Abstract
A plasmid-linked antimicrobial peptide, named coagulin, produced by Bacillus coagulans I4 has recently been reported (B. Hyronimus, C. Le Marrec and M. C. Urdaci, J. Appl. Microbiol. 85:42–50, 1998). In the present study, the complete, unambiguous primary amino acid sequence of the peptide was obtained by a combination of both N-terminal sequencing of purified peptide and the complete sequence deduced from the structural gene harbored by plasmid I4. Data revealed that this peptide of 44 residues has an amino acid sequence similar to that described for pediocins AcH and PA-1, produced by different Pediococcus acidilactici strains and 100% identical. Coagulin and pediocin differed only by a single amino acid at their C terminus. Analysis of the genetic determinants revealed the presence, on the pI4 DNA, of the entire 3.5-kb operon of four genes described for pediocin AcH and PA-1 production. No extended homology was observed between pSMB74 from P. acidilactici and pI4 when analyzing the regions upstream and downstream of the operon. An oppositely oriented gene immediately dowstream of the bacteriocin operon specifies a 474-amino-acid protein which shows homology to Mob-Pre (plasmid recombination enzyme) proteins encoded by several small plasmids extracted from gram-positive bacteria. This is the first report of a pediocin-like peptide appearing naturally in a non-lactic acid bacterium genus.
Bacteriocins are ribosomally synthesized antimicrobial polypeptides that are usually inhibitory only to strains closely related to the producing bacteria. These antimicrobial compounds are thought to provide the producer strain with a selective advantage over other strains. Bacteriocins produced by gram-positive bacteria are often membrane-permeabilizing cationic peptides with fewer than 60 amino acid residues (25, 29). In recent decades, the major advances in this field have been made in the lactic acid bacterium (LAB) family, due to the eminent economic importance of these microorganisms. Hence, the great structural diversity of LAB bacteriocins in combination with the fact that many bacteriocin producing LAB are present in a variety of naturally fermented food and feed products has led to a great interest in the potential of these bacteria as biopreservatives that could, at least partially, replace chemical preservatives (50). The bacteriocins of LAB have been divided into four distinct classes by biochemical and genetic means (28, 29). Bacteriocins of class I and II are by far the most studied because they are both the most abundant ones and the most prominent for industrial application (41). Class I bacteriocins called lantibiotics contain modified amino acid residues, lanthionine and methyllanthionine, which are formed posttranslationally (10). Class II consists of bacteriocins that lack modified residues. The pediocin-like bacteriocins constitute a large subgroup within class II: they are all small, heat-stable, membrane-active peptides that have a YGNGVXC consensus motif and are also characterized by their strong inhibitory effect on Listeria (29, 41). Representatives are widespread among LAB, including pediocins AcH and PA-1 produced by Pediococcus acidilactici (18); mesentericin Y105 produced by Leuconostoc mesenteroides (21); enterococin A produced by Enterococcus faecium (1); carnobacteriocins BM1, B2, and piscicocin V1a produced by Carnobacterium piscicola (2, 45); and divercin V41 produced by Carnobacterium divergens (35).
Antimicrobial compounds are also produced by other gram-positive bacteria (25), including the following Bacillus species: B. subtilis (26, 54), B. thuringiensis (8, 43), B. stearothermophilus (49), B. licheniformis (5), B. megaterium (24), B. thermoleovorans (42), and the food spoilage microorganisms B. cereus (39) and B. coagulans (23). Nevertheless, these reports usually suffer from limited biochemical characterization, often carried out on culture supernatant and not on the purified peptide, and a lack of genetic information. The only bacteriocins from Bacillus to be characterized at the amino acid and DNA sequence levels are subtilin (26, 30) and subtilosin (55) produced by B. subtilis. Subtilin is a member of linear lantibiotics (class I). Subtilosin is a cyclic bacteriocin, and the precursor peptide is reported to undergo several unique and unusual chemical posttranslational modifications, unlike those occurring during the synthesis of class I or some class II bacteriocins (55).
