Abstract
A global pandemic has erupted as a result of the new brand coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This pandemic has been consociated with widespread mortality worldwide. The antiviral immune response is an imperative factor in confronting the recent coronavirus disease 2019 (COVID-19) infections. Meantime, cytokines recognize as crucial components in guiding the appropriate immune pathways in the restraining and eradication of the virus. Moreover, SARS-CoV-2 can induce uncontrolled inflammatory responses characterized by hyper-inflammatory cytokine production, which causes cytokine storm and acute respiratory distress syndrome (ARDS). As excessive inflammatory responses are contributed to the severe stage of the COVID-19 disease, therefore, the pro-inflammatory cytokines are regarded as the Achilles heel during COVID-19 infection. Among these cytokines, interleukin (IL-) 1 family cytokines (IL-1, IL-18, IL-33, IL-36, IL-37, and IL-38) appear to have a strong inflammatory role in severe COVID-19. Hence, understanding the underlying inflammatory mechanism of these cytokines during infection is critical for reducing the symptoms and severity of the disease. Here, the possible mechanisms and pathways involved in inflammatory immune responses are discussed.
Keywords: Interleukin-1 family, Interleukin-18, Interleukin-33, Inflammation, COVID-19
Introduction
SARS-CoV-2, the causative agent of the ongoing pandemic of COVID-19, has become a health threat since December 2019 and is responsible for over 500 million infected cases and 6 million deaths as of 6th May 2022. This pandemic has posed catastrophic impacts on nations' healthcare resources, especially in regions with fragile healthcare infrastructures, such as Africa [1], Afghanistan [2], and Bangladesh [3], and overloaded healthcare systems due to the coexisting contagious disease, like Zika [4] and Tuberculosis [5].
SARS-CoV-2 belongs to the beta-Coronaviruses [6] and shares a similar genome with other members of this family, particularly Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV [7, 8], which were responsible for the outbreaks of CoVs in the past [9]. Similar to their clinical presentations [10], SARS-CoV-2 infection accompanies by plethoric manifestations such as fatigue, fever, dyspnea, cough, diarrhea, and in severe cases with ARDS and multi-systemic dysfunctions [11, 12]. For now, various therapeutic approaches, including antiviral therapeutics (Remdesivir, Opinavir/ritonavir, Tenofovir disoproxil fumarate, Chloroquine, etc.), and plasma therapies are consider as possible therapeutic options for COVID-19 treatment. Nevertheless, some antiviral drugs associated with adverse side effects, therefore, numerous researches to find appropriate therapeutic strategies for this diseases alongside comprehensive vaccination are ongoing. Hence, better perception of SARS-CoV-2 pathogenesis is contributed to the delineate proper therapeutic approaches [13].
SARS-CoV-2 utilizes angiotensin-converting enzyme 2 (ACE2) to enter the cells and induce cellular injury, predominantly in the respiratory system [14, 15]. Viral invasion by activating numerous immune mechanisms and immune cells such as monocytes (MOs), macrophages (MQs), dendritic cells (DCs), natural killer (NK) cells, B and T cells generates a disproportionate release of cytokines, known as cytokine storm [16, 17]. These altered cytokines include increased levels of both pro-and anti-inflammatory cytokines like interleukin (IL)-1 family, IL-6, interferons (IFNs), tumor necrosis factor (TNF), and C-reactive protein (CRP) [18, 19]. However, anti-inflammatory cytokines fail to preserve the pro‐/anti‐inflammatory proportion in the COVID-19 patients because of the overwhelming concentrations of pro-inflammatory cytokines, which are likely to trigger immune invasions, causing injuries to the healthy tissues [18, 20]. The IL-1 family consists of 11 cytokines, seven with agonistic features (IL-1α, IL-1β, IL-18, IL-33, IL-36α, IL-36β, IL-36γ) and four with antagonistic features (IL-1 receptor antagonist (IL-1Ra), IL-36Ra, IL-37, IL-38) [21]. IL-1 family cytokines activate signal transduction by the IL-1 receptor (IL-1R) family, which consists of 10 members: IL-1R1, IL-1R2, IL-1R accessory protein (IL-1RAcP), IL-18Rα, IL-18Rβ, ST2 (or IL-33R), and IL-36R [22]. Mounting evidence accumulates that IL-1 family cytokines have a distinct role in disease pathogenesis. To date, several studies have been published about the pivotal role of IL-1 family cytokines in the cytokine storm. However, to our knowledge, there is no data that explicitly study the immune-pathogenesis of IL-18, 1L-33, IL-36, IL-37, and IL-38 in the COVID-19. Therefore, here, we reviewed the immune-pathogenesis of the IL-1 family in COVID-19. Of note, we searched Web of Science, PubMed, Google Scholar, and conference/congress paper using the search terms “COVID-19” or “SARS-CoV-2” besides these terms “IL-1 family”, “inflammatory cytokines”, “IL-1”, “IL-18”, “IL-33”, “IL-36, IL-37, IL-38”, and “immunopathogenesis” of each cytokines in COVID-19. Using this search strategy, we conducted the screening process and selected relevant articles.
Genetic and structural features of COVID-19
The name Coronavirus is associated with its shape, which is coated the outer fringe with club-shaped glycoprotein, not sharp, causing disease in animals and humans and laboratory diseases [23]. The family Coronaviridae belongs to the order Nidovirales, which includes the other two families, Arteriviridae and Roniviridae, which cause disease in birds and insects, respectively. The family Coronaviridae includes two subfamilies, Coronavirinae and Torovirinae, and the subfamilies Torovirinae has not caused any human infection. The subfamily Coronavirinae includes four groups of Alphacoronavirus, Betacoronavirus, Gammacirnavirus, and Deltacoronavirus. In the genus Alphacoronavirus, human Coronavirus-229E (HCoV-229E) and HCoV-NL63 are human viruses. The genus Betacoronavirus contains the prototype mouse hepatitis virus (MHV), HCoV-OC43, HCoV-HKU1, SARS-HCoV, MERS-CoV, SARS-HCoV-2 (COVID-19), and another animal coronavirus. The Gammacoronavirus and Deltacoronavirus primarily infect birds (or poultry) [24, 25]. The human coronavirus was first isolated from the human in the 1960s, and it was studied as a Common Cold virus until 2003 when the upper respiratory tract was slightly affected, but since SARS-CoV caused severe respiratory disease in 2003, the coronaviruses have received more attention [26, 27].
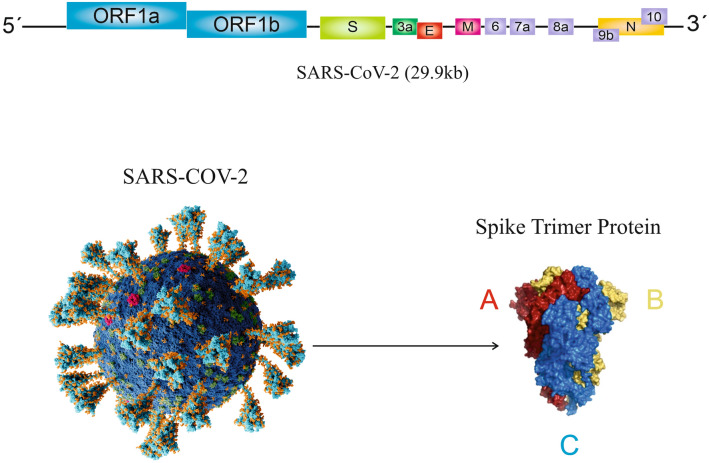
The coronavirus genome is single-stranded, positive-strand RNA (+ssRNA) that is designated as the longest RNA viral genome. They vary in length and expression in different species but share a common 3-end and a 5-leader sequence [10]. The 3-terminal region contains structural proteins, including spike protein (S), Membrane glycoprotein (M), nucleocapsid (N), and envelope protein (E); also several other proteins with hemagglutination and esterase functions, and these proteins may interfere with the immune system’s response (Fig. 1). The coronavirus genome contains 7–14 open reading frame (ORFs) such as SARS-CoV-2 genome includes 14 ORFs that translate into 27 proteins [24]. The majority of SARS-CoV through the S protein enters human cells using ACE2, which is expressed on lung and gastrointestinal cells [28]. Mutations in the S protein gene can alter receptor affinity and lead to a decrease or increase in its pathogenesis.
Fig. 1.
Coronaviruses and coronavirus Spike Glyco's genomic
Clinical, immunological and pathologic features of COVID-19
COVID-19 as an infectious disease rapidly transmitted among humans, and the mean incubation period after being exposed to the SARS-COV-2 and the onset of clinical symptoms is about 5.1 days (between 2 and 12 days) [29]. Medical indications of the COVID-19 patients vary from mild to severe symptoms [30]. Old age and underlying diseases, such as hypertension (HTN), diabetes mellitus (DM), and cardiovascular diseases, can consider risk factors for the severity of COVID-19 diseases [31, 32]. In the COVID-19 patients, fever (82.2%) with cough (61.7%) are main clinical symptom and followed by the fatigue (44.0%), dyspnea (41.0%), anorexia (40.0%), productive sputum (27.7%), myalgia (22.7%), sore throat (15.1%), nausea (9.4%), dizziness (9.4%), diarrhea (8.4%), vomiting (3.6%), abdominal pain (2.2%), myalgia (22.7%) and headache (6.7%),which are spectrum of other SARS-COV-2 clinical symptoms [31]. Accordingly, ACE2-expressing organs can be targeted by SARS-COV-2, such as the respiratory tract, gastrointestinal tract (GI) system, heart, kidney, and nervous system [33, 34]. Based on a meta-analysis evaluation with 6,022 COVID-19 patients (average age = 49.5 years), there is a likely strong association between COVID-19 severity with GI manifestations. Furthermore, this study represented the related symptoms with GI manifestations, such as diarrhea, nausea, abdominal pain, and vomiting [35]. A single-center study of 148 consecutive COVID-19 positive patients conducted by Fan et al. indicated that the duration of hospitalization of COVID-19 patients with the abnormal liver function was longer than other COVID-19 patients with normal liver function [36]. According to the available evidence, the COVID-19 can invade the nervous system and cause viral encephalitis, acute toxic encephalitis, and acute cerebrovascular disease [34]. Based on Mao L et al.’s investigation in a retrospective observational case series study with 214 patients admitted to the hospital, nearly 78 patients (36.4%) presented neurological manifestations, such as acute cerebrovascular diseases, impaired consciousness, and skeletal muscle injury [37]. Overall, among the clinical features of COVID-19, fever, cough, and fatigue consider common symptoms, and other symptoms like headache, sore throat, dizziness, and diarrhea are less common. Nevertheless, asymptomatic individuals have been identified who may not show any clinical manifestations and have been considered a challenge in controlling COVID-19 infection.
The induced antiviral response to SARS-CoV-2 by the host immunity is the determinant factor in controlling and eliminating COVID-19 infection [38]. The innate immune response is known as the primary line in recognizing and confrontation with infection following the virus’s entry. The onset of innate response is accompanied by hampering the viral replication through activation of phagocytic cells (MQs and neutrophils) and the secretion of pro-inflammatory cytokines. Moreover, pro-inflammatory cytokines have a decisive role in the stimulation of the adaptive arm of the immune system [39]. According to the knowledge obtained from the coronaviruses family, the viruses’ RNA is recognized by disparate pattern-recognition receptors (PRRs), such as the Toll-like receptors 3 (TLR3) and TLR7 as the endosomal PRR and retinoic acid-inducible gene 1 (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) as the cytosolic sensors. Because of inadequate data about the SARS-COV-2, these PRRs could be involved in identifying SARS-COV-2's RNA as well [40]. Subsequently, ligand binding induces the signaling cascades, which include several transcription factors, such as NF-κB, IRF3, and AP-1, that synergistically lead to type I interferon (IFN-I) production [41]. Furthermore, IFN-α/β cytokines trigger the expression of interferon-stimulated genes (ISGs) by activation of the downstream JAK-STAT signal pathway [38].
As aforementioned, the innate immune response triggers the adaptive immune response by pro-inflammatory cytokine secretion [39]. The adaptive immune response is characterized by two main components in COVID-19 infection, B cells as humoral immunity and T cells as cellular immunity [42]. When the immune system is stimulated, B cells, with the help of T cells, differentiate into plasma cells and then generate neutralizing antibodies (Abs) against viral antigens. Neutralizing Abs act as a barrier against the virus penetration into the host cell by blocking the virus [43]. Abs that produce through humoral immunity provide a protective role in restricting the viral infection and can prevent reinfection [44]. However, several primary reports illustrated that higher Ab titers are correlated to severe clinical symptoms in COVID-19 patients [45]. A prospective cohort study with 67 COVID-19 patients indicated that the anti-nucleocapsid protein (NCP) IgM began on day 7 and peaked on day 28, whereas the anti-NCP IgG was on day 10 and peaked on day 49. Moreover, the emergence of these Abs was earlier, and their titers are remarkably higher in severe patients in comparison with non-severe patients [46].
As for adaptive cellular immunity, naive CD4+ T cells activates by antigen‐presenting cells (APCs) that present the viral antigenic peptides. Then, activated CD4+ T cells enable the production of TNF‐α, IL‐2, and IFN‐γ [44]. During viral infection, cytotoxic T lymphocytes (CTLs) have the ability to kill the virally infected cells and contribute to viral elimination. CTLs induce apoptosis through several mechanisms like perforin or granzyme [47]. Accumulation evidence suggests that the dysregulation of immune response may play a critical role in SARS-CoV-2 pathogenesis [48]. One of the main features of severe COVID-19 infection is an increase in cytokine production. In COVID-19 disease, particularly in severe cases has been observed, an enhancement in the inflammatory cytokines' production, such as IL-1β, IL-2, IL-6, IL-7, IL-8, IL-10, IFN-γ, TNF-α, granulocyte-colony stimulating factor (G-CSF), granulocyte macrophage-colony stimulating factor (GM-CSF), interferon-inducible protein-10 (IP10), and monocyte chemotactic protein 1 (MCP-1) has been observed, which culminates leads to a “cytokine storm” or cytokine release syndrome (CRS) [40, 49]. During the immune response, the production of cytokines recruits pro-inflammatory cells, such as MQs and neutrophils, to the site of infection, which results in the induction of an inflammatory response. Despite the importance of these responses to clear the virus, they can also damage the healthy tissues of the host. Nevertheless, whether SARS CoV-2 stimulates cytokine storm via MOs/MQs requires further assessment [49]. Neutrophils, as reactive oxygen species (ROS) and neutrophil extracellular traps (NETs) producers, contribute to cell death [50]. Moreover, based on a study conducted by Veras et al., the concentration of NETs was increased in plasma, tracheal aspirate, and lung autopsies tissues of COVID-19 patients, and also were found the high levels of NETs produced by neutrophils. Notably, they reported that in the in vitro condition, the secreted NETs by neutrophils during SARS-CoV-2 infection promote lung epithelial cell death [51].
COVID-19 disease progression includes lymphopenia, elevated pro-inflammatory cytokines and also chemokines, accumulation of MQs and MOs in lungs, immune dysregulation, cytokine storms, and ARDS [39, 48, 50]. The development of ARDS or multi-organ damage often occurs due to secondary haemophagocytic lymphohistiocytosis (sHLH) in patients with a severe state of COVID-19. Both ARDS and sHLH are determined via excessive production of cytokine and hyperinflammation, which are hallmarks of cytokine storm [52]. Based on the evidence, lymphopenia (lymphocytopenia), a phenomenon characterized by a low count of lymphocytes in the blood, was observed in about 85% of patients with severe COVID-19 [44]. Consistently, the study illustrated that the number of T cells (TCD4+ and TCD8+) were remarkably diminished in COVID-19 patients, mainly in patients needing intensive care unit (ICU) care [53]. Therewith, recent evidence suggested that the exhaustion of T cells and NK cells during the COVID-19 infection, like what can happen in any chronic inflammation, may contribute to critically COVID-19 disease [39, 44]. Taken together, it can be concluded that just as the timely and correct response of the immune system inhibits infection, on the other side point, the dysregulated and excessive response of the immune system also causes the propagation of infection and damage to normal tissues (Fig. 2).
