Dear Editor,
The coronavirus pandemic has brought public health challenges worldwide. The main protease (Mpro), also called 3CLpro (3C-like protease), is encoded by the gene of non-structural protein 5 (nsp5). It is an attractive antiviral drug target to halt the progress of severe acute respiratory syndrome coronavirus-2 (SARS-COV-2), the causative pathogen of coronavirus disease 2019 (COVID-19) [1]. After viral infection in host cells, the replicase gene encodes two polyproteins, pp1a (486 kDa) and pp1ab (790 kDa). They are then cleaved by papain-like protease 2 (PL2pro) at 3 sites and Mpro at another 11 sites. Then, pp1a and pp1ab are processed to release a series of non-structural proteins (NSPs) that mediate viral replication and transcription [1]. The function of Mpro is indispensable to the viral life cycle. Thus, inhibiting the activity of Mpro can effectively impede the coronavirus replication. Over one 100 host-cell proteins that are involved in essential cellular processes have also been reported as substrates of SARS-COV-2 Mpro, suggesting that Mpro is a multifunctional viral factor [2].
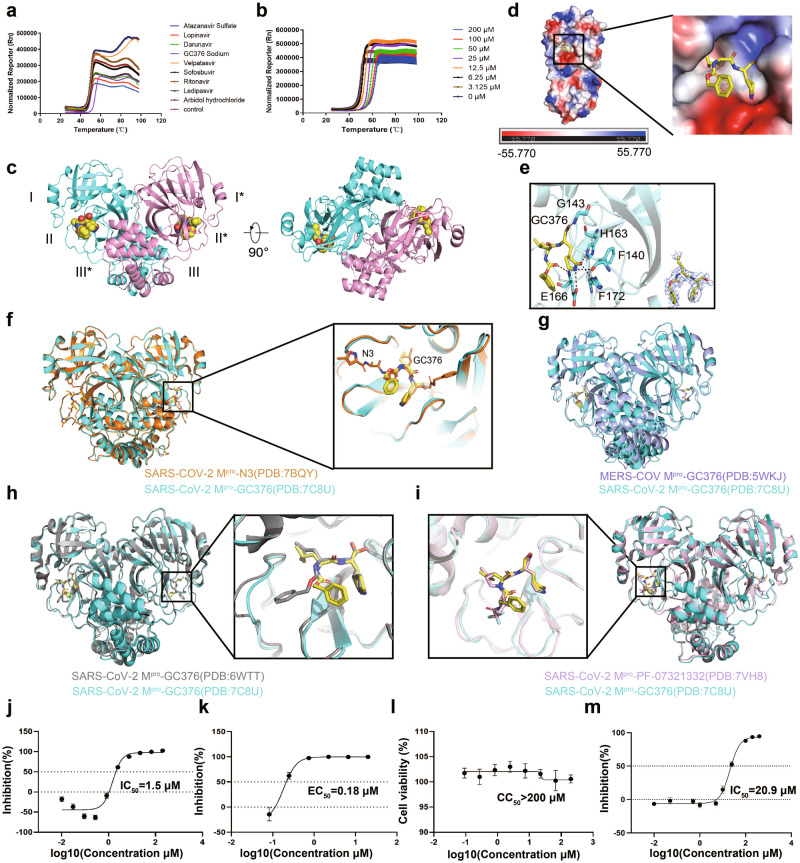
To evaluate drugs that may target SARS-COV-2 Mpro for COVID-19 treatment, we purified SARS-CoV-2 Mpro protein and attempted to validate certain compounds that have been reported to inhibit coronavirus Mpro effectively [3]. The SARS-COV-2 Mpro was expressed in Escherichia coli cells and then purified via affinity and size-exclusion chromatography. The purified SARS-COV-2 Mpro showed a monodispersed peak with a molecular weight of ~33.8 kDa (Supplementary information, Fig. S1a). We selected nine drugs and performed thermal shift assay (TSA) to assess the effect of each drug on SARS-CoV-2 Mpro. Each drug (20 μM) was incubated with SARS-COV-2 Mpro and the melting profile was monitored based on the SYBRO orange reaction in the range of 25–99 °C. Only GC376, an antiviral drug used to treat feline coronavirus disease [4], displayed an obvious effect, shifting the melting curve of SARS-CoV-2 Mpro from 50.9 °C to 55.2 °C (+4.3 °C). Other drugs showed little effect (Fig. 1a, Supplementary information, Table S2). These results show that GC376 may bind to SARS-COV-2 Mpro. To determine if the process is titratable, the TSA experiment was repeated with different concentrations of GC376 (0–20 μM). The melting temperature of Mpro was found to increase in a dose-dependent manner (Fig. 1b). Taken together, our results showed that GC376 interacts directly with SARS-COV-2 Mpro.
