Abstract
Abstract
Sex presents a vital determinant of a person’s physiology, anatomy, and development. Recent clinical studies indicate that sex is also involved in the differential manifestation of various diseases, affecting both clinical outcome as well as response to therapy. Genetic and epigenetic changes are implicated in sex bias and regulate disease onset, including the inactivation of the X chromosome as well as sex chromosome aneuploidy. The differential expression of X-linked genes, along with the presence of sex-specific hormones, exhibits a significant impact on immune system function. Several studies have revealed differences between the two sexes in response to infections, including respiratory diseases and COVID-19 infection, autoimmune disorders, liver fibrosis, neuropsychiatric diseases, and cancer susceptibility, which can be explained by sex-biased immune responses. In the present review, we explore the input of genetic and epigenetic interplay in the sex bias underlying disease manifestation and discuss their effects along with sex hormones on disease development and progression, aiming to reveal potential new therapeutic targets.
Key messages
Sex is involved in the differential manifestation of various diseases.
Epigenetic modifications influence X-linked gene expression, affecting immune response to infections, including COVID-19.
Epigenetic mechanisms are responsible for the sex bias observed in several respiratory and autoimmune disorders, liver fibrosis, neuropsychiatric diseases, and cancer.
Keywords: X-inactivation, Epigenetics, Sex bias, Hormones, COVID-19, X-linked genes, Autoimmune disorders, Liver fibrosis, Neuropsychiatric diseases, Cancer
Introduction
Gene expression is heavily influenced by epigenetic DNA changes, as well as post-translational modifications (PTMs) of histones which affect chromatin accessibility and structure [1–7]. DNA exists in a condensed form in the nucleus, wrapped around the nucleosomes, which are the functional units of chromatin, consisting of histone octamers (a pair of each H3, H4, H2A, H2B). Histone PTMs are mainly located at the N-terminal part of H3 and H4 and include methylation, acetylation, phosphorylation, ubiquitination, and sumoylation. The entirety of histone PTMs at different sites at each specific time point comprise the “histone code” and play a crucial role in the fate of gene expression by altering chromatin dynamics, including accessibility and transcription factor binding [8–12]. In this way, gene expression can be altered without any changes in the underlying nucleotide sequence, thus creating a link between genotype and phenotype. During development, a cross talk between epigenetic mechanisms, PTMs and hormones, such as estrogens and androgens acting upstream, have been shown to enhance a cascade of events that regulate mechanisms of gene expression inside the nucleus [13–16].
During embryogenesis in mammals, a single female X chromosome undergoes silencing, forming the Barr body. This developmental imprinting event ensures the proper dosage compensation of expression in the X-linked genes between males and females. Recent studies have revealed several genes which can evade X silencing process and are important for sex determination, immune response as well as developmental growth [17–19].
The choice of the silenced X chromosome creates a mosaic pattern, called lyonization in female cells. Although the inactivation of the X chromosome is random in humans, once the X chromosome gets inactivated, it remains inactive throughout the cell’s lifetime. On the contrary, the X inactivation process in marsupials and in mice is taking place exclusively at the paternally derived X chromosome [17–21].
Epigenetic mechanisms are highly implicated in gene regulation during the X-chromosome inactivation. In order to achieve proper dosage of X-chromosome–encoded proteins, a well-controlled fine-tuning of gene expression in females has been developed [19, 22]. During female mammal development, X chromosome inactivation (XCI) occurs through the action of Xist (X-inactive specific transcript), a long non-coding RNA (lncRNA) that is transcribed from the X chromosome. Each X chromosome contains an X inactivation center which contains both the Xist locus and its antisense transcription unit, Tsix [23]. Once XCI starts, Xist covers almost the entirety of one of the two X chromosomes and induces its silencing through Xist-mediated recruitment of chromatin-modifying, transcriptional-silencing, as well as other RNA-binding proteins [24]. In turn, this leads to epigenetic and structural modifications of the X chromosome, giving rise to one condensed Barr body covered by the Xist RNA [23]. In more detail, Xist promotes the aggregation of supramolecular complexes (SMACs) which include many copies of the Msx2-interacting protein (SPEN), critical for transcriptional repression. SMACs also favor the epigenetic regulators, polycomb group proteins (PcG) deposition inducing chromatin compaction. In this way, increased SMACs levels around genes propagate gene silencing along the X chromosome [24].
The polycomb repressive complex 2 (PRC2), one of the two PcG classes that is recruited by Xist, deposits H3K27me3 histone marks leading to further downregulation of target gene expression. Male cells contain one less chromosome X compared to females. This process aims to nullify the increased expression of X chromosome genes in females compared to males, in the absence of XCI. In combination, differential DNA methylation at CpG islands of the X-linked genes mediates the fine tuning of their expression [17–19]. This imprinting event in females is established through epigenetic mechanisms including DNA and histone methylation (especially H3K27 trimethylation, H3K27me3), mediated by the action of PCR2. It has been shown that the PRC2 subunit binds to a non-coding RNA (ncRNA) within the Xist RNA and is thereby targeted to the female X chromosome, mediating its inactivation [2, 20, 25–28].
Subsequently, the PRC2 complex plays an important role in establishing an inactive status of many genes on the X inactive chromosomes during development. The PRC2 group consists of four core components—the enhancer of zeste (EZH1/2 in mammals, E(z) in Drosophila); extra sexcombs (Eed in mammals, Esc in Drosophila); suppressor of zeste 12 (Suz12 in mammals, Su(z)12 in Drosophila); and a nucleosome remodeling factor (Rbbp7/4 in mammals, Nurf55 in Drosophila). The methyltransferases EZH1 and 2 are responsible for establishing and maintaining the inactive histone mark H3K27me3 on the female X chromosome [20, 25–28].
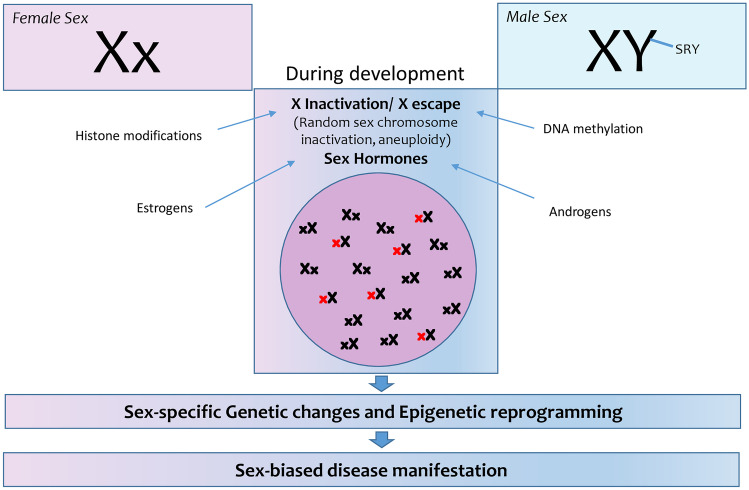
It is therefore evident that a better understanding of the epigenetic mechanisms that underlie the dosage compensation in different sexes may explain the differential response in disease progression such as chronic inflammation, autoimmune diseases, and liver fibrosis (Fig. 1).
Fig. 1.