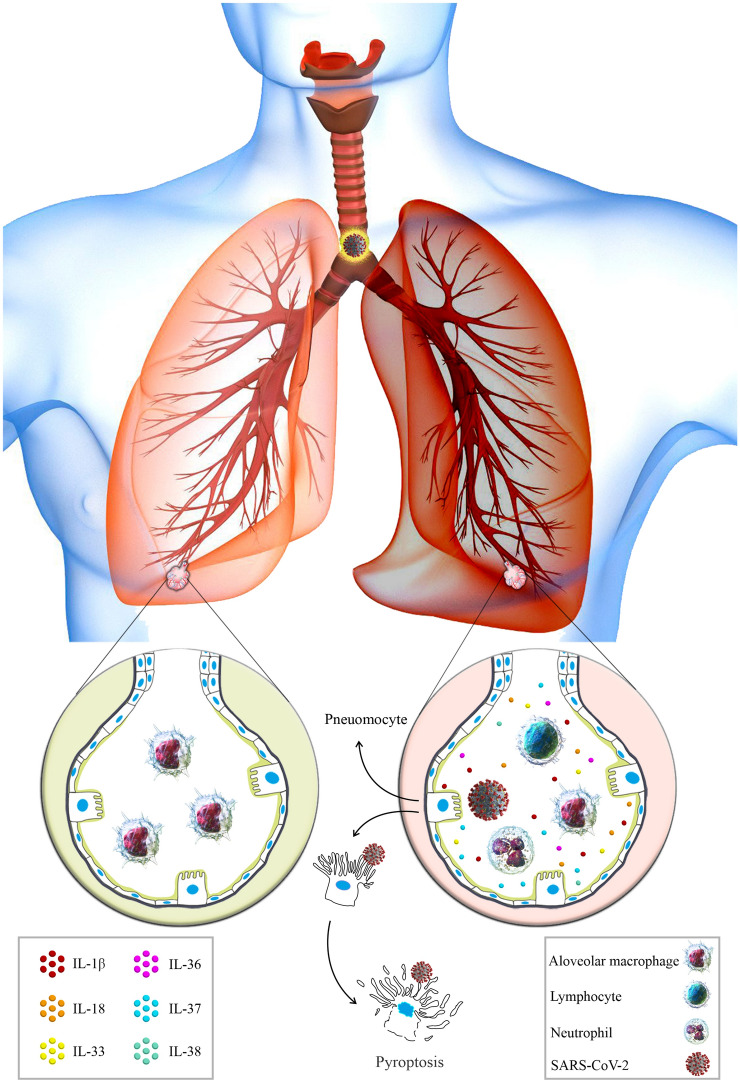
Fig. 2.
Healthy alveoli versus damaged alveoli in SARS-CoV-2 infection. Depicted the normal alveoli with coordinated functions (left), and damaged alveoli in SARS-CoV-2 infection (right). The wide cytokines production following immune activation through PAMPs or DAMPs, which recruited more immune cells to the infection site, such as neutrophils and lymphocytes, triggers an inflammatory cascade leading to asynchronized immune responses and ultimately to the pyroptosis of the pneumocytes
IL-1 and COVID-19
IL-1 structure and function
IL-1 is recognized as a pro-inflammatory cytokine [54] and belongs to the IL-1 cytokine’s family. IL-1 is composed of two cytokines, including IL-1α and IL- 1β. The IL-1α and IL-1β genes are located next to each other in the q14.1 band of chromosome 2 (2q14.1) and encode IL-1α and IL-1β, respectively [55]. Both IL-1α and IL-1β are primarily synthesized as precursor’s proteins, pro-IL-1α (271aa) and pro-IL-1β (269aa) by the innate immune cells in response to an inflammatory stimulus [55, 56]. After processing, pro-IL-1α and pro-IL-1β are converted to mature form with 158aa and 152aa, respectively [55]. Unlike pro-IL-1β, both pro-IL-1α and IL-1α (unprocessed and processed) are biologically active. Although, the biological activity and binding affinity of mature IL-1a is much higher than the immature (pro-IL-1α) form. Of note, various IL-1α and IL-1β proteases have been identified to convert and process the immature form into mature. Pro-IL-1α is cleaved by Calpain, granzyme B, neutrophil elastase, and chymase while, pro-IL-1β is processed by caspase-1 as the main pro-IL-1β protease, and then with neutrophil elastase, cathepsin G, chymase, and proteinase-3 [57]. In addition, IL-1α is produced by epithelial cells, endothelial cells, stromal cells, hematopoietic, and non-hematopoietic cells. Plus, it is presented by resident MQs and myeloid-derived cells [55]. IL-1 receptors are categorized based on their functions, IL-1R1 as activatory and both IL-1R2 and IL-1Ra as inhibitory receptors [57]. IL-1R1 (CD121a) as an activator receptor contains the intracellular Toll/interleukin-1 receptor (TIR) domains that it is essential for the formation of signaling complex [58]. IL-1R1, along with IL-1RAcP as a co-receptor [57] and TIR domains, cause the activation of NF-κB, p38, JNKs, and mitogen-activated protein kinases (MAPKs) pathways [55]. IL-1R2 (CD121b) is another receptor for IL-1 cytokines. Although IL-1R2 binds to IL-1, it is unable to initiate the signal transduction due to the lacking the TIR domains, thus known as a decoy receptor [58]. Furthermore, IL-1Ra is described as a third ligand in the IL-1 family that binds to IL-1R1 and IL-1R2, and acts as a physiological IL-1 blocking factor. IL-1Ra fails to activate signal transduction because it is incapable of recruiting the IL-1RAcP [55, 59].
Noteworthy, IL-1 is considered a key mediator of immunity and inflammation [60]. IL-1α and IL-1β recruited and activated neutrophils, MOs, and MQs with some differences in timing and activity. These cytokines (IL-1α and IL-1β) exert numerous biological effects, systemically and at sites of local inflammation, such as inflammatory response, promoting fever, vasodilation, angiogenesis, leukocyte recruitment, proliferation Abs production, and T cell differentiation, maturation antigen presentation, hematopoiesis, and the acute-phase response [57, 61]. Based on the findings, IL-1α has an alarming function, which is released from dying cells and initiates the primary phase of sterile inflammation. On the other hand, IL-1β is generated by inflammasomes in tissue infection areas or sterile injury [55, 61].
To date, straightforward evidence appears of the fundamental role of nucleotide-binding oligomerization domain (NOD)-like receptor family pyrin domain containing 3 (NLRP3) as a fundamental part of the inflammasome in the activation of IL-1β secretion [62, 63]. The enzymatic activity of caspase-1 within the cells is mostly responsible for the converting of pro–IL-1β to its active form [64, 65]. The inflammasome, a macromolecular complex, is formed by the NLRP3 scaffold, adaptor apoptosis speck-like protein (ASC), and the effector procaspase-1, which recruits and activates procaspase-1 to produce active caspase-1 [66]. The NLRP3 is one of the vital components of the inflammasome complex and acts as a sensor protein that consists of a NACHT, LRR, and PYD domains-containing protein 3 (NACHT, LRR, and PYD domains-containing protein 3) intracellular receptor that responds to intracellular or extracellular danger signals [62, 67]. Hoffman H M et al. are credited with discovering the involvement of NLRP3 [62, 68], whereas Agostini L et al. were the first to show that NLRP3 is a constitutive member of the inflammasome in the processing of IL-1 [62, 69].
IL-1 in diseases
Nowadays, more than any other cytokine family, the role of the IL-1 family, more importantly, the IL-1 subfamily (IL-1α and IL-1β), is well established in the spectrum of auto-inflammatory disorders, cardiovascular, and degenerative diseases [70–72]. The imbalance between IL-1 and IL-1Ra levels can result in overstated inflammatory responses and is contributed to several human diseases [70, 73]. IL-1β, as a crucial cytokine, is involved in promoting inflammation, especially in auto-inflammatory diseases, while IL-1α and the IL-36, as other members of the IL-1 family, are correlated with skin diseases. Also, IL-1β has been identified as a cardinal cytokine in the pathogenesis of numerous rheumatic disorders [74]. The observation implicates that the IL-1β gene expression is noticeably incremented in blood MOs in disease conditions, whereas it is low or invisible in health [62]. In this context, the IL-1α central role is described in the pathogenesis of several diseases, as systemic sclerosis (SS), myocarditis, osteoarthritis (OA), Kawasaki disease, and the role of IL-1β is certified in the pathogenesis of gout, OA, DM, and heart failure [74, 75]. Based on evidence, IL-1 pathway has prominent antiviral effect and helps to immune system in dealing with viral infection as well [76, 77]. In the next section, we focus on understanding the antiviral role of IL-1 in pathogenesis of COVID-19 disease and elucidating the possible mechanisms underlying disease for devising therapeutic approaches.
The pathogenesis of IL-1 in COVID-19
The findings point that during cytokine storms, the number of inflammatory mediators increases in critically ill patients [78]. Therefore, understanding the mechanisms and mediators involved in the cytokine storm assists in finding effective ways to treat and combat SARS-COV-2 infection.
The COVID-19 infection is the reason for epithelial injury leading to the secretion of IL-1α (cause to the recruitment of neutrophils and MOs to the infection’s site) and production of IL-1β in MOs/MQs [79]. IL-1β and IL-1Ra have been perceived in the peripheral blood and bronchoalveolar lavage fluid (BALF) of patients with COVID-19-induced pneumonia [80]. IL-1β as a pro-inflammatory cytokine is activated and released following activation of the inflammasome [81]. Intriguingly, according to other research results, patients with severe/critical COVID-19 infection had considerably higher levels of inflammatory cytokines in their BALFs than patients with mild COVID-19 infection, especially IL-8, IL-6, and IL-1β [82] (Table 1).
Table 1.
IL-1 family levels and its variation in the COVID-19 patients
| Cytokine | Findings | References |
|---|---|---|
| IL-1 | Elevated serum levels of IL-1 | [13] |
| Correlation of IL-1 levels with disease severity | [12] | |
| Elevated levels of IL-1β, and IL-1Ra in the BALF | [73] | |
| Not significantly increased in the fecal samples | [226] | |
| IL-18 | Elevated serum levels of NLRP3 inflammasome-derived products, IL-18, and 18BP, activated caspase-1, inflammatory factors, such as IL‐1, IL-4, IL-6, IL-7, IL-10, TNF-α, IFN-γ, and measurements of disease activity, such as LDH, CRP, ferritin | [130, 134, 136, 150–152] |
| Elevated levels of NLRP3 independent IL-18 processing enzymes, like granzyme B and proteinase 3 | [133, 134] | |
| Elevated levels of the IL-18BP | [129, 135, 136] | |
| Correlation between IL-18 levels and the activation of innate NK cells, γδ T cells, CD4 + , and CD8 + T cells | [139] | |
| Correlation of IL-18 levels with disease severity | [133, 153, 154] | |
| Identifying IL-18, IL-15 and Gal-9, as organ-failure-specific immunological markers of respiratory system failure | [154, 155] | |
| Correlations of the IL-18 with hematological parameters, renal and hepatic biochemical markers, and cardiac injury markers | [135, 153] | |
| Correlations of the IL-18 with increased levels of fibrinogen and D-dimer, and decreased platelet numbers | [153] | |
| Higher serum levels of IL-18, and LDH in patients with underlying liver diseases | [128] | |
| Correlations of IL-18 coupled with IL-6, IFN-γ, IP-10, and RANTES with LDL and HDL in the recovered patients | [165] | |
| Higher levels of IL-18, IL-8, and CCL5 and more robust induction of non-classical MOs in male patients | [156] | |
| Correlation of IL-18 and IL-18BP with patients’ age | [136] | |
| Positive associations of IL-18 levels with the saliva viral loads | [157] | |
| Elevated levels of IL-18 in children with MIS-C | [158–160] | |
| High levels of IL-18 consistent with CCR2, CX3CR1, CCL3, CXCL9, and macrophage inflammatory factors in the recovery stages of the infection | [154, 166] | |
| Higher concentrations of IL-18 as a differentiating marker between COVID-19 and influenza, AOSD, MAS, and sHLH | [152, 167–169] | |
| Negative correlation of IL-18 levels with anti-SARS-CoV-2 IgA and IgG Abs in the nasal fluids | [161] | |
| Increased IL-18 levels in the fecal samples | [164] | |
| Negative association between IL-18 gene expression and risk of the SARS-CoV-2 infection | [171] | |
| High gene expression of IL-18, IL-6, IL-8, TNF-α, IFN-γ, and CCL5 in the postmortem cardiac tissues | [162] | |
| IL-33 | Elevated serum levels of the IL-33 | [199, 200] |
| Correlation of the IL-33 with clinical and radiographic parameters of the patients and disease severity | [198, 199] | |
| Positive correlation of the IL-33 with TNF-α, IL-1β, IL-6, IL-12, and IL-23 levels | [200] | |
| Negative correlation of the IL-33 with O2 saturation | ||
| Correlation of IL-33 and IL-12p70 levels with seroconversion of IgG Abs | [200] | |
| Correlation of the IL-33 and IL-18 levels with renal toxicity markers such as GST and osteopontin | [196] | |
| Un-changed levels of the IL-33 accompanied by increased levels of the sST2 receptor | [201] | |
| Inverse correlation of the serum sST2 levels with the counts of CD4+ and CD8+ T cells | ||
| Positive correlation of the serum sST2 levels with disease severity | ||
| Correlation of the elevated expression of IL-33 in the cells isolated from the BALF samples with disease severity | [202, 203] | |
| IL-33 gene as one of the target genes of the SARS-CoV-2 encoded miRNAs | [204] | |
| IL-33 gene co-expression with ACE2, in human lung epithelial cells | [205] | |
| Increased serum levels of the IL-33, alongside the IL-1α, IL-6, IFNs, TNF-α, and chemokines in children with PIMS-TS compared to those of the COVID-19, seropositive for SARS-CoV-2 infection, and control children | [206] | |
| Higher concentration of IL-33 in the nasal mucosa of CRSwNP patients compared to COVID-19 patients | [207] | |
| Depletion of the IL-33 and AEC2 in the postmortem lung tissues of COVID-19 patients compared to those of the healthy subjects and patients with COPD or IPF | [176] | |
| Increased amounts of IL-33 and AEC2 in the fibrotic lung samples of the COVID-19 survived patients compared to patients with COPD and IPF | ||
| Heighten IL-33 levels in SARS-CoV-2 co-infected patients with LTBI | [208] | |
| IL-36 | Increased serum levels of the IL-36α and IL-38 | [220] |
| Positive correlation of the IL-36α with disease severity | ||
| IL-37 | Elevated plasma levels of IL-37 together with higher IFN-α and lower IL-6 and IL-8 levels | [222] |
| Lower levels of the IL-37 and higher IL-8 and CRP levels | ||
| IL-38 | Lower serum levels of the IL-37 and vitamin D | [223] |
| Negative correlation of the IL-37 with disease severity | [222, 223] | |
| Association of the IL-37 genes mutations with the COVID-19 susceptibility | [224] |
Ab, antibody; ACE2, angiotensin-converting enzyme 2; AEC2, alveolar epithelial cells type 2; AOSD, adult-onset still’s disease; BALF, bronchoalveolar lavage fluid; COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; CRSwNP, chronic rhinosinusitis with nasal polyps; GST, glutathione S-transferase; HDL, high-density lipoprotein; sHLH, secondary hemophagocytic lymphohistiocytosis; IFN, interferon; IL, interleukin; IL-18BP, IL-18 binding protein; IPF, idiopathic pulmonary fibrosis; LDH, lactate dehydrogenase; LDL, light-density lipoprotein; LTBI, latent Tuberculosis infection; MAS, macrophage activation syndrome; NLRP3, nucleotide-binding oligomerization domain-like receptor family pyrin domain containing 3; NK, natural killer; PIMS-TS, pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 infection; RANTES, regulated on activation; normal T expressed and secreted; miRNA, microRNA
Furthermore, the peripheral blood mononuclear cells (PBMCs) analysis of the COVID-19 patients indicated a significant reduction in CD4+ T cells and CD8+ T cells and enhancement in MOs in the early recovery stage. Moreover, in the erythrocyte sedimentation rate (ESR), there was a higher ratio of classical CD14++ MOs with strong inflammatory gene expression and a higher quantity of CD14++ IL-1β+ MOs (Table 2). In addition, the integrated analysis anticipated that IL-1β and macrophage colony-stimulating factor (M-CSF) could be new prospective target genes for inflammatory storms, as well as TNF Superfamily Member 13 (TNFSF13), IL-18, IL-2, and IL-4 could help COVID-19 patients recover [83]. Research conducted on PBMCs and plasma samples of 13 COVID-19 patients demonstrated a fourfold greater IL-6 production in COVID-19 convalescent patients. Besides, convalescent patients had an increased level of IL-1β and a comparable amount of IFN-γ. In addition, approximately 46% of COVID-19 convalescent patients had greater TNF-α and an elevated plasma level of CXCL11 [84]. The striking property of IL-1 is the ability to produce and secretion of more IL-1, a process called an auto-inflammatory loop. This character of IL-1 ushered in the production of more IL-1 and the activation and recall of more innate immune cells [79]. Therefore, several hypotheses and studies have been formed based on the blocking of IL-1, which may alleviate the critically of cytokines storm and improve COVID-19 patients' symptoms. We will mention some clinical trials in this section.
