extended Data Fig. 4c). RNA was isolated as mentioned in the ‘RNA isolation’ section from salispheres and cDNA was prepared in a similar manner to that used for the tissue samples. The cDNA obtained was run using the corresponding primers of EDIM, MNV-1 and CR6 (Supplementary Table 2) along with SYBR Green. The Ct values obtained were analysed along the standard curves obtained for each virus as mentioned in the ‘Standard curve preparation for EDIM, MNV-1, CR6, WU23, MNV-3 and MNV-4’ section and the viral genome copy number per millilitre was obtained for each sample.
Immunofluorescence labelling
SMGs or mammary glands were collected from euthanized adult mice or pups. Samples were then fixed overnight at 4 °C in 4% PFA (catalogue no. 15710; Electron Microscopy Sciences). The fixed samples were further processed at the NHLBI Pathology Core: samples were incubated for 24 h in 30% sucrose solution at 4 °C and embedded in optimal cutting temperature compound before freeze sectioning on a microtome (Leica Cryo-start, catalogue no. CM3050S; Leica Biosystems). For all immunofluorescence staining, antigen retrieval was performed by incubating slides in Universal HIER antigen retrieval reagent (catalogue no. ab208572; Abcam) in a boiling water bath for 20 min. Samples were then cooled and tissue sections were permeabilized for 2 h at room temperature with 0.1% Triton X100 (catalogue no. T9284; Sigma-Aldrich) in DPBS (catalogue no. 14190-144; Thermo Fisher Scientific) supplemented with 10% FCS. Then samples were incubated overnight at 4 °C in primary antibodies diluted in DPBS supplemented with 10% FBS. Next, samples were rinsed three times with DPBS and incubated for 2 h at room temperature in fluorescently labelled secondary antibodies diluted in DPBS supplemented with 10% FCS. Finally, samples were rinsed three times with DPBS and mounted using Fluormount-G containing 4′,6-diamidino-2-phenylindole (DAPI) stain (catalogue no. 17984-24; Electron Microscopy Sciences). Imaging was performed on a Zeiss LSM 780 confocal laser scanning microscope and analysed with Zen Blue software (Zeiss Zen 3.1 Blue edition). After the incubation period, NS-SV-TT-DC cells were fixed with 4% PFA at room temperature for 15 min. Thereafter, PFA was removed and washed with PBS three times. Cells were then blocked with BSA supplemented with the permeabilization agent saponin for 1 h. Cells were then incubated with anti-VP1 for 1 h followed by washes and adding appropriate secondary antibody for 1 h. All incubations were done at room temperature. After the final washes, cells were mounted by mounting agent with DAPI and imaged as mentioned earlier. Details of the antibodies are listed in Supplementary Table 1.
Immunoblots
All uncropped/unprocessed blots are supplied in Supplementary Fig. 1.
Saliva
For immunoblotting, 40 µl of saliva from uninfected and 2, 3 and 4 dpi samples were directly cooked in 4× Laemmli sample buffer (catalogue no. 161-0747; Bio-Rad Laboratories); cultured cells were lysed and then centrifuged, and the collected supernatants were mixed with 4× Laemmli sample buffer. SDS–PAGE was carried out in all samples using 10% SDS–polyacrylamide gel electrophoresis (catalogue no. 4561034; Bio-Rad Laboratories) and the sample transferred to a nitrocellulose membrane (catalogue no. 1704159; Bio-Rad Laboratories). Then the membranes were blocked in 5% bovine serum albumin (BSA) (catalogue no. BP1600-100; Thermo Fisher Scientific) in Tris-buffered saline buffer (catalogue no. 1706435; Bio-Rad Laboratories) containing Tween-20 (TBST) (catalogue no. 9005-64-5; Affymetrix) for 1 h at room temperature. Thereafter, membranes were incubated overnight with primary antibodies for EDIM VP6 (gift from J. Patton, Indiana University) and MNV-1 VP1 (gift from K. Green, NIAID). Membranes were washed five times with TBST and probed with specific horseradish peroxidase-tagged secondary antibodies for 1 h. This was followed by washing steps and development with Lumigen ECL Ultra (TMA-6) (catalogue no. TMA-100; Lumigen). Blots were imaged using the Amersham Imager 600 (catalogue no. 29083461; GE Healthcare). Details of the antibodies are listed in Supplementary Table 1.