According to some authors (41), nonlantibiotic bacteriocins, which are widespread in LAB, are probably produced by other gram-positive bacteria. Nevertheless, no class II bacteriocin has so far been characterized in the genus Bacillus. In this paper, we report on the detailed study of the antilisterial peptide produced by B. coagulans strain I4, isolated in the course of a survey of spore-forming LAB for novel antimicrobial compounds (23). Coagulin is described by the N-terminal sequencing of the purified peptide and sequencing of the dedicated operon. Analysis demonstrates that coagulin is a new member in the pediocin-like family of bacteriocins. A putative Mob-RsA module was identified in the vicinity of the coa operon. The implications of our data for the intergeneric transfer of the bacteriocin operon are discussed.
MATERIALS AND METHODS
Bacterial strains and media.
The bacteriocin producer B. coagulans I4 was previously isolated from cattle feces (23). The strain was propagated aerobically in MRS (de Man, Rogosa, and Sharpe) broth (Difco Laboratories, Detroit, Mich.) at 37°C and maintained as a frozen stock at −20°C in the same medium supplemented with 20% (vol/vol) glycerol. Escherichia coli NM522 [supE thi Δ(lac-proAB) Δ(hsdMS-mcrB)5 F′(proAB lacIq ΔlacZM15)] (33) was used for all genetic manipulations and propagated in Luria-Bertani broth or agar (15 g/liter; Difco Laboratories) at 37°C (46).
The vector pUC19 (Stratagene) was used in cloning experiments. The selective concentration of ampicillin for growing E. coli cells containing the various recombinant plasmids was 50 μg/ml.
Determination of bacteriocin activity.
Bacteriocin activity was determined by the well diffusion assay (deferred method) described by Tagg and McGiven (51) in tryptose agar (Difco) seeded with Listeria innocua. The overnight culture of the indicator strain was grown in tryptose broth. To quantitate inhibitory activity, the diameter of the inhibition zone (in millimeters) was measured. For strong activities, the titer (expressed in activity units [AU] per milliliter), corresponding to the reciprocal of the highest dilution showing definite inhibition of the indicator lawn, was determined.
DNA manipulations.
Plasmids from E. coli and B. coagulans were extracted and purified as previously described (23, 46). Plasmid DNA was digested with restriction enzymes (Eurogentec, Seraing, Belgium) according to the supplier's recommendations. For cloning purposes, EcoRI or BamHI fragments of pI4 DNA were ligated into EcoRI- or BamHI-digested pUC19 using T4 DNA ligase (Eurogentec). The ligated DNAs were transformed into competent E. coli NM522 cells as described previously (31). Analytical and preparative agarose gel electrophoresis in Tris-borate-EDTA (pH 8.3) was performed as described (46). DNA fragments were isolated and purified from 1% (wt/vol) agarose gels with the Nucleotrap kit (Clontech, Palo Alto, Calif.). Southern blot hybridizations (46) were performed using Hybond-N+ nucleic transfer membranes (Amersham, Little Chalfont, Buckinghamshire, United Kingdom). DNA probes were labeled and used for hybridization experiments with the enhanced chemiluminescence kit according to the high-stringency conditions specified by the supplier (hybridization and washing steps at 42°C and in 0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]; Amersham). Nucleotide sequencing, based on the chain termination method (47), was done using an Auto-read sequencing kit (Pharmacia Biotech, Uppsala, Sweden) with either standard or specific primers in an automated DNA sequencer (ALF; Pharmacia).
Sequence analyses and alignments.
DNA and protein homology searches (Genbank, EMBL and SWISS-PROT) and sequence analysis were performed with the programs of the Genetics Computer Group sequence analysis software package (University of Wisconsin). Multiple sequence alignment was carried out with CLUSTAL W 1.60 (52).
Bacteriocin purification. (i) Concentration.
The bacteriocin was purified from 1-liter cultures of B. coagulans I4 grown in MRS broth at 37°C, to late logarithmic phase (A600, approximately 108 CFU/ml). The cells were removed by centrifugation at 4,000 × g for 15 min at 4°C, after which the supernatant was adjusted to pH 6.5 and filtered through a 0.45-μm-pore size HVLP membrane (Millipore SA, Molsheim, France). The filtrate was concentrated 10-fold using a spiral cartridge concentrator (model CH 2; Amicon Division, W. R. Grace and Co., Beverly, Mass.) with a molecular size cutoff of the membrane of 10,000 Da.
(ii) Solid-phase extraction.