Table 2.
In vitro evaluations of the inflammatory pathways related to IL-1 family cytokines in the SARS-CoV-2 infection
| Cytokine | Findings | Reference |
|---|---|---|
| IL-1 | Increased expression of IL-1β following NLRP3 inflammasome and caspase-1 activation in the purified PBMCs, granulocyte subsets, and T cells | [128, 129] |
| Increased expressions of IL-1β following NLRP3 inflammasome and caspase-1 activation in direct infection of PBMCs with SARS-CoV-2 | [127, 130] | |
| Lower pro-IL-1β together with higher IL-18, and IFN-α, IFN-β, IFN-λ mRNA expression in the peripheral blood cells | [137] | |
| Higher production of IL-1β in the anti-CD3/CD28 Ab stimulated PBMCs of the COVID-19 patients compared to healthy controls | [140] | |
| Up-regulated expression IL-1β in the direct infection of LORGs, HSCs, HAECs, EPCs, and VSELs by SRAS-CoV-2 | [146–148] | |
| Increased IL-1β levels in the SARS-CoV-2 infected PBMCs with recombinant form of BPIFB4 | [170] | |
| IL-18 | Increased expression of IL-1β, IL-18, IL-18BPa, and GSDMD following NLRP3 inflammasome and caspase-1 activation in the purified PBMCs, granulocyte subsets, and T cells | [132, 142] |
| Increased expressions of ASC, IL-1β, IL‐18, IL‐6, IL‐12, IL‐15, IL‐16, IL‐36, TNF-α, and chemokine receptors following NLRP3 inflammasome and caspase-1 activation in direct infection of PBMCs with SARS-CoV-2 | [130, 131] | |
| Higher caspase-1 activation in T cells isolated from patients with underlying liver diseases | [128] | |
| Higher IL-18 together with IFN-α, IFN-β, IFN-λ, and lower pro-IL-1β mRNA expression in the peripheral blood cells | [137] | |
| Up-regulated gene expression of IL-18, amphiregulin, epiregulin, and ADAMTS2, and increased HIF-1 signaling pathway in the novel MO subsets | [138] | |
| Higher production of IL-18 and IL-1β in the anti-CD3/CD28 Ab stimulated PBMCs of the COVID-19 patients compared to healthy controls | [140] | |
| Partially decreased production of IL-18 in the co-culture of these cells with DPSCs | ||
| Overexpressed pro-inflammatory mediators like IL-32, down-regulated amphiregulin gene, and impeding anti-tumor responses in the IL-18 treated T-regs | [141] | |
| Increased IL-18 levels, activation of MAIT cells in the lungs and blood, increased IFN-γ production, decreased IFN-α2 levels, down-regulated type I IFN-responsive transcription factors, and up-regulated NLRP3 inflammasome, in the serum, plasma, and SARS-CoV-2 infected MOs/MQs and MAIT cells | [142] | |
| Up-regulated levels of IL-18 and IL-6 by classical MQs, inhibited growth and enhanced apoptosis of lung cells in the infection of HPSCs derived lung cells and MQs with SARS-CoV-2 pseudovirus | [145] | |
| Up-regulated expression of NLRP3 inflammasome, caspase-1, IL-1β, IL-18, GSDM, and ASC in the direct infection of LORGs, HSCs, HAECs, EPCs, and VSELs by SRAS-CoV-2 | [146–148] | |
| Increased IL-18 and IL-1β levels, down-regulated CD69 activating-marker for T cells (both CD4+ T and CD8+ T cells) and MCP-1, and limited cellular damages in the SARS-CoV-2 infected PBMCs with recombinant form of BPIFB4 | [170] | |
| IL-33 | Higher increase of IL-33 accompanied by IL-6, IFN-α2, and IL-23 secretion in the culture of the SARS-CoV-2 peptide stimulated PBMCs from seropositive patients than those of the seronegative patients | [203] |
| Positive correlation of enhanced production of the IL-33 with the expression of CD69 in CD4+ T cells | [203] | |
| Higher level of intracellular IL-33 in CD14+ MOs in the PBMCs | [203] | |
| Higher ST2 expression in the B cells, followed by MOs and DCs in the immune cells | ||
| Association of increased serum concentrations of IL-33 with increased bone marrow precursor cells in PBMCs | [209] | |
| Positive association between IL-33 production from SARS-CoV-2 peptide stimulated PBMCs of the seropositive patients and their IgG titers | [203] | |
| Increased IL-33 mRNA expression in the direct infection of human epithelial cell lines, Fadu and LS513, by SRAS-CoV-2 | [195] | |
| Impaired production of IL-33 and IL-1β, IL-6, and IL-8 together with up-regulated SIGIRR, down-regulated AEC2, and up-regulated IFN-β expression, and activation of various antiviral immune pathways following treatment of poly(I:C) stimulated HBECs with Imiquimod | [210] | |
| IL-36 | Increased expressions IL‐36 following NLRP3 inflammasome and caspase-1 activation in direct infection of the PBMCs with SARS-CoV-2 | [131] |
| IL-37 | Attenuated respiratory inflammation by IL-37 administration in the human transgenic ACE2 mice | [222] |
| IL-38 | N/A | N/A |
Ab, antibody; ACE2, angiotensin-converting enzyme 2; ADAMTS2, a disintegrin and metalloproteinase with thrombospondin motifs 2; AEC2, alveolar epithelial cells type 2; BPIFB4, bactericidal/permeability-increasing fold-containing family-B-member-4; DPSC, dental pulp stem cell; EPC, endothelial progenitor cell; GSDMD, gasdermin D; HAEC, human aortic endothelial cell; HBEC, human bronchial epithelial cells; HIF-1, hypoxia inducible factor 1; HPSC, human pluripotent stem cell; HSC, hematopoietic stem cell; IFN, interferon, IL, interleukin; IL-18BP, IL-18 binding protein; LORG, lung organoids; MAIT, mucosal-associated invariant T; MCP-1, monocyte chemoattractant protein 1, N/A, non-applicable; NLRP3, nucleotide-binding oligomerization domain-like receptor family pyrin domain containing 3; PBMC, peripheral blood mononuclear cell, SIGIRR, single immunoglobulin IL-1-related receptor; mRNA, messenger RNA; VSEL, very small embryonic-like stem cell
Anakinra is one of the known IL-1-blocking agents that acts like IL-1Ra and blocks the function of IL-1. Thus, it can inhibit the auto-inflammatory loop’s activity [79, 85]. Anakinra has also been shown to reduce mortality in patients with sHLH caused by a virus in both children and adults [86]. For example, a case report study of a COVID-19 + 50-year-old man with ADRS manifestation (occurred due to cytokine storm) found that anakinra ameliorates inflammatory markers and ferritin, decreasing the patient's need for oxygen. Moreover, they mentioned that although its clinical efficacy cannot be ascertained by studying one person, anakinra may have a positive effect on the inflammatory condition, which is correlated with cytokine storm in SARS-COV-2 infection [87]. In another case series, study with three acute leukemia patients with severe COVID-19 pneumonia illustrated that treatment with anakinra may be contributed to clinical improvement [88]. Moreover, Huet T et al. conducted a COVID-19 cohort study with 52 consecutive patients who received anakinra as treatment and 44 historical patients. They indicated that anakinra decreased both requirements for invasive mechanical ventilation in the ICU and mortality between patients with severe COVID-19, without critical adverse effects [89]. Based on Cavalli G et al. retrospective cohort study, anakinra therapy with high-dose intravenous in patients with COVID-19 and ARDS managed with non-invasive ventilation outside of the ICU was safe and correlated with clinical advantage in 72% of patients [90]. Besides, another investigation evaluated the effect of the anakinra along with methylprednisolone in acute COVID-19 patients with pneumonia and hyperinflammation. At 28 days, treated patients had a mortality rate of 13.9%, whereas controls had a mortality rate of 35.6%. Consequently, based on the study, outcome can be assumed that the combination of anakinra with methylprednisolone may be a good candidate for the treatment of COVID-19 patients with respiratory failure and hyper-inflammation [91].
Franzetti M et al., in the retrospective observational study, assayed the effect of anakinra on the survival of COVID-19 patients with ARDS. In this research, out of 112 patients, 56 individual’s received anakinra, and 56 composed controls. According to the results, anakinra-treated individuals had a much greater survival rate than the controls [92].
Collectively, according to the findings, it can be concluded that treatment with anakinra is safe, and due to its short half-life, side effects and undesirable effects can be prevented quickly. Conceivably, this treatment may be an appropriate option for COVID-19 patients, but confirmation requires further investigation (Fig. 3).
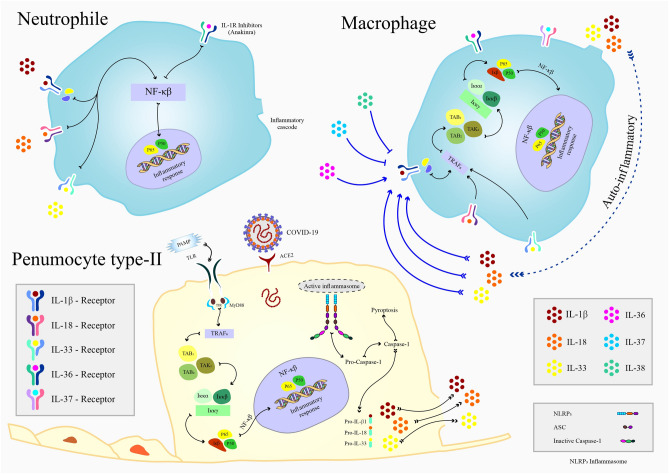
Fig. 3.
Schematic model of strategies for IL-1 family members’ intervention during SARS-CoV-2 infection. Following virus entry, detection of the viral PAMPs by TLRs leads to activated NLRP3 inflammasome in the type II pneumocyte cells of the respiratory tract and induction of IL-1β, IL-18, and IL-33 expression via NF-κB transcription factor signaling pathway. On the other side, the aggregated immune response against SARS-COV-2 by macrophages and neutrophils results in the production of IL-1 family cytokines
IL-18 and COVID-19
IL-18 structure and function
IL-18 is a 193 amino acids IL-1 family member that initially designated IFN-γ-inducing factor (IGIF) [93]. IL-18 has structural similarities with its sister cytokines, particularly IL-1β [94]. Like IL-1β, but unlike IL-1α or IL-33, it first synthesizes as an inactive precursor, pro-IL-18, in various hematopoietic and non-hematopoietic cells [95, 96]. To become a biologically active cytokine, pro-IL-18 requires subsequent proteolytic cleavage. Similar to pro-IL-1β, activated caspase-1 is the pivotal pro-IL-18 processing enzyme, which its activation is mainly dependent on the priming of inflammasomes, primarily the NLRP3 [95, 97]. In addition, IL-18 can be activated independently from caspase-1 by other enzymes, such as neutrophilic proteinase 3 [98], granzyme B (in NK cells and CTLs) [99], and meprin β (in intestinal cells) [100]. In contrast to IL‐1β, IL‐18 has constitutive expression in MOs, MQs, and DCs, and like IL‐1α and IL‐33, in endothelial and intestinal epithelial cells (IECs) of healthy subjects [95, 101]. Moreover, in the inflammatory states, recognition of pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) by TLRs culminates in the NLRP3 priming, followed by an overproduction of IL-18 and its secretion in a process called pyroptosis, in which IL-18 release from cell-membrane pores created by gasdermin D (GSDMD), another product of NLRP3 activation [102]. Although this mechanism is essential for releasing IL-18 from cells, it drives the disturbance of innate and adaptive immune responses.
IL-18 harbors pleiotropic features in both innate and adaptive arms of the immune system. It has fundamental roles in the clearance of intracellular pathogens, especially viruses, by promoting IFN-γ production and activation of NK cells and various subsets of T cells, such as T helper (Th)-1, Th-2, Th-17, CTLs, and regulatory T (T-reg) cells [103, 104]. IL‐18 facilitates IFN-γ production by CD4+ T cells, NK cells, B cells, DCs, and MQs with the help of IL‐12, since IL-12 significantly increases the expression of IL‐18R [105, 106]. Moreover, IL‐18, with synergistic effects of IL-12 and IL-15, up-regulates perforin and Fas ligand (FasL) expression and promotes FasL‐dependent cytotoxicity in NK cells and CD8+ T cells [107, 108]. Additionally, studies suggested that IL-18, in synergy with IL-23, enhances IL-17 production by Th-17 cells [104]. Second, IL-18 mediates subcapsular MQ-dependent expansion of B cells in the lymph nodes and their IgG Ab production [109, 110].
In addition to its crucial effects, mediating adaptive immune responses, IL-18 amplifies the innate immune response by regulating a complex network of cytokines. IL-18 with IL-2 enhance the production of Th-2 cytokines like IL-3, IL-4, IL-9, and IL-13 from CD4+ NK T (NKT) cells, NK cells, and even Th-1 cells [106, 111]. Accompanied by IL-3, IL-18 stimulates mast cells and basophils to produce IL-4, IL-13, and histamine but not IFN-γ. These activated basophils also could induce the development of Th-2 cells [106]. Moreover, it stimulates other cytokine production such as IL-1β, IL-6, IL-17, TNF-a, CRP, and chemokines, which are engaged in host innate defense and cytokine storm leading to ARDS [103, 112].
Second, IL-18 has pro-fibrotic features [70]. Apart from its direct inducer actions on fibroblasts, overexpressed IL-18 accumulates inflammatory cells (primarily CD8 + T cells, MQs, neutrophils, and eosinophils) in airway fibrosis and lung injury [113, 114]. Furthermore, IL-18, and in the second place, IL-33 induces the expression of amphiregulin, an epidermal growth factor-like cytokine necessary for tissue repair, in lung T-reg cells, the predominant amphiregulin source in the lungs [115]. However, its overproduction in the inflammatory states can be pathogenic by inducing lung fibrosis [116].
IL-18 activity is regulated by IL-18-binding protein (IL-18BP) [117]. IL-18BP is stimulated by IFN-γ and inhibits IL-18-induced cell responses like IFN-γ production by binding to IL-18, creating a negative feedback loop [70].
IL-18 in diseases
Numerous studies have described the implication of dysregulated IL‐18 in several inflammatory diseases, particularly in viral infections like influenza, Epstein–Barr virus (EBV), and poxviruses [118, 119]. Viral induction of IL-18 is profoundly mediated by NLRP3 inflammasome and subsequently caspase-1 activation in multiple cells like MOs, MQs, and epithelial cells of the respiratory system [120]. This pathway can be a defensive mechanism in the early stages of viral invasion since IL‐18 enhances host-viral defense and cell cytotoxicity [121, 122]. However, sustained NLRP3 stimulation induces an aberrant amount of IL‐18, detrimentally triggering the exuberant secretion of other downstream inflammatory mediators, such as IFN-γ, IL-6, TNF-α, which are involved in the cytokine storm formation [123, 124]. The abundant presence of IL-18R in the lungs than in other tissues makes the respiratory system more appealing for IL-18 to endorse a cytokine storm [125]. Prominent elevated serum and lung IL-18 levels have been correlated with increased inflammatory factors, disease pathogenesis, and severity in viral infections featured by cytokine storm and ARDS [126, 127].