Fig. 1. Structure of SARS-CoV-2 Mpro and GC376 activity.
a Thermal shift assay of SARS-CoV-2 Mpro with different drugs. b Thermal shift assay of SARS-CoV-2 Mpro with different concentrations of GC376. c Overall structure of SARS-CoV-2 Mpro in complex with GC376 in two different views. d Electrostatic surface of SARS-CoV-2 Mpro. Blue: positive charge potential; Red: negative charge potential. The value ranges from −55.77 (red) to 0 (white) to 55.77 (blue). e Binding of GC376 (yellow sticks) to the SARS-CoV-2 Mpro pocket with the electron density map for GC376. f Comparison between SARS-CoV-2 Mpro-GC376 and SARS-COV-2 Mpro-N3 crystal structures. g Comparison between SARS-CoV-2 Mpro-GC376 and MERS-COV Mpro-GC376. h Comparison of the SARS-COV-2-GC376 structure we reported (PDB: 7CBU) to the SARS-COV-2-GC376 structure reported by Ma et al. (PDB:6WTT). i Comparison of SARS-CoV-2 Mpro-GC376 and SARS-CoV-2 Mpro-PF-07321332. j IC50 of GC376 on the purified Mpro enzyme. k EC50 of GC376 on inhibition of SARS-Cov-2 in Vero E6 cells. l CC50 of GC376 on Vero E6 cells. m IC50 of GC376 on the purified Mpro P132H.
GC376 is a dipeptidyl bisulfite adduct salt and has been reported to inhibit Mpro [4], suggesting that GC376 may be a broad-spectrum antiviral drug. To determine the structural basis of SARS-COV-2 Mpro inhibition by GC376, we determined the structure of GC376 bound SARS-COV-2 Mpro at the resolution of 2.35 Å. Like SARS-CoV Mpro, the SARS-CoV-2 Mpro forms a dimer with the two protomers vertically packed into each other (Fig. 1c and Supplementary information Table S1). The SARS-COV-2 Mpro monomer has three domains with domains I (residues 8–101) and II (residues 102–184) forming a barrel structure of six antiparallel β-strands. Between domain I and domain II is a cleft that forms the catalytic dyad C145-H41 and the substrate binding site, which is composed of three highly conserved sub-pockets S1, S2 and S4. Domain III (residues 201–303) comprises a globular cluster of five antiparallel α-helices, which is involved in the dimerization of Mpro. Domains II and III are connected through a loop (residues 185–200), while domain I contains a stretched out “N-finger” (NH2-terminal) that is inserted into domain II of its adjacent protomer via interaction with F140 and G166. The surface charge distribution in the active site of SARS-CoV-2 Mpro displays a type of bipolar distribution that is suitable for the binding of peptide substrates (Fig. 1d). GC376 was found to occupy the Mpro substrate binding site (Fig. 1e) and form extensive hydrogen bonds with the surrounding residues of F140, G143, H163, E166, and H172 of Mpro. The glutamine surrogate ring and the leucine group of GC376 fit into the pockets in Mpro S1 and S2 and serve as conserved residents for substrate recognition. GC376 also uses an aldehyde bisulfite to covalently bind to the catalytic residue C145, suggesting its potentiality to block the catalytic dyad. This shows the potential of GC376 to act as a covalent inhibitor to prevent the binding and cleavage of substrates by SARS-CoV-2 Mpro. When we compared the structure of our GC376-bound Mpro complex to the previously reported SARS-CoV-2 Mpro structure [5] (Fig. 1f) and MERS-COV Mpro-GC376 