Genetic changes and epigenetic reprogramming during development underlie the dosage compensation in female and male cells, contributing to sex-biased disease (3.1) manifestation and progression
Differential manifestation of infectious diseases in male and female patients
It has long been observed that males and females harbor differences in their immune responses against most bacterial, viral, and parasitic infections, such as influenza, hepatitis, AIDS, tuberculosis, and malaria. The underlying pathways, which promote these sex-based immunologic differences in response to infectious diseases, have only recently been investigated. Among them, the influence of sex hormones on pathways regulating the immune system, along with the effect of X-chromosome inactivation on X-related immune genes, appears to play a central role.
Susceptibility differences to lower respiratory tract infections between the two sexes have been reported and extensively studied in different mice models. Intranasal inoculation of mice with Streptococcus pneumoniae, the most commonly isolated cause of bacterial pneumonia, demonstrated a higher susceptibility and decreased survival in males compared to females [29]. This was attributed to the differential immune response between the two sexes. It has been suggested that increased mortality from infectious diseases in males was related to testosterone-induced immunosuppression in post-pubertal males [26]. Moreover, a greater number of neutrophils was shown to infiltrate the pulmonary tissues, being accompanied by elevated inflammatory mediators, such as IL-17A, which has been linked to lower levels of female sex hormones [29, 30]. Yang et al. showed that female sex hormones exhibit a protective effect against bacterial pneumonia by enhancing the antimicrobial action of macrophages [31]. This was explained by the estrogen-mediated increase in the expression of nitric oxide synthase-3 (NOS-3), which enhances the killing of bacteria ingested by macrophages. Previous studies have demonstrated an association between estrogen receptors (ERs) with NOS-3 and the lncRNAs HOTAIR and MALAT1 in breast and prostate cancer cells [32]. These two lncRNAs were shown to be regulated by estrogens and further interact with NOS-3 and ERs [32], suggesting a potential role for sex hormones to control the macrophage bactericidal properties in an epigenetic manner.
A similar trend has been reported in mouse models treated with Mycoplasma pulmonis, where the mortality of males was increased compared to females and was correlated with dense inflammatory cell infiltrates within the pulmonary alveoli of males [33]. Interestingly, there were no big differences in IgM serum levels between male and female mice.
Effects of COVID-19 infection in male versus female patients
The COVID-19 pandemic has led to numerous deaths worldwide. In a recent study, an increased incidence of COVID-19 without respiratory failure was observed in males (57.7%) compared to females (42.3%), but a significantly increased incidence of COVID-19-related severe respiratory failure was detected in males (89.3%) compared to females (10.7%). Moreover, the death rates in the different age groups revealed that males were more likely to die of the infection in age groups below 90 years old [34].
In search of the underlying mechanisms that define the sex bias in infection severity and pathogenicity of the virus, several preliminary studies have proposed different biological mechanisms regulating immune responses in a sex-biased manner, contributing to viral defense mechanisms [29–35].
Studies in mice have shown that male mice demonstrate a greater susceptibility to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection compared to female mice of the same age, with the difference being far more prominent at increasing age. Males exhibited increased virus titers and vascular leakage, more alveolar edema as well as inflammatory monocyte and neutrophil lung accumulation, whereas no differences between the two sexes were associated with T or B cell activity. The increased susceptibility of male mice to SARS-CoV-2 infection was mainly attributed to the effect of estrogens and their contribution to viral defense, since ovariectomy or the use of estrogen receptor antagonists led to increased mortality in female mice, highlighting the protective effects of estrogen receptor signaling in SARS-CoV-2 infections [35].
On the other hand, several molecular factors induce the expression of key proteins in viral entry at the epithelial cells, which are also affecting the infectivity rate. One such entry protein expressed in host cells is angiotensin converting enzyme 2 (ACE2). ACE2 is a dipeptidyl carboxy dipeptidase of the angiotensin-converting enzyme family which cleaves angiotensin I into angiotensin 1-9 and angiotensin II into the vasodilator angiotensin 1-7. ACE2 is the entry point of the SARS-CoV-2 virus, and its levels define the magnitude of viral infection. Regulation of this protein is fine-tuned by genetic and epigenetic mechanisms [36–41]. The Ace2 gene in humans is located on the X chromosome, implying a potential sex-specific gene regulation. The Ace2 gene is highly expressed in a lot of tissues including the epithelial cells of the lung, heart, and testis. It is an X chromosome inactivation escaper gene and therefore its expression is not regulated by the dosage compensation machinery [42]. Male cells are expressing higher levels of the ACE2 protein, especially in the lungs, evidence that may explain potential sex differences that have been observed during the progression of the COVID-19 infection and its complications [43]. A study of 450 DNA methylation data from 244 fresh human lung tissues has detected a differential methylation profile for the Ace2 gene at cg23232263 and cg16734967 [44]. It was shown that the male lung tissue had decreased methylation levels of this gene compared to females. Furthermore, males and females who were smokers or had chronic obstructive pulmonary disease demonstrated increased levels of Ace2 gene methylation [44]. In addition, DNA methylation levels at a CpG island in the dataset related to the Ace2 gene exhibited a large degree of variability in both men and women suggesting that DNA methylation of Ace2 varies by individual. Of note, these datasets did not have comparable metadata for age and the differences in DNA methylation related to Ace2 may reflect the cell type differences in lung tissues [19, 45–47].
Sex-bias effects in respiratory diseases
Sex-based differences are also prevalent in respiratory diseases, with DNA methylation changes between the two sexes being observed as early as at birth [48]. In respect to asthma, during the early years of childhood, there is an uneven distribution of the disease between males and females, with exceeding incidence in males compared to females [49–52]. This distribution, however, changes at pre-adolescence, when females begin to get asthma more frequently than males, and continues at post-adolescence where the increased incidence in females leads to a “sex-reversal” [53] and a more severe phenotype [54]. Asthma appears to be associated with changes in the DNA methylome [55]. In the study by Patel et al., the DNA methylation patterns were observed in a variety of genes in pre-adolescent and post-adolescent subjects [56]. Their results pointed towards 13 CpG sites which demonstrate sex and age-related changes. They also detected a sex-related effect difference at 9 of those CpG islands, including an increase in CpG site cg03269757 methylation which led to decreased risk of asthma acquisition in males and increased risk in females. In a similar manner, 5 genes which were mapped to CpG islands showed a sex-related response to elevation of DNA methylation, indicating that the effect of DNA methylation was affected by the subject’s sex. Increased methylation of the cg11295724 CpG island at subjects of 10 years of age resulted in increased expression of the signal regulatory protein delta precursor (SIRPD) gene which regulates T cell activation in males and its decreased expression in females. Moreover, the interferon related developmental regulator 1 (IFRD1) gene, a transcription factor which regulates skeletal muscle differentiation in asthmatic responses of the airway, shows strong sex-specific effects on asthma transition.
Sex-related differences have also been observed in chronic obstructive pulmonary disease (COPD) [57] and many epigenetic pathways have been associated with smoking [58]. Koo et al. investigated tissue samples from adult patients and fetal lungs, as well as cord blood and detected differences in CpG sites, such as cg03691818 where the keratin 77 (KRT77) gene is mapped. Of note, keratins are involved in the structural integrity of epithelia cells and this site was found to be hypomethylated in males compared to females, an effect observed also in other CpG sites [59].