Noteworthy, IL-18 regulates a wide range of immunological functions, such as stimulating IFN-γ production, priming NK cells cytotoxicity, and promoting differentiation of Th-1 cells in the gastrointestinal (GI) system [128]. However, it has also been suggested to contribute to the aggregation of inflammation and amplifying damages elicited to the GI tract by viral pathogens like rotaviruses and human immunodeficiency virus (HIV) [128].
Moreover, the imbalance between IL-18 and IL-18BP has been implicated in the severity of many diseases such as myocardial infarction (MI), acute kidney injury (AKI), inflammatory bowel disease (IBD), and sepsis [129].
The pathogenesis of IL-18 in COVID-19
Interestingly, the mechanism mentioned above is applicable for the pathogenesis of ARDS induced by coronaviruses since both SARS-CoV and SAR-CoV-2 have exhibited NLRP3 activation [130, 131]. However, there are discrepancies in the literature regarding the role of IL-18 in SARS-CoV infection, as the demonstration of its markedly increased expression in the lung tissues of SARS-CoV patients with ARDS in some research [124, 132] was accompanied by reports of its low serum levels in other studies [133].
In contrast to SARS-CoV, mounting clinical evidence supplemented by recently in vitro investigations deciphered solid implication of IL-18 and its downstream cytokine cascade, predominantly through NLRP3 inflammasome priming in the COVID-19 pathogenesis (Table 2). These studies reported increased levels of IL-18 together with the up-regulated expression of NLRP3, ASC, activated caspase-1, and IL-1β secretion in PBMCs, granulocyte subsets, and T cells purified from COVID-19 patients that were correlated with disease severity [134–139]. Correspondingly, direct infection of these cells with SARS-CoV-2 spike (S) protein revealed its interplay with ACE2 as the principal trigger of the NLRP3 activation [134, 137, 138]. These results were coupled with NLRP3 and ASC expression in postmortem lung tissues in COVID-19 patients [137]. Plus, multiple investigations found elevated levels of NLRP3 independent IL-18 processing enzymes, particularly granzyme B and proteinase 3, suggesting roles of other inflammatory mechanisms in SARS-CoV-2-induced production of IL-18 [140, 141]. Second, several studies documented significantly increased levels of the IL-18BP alongside the IL-18 in COVID-19 patients [136, 142, 143]. However, it seems that enhanced production of IL-18BP in response to IFN-γ and other stimulators is insufficient for inhibiting the IL-18–IFN-γ axis, leading to immune dysregulation, particularly during the final stages of the disease.
Additionally, analysis of messenger RNA (mRNA) transcription displayed higher IL-18 together with IFN-α, IFN-β, IFN-λ, and lower pro-IL-1β mRNA expression in peripheral blood cells of COVID-19 patients in comparison to healthy subjects. Furthermore, there was a positive correlation between mRNA expression of IL-18 and pro-IL-1β with TLR4 [144].
Furthermore, Zhang Y et al., by analyzing PBMCs of COVID-19 patients with single-cell RNA sequencing (scRNA-Seq), discovered novel MO subsets with up-regulated gene expression of IL-18 and pro-fibrosis mediators such as amphiregulin, epiregulin, and a disintegrin and metalloproteinase with thrombospondin motifs 2 (ADAMTS2). In addition, there was an increased hypoxia-inducible factor-1 (HIF-1) signaling in these cells [145].
In agreement with these findings, several observations were consistent with the ability of IL-18/IL-18R signaling to enhance the activation of T cells [106], unifying the elevated levels of IL-18 and its signaling pathways with hyperactivation of T cells in COVID-19 disease [146].
Croci S et al. reported that compared to healthy controls, PBMCs of COVID-19 patients which stimulated with anti-CD3/CD28 Ab to mimic the activation of T lymphocytes that occurs in infected tissues displayed higher production of IL-18, highlighting T lymphocyte's role in the production of IL-18 from PBMCs [147]. Additionally, the co-culture of these cells with dental pulp stem cells (DPSCs) resulted in partially decreased production of IL-18 [147].
Moreover, a study done by Galván-Peña S et al. demonstrated the high similarity between IL-18 treated T-regs and T-regs purified from COVID-19 patients in terms of cytometric and transcriptomic profile addressing the contribution of IL-18 to perturbations of T-regs in COVID-19 patients [148]. These T-regs exhibited a distinct transcriptional signature, with overexpression of several pro-inflammatory mediators like IL-32, and a remarkable similarity to T-regs that impede anti-tumor responses, suggesting their nefarious roles in the disease by suppressing antiviral T cell responses and production of pro-inflammatory mediators [148]. However, in contrast to the previous study, they found down-regulated amphiregulin gene in IL-18 treated T-regs [148].
Likewise, Flament H et al. showed increased IL-18 levels associated with severe clinical outcomes and activation of mucosal-associated invariant T (MAIT) cells in the lungs and blood of COVID-19 patients [149]. These cells are innate-like T cells involved in mucosal viral defense and display a pronounced cytotoxic phenotype in the lungs using the IL-18–IFN-γ axis [150]. Additionally, plasma IFN-α2 levels were inversely correlated with elevated IL-18 levels. Co-culture study of SARS-CoV-2 infected MOs/MQs and MAIT cells determined the IFN-α–IL-18 imbalance due to the down-regulated type I IFN-responsive transcription factors and up-regulated NLRP3 inflammasome to be responsible for the altered functions of MAIT cells, as depicted by their overwhelming IFN-γ production [149]. Respectively, increased IL-18 production and MAIT cells activation during innate immune response to SARS-CoV-2 may thereby lead to B cell activation and production of the anti-SARS-CoV-2 IgG Abs [151]. Concomitantly, correlations of the IL-18 levels and the activation of innate NK cells, as well as γδ T cells, CD4 + , and CD8 + T cells, have been described in these patients [146].
In addition, multiple investigations using various cell lines displayed the capacity of SARS-CoV-2 in instigating IL-18 secretion following the NLRP3 inflammasome activation. Duan F et al., in a co-culture study of lung cells and MQs, both derived from human pluripotent stem cells (HPSCs), with SARS-CoV-2 pseudovirus, displayed significantly up-regulated levels of IL-18 and IL-6 by classical MQs, accompanied by their ability to inhibit growth and enhance apoptosis of lung cells [152]. A similar case has been reported in the interactions of SRAS-CoV-2 and ACE2 or TLR4 in human stem cell-derived lung organoids (LORGs), hematopoietic stem cells (HSCs), human aortic endothelial cells (HAECs), endothelial progenitor cells (EPCs), and very small embryonic-like stem cells (VSELs), with the resultant, up-regulated expression of NLRP3, caspase-1, IL-18, IL-1β, GSDMD, and ASC [153–156]. These pathways can be inhibited by recombinant human ACE2, TLR4 inhibitors like TAC-242 or MCC950 [154], mineralocorticoid receptor antagonists (MRAs) like spironolactone or eplerenone, and G3P-01, an inhibitor of Galectin (Gal)-3, which is a downstream mediator of MR-induced vascular, cardiac, and renal inflammation [156].
In parallel with these findings, mounting investigations have reported remarkably elevated serum or plasma levels of IL-18, positively correlated with increased other inflammatory cytokines such as IL‐1, IL-6, TNF-α, IFN-γ, and measures of disease activity such as lactate dehydrogenase (LDH), CRP, and ferritin, the prominent inflammation signature of SARS-CoV-2-induced cytokine storm and ARDS [20, 137, 141, 143, 157–159].
Furthermore, a wealth of clinical data determines the positive correlation of heightened IL-18 concentrations with the occurrence of the ARDS, lung fibrosis, and AKI and disease severity, contributing to the early identification of critical COVID-19 patients [19, 140, 160, 161] (Table 1). Concomitantly, some studies identified IL-18, alongside the IL-15 and Gal-9, as an organ-failure-specific immunological marker of the respiratory system [161, 162].
Notably, levels of IL-18, together with IL-8 and CCL5, were found higher in male patients along with more robust induction of non-classical MOs [163], and both IL-18 and IL-18BP were correlated with the age of COVID-19 patients [143]. Positive associations of IL-18 levels with the saliva viral loads have also been reported [164].
Consistently, IL-18 levels and other inflammatory factors such as IL-1 and IL-6 were higher in children with the multisystem inflammatory syndrome (MIS-C), a severe clinical form of SARS-CoV-2 infection [165–167]. Moreover, IL-18 level was inversely correlated with anti-SARS-CoV-2 IgA, and IgG was detected in the nasal fluids of both children and adult patients, suggesting its implementation in the mucosal immune response against SARS-CoV-2 [168].
Interestingly, numerous data extend the contribution of the IL-18 to the pathogenesis of the SARS-CoV-2 induced non-respiratory damages. Its concentrations have significant correlations with hematological parameters, renal and hepatic biochemical markers, and cardiac injury markers like troponin [142, 160]. Likewise, IL-18 gene expression as well as IL-6, TNF-α, and IFN-γ were higher in cardiac tissues of COVID-19 autopsy cases that detected positive for SARS-CoV-2, addressing IL-18 s’ participation in the cardiac involvements of SASR-CoV-2 infection [169]. Furthermore, IL-18 correlations with increased fibrinogen and D-dimer, and decreased platelet numbers, were indicative of its actions as a bridge between inflammation and coagulation [160, 170]. Additionally, COVID-19 patients with underlying liver diseases like non-alcoholic fatty liver disease (NAFLD) and cirrhosis in comparison to patients without comorbidities displayed higher serum levels of IL-18, LDH, and activated caspase-1 in T cells, suggesting the boosting effect of underlying inflammatory state on IL-18 production and COVID-19 severity [135]. Moreover, IL-18 levels were higher in the fecal samples of COVID-19 patients than in those of controls, addressing IL-18 produced by lymphocytes, myeloid and epithelial cells of the GI system as an indicator of SARS-CoV-2 elicited intestinal infection [128, 171]. Concomitantly, IL-18 coupled with IL-6, IFN-γ, and IP-10, and regulated on activation, normal T expressed and secreted (RANTES) exhibited significant correlations with lipoproteins like light-density lipoprotein (LDL) and high-density lipoprotein (HDL) in recovered COVID-19 patients, indicating the involvement of IL-18 in the interactions of the SARS CoV-2 infection with the lipoproteins and immunometabolic profile of the disease [172].
Importantly, IL-18 has been proposed as a regulator of immunity in the recovery stages of the COVID-19. A study done by Singh et al. showed sustained high levels of IL-18 consistent with CCR2, CX3CR1, and macrophage inflammatory factors even in the recovery stages of the infection, suggesting IL-18 s’ roles in the compromised immunity of recovered patients [173]. Likewise, in the recovery period from ARDS, the serum levels of IL-18, CCL3, and CXCL9 were higher in COVID-19 patients with dyspnea or renal failure, representing IL-18 as a reflecting marker of residual inflammation in these patients [161]. However, prediction analysis of cell-to-cell interactions utilizing data from the scRNA-Seq in a computational approach indicated that, in late recovery stage (LRS) patients, DC-derived IL-18 and TNF TNFSF13 accompanied by T cell-derived IL-2 and IL-4 might promote B cell survival, proliferation, and differentiation, culminating in the production of various SARS-CoV-2-specific Abs [83].
It is also noteworthy that various studies determine higher concentrations of IL-18 in SARS-CoV-2 infected patients as a differentiating marker between COVID-19 and influenza [159], adult-onset still’s a disease (AOSD), an auto-inflammatory disease [174, 175], and other cytokine storm conditions like MAS or sHLH [176].
Nevertheless, IL-18 can show protective features in the early stages of viral infections. A study done by Ciaglia E et al. demonstrated this fact in the initial stages of the SAR-CoV-2 infection by administrating a recombinant form of bactericidal/permeability-increasing fold-containing family-B-member-4 (BPIFB4), a protein involved in the homeostatic response to the inflammatory stimulation, which its plasma level was found significantly decreased in COVID-19 patients [177]. Treatment of SARS-CoV-2 infected PBMCs with this agent demonstrated an elevation in IL-18 and IL-1β levels accompanied by down-regulation of the CD69 activating marker for T cells (both CD4+ T and CD8+ T cells) and MCP-1 that culminated in the limited cellular damages [177]. Additionally, analyzing the genome sequence of the COVID-19 patients revealed a negative association between IL-18 and the risk of the SARS-CoV-2 infection, suggesting its protective role in the disease [178]. Collectively, these studies accentuated the undeniable pro-inflammatory role of IL-18 in disease progress and identified it as a prognostic factor for SARS-CoV-2-infection [160] (Fig. 3). As described, cytokine storm is a life-threatening factor in COVID-19 patients, therefore, blocking the inflammatory cascades represents a promising potential therapeutic target along with other candidate drugs. Consequently, the use of cytokines inhibitors such as anakinra, rilonacep, tocilizumab as immunomodulatory agents has been recommended, however, still have require comprehensive studies for find effective treatment for cytokines involved in severe form of COVID-19 including IL-18 [179].
As mentioned earlier, IL-18 secretion following the activation of NLRP3 inflammasome in SARS-CoV-2-infected cells is in conjunction with pyroptotic cell death [102]. Thus, NLRP3 activation has been proposed as a crucial mediator of leukopenia and lymphopenia generated in some COVID-19 patients [102, 135]. SARS-CoV-2-infected cells that undergo pyroptosis release PAMPs, like viral RNA and DAMPs, like DNA and ASC, which their recognition by TLRs initiates an inflammatory response. Thus, this phenomenon may lead to a secondary inflammatory response that can further exacerbate the aberrant primary inflammatory response to the viral invasion [180]. Additionally, pro-IL-1β, pro-IL-18, and NLRP3 gene transcription can be induced in response to the recognition of PAMPs by TLRs [78, 95]. Therefore, activation of these cytokines can initiate a sequence of events leading to amplification of the underlying disturbance of innate and adaptive immune responses. However, discussing the roles of NLRP3 inflammasome in SARS-CoV-2 pathogenesis falls outside the scope of this review and it has been extensively reviewed in [131].
IL-33 and COVID-19
IL-33 structure and function
IL-33 is a recently discovered member of the IL-1 superfamily that was initially identified as a nuclear factor from high endothelium venules (NF-HEV) [181]. Unlike most of the other members of the IL-1 superfamily, the IL-33 gene is located on chromosome 9 at 9p24.1 and contains eight exons (one non-coding and seven coding exons) that span more than 42 kb of genomic DNA. Human IL-33 mRNA encodes a 30 kD protein with 270 amino acids characterized by a conserved structure at the carboxyl-terminal named the molecule β-trefoil structure, through which represents its cytokine activity [182, 183]. IL-33 constitutively is produced as a 30-kDa pro-peptide (full-length IL-33; f-IL 33) and stored in the nucleus of cells. Like IL-1β and IL-18, it is cleaved by caspase-1 to generate mature 18 kDa IL-33 (m-IL-33). The f-IL-33 contains a nuclear localization sequence (NLS) and a homeodomain-like helix-turn-helix DNA-binding domain. Similar to pro-IL-1α, nuclear pro-IL-33 appeared to exert its biologic activities independent of caspase-1 cleavage and cell surface receptor binding [70, 184]. It is estimated that the bioactivity of m-IL-33 increases ~ 10–30 times compared to f-IL-33 [185].
IL-33, in both homeostasis and inflammation, is mainly produced at high levels in the endothelial cells, epithelial cells, fibroblast-like cells, and myofibroblasts [186]. Tissue or cellular damage like mechanical wounding epithelial cells may lead to the release of IL-33 and have essential roles in inducing type 2 immune response through ST2, an orphan receptor of IL-1, and IL-1RAcP [187]. The ST2 receptor is present in many immune cells, such as neutrophils, basophils, mast cells, Th-2 cells, NK cells, DCs, alternatively activated macrophages (AAMs), and invariant NKT (iNKT) cells [70]. IL-33 binds to the T1/ST2 receptor, and this complex engages the IL-1RAcP as a co-receptor. A soluble form of ST2 (sST2) (also known as IL-1RL1) may function as an inhibitor of IL-33 by binding IL-33 in the cell microenvironment and soluble IL-1RAcP may enhance the inhibitory effects of sST2 [70]. The structure of the IL-33/ST2 complex is composed of the very highly acidic C-terminal part of IL-33, which is similar to IL-1 and has a critical role in binding to the ST2. The studies have shown that single-point genetic mutation of the acidic C-terminal part of IL-33 at these two sites reduces the desire to bind to ST2 [186, 188]. IL-33, by binding to ST2, activates NF-kB and MAPK pathways in the target cells alters activity cells, and enhances secretion of Th-2-associated cytokines, like IL-5 IL-13 but not IL-4 [189].