The lyophilized retentate (concentrated crude extract) was solubilized in 5 mM NH4HCO3 buffer, pH 8.0, and loaded on a Sep-Pak Plus C18 cartridge (Waters, Division of Millipore Corp., Bedford, Mass.) for solid-phase extraction. Nonactive material was discarded after washing the Sep-Pak cartridge with 5 ml of NH4HCO3–2-propanol (80:20, vol/vol). Active fractions (i.e., those displaying anti listeria activity) were eluted using 5 ml of 5 mM NH4HCO3–2-propanol (30:70, vol/vol) and then dried under vacuum centrifugation.
(iii)RP chromatography.
The dry residue (fraction I) was dissolved in 5 mM NH4HCO3–2-propanol (30:70, vol/vol) and centrifuged at 10,000 × g for 5 min. The supernatant was further used for bacteriocin purification by reverse phase (RP) chromatography performed with an analytical high-performance liquid chromatography (HPLC) system (Kontron Instruments) with an automated gradient controller, 332 UV detector, and 425 integrator. The supernatant (0.5 ml) was loaded on a Delta-Pack C18 (Waters) semipreparative RP-HPLC column (8 by 100 mm; diameter, 15 μm; 100 Å), equilibrated at a flow rate of 2 ml/min with H2O-acetonitrile (90:10, vol/vol) containing 0.1% trifluoroacetic acid (solvent A). Activity was eluted with a linear gradient ranging from 15 to 60% acetonitrile containing 0.07% trifluoroacetic acid (solvent B). Active fractions were pooled, dried under vacuum (fraction II), dissolved in 5 mM NH4HCO3–2-propanol (30:70, vol/vol), and then rechromatographed on a Licrospher C18 (Merck, Darmstadt, Germany) analytical RP-HPLC column (4 by 125 mm; diameter, 5 μm; 100 Å). The column was equilibrated at a flow rate of 1 ml/min with solvent A. The bacteriocin was eluted with a gradient of solvent B. Elution conditions were as follows: 0 to 5 min, 100% A to 90% A–10% B (vol/vol); from 5 to 30 min, linear gradient to 75% A–25% B (vol/vol). Active fractions (1 ml) containing the purified bacteriocin were collected, dried under vacuum and stored at −20°C (fraction III).
Determination of protein.
Protein content was determined by the Bradford method (4) using the Bio-Rad assay reagent (Bio-Rad, Munich, Germany) with bovine serum albumin as the standard.
Amino acid sequencing, enzymatic cleavage, and MS of coagulin.
Amino acid sequencing was performed by Edman degradation with an automatic liquid phase sequence analyzer (model 473; Applied Biosystems, Foster City, Calif.). Presence of disulfide bonds was pointed out by hydrolysis of the purified bacteriocin with the Lysobacter enzymogenes Lys-C endoproteinase (Sigma Chemical Chimie, St Quentin Fallavier, France).
Mass measurements were performed using matrix-assisted laser desorption ionization–time of flight mass spectrometry. Analysis was performed by the PE Applied Biosystems Division of Perkin-Elmer, Courtabœuf, France, using a voyager-STR instrument (Perseptive Biosystems, Framingham, Mass.) equipped with a single-stage reflector and delayed extraction. The instrument has a linear and reflector flight path length of 2.0 and 3.0 m, respectively.
Analysis of coagulin C-terminal sequence hydrophobicity and hydrophobic moment.
The average sequence hydrophobicity and average sequence hydrophobic moments of coagulin sequence regions were calculated by using the normalized amino acid hydrophobicity scale and hydrophobic moment calculation method as described by Eisenberg et al. (12).
Nucleotide sequence accession number.
The nucleotide sequence corresponding to the coa operon and mob-RsA module has been deposited in the GenBank database under accession no. AF 300457.
RESULTS
Purification of antimicrobial peptide produced by B. coagulans I4.