Salisphere culture
After incubating for 48 h with MNV-1 or CR6, cells were spun down at 1,000 r.p.m. for 5 min; the collected pellet was resuspended in 100 μl of lysis buffer (catalogue no. 559759; BD Biosciences) supplemented with protease inhibitor (catalogue no. A32955; Invitrogen) and subjected to trichloroacetic acid (TCA) (catalogue no. T6399-100G; Sigma-Aldrich) precipitation (1:4 vol/vol, 15 min at 4 °C). The TCA precipitate was washed twice with acetone and air-dried. Samples were dissolved in 4× Laemmli sample buffer and run in a 4–20% gradient gel (catalogue no. 4561094; Bio-Rad Laboratories). Transfer and blocking were carried out as mentioned earlier. Blots were probed against anti-VP1 and GAPDH was used as the loading control.
NS-SV-TT-DC and NS-SV-TT-AC cell lines
After the incubation period was over, cells were collected and subjected to TCA precipitation as stated in the ‘Salisphere culture’ section. Samples were run in 4–20% gradient gels and transferred to a nitrocellulose membrane for probing against anti-HuNoV NS7 and VP1 and to a polyvinylidene fluoride membrane (catalogue no. 1704157; Bio-Rad Laboratories) for anti-HuNoV NS6 (all antibodies were a gift from K. Green, NIAID). β-Actin was used as the loading control. The rest of the procedure was same as mentioned in the ‘Salisphere culture’ section. Densitometry to analyse the expression levels of all proteins was done using ImageJ software (bundled with 64-bit Java, 1.8.0_172; National Institutes of Health). The measure intensities for the virus-infected samples were normalized over mock and over the corresponding loading control and represented on a graph.
Secretory IgA measurement
Small intestine content was extracted from mouse pups and the amount was measured and made into a 10% solution in PBS; 100 μl solution was directly transferred to an antibody-coated 96-well format of ELISA Kit for Secretory Immunoglobulin A (sIgA) (catalogue no. SEA641Mu; Cloud-Clone Corporation) and measurements were carried out according to the manufacturer’s instructions. Similarly, milk collected in-1 ml tubes was diluted to a 10% solution in PBS and processed for sIgA measurement with the kit. The concentrations for the samples were measured from the standard curve plotted using the standards provided in the kit in micrograms per millilitre.
Cell sorting from SMGs
SMGs were extracted from animals after euthanization and homogenized in ice-cold PBS (supplemented with 10% FCS). The homogenate was centrifuged at 1,000 r.p.m. for 5 min at 4 °C to pellet down cells; the homogenate was further incubated for 20 min at 37 °C in 3 ml of Gentle Collagenase/Hyaluronidase solution (catalogue no. 07919; Stemcell Technologies) with shaking. Thereafter, the solution was centrifuged again at 1,000 r.p.m. for 5 min at 4 °C to collect the pellet and discard the supernatant. The pellet was further trypsin-treated for 5 min at 37 °C and passed through a 70-μm filter to eliminate undissociated tissue. The filtrate was then treated with a 4:1 NH4Cl:PBS solution to eliminate blood cells and subjected to centrifugation at 1,000 r.p.m. for 5 min at 4 °C. After leaving the red layer of cells at bottom, the supernatant consisted of single cells isolated from tissue. Cells were counted and incubated with anti-EpCAM conjugated to allophycocyanin (catalogue No. 17-5791-82; Thermo Fisher Scientific) and anti-CD45 conjugated to phycoerythrin (catalogue no. 12-0451-82; Thermo Fisher Scientific) for 1 h at 4 °C. Cells were subsequently washed and stained with LIVE/DEAD Fixable Aqua Dead cell stain (catalogue no. L34957; Thermo Fisher Scientific). Resuspended cells were sorted on a FACS ARIA III (BD Biosciences) cell sorter equipped with 355 nM, 407 nM, 488 nM, 532 nM and 640 nM laser lines using the FACSDiva v.8.1 software at 70 psi pressure using a 70 μm nozzle. Debris was removed based on scattering properties using forward scatter and side scatter parameters. Live-gated cells were purified for leukocytes identified as CD45+ EpCAM− live cells whereas CD45−EpCAM+ cells were identified as epithelial cells (Extended Data Fig. 3). Purified populations were collected in 10% PBS supplemented with FCS for downstream analysis either by TCID50 or qPCR.