Critically, regulating IL-33 s’ activity is vital to inhibit its constitutive production and harmful multi-organ inflammation and damage. IL-33 activity is regulated by several mechanisms like; the maintenance of IL-33 in the nucleus of generating cells during homeostasis and cleavage in the IL-1-like cytokine domain by caspases during pyroptosis. Neutralization by the decoy receptor sST2 and rapid oxidation and formation of disulfide bridges (S–S) in IL-33 could be other central mechanisms to inhibit its excessive activity [182].
IL-33 in diseases
The documented role of IL‑33 has extended from infectious disease to inflammatory and metabolic disorders. IL‑33 is produced during cell death and stress or following the invasion of infectious agents and acts as an alarmin or self-molecules that alert the immune system to damage [190]. However, it can make inordinate inflammatory responses in the long term, resulting in parallel injury to local and systemic tissues[182]. Increasing in IL-33 was observed in numerous diseases, such as asthma, chronic obstructive pulmonary disease (COPD), ARDS, idiopathic pulmonary fibrosis (IPF), rheumatoid arthritis (RA), IBD, and cardiovascular disease [191]. The ST2 axis is usually more basic in all of these, which strengthens Th-2 or T-reg responses.
Thanks to IL-33 causes decreases in IFN-γ and increases IL-4, IL-5, and IL-13 by switching the immune response to Th-2 and also causes changes in plasma immunoglobulin levels, increasing IgG1, IgE, and IgA but decreasing IgG2a appears that IL-33 may help to prevent atherosclerosis [192]. Augmented IL-33 levels have also been documented in the RA that can exacerbate joint inflammation by increasing migration and activation of neutrophils and mast cells, which increase inflammatory cytokines like IL-6 and IL-1β. Respectively, studies have shown the therapeutic relevance of the IL-33 inhibitors in RA [193]. In the case of colitis, IL-33 can have both a moderating and exacerbating role, and the effect of IL-33 varies depending on whether it is in the acute or chronic phase and which type of immunity (Th-1 or Th-2) is dominant [194].
Since IL-33 is secreted from alveolar epithelial cells type 2 (AEC2) and other resident immune cells of the respiratory system, like alveolar macrophages (AMs) [195], it has critical functions in mediating immune responses and tissue damage in respiratory diseases. The elevation of IL-33 during respiratory disease may switch immune response to type 2 through stimulating mast cells, Th-2 cells, M2 MQs, eosinophils, and innate lymphoid cells (ILC2s) and elevating type 2 cytokines including IL-4, IL-9, IL-10, IL-13, and tumor growth factor β (TGF-β) [196]. For instance, in asthma, a type I hypersensitivity disease with an increase in IgE level and Th-2 responses, studies showed an increase in the expression of IL-33 in the airways, which can play an aggravating role in disease pathogenesis [192]. Moreover, despite IL-5 central role in the maturation and production of eosinophils in the bone marrow, IL-33 could induce the production of eosinophils from progenitor CD117+ cells, which may be a reason for asthma resistance to the anti-IL-5 drug. The activation of eosinophils can increase the expression of CCL7, which is an essential factor for the accumulation of lymphocytes in the airways, causing exacerbated asthma inflammation [197]. IL-33 could also increase inflammatory conditions by activating AAMs during airway inflammation, which has discrete roles in type 2 immune responses [198]. Furthermore, it was reported that a significant increase in serum levels of IL-33 in COPD has a mechanism almost similar to asthma and can be proposed as a risk factor for this disease [199].
However, the exact role of IL-33 in ARDS has not yet been entirely determined [195]. IL-33 can control ARDS inflammation through mediating T-reg secreted IL-13, which has reparative functions by activating tissue-repair MQs [190]. On the other hand, animal studies showed that the IL-33 neutralizing Abs has been useful in reducing inflammation in ARDS [200].
In addition, Jing Xu et al. indicated that IL-33 activated ILC2s could initiate early detrimental type 2 immune responses following severe trauma through the production of IL-5 and stimulation of the IL-5 expression in neutrophils, which creates the IL-33-ILC2-IL5-neutrophil axis in lung injury [196]. IL-33 can contribute to the development of neutrophils by inducing GM-CSF, CXCL1, CXCL2, E-selectin, intracellular adhesion molecule 1 (ICAM1), and vascular cell adhesion molecule 1 (VCAM1). In return, proteases released from neutrophils activate IL-33 [201]. Additionally, in chronic respiratory infection that leads to lung fibrosis, IL-33 by activating M2 MQs, ILC2, mast cells, and myofibroblasts and stimulating TGF-β, IL-13, CCL2, and CXCL6 secretion plays an important role in the fibrosis mechanism [202]. Interestingly, this pro-fibrotic feature of the IL-33 can be helpful in lung repair [184].
Notably, IL-33 has also been implicated in the pathogenesis of respiratory viral diseases like respiratory syncytial virus (RSV) induced respiratory inflammation [203]. RSV, by activating TLR3 or TLR7, can up-regulate the expression of IL-33 mRNA in AMs and DCs, and enhance the production of IL-33 in these cells [203].
Conclusively, like IL-1 and IL-18, IL-33 plays a dual role in the pathogenesis of the viral and respiratory inflammations, in which activating immune cells in the early stages potentiate immune responses, but in the late stages of the inflammation, drives the lung fibrosis [204].
The pathogenesis of IL-33 in COVID-19
Although there are data regarding IL-33 functions in the COVID-19, recent studies have begun to unravel its roles in the disease course. In one of the first studies, Burke H et al. revealed increased levels of the IL-33 as a significant predictive marker of adverse outcome in COVID-19 patients [205]. Following observations similarly demonstrated its elevated levels that were correlated with clinical and radiographic parameters of the disease and established its positive correlation with TNF-α, IL-1β, IL-6, IL-12, and IL-23 levels and its negative correlation with O2 saturation of the COVID-19 patients [206, 207] (Table 1). Additionally, elevated levels of the IL-33 together with IL-18 were coupled with higher concentrations of the renal toxicity markers such as glutathione S-transferase (GST) and osteopontin in COVID-19 patients, which confers its putative interference in multi-systemic damages of the SARS-CoV-2 infection [208]. Second, higher levels of IL-33 accompanied by IL-12p70 were correlated with seroconversion of IgG Abs in COVID-19 patients, indicating IL-33 s’ potential participation in regulating adaptive immunity [207].
Despite relatively abundant evidence representing its augmented levels in the COVID-19, a study done by Zeng et al. did not show altered levels of the IL-33, probably due to the increased levels of its decoy receptor sST2 [209]. Second, elevated serum sST2 levels of the patients had an inverse correlation with the counts of CD4+ and CD8+ T cells and a positive correlation with disease severity, which highlights the potential protective features of the IL-33 in the infected patients [209].
Interestingly, data provided by scRNA-seq of cells isolated from the BALF samples of COVID-19 patients have proven elevated expression of IL-33 [210]. A similar result has also been reported by Stanczak et al. that was concomitant with the correlation of its expression with disease severity [211]. Moreover, explorations revealed the IL-33 gene as one of the target genes of the SARS-CoV-2 encoded microRNAs (miRNAs) [212], which its co-expression with the entry receptor of the virus, ACE2, in human lung epithelial cells [213], further extending the implication of IL-33 in the disease.
Similar to IL-1 and IL-18, IL-33 also has a crucial role in the clinical phenotypes of the SARS-CoV-2 infection in children. Increased IL-33, alongside the IL-1α, IL-6, IFNs, TNF-α, and chemokines, was detected in children with pediatric inflammatory, multisystem syndrome temporally associated with SARS-CoV-2 infection (PIMS-TS), which was higher than those of the COVID-19, seropositive for SARS-CoV-2 infection, and control children [214].
Moreover, IL-33 can act as a differentiating marker. For instance, compared to COVID-19 patients, a higher concentration of IL-33 was detected in the nasal mucosa of chronic rhinosinusitis with nasal polyps (CRSwNP) patients, which can be explained by its chronic course [215]. In addition, postmortem lung tissues of COVID-19 patients were depleted of IL-33 and AEC2 compared to those of the healthy subjects and patients with COPD or IPF [184]. However, fibrotic lung samples of the COVID-19 survived patient exhibited increased amounts of IL-33 and AEC2 compared to COPD and IPF, indicating the protective features of the AEC2 secreted IL-33 in lung repair by eliciting fibrosis [184]. Noteworthy, latent Tuberculosis infection (LTBI) can heighten IL-33 levels in SARS-CoV-2 co-infected patients [216].
Align with the mentioned clinical studies, in vitro investigations further illustrate IL-33 interactions in the infection (Table 2). Stanczak M A et al. discovered a higher increase of IL-33 accompanied by IL-6, IFN-α2, and IL-23 secretion in the culture of the SARS-CoV-2 peptide stimulated PBMCs from seropositive patients than those of the seronegative patients [211]. Enhanced production of the IL-33 is likely due to T cell-mediated effects on IL-33-producing cells since its production had a strong positive correlation with the expression of CD69, a lymphocyte activation marker, in CD4+ T cells [211]. This outcome has almost been confirmed by evaluating CD4+ T cells counts in COVID-19 patients [209]. The additional analysis detected CD14+ MOs, as the major responders to T-cell-mediated IL-33 production since they had the highest level of intracellular IL-33 among PBMCs. Finding potential candidates for the regulation exerted by IL-33, identified B cells, followed by MOs and DCs, as the immune cells with the highest ST2 expression [211]. Noteworthy, increased serum concentrations of IL-33 have previously been associated with increased bone marrow precursor cells in PBMCs of patients infected with SARS-CoV-2 [217].
Moreover, they discovered a positive association between IL-33 production from SARS-CoV-2 peptide stimulated PBMCs of the seropositive patients and their IgG titers [211] that were in keeping with the data exhibited in [207]. Accommodating data imply the contribution of innate and both cellular and humoral arms of the adaptive immune system in regulating IL-33 s’ release in response to SARS-CoV-2 induced inflammation, which persists after the resolution of the inflammation.
In addition, infection of human epithelial cell lines, Fadu and LS513, with SARS-CoV-2 promoted IL-33 expression [202], providing more data for its potential implication in respiratory remodeling in the virus-induced lung inflammation [184]. Consistently, treatment of poly(I:C), a double-strand RNA (dsRNA) analogue and TLR3 agonist that mimics biological effects of viral infection, stimulated human bronchial epithelial cells (HBECs), with Imiquimod, a TLR7 agonist, impaired IL-33 and IL-1β, IL-6, and IL-8 production, potentially by up-regulation of the single immunoglobulin IL-1-related receptor (SIGIRR), the negative regulator of IL-1R signaling [218]. Consequently, impaired IL-33, together with down-regulated AEC2 expression, up-regulated IFN-β expression, and activation of various antiviral immune pathways following imiquimod treatment, contributed to the viral infection tolerance [218] (Fig. 3).
Accumulating data identify IL-33 as a pronounced factor in COVID-19 pathogenesis. Further probable IL-33 regulated pathways in SARS-CoV-2-induced infection have extensively been projected in [201]. Thus, there is an ongoing phase-2 clinical trial asserting the effects of its inhibition by using Astegolimab, an anti-IL-33 monoclonal Ab (mAb) that selectively inhibits the IL-33 receptor ST2 in patients with severe COVID-19 pneumonia [219]. Although existing evidence favors the deleterious roles of IL-33 in the COVID-19, it must be noted that by acting as an alarmin, IL-33 can stimulate antiviral CTL activity and Ab production. Therefore, it has been speculated that the persistence of cells producing IL-33 in response to T cell activation may benefit the antiviral defense against the virus [211].
The pathogenesis of IL-36, IL-37, and IL-38 in COVID-19
Recently discovered members of the IL-1 family, IL-36, IL-37, and IL-38, also have distinctive inflammatory or anti-inflammatory roles. These cytokines are secreted from various immune and non-immune cells, such as lymphocytes, myeloid and epithelial cells [220]. IL-36 is a pro-inflammatory cytokine and, through activation of MyD88, MAPK, mTOR, and NF-κB dependent signaling pathways, stimulates the generation of inflammatory factors such as IL-1β, IL-17, IFN-γ, and TNF-α [221, 222]. Additionally, IL-36 stimulates activation of the NLRP3 inflammasome [223]. Unlike IL-36, IL-37 and IL-38 have anti-inflammatory features and suppress the aforementioned inflammatory pathways and mediators [221, 222]. These cytokines have distinctive roles in different diseases, such as IBD, RA, and systemic lupus erythematosus (SLE) [224–226]. Additionally, IL-36 by inducing the secretion of inflammatory cytokines, and IL-37 and IL-38 by mitigating the inflammation in the respiratory milieu have been implicated in the pathogenesis of inflammatory respiratory disease and ARDS [227–229].
Likewise, recent studies have determined the functions of these cytokines in the SARS-CoV-2 infection (Fig. 3). Gao et al. revealed increased levels of the IL-36α accompanied by IL-38 in the COVID-19 patients correlated positively and negatively with disease severity [228]. Moreover, a study done by Li et al. exhibited elevated plasma levels of IL-37 together with higher IFN-α and lower IL-6 and IL-8 levels in the COVID-19 patients. Furthermore, low IL-37 levels combined with high IL-8 and CRP levels predicted severe clinical prognosis in these patients [230]. In addition, IL-37 administration attenuated respiratory inflammation in SARS-CoV-2 infected mice [230]. However, in another study, Ahmed AA and Ad'hiah AH demonstrated lower serum levels of IL-37 and its possible association with a higher risk of disease in COVID-19 patients. Additionally, IL-37 was significantly correlated with vitamin D levels [231]. Second, they discovered that mutations of the IL37 genes are also associated with the susceptibility to the COVID-19 [232]. Accumulation data suggest the inflammatory role of IL-36 and protective features of the IL-37 and IL-38 in SARS-CoV-2 induced inflammation. Noteworthy, Bozonnat A et al. reported a case of deficiency of interleukin thirty-six receptor antagonist (DITRA), an auto-inflammatory disease, flare triggered by SARS-CoV-2 infection [233].
Conclusion
The well-adjusted inflammatory cascade is the main piece of immune response in constraining COVID-19 infection. The over-activity or reduction in immune response activity are both unfavorable during infection and lead to detrimental outcomes. According to the literature, hyper-inflammatory IL-1 cytokines family such as IL-1α, IL-1β, IL-18, IL-33, and IL-36 act as the driving force of inflammatory activities, and the high levels of these cytokines are directly associated with the severity of the disease (Table 3). These cytokines exert the inflammatory response via triggering the NLRP3 inflammasomes. Hereupon, scientists have made significant progress in uncovering molecular insights into SARS-CoV-2 pathogenesis and development of vaccines, but there is still no viable treatment for SARS-CoV-2-infected patients. Therefore, a better understanding of the pathogenesis underlying severe forms of COVID-19 permits the design of novel immunotherapies and effective therapeutic strategies for targeting the hyperinflammatory key molecules during the COVID-19 inflammatory response. In this study, we aimed to evaluate and summarize the role of IL-1 family (IL-1, IL-18, IL-33, IL-36, IL-37, and IL-38) in COVID-19. We described the available therapeutic approaches based on the inhibitory mechanisms of these cytokines. Ultimately, IL-1 family blockers could be considered as efficient therapeutic agents in the treatment of COVID-19. However, further studies are required to provide detailed insights into the role of IL-1 family blockers in the alleviation and improvement of inflammatory response in COVID-19.
Table 3.