Furthermore, females demonstrated a predisposition for the development of pulmonary arterial hypertension (PAH) which has been partly attributed to the epigenetic regulation of the Ephx2 gene [60]. This gene codes for the soluble epoxide hydrolase, an enzyme which degrades epoxyeicosatrienic acids (EETs), regulating pulmonary circulation. The increased amount of estrogens observed in females is responsible for the downregulation of this gene expression through an epigenetic pathway, leading to increased EETs and subsequently, high PAH susceptibility [60].
Sex bias in autoimmune diseases
Autoimmune diseases demonstrate a high prevalence, affecting an estimated 5–10% of the population, with a female predominance. However, differences have been observed in female to male ratios of each autoimmune disease. For instance, the sex ratio in inflammatory bowel disease and diabetes mellitus type I exceeds 1:1 by a slight amount, but increases to almost 2:1 in multiple sclerosis (MS), to 3:1 in rheumatoid arthritis (RA), and to 9:1–10:1 in systemic lupus erythematosus (SLE) [22]. The molecular mechanisms underlying these differences are not well-understood. However, recent studies suggest that sex hormones such as estrogens and the X chromosome silencing are important drivers of sex-biased manifestations in autoimmune diseases [61].
Estrogens act through four different pathways, including the classical receptor-mediated, the non-classical, the non-ligand-mediated genomic (nuclear), and the non-genomic (extranuclear) pathway, to influence downstream gene expression, protein modifications, and signaling. The molecular actions of estrogens are mediated through the genomic and non-genomic pathways [62]. In the genomic pathway, estrogens bind to intracellular receptors (estrogen receptor, ERα, and ERβ). Upon binding, ERs are dimerized and translocated to the nucleus where they bind to specific motifs, the estrogen response elements (ERE), present in the target DNA. The consensus ERE site is 5′-GGTCAnnnTGACC-3′ and is located in promoters as well as in the distal regulatory elements of genes, affecting immune response and cell signaling pathways. On the other hand, in the non-classical genomic pathways, ER directly bound to DNA, cross talks with various transcription factors, or it may act in a tether-mediated manner as a co-factor of transcription factors, including the activating protein 1 (AP-1), NF-κB, and p300 proteins. In this way, estrogens are able to molecularly fine-tune adaptive and immune cell responsive pathways [62]. For example, activated T cells demonstrate estrogen receptor expression. At the same time, both ER mRNA and protein levels have been found in T and B cells, monocytes, and dendritic cells showing the importance of the hormone in immune response activation [63].
Sex hormone receptors are widely expressed in immune cells, while androgen and estrogen response elements are present in several genes involved in immune response, indicating a major effect of sex hormones on the inflammatory response pathways. Furthermore, a lot of variations have been detected during puberty, pregnancy, and menopause demonstrating the complex regulation of immunity by sex hormones [64].
The X chromosome contains a wide variety of genes which are related to the immune response [65]. In this way, immune function and dysregulation may stem from abnormal silencing of the X chromosome as well as from major defects on X chromosome genes, such as IL-1R associated kinase 1 (IRAK1), IL-2R γ chain, IL-3R α chain, IL-13 α chain, GATA1, TLR7, CD40L, and FOXP3 [61]. The involvement of the X chromosome in sex-biased immune responses is demonstrated in inherited disorders, including Klinefelter syndrome in males with XXY and Turner syndrome in females with XO, both characterized by hormonal and immune abnormalities [66, 67].
The effects of microRNAs (miRNA) in a sex-specific manner could potentially further promote differences in the immune response between the two sexes. It has been previously shown that miRNAs could play an inflammatory role by controlling the expression of important genes, such as IL-1 and TNF-α which are involved in systemic inflammation [68]. Differences between the two sexes based on the expression of the X-linked miRNA could explain the stronger immune system in females [61]. Despite the vast gap in knowledge concerning the differences between male and female miRNA expression, it has been found that the X chromosome contains nearly 800 miRNAs [69]. Some of the X-linked miRNAs have been associated with immune regulation. More specifically, miR106a, which is located on the X chromosome, has been shown to downregulate the anti-inflammatory cytokine IL-10 [70]. Furthermore, the X chromosome-contained miR-17-92 cluster was shown to be a crucial factor for B and T cell maturation [71, 72]. The other side of this phenomenon is that females are more likely to develop autoimmune diseases [67].
The X chromosome contains the information coding for 10% of the human genome miRNAs versus 2% of miRNAs on chromosome Y. The differential expression of miRNAs (for example, miR-18b, miR-223) from X chromosomes due to female mosaicism, X inactivation, and silencing escape may explain the higher responses of the immune system upon infection that are observed in females compared to males [68, 73].
A recent study by Moeser et al. showed that perinatal sex hormones during the embryonic development are responsible for mast cell-associated disorders later in life. Naturally, a high level of perinatal androgens is associated to reduced severity of mast cell-mediated anaphylaxis in male mice [74–76]. Mast cells recognized as effector cells of the immune system represent the first line of defense against various toxins. Their production and function are orchestrated by the activity of sex hormones, such as androgens and testosterone, regulating immune responses. Overactivation of mast cells has been connected to immunological disorders, chronic inflammatory diseases, and death. As part of their defense mechanism, mast cells release proteases, histamine, and serotonin [75]. Previous studies have shown that female mast cells produce, store, and release more proteases and amines than males. Thus, female mast cells are more capable to induce aggressive immune responses, leading to inflammatory and autoimmune diseases than the respective male cells [75, 77].
Current research has unraveled the mechanisms of action of perinatal hormones and the way sex may interfere with the severity of anaphylactic responses. In male embryos, high levels of perinatal androgens stimulate mast stem cells in the bone marrow to acquire a “masculinized” phenotype with reduced production and storage of granule mediators. As a result, they produce and release lower levels of inflammatory substances, leading to reduced possibility of anaphylactic responses and manifestation of chronic inflammatory disorders during the adult life. On the other hand, female embryos exposed to perinatal androgens exhibit reduced histamine levels and less-severe anaphylactic responses as adults [75].
Changes in chromatin configuration have been associated with the relaxation of XCI in female patients and the manifestation of autoimmune diseases. Notably, both naive T and B lymphocytes are characterized by an uncommon pattern of the inactive X chromosome at the level of chromosomal organization. Xist RNA and histone repressive marks are delocalized and reversed, leading to a minor biallelic transcription of autoimmune disease-associated X-linked genes, such as CD40L and CXCR3 [78]. Following lymphocyte activation, such epigenetic changes on the X chromosome are normally reversed in females but not in SLE patients. Biallelic expression of genes connected to the manifestation of the disease is higher in female patients compared to healthy individuals [79–81].
The Y chromosome in mammals is responsible for carrying genes contributing to sex differences and especially those related to testis maturation and spermatogenesis. Several studies in mice have revealed the importance of this chromosome in immune responses and manifestations of autoimmune diseases. Gene expression profiling in these mice demonstrated that Y variants control many immune response genes found on other chromosomes, functioning as trans-expression quantitative trait loci (eQTLs) [82].
In females, several genetic variants and X-linked genes are associated to the risk of autoimmune diseases, such as SLE and MS. One of these important genes, forkhead box p3 (FOXP3a), encodes a pioneer factor controlling T lymphocyte development into regulatory cells and suppressing self-reactivity [83].