TCID50 calculation
Saliva, SMGs from pups, proximal colon from adults, cells obtained from single-cell sorting from SMGs and supernatant from cultures inoculated with various murine norovirus strains were subjected to TCID50 analysis for viral titre calculation. The collection and processing methods for each sample were mentioned earlier. In brief, samples were added to RAW264.7 cells microplated on 96-well plates (3 × 104 cells per well) and serially diluted in DMEM by tenfold from 100 to 10−10. Plates were incubated until CPE was observed (day 7). Supernatant was removed and stained with crystal violet (catalogue no. HT901; Sigma-Aldrich) for 10 min. After rinsing with water twice, CPE wells were counted. TCID50 per millilitre was calculated as per refs. 39,40. The LOD was determined as the lowest concentration of virus where at least 50% CPE was observed. The detailed calculations are supplied in the source files for the corresponding figures.
FISH
The assay was performed using RNAscope (Advanced Cell Diagnostics) Multiplex Fluorescent Detection Kit v2 (catalogue no. 323110) and RNAscope probes custom-synthesized to detect negative sense human norovirus RNA gene region 5349–6428 of norovirus GII strain Hu/USA/2015/GII.P16-GII.4 Sydney/Pasadena. Cells adhered to coverslips were fixed 96 hpi in 4% PFA for 30 min. Pretreatment of the samples was performed by serially dehydrating the samples with ethanol followed by permeabilization with RNAscope protease III (catalogue no. 322340) at 1:5 dilution in PBS for 30 min at room temperature before hybridizing for 2 h at 40 °C with the FISH probe. The amplification and dye labelling steps were performed according to the RNAscope v2 assay protocol. Opal 570 dye (catalogue no. FP1488001KT; Akoya Biosciences) at 1:1,500 dilution in RNAscope TSA buffer (catalogue no. 322810) was used to visualize the FISH staining. Coverslips were mounted with Fluoromount-G mounting medium with DAPI (catalogue no. 00-4959-52; Thermo Fisher Scientific) and imaging was done using a Zeiss LSM 780 confocal microscope.
Statistics and reproducibility
Prism v.8 (GraphPad Software) was used for the statistical analysis. Error bars denote the s.e.m. in all figures. All statistical tests were done by two-tailed unpaired t-test. The number of experiments performed including the number of animals involved in each experiment are stated in the corresponding to figure legends. Statistical information can be found in Supplementary Table 4.
Reproducibility of micrographs
In Fig. 1g–n, representative images were obtained from five independent experiments for each virus infections (EDIM or MNV-1). Two mammary glands per animals were extracted from a total of five animals per virus and subjected to tissue sectioning and immunofluorescence analysis.
In Fig. 3c–f,i,j and Extended Data Fig. 1e,f, representative images were obtained from four independent experiments for each virus infections (EDIM or MNV-1). SMGs from a total of four pups per virus were subjected to tissue sectioning and immunofluorescence analysis.
In Fig. 4l,m, representative images were obtained from six biologically independent experiments from NS-SV-TT-DC and NS-SV-TT-AC infection with vesicle-cloaked HuNoV. After the incubation period, fixed cells were subjected to FISH analysis.
The scale bars for each micrograph panel are stated in the individual figure panels.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-022-04895-8.
Supplementary information
Source gels for Figs. 2b,c and 4f,k and Extended Data Fig. 4e–g.
Supplementary Table 1: antibody list; Supplementary Table 2: primer list; Supplementary Table 3: amplicon list; Supplementary Table 4: statistical analysis.
Acknowledgements
We thank M. Baldridge (Washington University in St Louis), S. Karst (University of Florida), C. Wobus (University of Michigan) and K. Green (NIAID, National Institutes of Health (NIH)) for providing the murine norovirus strains; S. Compton (Yale University) for providing the murine astrovirus; H. Greenberg (Stanford University) for providing the EDIM; C. B. Wilen (Yale University) for providing the Cd300lf−/− mice; P. Matzinger (NIAID), C. B. Wilen, S. Compton, C. Wobus, K. Green, B. Kelsall (NIAID), G. Parra (FDA) and G. Altan-Bonnet (National Cancer Institute) for discussion and critical reading of the manuscript; H. Vishwasrao and H. Shroff (Advanced Imaging and Microscopy Resource, National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH) for support with imaging the SGs; the NHLBI flow cytometry, pathology and animal cores for technical support; and A. Hoofring (NIH Medical Arts) for the illustrations. N.A.-B., S.G., M.K., M.Z., H.L. and M.S. were supported by the NHLBI and NIH. E.L. and Y.B. were supported by NIAID. Y.S. was supported by the NIBIB. M.P.H., J.A.C., H.N. and T.T. were supported by National Institute of Dental and Craniofacial Research (NIDCR)/NIH intramural funds. J.A.C. was also supported by an NIDCR/NIH intramural grant no. 1ZIADE000695.