Overview of IL-1, IL-18, IL-33, IL-36, IL-37, and IL-38 changes in different samples of the COVID-19 patients and their associated clinical or para-clinical features
| Sample/clinical or para-clinical feature | IL-1 | IL-18 | IL-33 | IL-36 | IL-37 | IL-38 | Reference |
|---|---|---|---|---|---|---|---|
| Serum/plasma | ↑ | ↑ | ↑ | ↑ | ↑/↓ | ↑ | [13, 43, 199, 220, 222, 223] |
| BALF | ↑ | N/A | ↑ | N/A | N/A | N/A | [73, 202, 203] |
| Feces | NS | ↑ | N/A | N/A | N/A | N/A | [164, 226] |
| Genome | N/A | * | * | N/A | * | N/A | [171, 204, 224] |
| Postmortem lung tissue | N/A | N/A | ↓ | N/A | N/A | N/A | [176] |
| Postmortem cardiac tissue | N/A | ↑ | N/A | N/A | N/A | N/A | [162] |
| Clinical phenotypes of the COVID-19 in children | ↑ | ↑ | ↑ | N/A | N/A | N/A | [158–160, 206] |
| Disease severity | + | + | + | + | - | - | [42, 153, 199, 220, 223] |
| Organ damage markers | + | + | + | N/A | N/A | N/A | [42, 135, 196] |
| Seroconversion | N/A | N/A | + | N/A | N/A | N/A | [200, 203] |
↑: Increase; ↓: Decrease; + : Positive correlation; − : Negative correlation; *: Association; N/A: not applicable; NS: not-significant
Abbreviations
- AAM
Alternative activated macrophage
- Ab
Antibody
- mAb
Monoclonal Ab
- ACE2
Angiotensin-converting enzyme 2
- ADAMTS2
A disintegrin and metalloproteinase with thrombospondin motifs 2
- AEC2
Alveolar epithelial cells type 2
- AKI
Acute kidney injury
- AM
Alveolar macrophage
- AOSD
Adult-onset still’s disease
- APC
Antigen‐presenting cell
- ARDS
Acute respiratory distress syndrome
- ASC
Apoptosis speck-like protein
- BALF
Bronchoalveolar lavage fluid
- BPIFB4
Bactericidal/permeability-increasing fold-containing family-B-member-4
- COPD
Chronic obstructive pulmonary disease
- COVID-19
Coronavirus disease 2019
- CRP
C-reactive protein
- CRS
Cytokine release syndrome
- CRSwNP
Chronic rhinosinusitis with nasal polyps
- CTL
Cytotoxic T lymphocyte
- DCs
Dendritic cell
- DAMP
Damage-associated molecular pattern
- DITRA
Deficiency of interleukin 36 receptor antagonist
- DM
Diabetes mellitus
- DPP4
Dipeptidyl peptidase 4
- DPSC
Dental pulp stem cell
- EBV
Epstein–Barr virus
- EPC
Endothelial progenitor cell
- ESR
Erythrocyte sedimentation rate
- FasL
Fas ligand
- G-CSF
Granulocyte-colony stimulating factor
- Gal
Galectin
- GI
Gastrointestinal
- GM-CSF
Granulocyte macrophage colony-stimulating factor
- GSDMD
Gasdermin D
- GST
Glutathione S-transferase
- HAEC
Human aortic endothelial cell
- HBEC
Human aortic endothelial cell, human bronchial epithelial cells
- HCoV
Human Coronavirus
- HDL
High-density lipoprotein
- HIF-1
Hypoxia-inducible factor 1
- HIV
Human immunodeficiency virus
- sHLH
Secondary hemophagocytic lymphohistiocytosis
- HPSC
Human pluripotent stem cell
- HSC
Hematopoietic stem cell
- HTN
Hypertension
- IBD
Inflammatory bowel disease
- ICAM1
Intracellular adhesion molecule 1
- ICU
Intensive care unit
- IEC
Intestinal epithelial cell
- IGIF
IFN-γ-inducing factor
- IFN
Interferon
- IL
Interleukin
- IL-18BP
IL-18-binding protein
- IL-1R
IL-1 receptor
- IL-1RAcP
IL-1R accessory protein
- IL-Ra
IL-1 receptor antagonist
- ILC2
Innate lymphoid cell
- IP10
Interferon-inducible protein-10
- IRAK1
IL-1 receptor-associated kinase
- ISG
Interferon-stimulated gene
- IPF
Idiopathic pulmonary fibrosis
- LDH
Lactate dehydrogenase
- LDL
Light-density lipoprotein
- LORG
Lung organoids
- LRS
Late recovery stage
- LTBI
Latent tuberculosis infection
- M-CSF
Macrophage colony-stimulating factor
- MAIT
Mucosal-associated invariant T
- MAPK
Mitogen-activated protein kinase
- MAS
Macrophage activation syndrome
- MCP-1
Monocyte chemo attractant protein 1
- MDA5
Melanoma differentiation-associated gene 5
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MHV
Mouse hepatitis virus
- MI
Myocardial infarction
- MIS-C
Multisystem inflammatory syndrome in children
- MO
Monocyte
- MQ
Macrophage
- MRA
Mineralocorticoid receptor antagonist
- NAFLD
Non-alcoholic fatty liver disease
- NCP
Nucleocapsid protein
- NET
Neutrophil extracellular trap
- NK
Natural killer
- NKT
Natural killer T
- iNKT
Invariant NKT
- NLRP3
Nucleotide-binding oligomerization domain (NOD)-like receptor family pyrin domain containing 3
- NLS
Nuclear localization sequence
- OA
Osteoarthritis
- ORF
Open reading frame
- PAMP
Pathogen-associated molecular pattern
- PRR
Pattern-recognition receptor
- PBMC
Peripheral blood mononuclear cell
- PIMS-TS
Pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 infection
- RA
Rheumatoid arthritis
- RANTES
Regulated on activation, normal T expressed and secreted
- RIG-1
Retinoic acid-inducible gene 1
- dsRNA
Double strand RNA
- messenger RNA
MRNA
- miRNA
MicroRNA
- scRNA-Seq
Single-cell RNA transcriptomic sequencing
- ssRNA
Single-stranded RNA
- ROS
Reactive oxygen species
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SIGIRR
Single immunoglobulin IL-1-related receptor
- SLE
Systemic lupus erythematosus
- Th
T helper
- T-reg
Regulatory T
- TGF
Tumor growth factor
- SS
Systemic sclerosis
- sSt2
Soluble ST2
- TIR
Toll/interleukin-1 receptor
- TLR
Toll-like receptor
- TNF
Tumor necrosis factor
- TNFSF13
TNF Superfamily member 13
- VCAM1
Vascular cell adhesion molecule 1
- VSEL
Very small embryonic-like stem cell
Author contributions
ES was involved in the study conception and design and supervised the work. Sh.M, AA, HK and VA wrote the manuscript in consultation with ES. VA and AM designed the pictures. All authors reviewed the results and approved the final version of the manuscript.
Declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shima Makaremi and Ali Asgarzadeh contributed equally to this work.
References
- 1.Aborode AT, et al. Impact of poor disease surveillance system on COVID-19 response in Africa: time to rethink and rebuilt. Clin Epidemiol Global Health. 2021;12:100841. doi: 10.1016/j.cegh.2021.100841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Essar MY, et al. Mucormycosis, conflicts and COVID-19: a deadly recipe for the fragile health system of Afghanistan. Int J Health Plan Manag. 2021 doi: 10.1002/hpm.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasan MM, et al. Emergence of highly infectious SARS-CoV-2 variants in Bangladesh: the need for systematic genetic surveillance as a public health strategy. Trop Med Health. 2021;49(1):1–3. doi: 10.1186/s41182-021-00360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardhan M, et al. Dual burden of Zika and COVID-19 in India: challenges, opportunities and recommendations. Trop Med Health. 2021;49(1):1–4. doi: 10.1186/s41182-021-00378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Awan HA, et al. Tuberculosis amidst COVID-19 in Pakistan: a massive threat of overlapping crises for the fragile healthcare systems. Epidemiol Infect. 2022 doi: 10.1017/S0950268822000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oberemok VV, et al. SARS-CoV-2 will continue to circulate in the human population: an opinion from the point of view of the virus-host relationship. Inflamm Res. 2020;69(7):635–640. doi: 10.1007/s00011-020-01352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren L-L, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133(9):1015. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim GU, et al. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin Microbiol Infect. 2020;26(7):948.e1–948.e3. doi: 10.1016/j.cmi.2020.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Morales AJ, et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis. 2020;34:101623. doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas P, et al. Candidate antiviral drugs for COVID-19 and their environmental implications: a comprehensive analysis. Environ Sci Pollut Res. 2021;28(42):59570–59593. doi: 10.1007/s11356-021-16096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohn MK, et al. Pathophysiology of COVID-19: mechanisms underlying disease severity and progression. Physiol (Bethesda) 2020;35(5):288–301. doi: 10.1152/physiol.00019.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellucci G, et al. SARS-CoV-2 meta-interactome suggests disease-specific, autoimmune pathophysiologies and therapeutic targets. F1000Res. 2020;9:992. doi: 10.12688/f1000research.25593.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta P, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. The lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan Y, Tang F. SARS-CoV-2-mediated immune system activation and potential application in immunotherapy. Med Res Rev. 2021;41(2):1167–1194. doi: 10.1002/med.21756. [DOI] [PubMed] [Google Scholar]
- 19.Chi Y, et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis. 2020;222(5):746–754. doi: 10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerget B, et al. Evaluation of alpha defensin, IL-1 receptor antagonist, and IL-18 levels in COVID-19 patients with macrophage activation syndrome and acute respiratory distress syndrome. J Med Virol. 2021;93(4):2090–2098. doi: 10.1002/jmv.26589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu D, Mu R, Wei X. The roles of IL-1 family cytokines in the pathogenesis of systemic sclerosis. Front Immunol. 2019;10:2025. doi: 10.3389/fimmu.2019.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinarello CA, Bufler P. Interleukin-37. In: Seminars in immunology. Elsevier. 2013 [DOI] [PubMed]
- 23.Haake C, et al. Coronavirus infections in companion animals: virology, epidemiology, clinical and pathologic features. Viruses. 2020;12(9):1023. doi: 10.3390/v12091023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burrell CJ, Howard CR, Murphy FA. Coronaviruses. In: Fenner and White's Medical Virology. 2017; 437–446.
- 25.Khan S, et al. Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: biology and therapeutic options. J Clin Microbiol. 2020;58(5):e00187–e220. doi: 10.1128/JCM.00187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Hoek L, et al. Identification of a new human coronavirus. Nat Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hon KL, et al. Overview: the history and pediatric perspectives of severe acute respiratory syndromes: novel or just like SARS. Pediatr Pulmonol. 2020;55(7):1584–1591. doi: 10.1002/ppul.24810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrosillo N, et al. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020;26(6):729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascarella G, et al. COVID-19 diagnosis and management: a comprehensive review. J Intern Med. 2020;288(2):192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tahmasebi S, Khosh E, Esmaeilzadeh A. The outlook for diagnostic purposes of the 2019-novel coronavirus disease. J Cell Physiol. 2020;235(12):9211–9229. doi: 10.1002/jcp.29804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siordia JA., Jr Epidemiology and clinical features of COVID-19: a review of current literature. J Clin Virol. 2020;127:104357. doi: 10.1016/j.jcv.2020.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao W, Li T. COVID-19: towards understanding of pathogenesis. Cell Res. 2020;30(5):367–369. doi: 10.1038/s41422-020-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, et al. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun. 2020;87:18–22. doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menon T, et al. Association of gastrointestinal system with severity and mortality of COVID-19: a systematic review and meta-analysis. Cureus. 2021 doi: 10.7759/cureus.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Z, et al. Clinical features of COVID-19-related liver functional abnormality. Clin Gastroenterol Hepatol. 2020;18(7):1561–1566. doi: 10.1016/j.cgh.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao L, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKechnie JL, Blish CA. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27(6):863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultze JL, Aschenbrenner AC. COVID-19 and the human innate immune system. Cell. 2021 doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taefehshokr N, et al. Covid-19: perspectives on innate immune evasion. Front Immunol. 2020 doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molaei S, et al. The immune response and immune evasion characteristics in SARS-CoV, MERS-CoV, and SARS-CoV-2: vaccine design strategies. Int Immunopharmacol. 2021;92:107051. doi: 10.1016/j.intimp.2020.107051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury MA, et al. Immune response in COVID-19: a review. J Infect Public Health. 2020 doi: 10.1016/j.jiph.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fathi N, Rezaei N. Lymphopenia in COVID-19: therapeutic opportunities. Cell Biol Int. 2020;44(9):1792–1797. doi: 10.1002/cbin.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee E, Oh JE. Humoral immunity against SARS-CoV-2 and the impact on COVID-19 pathogenesis. Mol Cells. 2021;44(6):392. doi: 10.14348/molcells.2021.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan W, et al. Viral kinetics and antibody responses in patients with COVID-19. MedRxiv. 2020;25:753. [Google Scholar]
- 47.Spicer BA, et al. Perforin—a key (shaped) weapon in the immunological arsenal. In: Seminars in cell and developmental biology. Elsevier. 2017 [DOI] [PubMed]
- 48.Zhou T, et al. Immune asynchrony in COVID-19 pathogenesis and potential immunotherapies. J Exp Med. 2020;217(10):e20200674. doi: 10.1084/jem.20200674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L, et al. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct Target Ther. 2020;5(1):1–8. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowery SA, Sariol A, Perlman S. Innate immune and inflammatory responses to SARS-CoV-2: implications for COVID-19. Cell Host Microbe. 2021 doi: 10.1016/j.chom.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veras FP, et al. SARS-CoV-2–triggered neutrophil extracellular traps mediate COVID-19 pathology. J Exp Med. 2020;217(12):e20201129. doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acharya D, Liu G, Gack MU. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol. 2020;20(7):397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diao B, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parisi V, et al. Imbalance between interleukin-1β and interleukin-1 receptor antagonist in epicardial adipose tissue is associated with non ST-segment elevation acute coronary syndrome. Front Physiol. 2020;11:42. doi: 10.3389/fphys.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galozzi P, et al. The revisited role of interleukin-1 alpha and beta in autoimmune and inflammatory disorders and in comorbidities. Autoimmun Rev. 2021 doi: 10.1016/j.autrev.2021.102785. [DOI] [PubMed] [Google Scholar]
- 56.Rivers-Auty J, et al. Redefining the ancestral origins of the interleukin-1 superfamily. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-03362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afonina IS, et al. Proteolytic processing of interleukin-1 family cytokines: variations on a common theme. Immunity. 2015;42(6):991–1004. doi: 10.1016/j.immuni.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Boraschi D, et al. The family of the interleukin-1 receptors. Immunol Rev. 2018;281(1):197–232. doi: 10.1111/imr.12606. [DOI] [PubMed] [Google Scholar]
- 59.Arend WP. The balance between IL-1 and IL-1Ra in disease. Cytokine Growth Factor Rev. 2002;13(4–5):323–340. doi: 10.1016/S1359-6101(02)00020-5. [DOI] [PubMed] [Google Scholar]
- 60.Litmanovich A, Khazim K, Cohen I. The role of interleukin-1 in the pathogenesis of cancer and its potential as a therapeutic target in clinical practice. Oncol Therapy. 2018;6(2):109–127. doi: 10.1007/s40487-018-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan AH, Schroder K. Inflammasome signaling and regulation of interleukin-1 family cytokines. J Exp Med. 2020 doi: 10.1084/jem.20190314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abbate A, et al. Interleukin-1 and the inflammasome as therapeutic targets in cardiovascular disease. Circ Res. 2020;126(9):1260–1280. doi: 10.1161/CIRCRESAHA.120.315937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, et al. Crosstalk between ER stress, NLRP3 inflammasome, and inflammation. Appl Microbiol Biotechnol. 2020;104:6129–6140. doi: 10.1007/s00253-020-10614-y. [DOI] [PubMed] [Google Scholar]
- 64.Mao L, et al. The role of NLRP3 and IL-1β in the pathogenesis of inflammatory bowel disease. Front Immunol. 2018;9:2566. doi: 10.3389/fimmu.2018.02566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanneganti T-D. Central roles of NLRs and inflammasomes in viral infection. Nat Rev Immunol. 2010;10(10):688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, et al. NLRP3 inflammasome and inflammatory diseases. Oxid Med Cell Long. 2020 doi: 10.1155/2020/4063562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grebe A, Hoss F, Latz E. NLRP3 inflammasome and the IL-1 pathway in atherosclerosis. Circ Res. 2018;122(12):1722–1740. doi: 10.1161/CIRCRESAHA.118.311362. [DOI] [PubMed] [Google Scholar]
- 68.Hoffman HM, et al. Fine structure mapping of CIAS1: identification of an ancestral haplotype and a common FCAS mutation, L353P. Hum Genet. 2003;112(2):209–216. doi: 10.1007/s00439-002-0860-x. [DOI] [PubMed] [Google Scholar]