An interesting observation comes from the studies in men with Klinefelter syndrome (XXY) and in females with Turner syndrome (X0). The presence of two X chromosomes increases the incidence of severe autoimmune diseases, indicating that X chromosome imbalance rather than hormonal differences is likely to contribute to immune disorders in a sex-specific manner [84]. This imbalance could come from both epigenetic and genetic factors that misregulate the X inactivation center across the single female chromosomes. Supporting this hypothesis, two escape genes, TLR7 and CD40L, are characterized by elevated expression in XX females, as well as XXY males versus XY males and X0 females [85].
Epigenetic mechanisms regulating liver fibrosis in a sex-specific manner
Transcriptional silencing of targeted genes is mainly epigenetically regulated though specific histone post-translational modifications, changes in DNA chemical modifications, and interplay with miRNAs that influence chromatin accessibility and gene expression patterns. Dysregulation of epigenetic mechanisms is often linked to certain diseases, such as cancer, liver fibrosis, and autoimmune disorders [86].
One of the most important protein complexes mediating gene silencing is PRC2 and its components EZH1 and EZH2 which deposit H3K27me3, inducing chromatin compaction. EZH1 and EZH2 possess complementary functions. EZH1 is mainly expressed in adult tissues and EZH2 dominates in embryonic cells, while both enzymes regulate a small set of the same genes through their activity [87–90].
Although the importance of chromatin states in the sex-dependent regulation of genes in female and male mouse livers was previously reported [91], the recent study of Lau-Corona et al. elucidated the sex-biased action of the two methyltransferases EZH1 and EZH2 in liver fibrosis and liver metabolism [92]. They detected that the trimethylation of histone H3K27 (H3K27me3) is a significant sex-biased repressive mark localized on many female-specific expressed genes, including cytochrome P450 (Cyp genes) and sulfotransferase (Sult genes) of male mouse livers, but not at male-biased genes of female mouse livers. In support to these findings, RNAseq data showed that various female-biased genes were upregulated (derepressed) in E1/E2-KO male livers and only a few male-biased genes in E1/E2-KO female livers [91, 92].
Sex-biased expression of related liver genes could explain why males exhibit increased susceptibility to liver fibrosis and showed increased male incidence, as well as progression of HCC [91]. Thus, several studies evaluated the implementation of the PRC2 complex in the progression of the disease. By using double knock-out mice for Ezh1 and Ezh2, they evaluated the epigenetic effects of these enzymes on a genome-wide level and the differences between the two sexes in the liver. As a result, more HCC and fibrosis-related genes were found to be dysregulated in E1/E2-KO male versus E1/E2-KO female liver. Thirty-two (32) sex-independent genes were identified as more highly upregulated in E1/E2-KO female than in E1/E2-KO male mouse livers. Moreover, in the double knock out mice, three genes which are strictly associated with HCC, including the lncRNA gene H19, Igf2, and miR675, were further characterized by accumulation of active marks on their promoters and open chromatin configurations that led to their overexpression in female mice [91].
Sex bias in neuropsychiatric disorders
Genes usually upregulated in autism spectrum disorder (ASD) were found to be located in hypermethylated regions in females. Hypermethylation could be responsible for decreased ASD-associated gene expression in females compared to males, explaining the difference in prevalence between the two sexes [93]. On the other hand, genes that are downregulated in major depressive disorder (MDD) were also found to be hypermethylated in females, which could explain the higher disease predisposition in females [93].
XY homologous genes escape X-inactivation, and the homolog genes of the Y-chromosome non-recombining region are of special interest in males. Differences in the X and Y homolog gene pairs contribute to sex differences, for example, in the ages of onset in psychosis. One example is the KDM5C gene which escapes X-inactivation and the NLGN4 gene which has been linked to autism, as well as the Protocadherin11 XY gene-pair which has been associated with psychosis [94].
Sex chromosomes also seem to contribute to sex differences in Alzheimer’s disease (AD) [95]. An additional X chromosome may result in a protective effect against AD by promoting the transcription of genes that would normally escape X inactivation. Around 15% of X-chromosome genes avoid this inactivation, leading to their increased expression in females compared to males [96]. For example, loss-of-function mutations of KDM6A which encodes for a histone demethylase have been associated with intellectual disability [97]. In this context, a recent study demonstrated that adding an extra X chromosome to mice makes them resilient to AD, likely due to increased KDM6A expression [98]. Another example is protocadherin 11 X-linked (PCDH11X), which also escapes X inactivation via epigenetic mechanisms [99]. A single nucleotide polymorphism in the PCDH11X gene (rs5984894) has been linked to higher AD risk in women [100]. Moreover, men with higher levels of loss of chromosome Y (LOY), the most common acquired somatic mutation associated with aging, had greater risks of AD. It has been suggested that LOY might cause a dysfunctional immune system and contribute to abnormal neuronal differentiation as well as early cell death [101, 102]. Moreover, hypomethylated CpG islands were found in the aurora kinase C gene promoter in males with AD, which were hypermethylated in female AD patients. Females are also characterized by reduced histone deacetylase 2 levels, when compared to males [103] and long noncoding RNA levels were also found to differ between the two sexes in AD [104].
Cancer and sex biases rely on epigenetics
Recent studies have linked the gene dosage imbalance on the X chromosome with the manifestation of cancer in a sex-biased mode. Several associations and whole genome studies have shown that the X chromosome contains a wide variety of genes which demonstrate differential expression between the two sexes. Moreover, XCI-evading genes have been correlated with cancer in humans. More specifically, six genes which belong to the tumor suppressor family that evade the XCI (i.e., CNKSR2, ATRX, DDX3X, KDM6A, KDM5C, and MAGEC3) have been shown to play a protective role in females when loss-of-function mutations are present, and partially explain the increased incidence of 23 cancer types in males compared to females [105]. Among them, KDM6A codes for a histone lysine demethylase which demonstrates functional variations from its Y homolog. A recent pioneer study in mice has associated the effects of this enzyme with the manifestation of bladder cancer, uncovering genetic and epigenetic alterations from hormonal effects associated with cancer [106].
Another interesting example is the case of increased male to female ratio in the incidence and mortality of glioblastoma which contradicts the increased mutation burden observed in females, but a higher incidence and mortality in men.
Many genes that are mutated in cancer cells are located on the X chromosome which has been shown to contain several areas which possess tumor suppressor genes [107]. This means that males are more susceptible to single hits in these loci, leading to increased propensity for cancer. These loci, however, are also commonly mutated in female cancers, such as breast and ovarian cancer [108, 109]. The alpha thalassemia/mental retardation syndrome X-linked (ATRX) gene which is responsible for DNA repair is also frequently found to be mutated [110]. The expression of this gene is tightly regulated in females by the Xist RNA and repressive epigenetic marks, to achieve a monoallelic expression and proper dosage balance. Upon misregulation of the Xist RNA expression, incomplete DNA repair and tumor progression occur. Interestingly, this gene escapes the X inactivation center (XIC) only in the brain of female individuals, resulting in its biallelic expression, protecting females against brain tumors, and more specifically glioblastomas [111].