structure [6] (Fig. 1g), we saw a good agreement. Comparison with the SARS-CoV-2 Mpro structure shows considerable conservation of domains I and II, with small discrepancies in domain III. Comparison with the MERS-COV Mpro-GC376 structure shows nearly overlapped conformation of the two GC376 molecules, indicating that the inhibition of these viruses by GC376 is mediated through a similar mechanism. We also compared our structure to the GC376-SARS-CoV-2 Mpro structure reported by Ma et al. [7]. (Fig. 1h) and found that the phenylmethyl group of GC376 in Ma et al.’s structure points deeper to S4 site in the pocket. The conformation of SARS-CoV-2 Mpro-GC376 reported by another group [8] is similar to our structure, with the phenylmethyl groups at slightly different positions (Supplementary information, Fig. S1b). We also compared our structure to GC376-SARS- CoV-2 Mpro structure reported by Fu et al. [1] and Vuong et al. [2] and also found a similar binding mode. These conformational differences may be caused by flexibility in the loop (Q189-A193) region near the site 4 (Supplementary information, Fig. S1c). We also compared our structure to the structure of the SARS-COV-2 Mpro in complex with the PF-07321332 [9], a SARS-CoV-2 Mpro inhibitor, which has been approved for emergency treatment of COVID-19, in combination with Ritonavir (brand name: Paxlovid) [10]. Comparison to the SARS-COV-2 Mpro-PF-07321332 structure showed that the glutamine surrogate rings of the GC376 and PF-07321332 fit into the S1 pocket well and form hydrogen bonds with the neighboring residues in the binding pocket, indicating that the GC376 and PF-07321332 act through a similar mechanism.
In addition to GC376, non-covalent SARS-COV-2 Mpro inhibitors have also been developed [11]. To verify the effectiveness of GC376, we tested the inhibitory activity of GC376 for SARS-CoV-2 Mpro, with the 50% inhibition concentration (IC50) of 1.5 μM (Fig. 1j). We also tested the ability of GC376 to inhibit SARS-CoV-2 infection in pre-seeded Vero E6 cells exposed to the authentic virus. GC376 showed a 50% maximal effect concentration (EC50) of 0.18 μM (Fig. 1k). Meanwhile, GC376 showed a 50% cytotoxic concentration (CC50) of more than 200 μM, indicating a strong safety profile (Fig. 1l). It has been reported that GC376 treatment showed little toxicity in K18-hACE2 mice, although it also had clearly beneficial effect against SARS-CoV-2 in vivo [12]. The Omicron sub-variants BA.1 and BA.2 of SARS-CoV-2 have become the dominant infective strains. The missense mutation P132H in Mpro is >95% prevalent in both Omicron sub-variants [13]. We also tested the ability of GC376 to inhibit Mpro P132H enzymatic activity with IC50 of 20 μM (Fig. 1m), suggesting that GC376 may act as a broad-spectrum drug for different variants of SARS-COV-2. A pre-IND (Investigational New Drug application) about usage of GC376 in COVID treatment has been submitted to the FDA [14]. All of these data suggest that GC376 is worthy of further testing in clinical trials. Both our structural analysis and our in vitro assays in this study indicate that GC376 is an effective and safe drug candidate suitable for use against COVID-19 by inhibiting SARS-CoV-2 Mpro.