- 69.Agostini L, et al. NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20(3):319–325. doi: 10.1016/S1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 70.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223(1):20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 71.Buckley LF, Abbate A. Interleukin-1 blockade in cardiovascular diseases: a clinical update. Eur Heart J. 2018;39(22):2063–2069. doi: 10.1093/eurheartj/ehy128. [DOI] [PubMed] [Google Scholar]
- 72.Gabay C, Lamacchia C, Palmer G. IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol. 2010;6(4):232–241. doi: 10.1038/nrrheum.2010.4. [DOI] [PubMed] [Google Scholar]
- 73.Palomo J, et al. The interleukin (IL)-1 cytokine family–Balance between agonists and antagonists in inflammatory diseases. Cytokine. 2015;76(1):25–37. doi: 10.1016/j.cyto.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 74.Dinarello CA. The IL-1 family of cytokines and receptors in rheumatic diseases. Nat Rev Rheumatol. 2019;15(10):612–632. doi: 10.1038/s41584-019-0277-8. [DOI] [PubMed] [Google Scholar]
- 75.Cavalli G, et al. Interleukin 1α: a comprehensive review on the role of IL-1α in the pathogenesis and targeted treatment of autoimmune and inflammatory diseases. Autoimmun Rev. 2021;20:102763. doi: 10.1016/j.autrev.2021.102763. [DOI] [PubMed] [Google Scholar]
- 76.Guidotti LG, Chisari FV. Cytokine-mediated control of viral infections. Virology. 2000;273(2):221–227. doi: 10.1006/viro.2000.0442. [DOI] [PubMed] [Google Scholar]
- 77.Orzalli MH, et al. An antiviral branch of the IL-1 signaling pathway restricts immune-evasive virus replication. Mol Cell. 2018;71(5):825–840.e6. doi: 10.1016/j.molcel.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu L, et al. Preventing mortality in COVID-19 patients: which cytokine to target in a raging storm? Front Cell Dev Biol. 2020;8:677. doi: 10.3389/fcell.2020.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van de Veerdonk FL, Netea MG. Blocking IL-1 to prevent respiratory failure in COVID-19. Crit Care. 2020;24(1):1–6. doi: 10.1186/s13054-020-03166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jamilloux Y, et al. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun Rev. 2020;19(7):102567. doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell. 2002;10(2):417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 82.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 83.Wen W, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discovery. 2020;6(1):1–18. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gong F, et al. Peripheral CD4+ T cell subsets and antibody response in COVID-19 convalescent individuals. J Clin Investig. 2020 doi: 10.1172/JCI141054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mantovani A, et al. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity. 2019;50(4):778–795. doi: 10.1016/j.immuni.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Monteagudo LA, Boothby A, Gertner E. Continuous intravenous anakinra infusion to calm the cytokine storm in macrophage activation syndrome. ACR open Rheumatol. 2020;2(5):276–282. doi: 10.1002/acr2.11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nemchand P, et al. Cytokine storm and use of anakinra in a patient with COVID-19. BMJ Case Rep CP. 2020;13(9):e237525. doi: 10.1136/bcr-2020-237525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Day JW, et al. IL-1 blockade with anakinra in acute leukaemia patients with severe COVID-19 pneumonia appears safe and may result in clinical improvement. Br J Haematol. 2020;190(2):e80–e83. doi: 10.1111/bjh.16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huet T, et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2(7):e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cavalli G, et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2(6):e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bozzi G, et al. Anakinra combined with methylprednisolone in patients with severe COVID-19 pneumonia and hyperinflammation: an observational cohort study. J Allergy Clin Immunol. 2021;147(2):561–566.e4. doi: 10.1016/j.jaci.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Franzetti M, et al. IL-1 receptor antagonist anakinra in the treatment of COVID-19 acute respiratory distress syndrome: a retrospective, observational study. J Immunol. 2021;206(7):1569–1575. doi: 10.4049/jimmunol.2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Okamura H, et al. Cloning of a new cytokine that induces IFN-γ production by T cells. Nature. 1995;378(6552):88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 94.Okamura H, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378(6552):88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 95.Kaplanski G. Interleukin-18: biological properties and role in disease pathogenesis. Immunol Rev. 2018;281(1):138–153. doi: 10.1111/imr.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li YF, et al. Analyses of caspase-1-regulated transcriptomes in various tissues lead to identification of novel IL-1β-, IL-18- and sirtuin-1-independent pathways. J Hematol Oncol. 2017;10(1):40. doi: 10.1186/s13045-017-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Platnich JM, Muruve DA. NOD-like receptors and inflammasomes: a review of their canonical and non-canonical signaling pathways. Arch Biochem Biophys. 2019;670:4–14. doi: 10.1016/j.abb.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 98.Fu Z, et al. Potent and broad but not unselective cleavage of cytokines and chemokines by human neutrophil elastase and proteinase 3. Int J Mol Sci. 2020;21(2):651. doi: 10.3390/ijms21020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Akeda T, et al. CD8+ T cell granzyme B activates keratinocyte endogenous IL-18. Arch Dermatol Res. 2014;306(2):125–130. doi: 10.1007/s00403-013-1382-1. [DOI] [PubMed] [Google Scholar]
- 100.Li YJ, et al. Meprin-β regulates production of pro-inflammatory factors via a disintegrin and metalloproteinase-10 (ADAM-10) dependent pathway in macrophages. Int Immunopharmacol. 2014;18(1):77–84. doi: 10.1016/j.intimp.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 101.Dinarello C, et al. Interleukin-18 and IL-18 binding protein. Front Immunol. 2013;4:289. doi: 10.3389/fimmu.2013.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferreira AC, et al. SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021;7(1):43. doi: 10.1038/s41420-021-00428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Slaats J, et al. IL-1β/IL-6/CRP and IL-18/ferritin: distinct inflammatory programs in infections. PLoS Pathog. 2016;12(12):e1005973. doi: 10.1371/journal.ppat.1005973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lim HX, et al. Principal role of IL-12p40 in the decreased Th1 and Th17 responses driven by dendritic cells of mice lacking IL-12 and IL-18. Cytokine. 2013;63(2):179–186. doi: 10.1016/j.cyto.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 105.Nakanishi K, et al. Interleukin-18 regulates both Th1 and Th2 responses. Annu Rev Immunol. 2001;19(1):423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 106.Nakanishi K. Unique action of Interleukin-18 on T cells and other immune cells. Front Immunol. 2018;9:763. doi: 10.3389/fimmu.2018.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lutshumba J, et al. Selective upregulation of transcripts for six molecules related to T cell costimulation and phagocyte recruitment and activation among 734 immunity-related genes in the brain during perforin-dependent, CD8(+) T cell-mediated elimination of toxoplasma gondii cysts. nSystems. 2020 doi: 10.1128/mSystems.00189-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oka N, et al. IL-12 regulates the expansion, phenotype, and function of murine NK cells activated by IL-15 and IL-18. Cancer Immunol Immunother. 2020;69(9):1699–1712. doi: 10.1007/s00262-020-02553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kunikata T, et al. Constitutive and induced IL-18 receptor expression by various peripheral blood cell subsets as determined by anti-hIL-18R monoclonal antibody. Cell Immunol. 1998;189(2):135–143. doi: 10.1006/cimm.1998.1376. [DOI] [PubMed] [Google Scholar]
- 110.Desbien AL, et al. IL-18 and subcapsular lymph node macrophages are essential for enhanced B cell responses with TLR4 agonist adjuvants. J Immunol. 2016;197(11):4351–4359. doi: 10.4049/jimmunol.1600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blom L, Poulsen LK. IL-1 family members IL-18 and IL-33 upregulate the inflammatory potential of differentiated human Th1 and Th2 cultures. J Immunol. 2012;189(9):4331–4337. doi: 10.4049/jimmunol.1103685. [DOI] [PubMed] [Google Scholar]
- 112.Szczuciński A, Kalinowska A, Losy J. CXCL11 (Interferon-inducible T-cell alpha chemoattractant) and interleukin-18 in relapsing-remitting multiple sclerosis patients treated with methylprednisolone. Eur Neurol. 2007;58(4):228–232. doi: 10.1159/000107945. [DOI] [PubMed] [Google Scholar]
- 113.Borthwick L. The IL-1 cytokine family and its role in inflammation and fibrosis in the lung. In: Seminars in immunopathology. Springer. 2016. [DOI] [PMC free article] [PubMed]
- 114.Kitasato Y, et al. Enhanced expression of interleukin-18 and its receptor in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2004;31(6):619–625. doi: 10.1165/rcmb.2003-0306OC. [DOI] [PubMed] [Google Scholar]
- 115.Arpaia N, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162(5):1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hirahara K, et al. The immunopathology of lung fibrosis: amphiregulin-producing pathogenic memory T helper-2 cells control the airway fibrotic responses by inducing eosinophils to secrete osteopontin. Semin Immunopathol. 2019;41(3):339–348. doi: 10.1007/s00281-019-00735-6. [DOI] [PubMed] [Google Scholar]
- 117.Novick D, et al. Interleukin-18 binding protein: a novel modulator of the Th1 cytokine response. Immunity. 1999;10(1):127–136. doi: 10.1016/S1074-7613(00)80013-8. [DOI] [PubMed] [Google Scholar]
- 118.Zhao C, Zhao W. NLRP3 inflammasome—a key player in antiviral responses. Front Immunol. 2020;11:211. doi: 10.3389/fimmu.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Y, et al. IL-12p40 and IL-18 play pivotal roles in orchestrating the cell-mediated immune response to a poxvirus infection. J Immunol. 2009;183(5):3324–3331. doi: 10.4049/jimmunol.0803985. [DOI] [PubMed] [Google Scholar]
- 120.Yasuda K, Nakanishi K, Tsutsui H. Interleukin-18 in health and disease. Int J Mol Sci. 2019 doi: 10.3390/ijms20030649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kuriakose T, Kanneganti TD. Regulation and functions of NLRP3 inflammasome during influenza virus infection. Mol Immunol. 2017;86:56–64. doi: 10.1016/j.molimm.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pinkerton JW, et al. Inflammasomes in the lung. Mol Immunol. 2017;86:44–55. doi: 10.1016/j.molimm.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 123.Makabe H, et al. Interleukin-18 levels reflect the long-term prognosis of acute lung injury and acute respiratory distress syndrome. J Anesth. 2012;26(5):658–663. doi: 10.1007/s00540-012-1409-3. [DOI] [PubMed] [Google Scholar]
- 124.Huang KJ, et al. An interferon-γ-related cytokine storm in SARS patients. J Med Virol. 2005;75(2):185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cero FT, et al. IL-18 and IL-12 synergy induces matrix degrading enzymes in the lung. Exp Lung Res. 2012;38(8):406–419. doi: 10.3109/01902148.2012.716903. [DOI] [PubMed] [Google Scholar]
- 126.Guo J, et al. The serum profile of hypercytokinemia factors identified in H7N9-infected patients can predict fatal outcomes. Sci Rep. 2015;5:10942. doi: 10.1038/srep10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dong G, et al. Serum interleukin-18: a novel prognostic indicator for acute respiratory distress syndrome. Med (Baltim) 2019;98(21):e15529. doi: 10.1097/MD.0000000000015529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Williams MA, O'Callaghan A, Corr SC. IL-33 and IL-18 in inflammatory bowel disease etiology and microbial interactions. Front Immunol. 2019;10:1091–1091. doi: 10.3389/fimmu.2019.01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wawrocki S, et al. Interleukin 18 (IL-18) as a target for immune intervention. Acta Biochim Pol. 2016;63(1):59–63. doi: 10.18388/abp.2015_1153. [DOI] [PubMed] [Google Scholar]
- 130.Zalinger ZB, Elliott R, Weiss SR. Role of the inflammasome-related cytokines Il-1 and Il-18 during infection with murine coronavirus. J Neurovirol. 2017;23(6):845–854. doi: 10.1007/s13365-017-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shah A. Novel coronavirus-induced NLRP3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baas T, et al. SARS-CoV virus-host interactions and comparative etiologies of acute respiratory distress syndrome as determined by transcriptional and cytokine profiling of formalin-fixed paraffin-embedded tissues. J Interferon Cytokine Res. 2006;26(5):309–317. doi: 10.1089/jir.2006.26.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chin Med J (Engl), 2003. 116(9): 1283–7. [PubMed]
- 134.Plassmeyer M, et al. Caspases and therapeutic potential of caspase inhibitors in moderate-severe SARS CoV2 infection and long COVID. Allergy. 2021 doi: 10.1111/all.14907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kroemer A, et al. Inflammasome activation and pyroptosis in lymphopenic liver patients with COVID-19. J Hepatol. 2020;73(5):1258–1262. doi: 10.1016/j.jhep.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Courjon J, et al. Heterogeneous NLRP3 inflammasome signature in circulating myeloid cells as a biomarker of COVID-19 severity. Blood Adv. 2021;5(5):1523–1534. doi: 10.1182/bloodadvances.2020003918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rodrigues TS, et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2021. 218(3). [DOI] [PMC free article] [PubMed]
- 138.Theobald SJ, et al. Long-lived macrophage reprogramming drives spike protein-mediated inflammasome activation in COVID-19. EMBO Mol Med. 2021;13(8):e14150. doi: 10.15252/emmm.202114150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Junqueira C, et al. SARS-CoV-2 infects blood monocytes to activate NLRP3 and AIM2 inflammasomes, pyroptosis and cytokine release. Res Square 2021; p. rs.3.rs-153628.
- 140.Huang W, et al. The inflammatory factors associated with disease severity to predict COVID-19 progression. J Immunol. 2021;206(7):1597–1608. doi: 10.4049/jimmunol.2001327. [DOI] [PubMed] [Google Scholar]
- 141.Fraser DD, et al. Inflammation profiling of critically Ill coronavirus disease 2019 patients. Crit Care Explorat. 2020;2(6):e0144–e0144. doi: 10.1097/CCE.0000000000000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hoel H, et al. Elevated markers of gut leakage and inflammasome activation in COVID-19 patients with cardiac involvement. J Intern Med. 2021;289(4):523–531. doi: 10.1111/joim.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Luft T, et al. EASIX for prediction of outcome in hospitalized SARS-CoV-2 infected patients. Front Immunol. 2021;12:2425. doi: 10.3389/fimmu.2021.634416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Menezes MCS, et al. Lower peripheral blood Toll-like receptor 3 expression is associated with an unfavorable outcome in severe COVID-19 patients. Sci Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-94624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang Y, et al. Identification of monocytes associated with severe COVID-19 in the PBMCs of severely infected patients through single-cell transcriptome sequencing. Engineering. 2021 doi: 10.1016/j.eng.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koutsakos M, et al. Integrated immune dynamics define correlates of COVID-19 severity and antibody responses. Cell Rep Med. 2021;2(3):100208. doi: 10.1016/j.xcrm.2021.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Croci S, et al. Human dental pulp stem cells modulate cytokine production in vitro by peripheral blood mononuclear cells from coronavirus disease 2019 patients. Front Cell Dev Biol. 2021;8:1876. doi: 10.3389/fcell.2020.609204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Galván-Peña S, et al. Profound treg perturbations correlate with COVID-19 severity. Proc Natl Acad Scie. 2021;118(37):e1006507. doi: 10.1073/pnas.2111315118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Flament H, et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat Immunol. 2021;22(3):322–335. doi: 10.1038/s41590-021-00870-z. [DOI] [PubMed] [Google Scholar]
- 150.Toubal A, et al. Mucosal-associated invariant T cells and disease. Nat Rev Immunol. 2019;19(10):643–657. doi: 10.1038/s41577-019-0191-y. [DOI] [PubMed] [Google Scholar]
- 151.Seddiki N, French M. COVID-19 and HIV-associated immune reconstitution inflammatory syndrome: emergence of pathogen-specific immune responses adding fuel to the fire. Front Immunol. 2021 doi: 10.3389/fimmu.2021.649567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Duan F, et al. Modeling COVID-19 with human pluripotent stem cell-derived cells reveals synergistic effects of anti-inflammatory macrophages with ACE2 inhibition against SARS-CoV-2. Res Square 2020; p. rs.3.rs-62758.