Moreover, the E1A-binding protein p400, EP400, a component of the nucleosome-acetyltransferase of histone H4 (NuA4) complex, is responsible for acetylating histones H4 and H2A, leading to transcription and high expression of targeted genes. It interacts with the Myc protein to regulate target gene expression during proliferation. Recently, a functional interaction of EP400 with the ataxia-telangiectasia mutated (ATM) protein kinase has been reported, which works to achieve efficient DNA damage response and repair [112]. Taken together, these findings shed light to the importance of mutations on EP400 as tumor drivers. A study of male and female patients with stomach and esophageal cancers has revealed an association between the EP400 mutation and an increased mutation load in female-derived samples when compared to male ones [113].
Lastly, there is a hypothesis in the field of cancer biology that chromatin configuration and genome architecture could be manipulated in a sex-specific manner. This change can further contribute to the manifestation of several diseases, including cancer. Supporting this working model, recent studies have shown that two major factors of genome topology and genome organization (i.e., COHESIN and CTCF) bind via a sex-related manner to chromatin and that mutated binding sites of CTCF may induce tumorigenic effects [79, 114, 115]. Moreover, sex-specific chromatin quaternary structure and accessibility could influence the efficacy or efficiency of the DNA damage response. This evidence provides a better understanding of the mechanistic aspect of the DNA damage response and its effect in the sex bias observed in tumor development. Further research addressing both genetic as well as epigenetic alterations in normal and precancerous tissues is expected to shed more light into the mechanisms underlying sex-specific differences in genomic mutations and their repair.
Conclusion—future perspectives
Taken all together, it is evident that several disease manifestations are attributed to sex bias mediated by epigenetics mechanisms and are not always related to sex hormones (Table 1, Fig. 2). The unique genomic and epigenomic profiling of the two sexes could further define the respective immune response. One of the main drivers for this differentiation is the dosage compensation machinery between the two sexes. Through X inactivation and chromosomal organization, which occur very early in development, female cells regulate a series of critical genes in their lifespan. Misregulation of X inactivation process leads to a sex-biased manifestation of several immunological disorders. The underlying regulatory mechanism is based on epigenetic factors, such as DNA and histone methylation, which drives gene silencing on the inactive female X chromosome.
Table 1.
Epigenetic mechanisms and sex bias in various diseases
| Disease | Epigenetic mechanism | Affected gene | References |
|---|---|---|---|
| Respiratory diseases | |||
| LRTIs | Increased susceptibility and mortality of males, since estrogens regulate lncRNAs, which interact with NOS-3 that enhances macrophage-mediated killing of bacteria | [32] | |
| COVID-19 | Increased susceptibility and mortality of males, due to estrogen contribution to viral defense and decreased gene methylation in males | ACE2 | [44] |
| Asthma | Increased incidence in pre-adolescent males and post-adolescent females, due to sex and age-related changes in gene methylation | SIRPD, IFRD1 | [55, 56] |
| COPD | Decreased gene methylation in males | KRT77 | [59] |
| PAH | Increased susceptibility in females, due to estrogen-mediated increase in gene methylation | Ephx2 | [60] |
| Autoimmune diseases | |||
| IBD, MS, RA, SLE | Increased susceptibility in females, due to estrogen-mediated control of immune responses, abnormal silencing of X-linked genes and X-linked miRNA expression (miR106A, miR-17-92, miR-18b, miR-223) | IL-1R/2R/3R, IL-13, TLR7, IRAK1, FOXP3, GATA1, CD40L, IL-10, CXCR3 | [61] |
| Gastrointestinal diseases | |||
| HCC and liver fibrosis | Increased susceptibility in males due to increased histone methylation in males, causing gene silencing | Cyp, Sult, H19, Igf2, miR675 | [91] |
| Neuropsychiatric diseases | |||
| ASD |
Increased incidence in males due to: Decreased ASD-related gene methylation and thus increased expression |
[93] | |
| Expression of Y-linked genes escaping inactivation | KDM5C, NLGN4 | [94] | |
| MDD | Increased incidence in females, due to increased gene methylation and thus decreased gene expression of anti-MDD genes | [93] | |
| AD |
Increased incidence in males due to: Decreased gene inactivation in females |
KDM6A, PCDH11X | [98] |
| Hypomethylation of genes in males | Aurora kinase C | [103] | |
| Reduced HDAC2 levels in females | [103] | ||
| Cancer | |||
| 43 cancer types including glioblastoma | Decreased incidence and mortality of some cancer types, e.g., glioblastoma in females due to tumor suppressor genes escaping XCI | CNKSR2, ATRX, DDX3X, KDM6A, KDM5C, MAGEC3 | [105] |
| Breast and ovarian cancer | Increased susceptibility in females due to dysregulation of the Xist RNA expression and repressive marks, causing gene expression imbalances | ATRX | [111] |
| Stomach and esophageal cancer | Increased susceptibility in females due to gene mutations, hindering interaction with ATM protein kinase to ensure efficient DNA damage response and repair | EP400 | [112] |
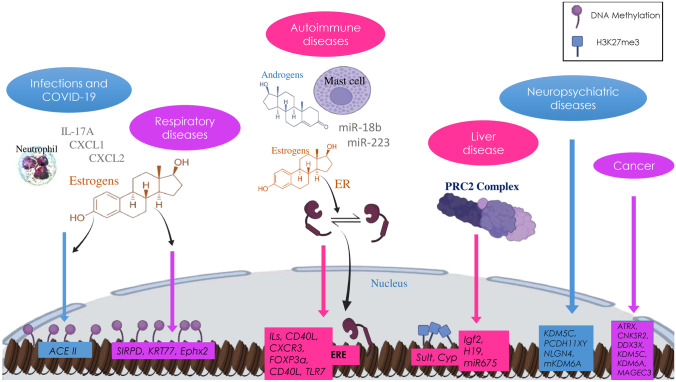
Fig. 2.
Sex-bias epigenetic mechanisms underlie the manifestation of various diseases. Women and men differ in the susceptibility and manifestations of various diseases due to epigenetic mechanisms regulating the effects of sex hormones, as well as the differential expression of X-chromosome–encoded genes
Several studies are currently elucidating the critical role of epigenetic enzymes, such as EZH2 and respective histone modifications to the sex-biased manifestation of diseases. Recently, COVID-19 susceptibility and progression were connected to epigenetic factors and mechanisms that act in a different way between the two sexes. Further studies are, however, needed to fully uncover the sex-biased epigenome in different disease states, opening a new window to personalized medicine and treatment, with the use of drugs targeting important epigenetic mechanisms.