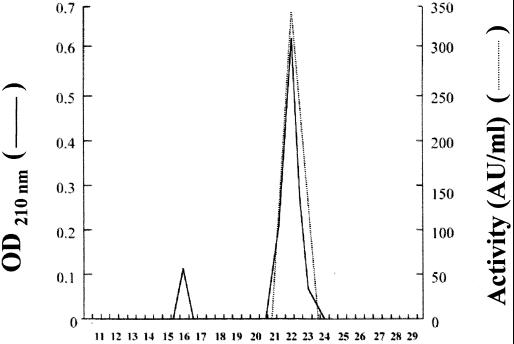
The antimicrobial peptide produced by B. coagulans I4 was purified to homogeneity from a late-logarithmic-phase culture, grown at 37°C in MRS broth. Results are summarized in Table 1. Ultrafiltration was used successfully to concentrate the activity from the growth medium. Approximately a two-fold increase in the specific activity and a 65% recovery of the peptide were obtained (Table 1). Ultrafiltration was followed by solid-phase extraction on a silica-based bonded phase with strong hydrophobicity. The recovery, 52%, remained high after binding the peptide contained in the concentrated crude extract and eluting with 5 mM NH4HCO3–2-propanol (Table 1, fraction I). The largest increase in specific activity, about 109 times, was obtained after the final analytical RP-HPLC steps (Table 1, fraction III). A single absorbance peak, corresponding with the activity peak, was obtained (Fig. 1). The final specific activity of the purified peptide was evaluated to be about 140,000 AU/mg.
TABLE 1.
Summary of coagulin purification
| Step | Vol (ml) | Total protein (mg)a | Total activity (AU)b | Sp. act. (AU/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Culture supernatant (crude extract) | 1,000 | 250 | 320,000 | 1,280 | 100 | 1.0 |
| UF concentrate (concentrated crude extract) | ||||||
| Solid-phase extract (fraction I) | 50 | 93 | 280,000 | 2,240 | 65 | 1.8 |
| RP chromatography | 50 | 16.5 | 166,000 | 10,080 | 52 | 7.9 |
| Semiprep run (fraction II) | 8 | 1.16 | 58,200 | 67,479 | 18 | 52.7 |
| Analytical run (fraction III) | 2 | 0.23 | 32,000 | 139,130 | 10 | 108.7 |
Protein content was estimated by the Bradford method (4).
Total bacteriocin activity (AU) was determined against L. innocua as described (23).
FIG. 1.
Reverse-phase chromatography of the antimicrobial peptide produced by B. coagulans I4. The amount applied to the column was obtained from a 1-liter culture.
The antimicrobial peptide produced by B. coagulans I4 is a pediocin-like bacteriocin.
The purified peptide was analyzed by mass spectrometry, which confirmed the purity of the sample and showed a molecular mass of 4,612 Da. Amino acid sequencing enabled the partial determination of the first 39 amino acid (aa) residues. Seven positions (9, 14, 19, 24, 29, 33, and 34) could not be identified. Comparisons with protein sequences contained in the data banks demonstrated a total identity of the 32 identified amino acid residues with the sequence of pediocin PA-1 (22). This 44-aa bacteriocin, with a molecular mass of 4,624 Da, contains four cysteine residues forming two disulfide bridges, between C-9 and C-14 and between C-24 and C-44, which are required for the antibacterial activity (6, 13, 22). Digestion of the newly purified bacteriocin by Lys-C endoprotease produced only two RP-HPLC peaks (results not shown) as has been previously observed for pediocin when submitted to the same treatment (15, 22). These results demonstrate the presence of disulfide bridge(s) in the structure of the peptide. In spite of the observed similarities between the partial sequence of the two bacteriocins, the difference between the molecular masses might be suggestive of a slight variation in the primary structure.
Cloning and nucleotide sequence analysis of the bacteriocin locus.