Supplementary information
Acknowledgements
This work was supported by Beijing Natural Science Foundation (M21016 to SZ); CAMS Innovation Fund for Medical Sciences (2021-I2M-1-003 to SZ); CAMS Innovation Fund for Medical Sciences (2021-CAMS-JZ004 to SZ); Tsinghua University-Peking University Center for Life Sciences (045-61020100122 to SYZ) and Institute of biomedicine, Tsinghua University. We thank the assistance of Protein Preparation and Identification Facility at Technology Center for Protein Science of Tsinghua University. We appreciate the help from Genscript Biotech Co. Ltd for DNA synthesis, Vazyme Biotech Co. Ltd for offering ClonExpress II One Step Cloning Kit, NEST Biotechnology Co. Ltd for offering centrifugal tube and Topscience for offering GC376.
Author contributions
XDL performed the protein purification and structure analysis of Mpro and wrote the paper under the supervision of SYZ; BXC assisted in the work of XDL; WJS performed cell experiments under supervision of LKZ; WCY, YJ, and BXC participated in writing the manuscript and preparing figures; SYZ and HEX helped design the study and wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: due two errors in the text.
These authors contributed equally: Xiao-dong Luan, Bin-xian Chen, Wei-juan Shang, Lei-ke Zhang, H. Eric Xu, Shu-yang Zhang
Change history
8/10/2022
A Correction to this paper has been published: 10.1038/s41401-022-00964-w
Contributor Information
Lei-ke Zhang, Email: zhangleike@wh.iov.cn.
H. Eric Xu, Email: eric.xu@simm.ac.cn.
Shu-yang Zhang, Email: shuyangzhang103@nrdrs.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-022-00929-z.
References
- 1.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–7. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 2.Pablos I, Machado Y, de Jesus HCR, Mohamud Y, Kappelhoff R, Lindskog C, et al. Mechanistic insights into COVID-19 by global analysis of the SARS-CoV-2 3CL(pro) substrate degradome. Cell Rep. 2021;37:109892. doi: 10.1016/j.celrep.2021.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2012;19:149–50. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y, Lovell S, Tiew KC, Mandadapu SR, Alliston KR, Battaile KP, et al. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J Virol. 2012;86:11754–62. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–93. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 6.Galasiti Kankanamalage AC, Kim Y, Damalanka VC, Rathnayake AD, Fehr AR, Mehzabeen N, et al. Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element. Eur J Med Chem. 2018;150:334–46. doi: 10.1016/j.ejmech.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, Szeto T, et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–92. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Shuai L, Wen Z, Wang C, Yan Y, Jiao Z, et al. The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract. Emerg Microbes Infect. 2021;10:481–92. doi: 10.1080/22221751.2021.1899770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, Fang C, Zhang Q, Zhang RX, Zhao XB, Duan YK, et al. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 2022. 10.1007/s13238-021-00883-2 (in press). [DOI] [PMC free article] [PubMed]
- 10.FDA authorizes paxlovid for treatment of COVID-19. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 11.Rossetti GG, Ossorio MA, Rempel S, Kratzel A, Dionellis VS, Barriot S, et al. Non-covalent SARS-CoV-2 M(pro) inhibitors developed from in silico screen hits. Sci Rep. 2022;12:2505. doi: 10.1038/s41598-022-06306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caceres CJ, Cardenas-Garcia S, Carnaccini S, Seibert B, Rajao DS, Wang J, et al. Efficacy of GC-376 against SARS-CoV-2 virus infection in the K18 hACE2 transgenic mouse model. Sci Rep. 2021;11:9609. doi: 10.1038/s41598-021-89013-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ullrich S, Ekanayake KB, Otting G, Nitsche C. Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir. Bioorg Med Chem Lett. 2022;62:128629. doi: 10.1016/j.bmcl.2022.128629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anivive Repurposes Veterinary Drug GC376 for COVID-19 And Submits Pre-IND to FDA. https://www.anivive.com/news/anivive-repurposes-veterinary-drug-gc376-for-covid-19.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.