Abbreviations
- ACE2
Angiotensin converting enzyme 2
- AD
Alzheimer’s disease
- ASD
Autism spectrum disorder
- ATRX
ATRX chromatin remodeler
- CD40L
Cluster of differentiation 40 ligand
- CXCL1
C-X-C motif chemokine ligand 1
- CXCL2
C-X-C motif chemokine ligand 2
- COPD
Chronic obstructive pulmonary disease
- CNKSR2
Connector enhancer of kinase suppressor of Ras 2
- Cyp
Cytochrome P family
- DDX3X
DEAD-box helicase 3 X-linked
- Ephx2
Epoxide hydrolase 2
- ER
Estrogen receptor
- ERE
Estrogen response element
- FOXP3
Forkhead box P3
- H3K27me3
Histone 3 lysine 27 trimethylation
- HCC
Hepatocellular carcinoma
- H19
H19 imprinted maternally expressed transcript
- IGF2
Insulin like growth factor 2
- IL-17A
Interleukin 17 alpha
- IFRD1
Interferon related developmental regulator 1
- KRT77
Keratin 77
- KDM5C
Lysine demethylase 5C
- lncRNAs
Long non-coding RNAs
- LRTIs
Lower respiratory tract infections
- MAGEC3
MAGE family member C3
- MDD
Major depressive disorder
- miR
microRNA
- mKDM6A
Mutated lysine demethylase 6 alpha
- NLGN4
Neuroligin 4 X-linked
- NOS-3
Nitric oxide synthase-3
- PRC2
Polycomb repressive complex 2
- PAH
Pulmonary arterial hypertension
- PCDH11XY
Protocadherin 11 X/Y-linked
- SIRPD
Signal regulatory protein delta
- Sult
Sulfotransferase
- XCL
X-C motif chemokine ligand 1
Author contribution
SC, MM, and DS performed the literature survey and wrote the first draft of the manuscript. CP conceived, revised, and edited the final manuscript. All authors read and approved the final manuscript.
Data availability
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trotman JB, Calabrese JM. How to silence an X chromosome. Nature. 2020;578:365–366. doi: 10.1038/d41586-020-00207-0. [DOI] [PubMed] [Google Scholar]
- 3.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 4.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luger K, Mäder AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 7.Xhemalce B, Dawson MA, Bannister AJ (2011) Histone modifications. Rev Cell Biol Mol Med
- 8.Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed]
- 9.Tchasovnikarova IA, Kingston RE. Beyond the histone code: a physical map of chromatin states. Mol Cell. 2018;69:5–7. doi: 10.1016/j.molcel.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 10.Peterson CL, Laniel M-A. Histones and histone modifications. Curr Biol. 2004;14:R546–51. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Ng MK, Cheung P. A brief histone in time: understanding the combinatorial functions of histone PTMs in the nucleosome context. Biochem Cell Biol. 2015;94:33–42. doi: 10.1139/bcb-2015-0031. [DOI] [PubMed] [Google Scholar]
- 12.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Snyder EM, Small CL, Li Y, Griswold MD. Regulation of gene expression by estrogen and testosterone in the proximal mouse reproductive tract1. Biol Reprod. 2009;81:707–716. doi: 10.1095/biolreprod.109.079053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 15.Parker MG. Transcriptional activation by oestrogen receptors. Biochem Soc Symp. 1998;63:45–50. [PubMed] [Google Scholar]
- 16.Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–3054. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- 17.Galupa R, Heard E. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu Rev Genet. 2018;52:535–566. doi: 10.1146/annurev-genet-120116-024611. [DOI] [PubMed] [Google Scholar]
- 18.Żylicz JJ, Bousard A, Žumer K et al (2019) The implication of early chromatin changes in X chromosome inactivation. Cell 176:182-197.e23. 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed]
- 19.Tukiainen T, Villani A-C, Yen A, et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Żylicz JJ, Heard E. Molecular mechanisms of facultative heterochromatin formation: an X-chromosome perspective. Annu Rev Biochem. 2020;89:255–282. doi: 10.1146/annurev-biochem-062917-012655. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Soloway PD, Clark AG. Paternally biased X inactivation in mouse neonatal brain. Genome Biol. 2010;11:R79–R79. doi: 10.1186/gb-2010-11-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Invernizzi P, Pasini S, Selmi C, et al. Female predominance and X chromosome defects in autoimmune diseases. J Autoimmun. 2009;33:12–16. doi: 10.1016/j.jaut.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Fang H, Disteche CM, Berletch JB. X inactivation and escape: epigenetic and structural features. Front cell Dev Biol. 2019;7:219. doi: 10.3389/fcell.2019.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markaki Y, Gan Chong J, Wang Y et al (2021) Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell 184:6174-6192.e32. 10.1016/j.cell.2021.10.022 [DOI] [PMC free article] [PubMed]
- 25.Collombet S, Ranisavljevic N, Nagano T, et al. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature. 2020;580:142–146. doi: 10.1038/s41586-020-2125-z. [DOI] [PubMed] [Google Scholar]
- 26.Loda A, Heard E. Xist RNA in action: past, present, and future. PLoS Genet. 2019;15:e1008333–e1008333. doi: 10.1371/journal.pgen.1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto I, Patrat C, Thépot D, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- 28.Kalantry S, Magnuson T. The polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLOS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadioglu A, Cuppone AM, Trappetti C, et al. Sex-based differences in susceptibility to respiratory and systemic pneumococcal disease in mice. J Infect Dis. 2011;204:1971–1979. doi: 10.1093/infdis/jir657. [DOI] [PubMed] [Google Scholar]
- 30.Tyagi AM, Srivastava K, Mansoori MN, et al. Estrogen deficiency induces the differentiation of IL-17 secreting Th17 cells: a new candidate in the pathogenesis of osteoporosis. PLoS One. 2012;7:e44552–e44552. doi: 10.1371/journal.pone.0044552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z, Huang Y-CT, Koziel H, et al. Female resistance to pneumonia identifies lung macrophage nitric oxide synthase-3 as a therapeutic target. Elife. 2014;3:e03711. doi: 10.7554/eLife.03711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aiello A, Bacci L, Re A, et al. MALAT1 and HOTAIR long non-coding RNAs play opposite role in estrogen-mediated transcriptional regulation in prostate cancer cells. Sci Rep. 2016;6:38414. doi: 10.1038/srep38414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yancey AL, Watson HL, Cartner SC, Simecka JW. Gender is a major factor in determining the severity of mycoplasma respiratory disease in mice. Infect Immun. 2001;69:2865–2871. doi: 10.1128/IAI.69.5.2865-2871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Jerkic M, Slutsky AS, Zhang H. Molecular mechanisms of sex bias differences in COVID-19 mortality. Crit Care. 2020;24:405. doi: 10.1186/s13054-020-03118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Channappanavar R, Fett C, Mack M, et al. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D, Lee J-Y, Yang J-S, et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J-M, Chung Y-S, Jo HJ et al (2020) Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong public Heal Res Perspect 11:3–7. 10.24171/j.phrp.2020.11.1.02 [DOI] [PMC free article] [PubMed]
- 38.Jia HP, Look DC, Shi L, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan Y, Shang J, Graham R, et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gemmati D, Bramanti B, Serino ML, et al (2020) COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int J Mol Sci 21:3474. 10.3390/ijms21103474 [DOI] [PMC free article] [PubMed]