- 153.Tiwari SK, et al. Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell-derived lung and cerebral organoids. Stem Cell Rep. 2021;16(3):437–445. doi: 10.1016/j.stemcr.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kucia M, et al. An evidence that SARS-Cov-2/COVID-19 spike protein (SP) damages hematopoietic stem/progenitor cells in the mechanism of pyroptosis in Nlrp3 inflammasome-dependent manner. Leukemia. 2021 doi: 10.1038/s41375-021-01332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ratajczak MZ, et al. SARS-CoV-2 entry receptor ACE2 Is expressed on very small CD45(-) precursors of hematopoietic and endothelial cells and in response to virus spike protein activates the Nlrp3 inflammasome. Stem Cell Rev Rep. 2021;17(1):266–277. doi: 10.1007/s12015-020-10010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jover E, et al. Beneficial effects of mineralocorticoid receptor pathway blockade against endothelial inflammation induced by SARS-CoV-2 spike protein. Biomedicines. 2021;9(6):639. doi: 10.3390/biomedicines9060639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gibellini L, et al. Altered bioenergetics and mitochondrial dysfunction of monocytes in patients with COVID-19 pneumonia. EMBO Mol Med. 2020;12(12):e13001–e13001. doi: 10.15252/emmm.202013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Karaba AH, et al. Differential cytokine signatures of SARS-CoV-2 and influenza infection highlight key differences in pathobiology. MedRxiv. 2021;182:1401. doi: 10.1093/cid/ciab376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Satış H, et al. Prognostic value of interleukin-18 and its association with other inflammatory markers and disease severity in COVID-19. Cytokine. 2021;137:155302–155302. doi: 10.1016/j.cyto.2020.155302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hasegawa T, et al. Type 1 inflammatory endotype relates to low compliance, lung fibrosis, and severe complications in COVID-19. Cytokine. 2021 doi: 10.1016/j.cyto.2021.155618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Laudanski K, et al. Unbiased analysis of temporal changes in immune serum markers in acute COVID-19 infection with emphasis on organ failure, anti-viral treatment, and demographic characteristics. Front Immunol. 2021 doi: 10.3389/fimmu.2021.650465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Takahashi T, et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588(7837):315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Silva J, et al. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. medRxiv. 2021.
- 165.Peart Akindele N, et al. Distinct cytokine and chemokine dysregulation in hospitalized children with acute COVID-19 and multisystem inflammatory syndrome with similar levels of nasopharyngeal SARS-CoV-2 shedding. J Infect Dis. 2021 doi: 10.1093/infdis/jiab285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gruber CN, et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C) Cell. 2020;183(4):982–995.e14. doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Esteve-Sole A, et al. Similarities and differences between the immunopathogenesis of COVID-19–related pediatric multisystem inflammatory syndrome and Kawasaki disease. J Clin Investig. 2021 doi: 10.1172/JCI144554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pierce CA, et al. Natural Mucosal Barriers and COVID-19 in Children. medRxiv. 2021;32:119. doi: 10.1172/jci.insight.148694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lindner D, et al. Association of cardiac infection With SARS-CoV-2 in confirmed COVID-19 autopsy cases. JAMA Cardiol. 2020;5(11):1281–1285. doi: 10.1001/jamacardio.2020.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Piazza O, et al. Thrombin antithrombin complex and IL-18 serum levels in stroke patients. Neurol Int. 2010;2(1):e1. doi: 10.4081/ni.2010.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tao W, et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med Microecol. 2020;5:100023. doi: 10.1016/j.medmic.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lodge S, et al. NMR spectroscopic windows on the systemic effects of SARS-CoV-2 infection on plasma lipoproteins and metabolites in relation to circulating cytokines. J Proteome Res. 2021;20(2):1382–1396. doi: 10.1021/acs.jproteome.0c00876. [DOI] [PubMed] [Google Scholar]
- 173.Singh R, et al. Sustained expression of inflammatory monocytes and activated T cells in COVID-19 patients and recovered convalescent plasma donors. Immun Inflam Dis. 2021 doi: 10.1002/iid3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Meng J, et al. Cytokine storm in coronavirus disease 2019 and adult-onset Still’s disease: similarities and differences. Front Immunol. 2021;11:3625. doi: 10.3389/fimmu.2020.603389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chen P-K, et al. Interleukin-18 Is a potential biomarker to discriminate active adult-onset still’s disease From COVID-19. Front Immunol. 2021 doi: 10.3389/fimmu.2021.719544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kessel C, et al. Discrimination of COVID-19 from inflammation-induced cytokine storm syndromes by disease-related blood biomarkers. Arthritis Rheumatol. 2021 doi: 10.1002/art.41763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ciaglia E, et al. BPIFB4 circulating levels and its prognostic relevance in COVID-19. J Gerontol Ser A. 2021;76(10):1775–1783. doi: 10.1093/gerona/glab208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schooling C, Li M, Yeung SA. Interleukin-18 and COVID-19. Epidemiol Infect. 2022 doi: 10.1017/S0950268821002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu B, et al. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J Autoimmun. 2020;111:102452. doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tay MZ, et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Carriere V, et al. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci. 2007;104(1):282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Liew FY, Girard J-P, Turnquist HR. Interleukin-33 in health and disease. Nat Rev Immunol. 2016;16(11):676–689. doi: 10.1038/nri.2016.95. [DOI] [PubMed] [Google Scholar]
- 183.Tsuda H, et al. Identification of the promoter region of human IL-33 responsive to induction by IFNγ. J Dermatol Sci. 2017;85(2):137–140. doi: 10.1016/j.jdermsci.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 184.Gaurav R, et al. IL-33 depletion in COVID-19 lungs. Chest. 2021;160(5):1656–1659. doi: 10.1016/j.chest.2021.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lefrançais E, et al. IL-33 is processed into mature bioactive forms by neutrophil elastase and cathepsin G. Proc Natl Acad Sci. 2012;109(5):1673–1678. doi: 10.1073/pnas.1115884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cayrol C, Girard JP. Interleukin-33 (IL-33): a nuclear cytokine from the IL-1 family. Immunol Rev. 2018;281(1):154–168. doi: 10.1111/imr.12619. [DOI] [PubMed] [Google Scholar]
- 187.Schmitz J, et al. IL-33, an Interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper Type 2-associated cytokines. Immunity. 2005;23(5):479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 188.Liu X, et al. Structural insights into the interaction of IL-33 with its receptors. Proc Natl Acad Sci. 2013;110(37):14918–14923. doi: 10.1073/pnas.1308651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Oboki K, et al. IL-33 and IL-33 receptors in host defense and diseases. Allergol Int. 2010;59(2):143–160. doi: 10.2332/allergolint.10-RAI-0186. [DOI] [PubMed] [Google Scholar]
- 190.Liu Q, et al. IL-33-mediated IL-13 secretion by ST2+ Tregs controls inflammation after lung injury. JCI insight. 2019;4(6):e123919. doi: 10.1172/jci.insight.123919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Miller AM. Role of IL-33 in inflammation and disease. J Inflamm. 2011;8(1):22. doi: 10.1186/1476-9255-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fulgheri G, Malinowski B. The Role of IL-33 in the inflammation process of asthma and atherosclerosis. EJIFCC. 2011;22(3):79–91. [PMC free article] [PubMed] [Google Scholar]
- 193.Macedo RBV, AM Kakehasi, MV Melo de Andrade. IL33 in rheumatoid arthritis: potential contribution to pathogenesis. Revista Brasileira de Reumatologia 2016; 56: 451–457. [DOI] [PubMed]
- 194.Groβ P, et al. IL-33 attenuates development and perpetuation of chronic intestinal inflammation. Inflamm Bowel Dis. 2012;18(10):1900–1909. doi: 10.1002/ibd.22900. [DOI] [PubMed] [Google Scholar]
- 195.Lin S-H, et al. Inflammation elevated IL-33 originating from the lung mediates inflammation in acute lung injury. Clin Immunol. 2016;173:32–43. doi: 10.1016/j.clim.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 196.Xu J, et al. IL33-mediated ILC2 activation and neutrophil IL5 production in the lung response after severe trauma: a reverse translation study from a human cohort to a mouse trauma model. PLoS Med. 2017;14(7):e1002365. doi: 10.1371/journal.pmed.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stolarski B, et al. IL-33 exacerbates eosinophil-mediated airway inflammation. J Immunol. 2010;185(6):3472–3480. doi: 10.4049/jimmunol.1000730. [DOI] [PubMed] [Google Scholar]
- 198.Kurowska-Stolarska M, et al. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183(10):6469–6477. doi: 10.4049/jimmunol.0901575. [DOI] [PubMed] [Google Scholar]
- 199.Xia J, et al. Increased IL-33 expression in chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol. 2015;308(7):L619–L627. doi: 10.1152/ajplung.00305.2014. [DOI] [PubMed] [Google Scholar]
- 200.Lee HY, et al. Blockade of IL-33/ST2 ameliorates airway inflammation in a murine model of allergic asthma. Exp Lung Res. 2014;40(2):66–76. doi: 10.3109/01902148.2013.870261. [DOI] [PubMed] [Google Scholar]
- 201.Zizzo G, Cohen PL. Imperfect storm: is interleukin-33 the Achilles heel of COVID-19? Lancet Rheumatol. 2020;2(12):e779–e790. doi: 10.1016/S2665-9913(20)30340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liang Y, Ge Y, Sun J. IL-33 in COVID-19: friend or foe? Cell Mol Immunol. 2021;18(6):1602–1604. doi: 10.1038/s41423-021-00685-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Qi F, et al. Respiratory macrophages and dendritic cells mediate respiratory syncytial virus-induced IL-33 production in TLR3- or TLR7-dependent manner. Int Immunopharmacol. 2015;29(2):408–415. doi: 10.1016/j.intimp.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 204.Zizzo G, Cohen PL. Imperfect storm: is interleukin-33 the Achilles heel of COVID-19? Lancet Rheumatol. 2020 doi: 10.1016/S2665-9913(20)30340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Burke H, et al. Inflammatory phenotyping predicts clinical outcome in COVID-19. Respir Res. 2020;21(1):245. doi: 10.1186/s12931-020-01511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Markovic SS, et al. IL 33 Correlates With COVID-19 Severity, Radiographic and Clinical Finding. Front Med. 2021 doi: 10.3389/fmed.2021.749569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Munitz A, et al. Rapid seroconversion and persistent functional IgG antibodies in severe COVID-19 patients correlates with an IL-12p70 and IL-33 signature. Sci Rep. 2021;11(1):3461. doi: 10.1038/s41598-021-83019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fonseca W, et al. COVID-19 modulates inflammatory and renal markers that may predict hospital outcomes among African American males. Viruses. 2021;13(12):2415. doi: 10.3390/v13122415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zeng Z, et al. Serum-soluble ST2 as a novel biomarker reflecting inflammatory status and illness severity in patients with COVID-19. Biomark Med. 2020;14(17):1619–1629. doi: 10.2217/bmm-2020-0410. [DOI] [PubMed] [Google Scholar]
- 210.Xiong Y, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microb Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Stanczak MA, et al. IL-33 expression in response to SARS-CoV-2 correlates with seropositivity in COVID-19 convalescent individuals. medRxiv. 2020;26:453. doi: 10.1038/s41467-021-22449-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Aydemir MN, et al. Computationally predicted SARS-COV-2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Rep. 2021;22:101012. doi: 10.1016/j.genrep.2020.101012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wruck W, Adjaye J. SARS-CoV-2 receptor ACE2 is co-expressed with genes related to transmembrane serine proteases, viral entry, immunity and cellular stress. Sci Rep. 2020;10(1):1–14. doi: 10.1038/s41598-020-78402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Venkataraman A, et al. Plasma biomarker profiling of PIMS-TS, COVID-19 and SARS-CoV2 seropositive children—a cross-sectional observational study from southern India. EBioMedicine. 2021;66:103317. doi: 10.1016/j.ebiom.2021.103317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jeican II, et al. Respiratory nasal mucosa in chronic rhinosinusitis with nasal polyps versus COVID-19: histopathology, electron microscopy analysis and assessing of tissue interleukin-33. J Clin Med. 2021;10(18):4110. doi: 10.3390/jcm10184110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rajamanickam A, et al. Latent tuberculosis co-infection is associated with heightened levels of humoral, cytokine and acute phase responses in seropositive SARS-CoV-2 infection. J Infect. 2021;83(3):339–346. doi: 10.1016/j.jinf.2021.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bernardes JP, et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity. 2020;53(6):1296–1314.e9. doi: 10.1016/j.immuni.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nieto-Fontarigo JJ, et al. Imiquimod boosts interferon response, and decreases ACE2 and pro-inflammatory response of human bronchial epithelium in asthma. Front Immunol. 2021 doi: 10.3389/fimmu.2021.743890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Patel S, Saxena B, Mehta P. Recent updates in the clinical trials of therapeutic monoclonal antibodies targeting cytokine storm for the management of COVID-19. Heliyon. 2021 doi: 10.1016/j.heliyon.2021.e06158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Akdis M, et al. Interleukins (from IL-1 to IL-38), interferons, transforming growth factor β, and TNF-α: receptors, functions, and roles in diseases. J Allergy Clin Immunol. 2016;138(4):984–1010. doi: 10.1016/j.jaci.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 221.Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281(1):8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Boutet M-A, Nerviani A, Pitzalis C. IL-36, IL-37, and IL-38 cytokines in skin and joint inflammation: a comprehensive review of their therapeutic potential. Int J Mol Sci. 2019;20(6):1257. doi: 10.3390/ijms20061257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chi H-H, et al. IL-36 signaling facilitates activation of the NLRP3 inflammasome and IL-23/IL-17 axis in renal inflammation and fibrosis. J Am Soc Nephrol. 2017;28(7):2022–2037. doi: 10.1681/ASN.2016080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ding L, et al. IL-36 cytokines in autoimmunity and inflammatory disease. Oncotarget. 2018;9(2):2895. doi: 10.18632/oncotarget.22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wang X, et al. Role of interleukin-37 in inflammatory and autoimmune diseases. Iran J Immunol. 2018;15(3):165–174. doi: 10.22034/IJI.2018.39386. [DOI] [PubMed] [Google Scholar]
- 226.van de Veerdonk FL, et al. Biology of IL-38 and its role in disease. Immunol Rev. 2018;281(1):191–196. doi: 10.1111/imr.12612. [DOI] [PubMed] [Google Scholar]
- 227.Kovach MA, et al. IL-36γ is a crucial proximal component of protective type-1-mediated lung mucosal immunity in Gram-positive and-negative bacterial pneumonia. Mucosal Immunol. 2017;10(5):1320–1334. doi: 10.1038/mi.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gao X, et al. Interleukin-38 ameliorates poly (I: C) induced lung inflammation: therapeutic implications in respiratory viral infections. Cell Death Dis. 2021;12(1):1–18. doi: 10.1038/s41419-020-03283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Li B, et al. Interleukin-37 attenuates lipopolysaccharide (LPS)-induced neonatal acute respiratory distress syndrome in young mice via inhibition of inflammation and cell apoptosis. Med Scie Monit. 2020;26:e920365–e920371. doi: 10.12659/MSM.920365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li A, et al. Correlation between early plasma interleukin 37 responses with low inflammatory cytokine levels and benign clinical outcomes in severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis. 2021;223(4):568–580. doi: 10.1093/infdis/jiaa713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ahmed AA, Ad'hiah AH. Interleukin-37 is down-regulated in serum of patients with severe coronavirus disease 2019 (COVID-19) Cytokine. 2021;148:155702. doi: 10.1016/j.cyto.2021.155702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ahmed AA, Ad'hiah AH. Interleukin-37 gene polymorphism and susceptibility to coronavirus disease 19 among Iraqi patients. Meta Gene. 2022;31:100989. doi: 10.1016/j.mgene.2021.100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bozonnat A, et al. SARS-CoV-2 infection inducing severe flare up of deficiency of interleukin thirty-six (IL-36) receptor antagonist (DITRA) resulting from a mutation invalidating the activating cleavage site of the IL-36 receptor antagonist. J Clin Immunol. 2021;41(7):1511–1514. doi: 10.1007/s10875-021-01076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]