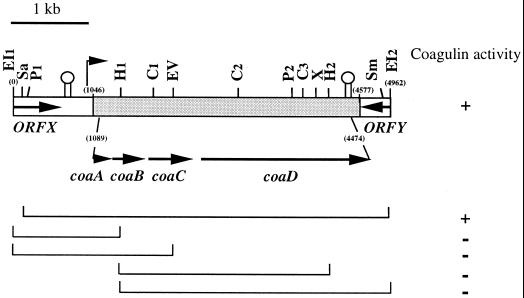
Evidence for plasmid-encoded bacteriocin was obtained previously by plasmid curing (23), suggesting that production and immunity are associated with the 14-kb plasmid, named pI4, present in B. coagulans I4. In an attempt to express the bacteriocin production gene in E. coli, restriction fragments from plasmid pI4 DNA were cloned in pUCI9 (see Materials and Methods), and the bacteriocin production of transformants was tested using the agar well diffusion method. L. innocua was used as the indicator strain. Cells carrying the recombinant clone pBC8 showed a zone of inhibition in the indicator lawn, while the control strain E. coli NM522 containing pUC19 did not. This observation indicated that the bacteriocin was produced and, most likely, secreted into the medium. Southern blot hybridization analysis showed that this clone contained a 5-kb EcoRI fragment of plasmid pI4 from B. coagulans I4. To localize the bacteriocin production gene, the 5-kb EcoRI1–2 fragment was mapped and the identified restriction sites were used to obtain deletion derivatives. An overview is given in Fig. 2. The SacI deletion derivative retained activity while neither of the other derivatives obtained displayed zones of inhibition against the indicator strain. Since the SacI site was observed to be close to the EcoRI1 cloning site (Fig. 2), the sequence of the entire 5-kb EcoRI fragment from pI4 was determined.
FIG. 2.
Schematic representation of the cloned region of pI4 that mediates expression of coagulin in construction pBC8 and its deletion derivatives. The shaded area spanning coordinates 1046 to 4577 represents the section homologous to the ped-pap operon, present on the P. acidilactici pSMB 74 plasmid, and required for pediocin PA-1–AcH expression. The coding region containing coaA, coaB, coaC, and coaD spans position 1089 to 4474. The structures resembling rho-independent terminators are indicated. The vertical arrow represents the putative promoter for coagulin expression in B. coagulans. Abbreviations for restriction enzymes: C, ClaI; EI, EcoRI; EV, EcoRV; H, HindIII; P, PvuII; Sa, SacI; Sm, SmaI; X, XbaI. +, coagulase expressed; −, coagulase not expressed.
The sequences of the EcoRI1-HindIII1, HindIII1–2, and HindIII2-EcoRI2 fragments (Fig. 2) were determined and assembled, yielding a 4.962-kb sequence. The G+C content of this region, 37%, is lower than that of the B. coagulans chromosome (45 to 47%) (40). The internal 3.532-kb zone localized between nucleotide positions 1046 and 4577 displayed a total homology with the sequence of the operons ped (32) and pap (37) encoding the similar pediocins PA-1 and AcH, respectively (Fig. 2). Only six mismatches were identified between the Pediococcus and Bacillus 3.532-kb sequences. When translated in all possible reading frames, this sequence revealed the same organization as previously reported for the ped-pap operons, i.e., the presence of a 3.386-kb coding region (positions 1089 to 4474) organized in four open reading frames (ORFs) (ORF I to IV), each preceded by a putative ribosome binding site (Fig. 2). These ORFs encode proteins which consist of 62, 112, 174, and 724 aa, showing homology with the corresponding Ped-PapA, Ped-PapB, Ped-PapC, and Ped-PapD proteins, respectively. As for the pediocin operon, bacteriocin production was assigned to ORF I and immunity to ORF II. Transport was assigned to ORF III and ORF IV, encoding the so-called accessory protein and the ATP-binding cassette transporter, respectively (38).
A region of dyad symmetry was found downstream of coaD, between positions 4478 and 4515, i.e., within the homologous region. It is identical to the proposed rho-independent stem-loop terminator identified on the plasmid pSMB 74 that encodes pediocin AcH (38).
The bacteriocin operons from P. acidilactici and B. coagulans differ by six mismatches and display different promoter sequences.
As mentioned above, six mismatches were identified between the Pediococcus and Bacillus 3.532-kb homologous sequences. Five mapped in the transport genes ORF III and IV. They resulted in three substitutions in ORF IV, encoding the ATP-binding cassette transporter (V488F, F670L, and Y712H), and two in ORF III, encoding the accessory protein (R54K and V151I). The last of these mismatches (V151I) resulted in the presence of an EcoRV restriction site, demonstrated during the mapping of the 5-kb EcoRI fragment of pI4, and absent in pSMB74 (38). Hence, these five amino acids are not essential for transport activity since active coagulin was recovered in the supernatent of B. coagulans I4 cultures. The sixth mismatch was identified in ORF I, the bacteriocin production gene, and resulted in a single substitution at the C terminus of the peptide. Hence, a threonine residue was present at position 41 (T41) in coagulin while pediocins AcH and PA-1 contain an asparagine residue (N41) (22, 38).