- 43.Zhao Y, Zhao Z, Wang Y, et al. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beacon TH, Delcuve GP, Davie JR. Epigenetic regulation of ACE2, the receptor of the SARS-CoV-2 virus. Coronavirus Relat Res Collect. 2020;1:386–399. doi: 10.1139/gen-2020-0124@cfac.issue1. [DOI] [PubMed] [Google Scholar]
- 45.Corley MJ, Ndhlovu LC (2020) DNA methylation analysis of the COVID-19 host cell receptor, angiotensin I converting enzyme 2 gene (ACE2) in the respiratory system reveal age and gender differences. Preprints. 10.20944/preprints202003.0295.v1
- 46.Sawalha AH, Zhao M, Coit P, Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin Immunol. 2020;215:108410. doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chlamydas S, Papavassiliou AG, Piperi C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics. 2021;16:263–270. doi: 10.1080/15592294.2020.1796896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee N, Arathimos R, Chen S, et al. DNA methylation at birth is associated with lung function development until age 26 years. Eur Respir J. 2021 doi: 10.1183/13993003.03505-2020. [DOI] [PubMed] [Google Scholar]
- 49.Arathimos R, Granell R, Henderson J, et al. Sex discordance in asthma and wheeze prevalence in two longitudinal cohorts. PLoS One. 2017;12:e0176293. doi: 10.1371/journal.pone.0176293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuseini H, Newcomb DC. Mechanisms driving gender differences in asthma. Curr Allergy Asthma Rep. 2017;17:19. doi: 10.1007/s11882-017-0686-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Postma DS (2007) Gender differences in asthma development and progression. Gend Med 4 Suppl B:S133-46. 10.1016/s1550-8579(07)80054-4 [DOI] [PubMed]
- 52.Hohmann C, Keller T, Gehring U, et al. Sex-specific incidence of asthma, rhinitis and respiratory multimorbidity before and after puberty onset: individual participant meta-analysis of five birth cohorts collaborating in MeDALL. BMJ open Respir Res. 2019;6:e000460–e000460. doi: 10.1136/bmjresp-2019-000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vink NM, Postma DS, Schouten JP, et al. Gender differences in asthma development and remission during transition through puberty: the TRacking Adolescents’ Individual Lives Survey (TRAILS) study. J Allergy Clin Immunol. 2010;126:496–498. doi: 10.1016/j.jaci.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 54.Zein JG, Erzurum SC. Asthma is different in women. Curr Allergy Asthma Rep. 2015;15:28. doi: 10.1007/s11882-015-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.den Dekker HT, Burrows K, Felix JF, et al. Newborn DNA-methylation, childhood lung function, and the risks of asthma and COPD across the life course. Eur Respir J. 2019;53:1801795. doi: 10.1183/13993003.01795-2018. [DOI] [PubMed] [Google Scholar]
- 56.Patel R, Solatikia F, Zhang H, et al. Sex-specific associations of asthma acquisition with changes in DNA methylation during adolescence. Clin Exp allergy J Br Soc Allergy Clin Immunol. 2021;51:318–328. doi: 10.1111/cea.13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeMeo DL, Ramagopalan S, Kavati A, et al. Women manifest more severe COPD symptoms across the life course. Int J Chron Obstruct Pulmon Dis. 2018;13:3021–3029. doi: 10.2147/COPD.S160270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCartney DL, Stevenson AJ, Hillary RF, et al. Epigenetic signatures of starting and stopping smoking. EBioMedicine. 2018;37:214–220. doi: 10.1016/j.ebiom.2018.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koo H-K, Morrow J, Kachroo P, et al. Sex-specific associations with DNA methylation in lung tissue demonstrate smoking interactions. Epigenetics. 2021;16:692–703. doi: 10.1080/15592294.2020.1819662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang A, Kandhi S, Sun D. Roles of genetic predisposition in the sex bias of pulmonary pathophysiology, as a function of estrogens : sex matters in the prevalence of lung diseases. Adv Exp Med Biol. 2021;1303:107–127. doi: 10.1007/978-3-030-63046-1_7. [DOI] [PubMed] [Google Scholar]
- 61.Taneja V. Sex hormones determine immune response. Front Immunol. 2018;9:1931. doi: 10.3389/fimmu.2018.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai Y-J, Yu D, Zhang JH, Chen G-J. Cooperation of genomic and rapid nongenomic actions of estrogens in synaptic plasticity. Mol Neurobiol. 2017;54:4113–4126. doi: 10.1007/s12035-016-9979-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ortona E, Pierdominici M, Rider V. Editorial: sex hormones and gender differences in immune responses. Front Immunol. 2019;10:1076. doi: 10.3389/fimmu.2019.01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kadel S, Kovats S. Sex hormones regulate innate immune cells and promote sex differences in respiratory virus infection. Front Immunol. 2018;9:1653. doi: 10.3389/fimmu.2018.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bianchi I, Lleo A, Gershwin ME, Invernizzi P. The X chromosome and immune associated genes. J Autoimmun. 2012;38:J187–92. doi: 10.1016/j.jaut.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 66.Abramowitz LK, Olivier-Van Stichelen S, Hanover JA. Chromosome imbalance as a driver of sex disparity in disease. J genomics. 2014;2:77–88. doi: 10.7150/jgen.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quintero OL, Amador-Patarroyo MJ, Montoya-Ortiz G, et al. Autoimmune disease and gender: plausible mechanisms for the female predominance of autoimmunity. J Autoimmun. 2012;38:J109–19. doi: 10.1016/j.jaut.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays. 2011;33:791–802. doi: 10.1002/bies.201100047. [DOI] [PubMed] [Google Scholar]
- 69.Guo X, Su B, Zhou Z, Sha J. Rapid evolution of mammalian X-linked testis microRNAs. BMC Genomics. 2009;10:97. doi: 10.1186/1471-2164-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharma A, Kumar M, Aich J, et al. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci U S A. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khalifa O, Pers Y-M, Ferreira R, et al. X-linked miRNAs associated with gender differences in rheumatoid arthritis. Int J Mol Sci. 2016;17:1852. doi: 10.3390/ijms17111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rubtsova K, Marrack P, Rubtsov AV. Sexual dimorphism in autoimmunity. J Clin Invest. 2015;125:2187–2193. doi: 10.1172/JCI78082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mackey E, Thelen KM, Bali V, et al. Perinatal androgens organize sex differences in mast cells and attenuate anaphylaxis severity into adulthood. Proc Natl Acad Sci U S A. 2020;117:23751–23761. doi: 10.1073/pnas.1915075117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 77.Gupta K, Harvima IT. Mast cell-neural interactions contribute to pain and itch. Immunol Rev. 2018;282:168–187. doi: 10.1111/imr.12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J, Syrett CM, Kramer MC, et al. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci U S A. 2016;113:E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Credendino SC, Neumayer C, Cantone I. Genetics and epigenetics of sex bias: insights from human cancer and autoimmunity. Trends Genet. 2020;36:650–663. doi: 10.1016/j.tig.2020.06.016. [DOI] [PubMed] [Google Scholar]
- 80.El-Osta A. Introduction: understanding the consequences of epigenetic mechanisms and its effects on transcription in health and disease. Cancer Biol Ther. 2004;3:816–818. doi: 10.4161/cbt.3.9.1100. [DOI] [PubMed] [Google Scholar]