Extended data figures and tables
Source data
Source Data Extended Data Figs. 1, 2 and 4.
Author contributions
S.G. and N.A.-B. designed the experiments and wrote the manuscript. S.G., M.K., M.S., A.M., M.Z., H.L., Y.S., E.L., H.N., T.T. and A.M.C. performed the experiments; W.H., O.V., Y.B., J.A.C. and M.P.H provided critical reagents and contributed to the design of experiments.
Peer review
Peer review information
Nature thanks Ian Goodfellow and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The data generated in this study are provided as source data files corresponding to each figure. All statistics performed and the exact P values are presented in Supplementary Table 4. Primary data associated with figures are available in the source files. Any additional datasets generated and/or analysed are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
is available for this paper at 10.1038/s41586-022-04895-8.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-022-04895-8.
References
- 1.Petri WA, Jr, et al. Enteric infections, diarrhea, and their impact on function and development. J. Clin. Invest. 2008;118:1277–1290. doi: 10.1172/JCI34005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Posse, J. L., Dios, P. D. & Scully, C. in Saliva Protection and Transmissible Diseases (eds Posse, J. L., Dios, P. D. & Scully, C) 53–92 (Academic Press, 2017).
- 3.Huang N, et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021;27:892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez JM, Fernández L, Verhasselt V. The gut–breast Axis: programming health for life. Nutrients. 2021;13:606. doi: 10.3390/nu13020606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogier EW, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl Acad. Sci. USA. 2014;111:3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blutt SE, Miller AD, Salmon SL, Metzger DW, Conner ME. IgA is important for clearance and critical for protection from rotavirus infection. Mucosal Immunol. 2012;5:712–719. doi: 10.1038/mi.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vining KH, et al. Neurturin-containing laminin matrices support innervated branching epithelium from adult epithelial salispheres. Biomaterials. 2019;216:119245. doi: 10.1016/j.biomaterials.2019.119245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azuma M, Tamatani T, Kasai Y, Sato M. Immortalization of normal human salivary gland cells with duct-, myoepithelial-, acinar-, or squamous phenotype by transfection with SV40 ori- mutant deoxyribonucleic acid. Lab. Invest. 1993;69:24–42. [PubMed] [Google Scholar]
- 9.Lee JM, Garon E, Wong DT. Salivary diagnostics. Orthod. Craniofac. Res. 2009;12:206–211. doi: 10.1111/j.1601-6343.2009.01454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lima DP, Diniz DG, Moimaz SAS, Sumida DH, Okamoto AC. Saliva: reflection of the body. Int. J. Infect. Dis. 2010;14:e184–e188. doi: 10.1016/j.ijid.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 11.Corstjens PL, Abrams WR, Malamud D. Saliva and viral infections. Periodontol. 2000. 2016;70:93–110. doi: 10.1111/prd.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pisanic N, et al. Minimally invasive saliva testing to monitor norovirus infection in community settings. J. Infect. Dis. 2019;219:1234–1242. doi: 10.1093/infdis/jiy638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirby A, Dove W, Ashton L, Hopkins M, Cunliffe NA. Detection of norovirus in mouthwash samples from patients with acute gastroenteritis. J. Clin. Virol. 2010;48:285–287. doi: 10.1016/j.jcv.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Anfruns-Estrada, et al. Detection of norovirus in saliva samples from acute gastroenteritis cases and asymptomatic subjects: association with age and higher shedding in stool. Viruses. 2020;12:1369. doi: 10.3390/v12121369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Graaf M, van Beek J, Koopmans MP. Human norovirus transmission and evolution in a changing world. Nat. Rev. Microbiol. 2016;14:421–433. doi: 10.1038/nrmicro.2016.48. [DOI] [PubMed] [Google Scholar]