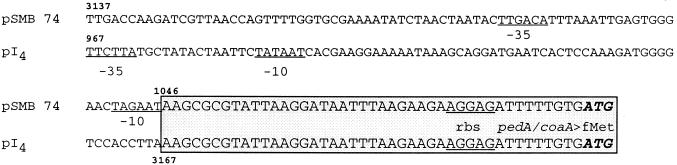
Interestingly, the common promoter sequence proposed for the pap operon in the P. acidilactici sequence was not present upstream of coaA in B. coagulans plasmid DNA. Comparison of the nucleotide sequences pI4 and pSMB 74 revealed that the first nucleotide of the homologous region, i. e., at position 1046 in pI4, corresponded to the nucleotide immediately downstream of the −10 box reported in P. acidilactici (Fig. 3). A putative promoter sequence for the coa locus in B. coagulans was present in the upstream 1.045-kb nonhomologous region (Fig. 3).
FIG. 3.
Translational and transcriptional signals upstream of coaA in pI4 from B. coagulans and pedA in pSMB 74 from P. acidilactici. Numbering for the pSMB74 DNA sequence is that reported for the complete sequence of the plasmid (38). The boxed area denotes the homologous sequences, starting at position 1046 in the EcoRI fragment from pI4. −35 and −10 boxes and the ribosome binding sequence (rbs) sequence are underlined.
A putative mobilization module is present immediately downstream of the coa operon.
The 3.532-kb homologous region containing the coa locus is located between two sequences of 1.045-kb and 0.384-kb, with G+C contents of 36 and 44%, respectively. Both sequences lack homology with pSMB 74 DNA and sequences found in the nucleotide sequence databases (EMBL and GenBank). When translated, each nonhomologous region revealed a truncated ORF (ORF X and ORF Y, respectively) (Fig. 2). ORF X, is followed by a region of dyad symmetry with a calculated free energy (ΔG0) of −32.6 kcal/mol, representing a putative rho-independent terminator. Complete elucidation of the sequences of ORF X and ORF Y was achieved by sequencing the restriction fragments contiguous to the 5-kb EcoRI fragment containing coa, which were positioned and cloned during the mapping of pI4 (C. Le Marrec, B. Hyronimus, P. Bressollier, and M. C. Urdaci, unpublished data). The additional 1.5-kb sequence containing the 5′ part of ORF X has a G+C content of 46%. Analysis did not reveal any homology to pSMB 74 DNA or to other known sequences (data not shown). Data revealed that ORF X specified a 181-aa protein with no homology between it and other protein sequences contained in the databases.
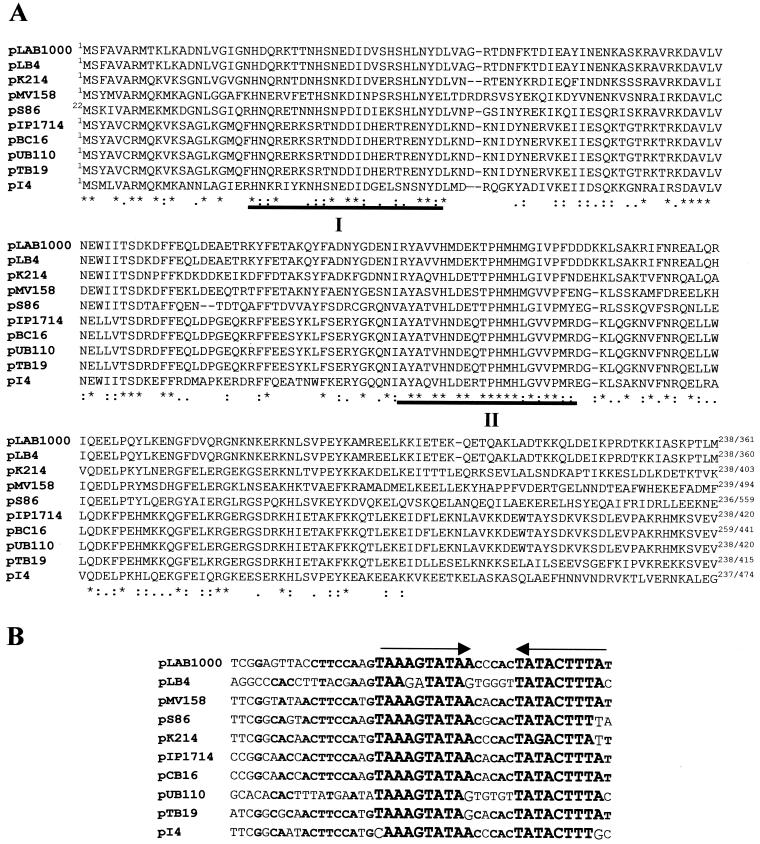
Complete characterization of ORF Y was achieved by sequencing a 1.5-kb downstream region (Le Marrec et al., unpublished data) which has a G+C content of 42%. As observed for ORF X, this oppositely oriented gene immediately downstream of coa did not exhibit any nucleotide homology to known sequences. Surprisingly, comparison with the databases indicated that the deduced 474-aa protein displayed a high degree of similarity (40 to 74%) to corresponding regions deduced from mob-pre (plasmid recombination enzyme) genes present on several plasmids, which were found in strains belonging to various gram-positive genera (including Bacillus, Lactococcus, Streptococcus, Lactobacillus, Enterococcus, and Staphylococcus). In