- 81.Long H, Yin H, Wang L, et al. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J Autoimmun. 2016;74:118–138. doi: 10.1016/j.jaut.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 82.Case LK, Wall EH, Dragon JA, et al. The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Res. 2013;23:1474–1485. doi: 10.1101/gr.156703.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hori S, Sakaguchi S. Foxp3: a critical regulator of the development and function of regulatory T cells. Microbes Infect. 2004;6:745–751. doi: 10.1016/j.micinf.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 84.Syrett CM, Anguera MC. When the balance is broken: X-linked gene dosage from two X chromosomes and female-biased autoimmunity. J Leukoc Biol. 2019;106:919–932. doi: 10.1002/JLB.6RI0319-094R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarmiento L, Svensson J, Barchetta I, et al. Copy number of the X-linked genes TLR7 and CD40L influences innate and adaptive immune responses. Scand J Immunol. 2019;90:e12776. doi: 10.1111/sji.12776. [DOI] [PubMed] [Google Scholar]
- 86.Murata M. Inflammation and cancer. Environ Health Prev Med. 2018;23:50. doi: 10.1186/s12199-018-0740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schuettengruber B, Bourbon H-M, Di Croce L, Cavalli G (2017) Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171:34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed]
- 88.Steffen PA, Ringrose L. What are memories made of? How polycomb and trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15:340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 89.Piunti A, Shilatifard A (2016) Epigenetic balance of gene expression by polycomb and COMPASS families. Science 352:aad9780. 10.1126/science.aad9780 [DOI] [PubMed]
- 90.Geisler SJ, Paro R. Trithorax and polycomb group-dependent regulation: a tale of opposing activities. Development. 2015;142:2876–2887. doi: 10.1242/dev.120030. [DOI] [PubMed] [Google Scholar]
- 91.Sugathan A, Waxman DJ. Genome-wide analysis of chromatin states reveals distinct mechanisms of sex-dependent gene regulation in male and female mouse liver. Mol Cell Biol. 2013;33:3594–3610. doi: 10.1128/MCB.00280-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lau-Corona D, Bae WK, Hennighausen L, Waxman DJ. Sex-biased genetic programs in liver metabolism and liver fibrosis are controlled by EZH1 and EZH2. PLoS Genet. 2020;16:e1008796–e1008796. doi: 10.1371/journal.pgen.1008796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xia Y, Dai R, Wang K, et al. Sex-differential DNA methylation and associated regulation networks in human brain implicated in the sex-biased risks of psychiatric disorders. Mol Psychiatry. 2021;26:835–848. doi: 10.1038/s41380-019-0416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crow TJ (2015) Is psychosis a disorder of XY epigenetics?. eBioMedicine 2:794–795. 10.1016/j.ebiom.2015.06.015 [DOI] [PMC free article] [PubMed]
- 95.Guo L, Zhong MB, Zhang L et al (2022) Sex differences in Alzheimer’s disease: insights from the multiomics landscape. Biol Psychiatry 91:61–71. 10.1016/j.biopsych.2021.02.968 [DOI] [PMC free article] [PubMed]
- 96.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 97.Miyake N, Mizuno S, Okamoto N et al (2013) KDM6A point mutations cause Kabuki syndrome. Hum Mutat 34:108–110. 10.1002/humu.22229 [DOI] [PubMed]
- 98.Davis EJ, Broestl L, Abdulai-Saiku S et al (2020) A second X chromosome contributes to resilience in a mouse model of Alzheimer’s disease. Sci Transl Med 12:eaaz5677. 10.1126/scitranslmed.aaz5677 [DOI] [PMC free article] [PubMed]
- 99.Lopes AM, Ross N, Close J, et al. Inactivation status of PCDH11X: sexual dimorphisms in gene expression levels in brain. Hum Genet. 2006;119:267–275. doi: 10.1007/s00439-006-0134-0. [DOI] [PubMed] [Google Scholar]
- 100.Carrasquillo MM, Zou F, Pankratz VS, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dumanski JP, Lambert J-C, Rasi C et al (2016) Mosaic loss of chromosome Y in blood is associated with Alzheimer disease. Am J Hum Genet 98:1208–1219. 10.1016/j.ajhg.2016.05.014 [DOI] [PMC free article] [PubMed]
- 102.Mendivil-Perez M, Velez-Pardo C, Kosik KS et al (2019) iPSCs-derived nerve-like cells from familial Alzheimer’s disease PSEN 1 E280A reveal increased amyloid-beta levels and loss of the Y chromosome. Neurosci Lett 703:111–118. 10.1016/j.neulet.2019.03.032 [DOI] [PMC free article] [PubMed]
- 103.Mahady L, Nadeem M, Malek-Ahmadi M et al (2019) HDAC2 dysregulation in the nucleus basalis of Meynert during the progression of Alzheimer’s disease. Neuropathol Appl Neurobiol 45:380–397. 10.1111/nan.12518 [DOI] [PMC free article] [PubMed]
- 104.Cao M, Li H, Zhao J et al (2019) Identification of age- and gender-associated long noncoding RNAs in the human brain with Alzheimer’s disease. Neurobiol Aging 81:116–126. 10.1016/j.neurobiolaging.2019.05.023 [DOI] [PMC free article] [PubMed]
- 105.Dunford A, Weinstock DM, Savova V, et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat Genet. 2017;49:10–16. doi: 10.1038/ng.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaneko S, Li X (2018) X chromosome protects against bladder cancer in females via a KDM6A-dependent epigenetic mechanism. Sci Adv 4:eaar5598–eaar5598. 10.1126/sciadv.aar5598 [DOI] [PMC free article] [PubMed]
- 107.Liu R, Kain M, Wang L. Inactivation of X-linked tumor suppressor genes in human cancer. Future Oncol. 2012;8:463–481. doi: 10.2217/fon.12.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buekers TE, Lallas TA, Buller RE. Xp22.2-3 loss of heterozygosity is associated with germline BRCA1 mutation in ovarian cancer. Gynecol Oncol. 2000;76:418–422. doi: 10.1006/gyno.1999.5713. [DOI] [PubMed] [Google Scholar]
- 109.Sirchia SM, Ramoscelli L, Grati FR, et al. Loss of the inactive X chromosome and replication of the active X in BRCA1-defective and wild-type breast cancer cells. Cancer Res. 2005;65:2139–2146. doi: 10.1158/0008-5472.CAN-04-3465. [DOI] [PubMed] [Google Scholar]
- 110.Ren W, Medeiros N, Warneford-Thomson R, et al. Disruption of ATRX-RNA interactions uncovers roles in ATRX localization and PRC2 function. Nat Commun. 2020;11:2219. doi: 10.1038/s41467-020-15902-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang H, Liao J, Zhang X, et al. Sex difference of mutation clonality in diffuse glioma evolution. Neuro Oncol. 2019;21:201–213. doi: 10.1093/neuonc/noy154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith RJ, Savoian MS, Weber LE, Park JH. Ataxia telangiectasia mutated (ATM) interacts with p400 ATPase for an efficient DNA damage response. BMC Mol Biol. 2016;17:22. doi: 10.1186/s12867-016-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li CH, Haider S, Shiah Y-J, et al. Sex differences in cancer driver genes and biomarkers. Cancer Res. 2018;78:5527–5537. doi: 10.1158/0008-5472.CAN-18-0362. [DOI] [PubMed] [Google Scholar]
- 114.Liu EM, Martinez-Fundichely A, Diaz BJ et al (2019) Identification of cancer drivers at CTCF insulators in 1,962 whole genomes. Cell Syst 8:446-455.e8. 10.1016/j.cels.2019.04.001 [DOI] [PMC free article] [PubMed]
- 115.Matthews BJ, Waxman DJ. Impact of 3D genome organization, guided by cohesin and CTCF looping, on sex-biased chromatin interactions and gene expression in mouse liver. Epigenetics Chromatin. 2020;13:30. doi: 10.1186/s13072-020-00350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.