- 16.Crawford SE, et al. Rotavirus infection. Nat. Rev. Dis. Primers. 2017;3:17083. doi: 10.1038/nrdp.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. Advances in norovirus biology. Cell Host Microbe. 2014;15:668–680. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santiana M, et al. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe. 2018;24:208–220.e8. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf JL, Cukor G, Blacklow NR, Dambraiuskas R, Trier JS. Susceptibility of mice to rotavirus infection: effect of age and administration of corticosteroids. Infect. Immun. 1981;33:565–574. doi: 10.1128/iai.33.2.565-574.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth AN, et al. Norovirus infection causes acute self-resolving diarrhea in wild-type neonatal mice. Nat. Commun. 2020;11:2968. doi: 10.1038/s41467-020-16798-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagavant H, et al. A method for the measurement of salivary gland function in mice. J. Vis. Exp. 2018;131:57203. doi: 10.3791/57203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arias A, Bailey D, Chaudhry Y, Goodfellow I. Development of a reverse-genetics system for murine norovirus 3: long-term persistence occurs in the caecum and colon. J. Gen. Virol. 2012;93:1432–1441. doi: 10.1099/vir.0.042176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graziano VR, et al. CD300lf conditional knockout mouse reveals strain-specific cellular tropism of murine norovirus. J. Virol. 2021;95:e01652-–20. doi: 10.1128/JVI.01652-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu CC, Riley LK, Wills HM, Livingston RS. Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp. Med. 2006;56:247–251. [PubMed] [Google Scholar]
- 25.Walker FC, et al. Norovirus evolution in immunodeficient mice reveals potentiated pathogenicity via a single nucleotide change in the viral capsid. PLoS Pathog. 2021;17:e1009402. doi: 10.1371/journal.ppat.1009402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knosp WM, Knox SM, Hoffman MP. Salivary gland organogenesis. Wiley Interdiscip. Rev. Dev. Biol. 2012;1:69–82. doi: 10.1002/wdev.4. [DOI] [PubMed] [Google Scholar]
- 27.Patel VN, Hoffman MP. Salivary gland development: a template for regeneration. Semin. Cell Dev. Biol. 2014;25–26:52–60. doi: 10.1016/j.semcdb.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocha-Pereira J, et al. The viral polymerase inhibitor 2′-C-methylcytidine inhibits Norwalk virus replication and protects against norovirus-induced diarrhea and mortality in a mouse model. J. Virol. 2013;87:11798–11805. doi: 10.1128/JVI.02064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortez V, et al. Characterizing a murine model of astrovirus using viral isolates from persistently infected immunocompromised mice. J. Virol. 2019;93:e00223-–19. doi: 10.1128/JVI.00223-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nice TJ, Strong DW, McCune BT, Pohl CS, Virgin HW. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J. Virol. 2013;87:327–334. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orchard RC, et al. Discovery of a proteinaceous cellular receptor for a norovirus. Science. 2016;353:933–936. doi: 10.1126/science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones MK, et al. Human norovirus culture in B cells. Nat. Protoc. 2015;10:1939–1947. doi: 10.1038/nprot.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ettayebi K, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y-H, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerviel A, Zhang M, Altan-Bonnet N. A new infectious unit: extracellular vesicles carrying virus populations. Annu. Rev. Cell Dev. Biol. 2021;37:171–197. doi: 10.1146/annurev-cellbio-040621-032416. [DOI] [PubMed] [Google Scholar]
- 36.Sosnovtsev SV, et al. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 2006;80:7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Compton SR, Booth CJ, Macy JD. Murine astrovirus infection and transmission in neonatal CD1 mice. J. Am. Assoc. Lab. Anim. Sci. 2017;56:402–411. [PMC free article] [PubMed] [Google Scholar]
- 38.Willingham K, et al. Milk collection methods for mice and Reeves’ muntjac deer. J. Vis. Exp. 2014;89:51007. doi: 10.3791/51007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh S, et al. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source gels for Figs. 2b,c and 4f,k and Extended Data Fig. 4e–g.
Supplementary Table 1: antibody list; Supplementary Table 2: primer list; Supplementary Table 3: amplicon list; Supplementary Table 4: statistical analysis.
Data Availability Statement
The data generated in this study are provided as source data files corresponding to each figure. All statistics performed and the exact P values are presented in Supplementary Table 4. Primary data associated with figures are available in the source files. Any additional datasets generated and/or analysed are available from the corresponding author on reasonable request.