several cases, the corresponding mob genes have been shown to be required for conjugative mobilization and site-specific recombination (34). Much higher levels of identity were observed within the N-terminal parts of these proteins. In Fig. 4A, a selection of the N-terminal part (∼250 aa) of representative Mob proteins were compared. They display 49 to 55% similarity and 63 to 74% identity with the putative Mob product from pI4. It was suggested that this N-terminal part could constitute an important functional region of these proteins (34). The two conserved motifs previously described for Mob proteins (20, 34) are present in the aligned sequences (Fig. 4A, boxes I and II). Motif I contains a putative DNA-binding tyrosine residue involved in DNA nicking activity (34).
FIG. 4.
Comparison of mobilization modules (Mob-RsA) from plasmids isolated from gram-positive bacteria. EMBL-GenBank accession numbers, given between square brackets, for the following plasmids are indicated pLAB1000 (Lactobacillus hilgardii [P35856]), pLB4 (L. plantarum [P20046]), pK214 (L. lactis [WX92946]), pMV158 (Streptococcus agalactiae [P13925]), pS86 (E. faecalis [AJ223161]), pIP1714 (Staphylococcus cohnii [AF015628]), pCB16 (B. cereus [U32369]), pUB110 (S. aureus [P22490]), and pTB19 (thermophilic Bacillus, [JQ1212]). (A) Partial alignments of Mob proteins (generated by using the CLUSTAL W program [52]). Conserved residues are marked with an asterisk. Closely related residues are indicated by colons, while less related residues are indicated by single dots. The two conserved regions I and II (20, 34) are indicated with filled bars. (B) Comparisons of RSA sites found upstream of Mob family members. The inverted repeats are indicated by arrows. Conserved nucleotides are printed in boldface type.
The corresponding structural features of reported mobilization modules are the oriT-RSA sites, the cognate sequences recognized and nicked by the Mob proteins (17, 34). The putative pI4 oriT-RSA site could be located through alignment immediately upstream of pI4 mob gene, as it is almost identical to corresponding sites present in plasmids encoding the Mob proteins presented Fig. 4A. It consists of a 23-bp inverted repeat, detected 43 -bp upstream of the ORF Y potential start codon (Fig. 4B). This 23-bp sequence is part of a larger 43-bp region conserved among the 10 plasmid sequences represented in Fig. 4.
DISCUSSION
In order to gain insight into the production of bacteriocins among Bacillus species, we characterized the antimicrobial compound produced by B. coagulans I4, using biochemical and genetic analyses. Purification of the bacteriocin was achieved using a four-step procedure that included ultrafiltration (UF) and RP chromatography. Good recovery of activity (65%) after the UF step is promising, because this technique is relevant for large-scale production. The purification level (109-fold) appears to be lower than those obtained for other structurally related bacteriocins (14, 15, 22). This was mainly the result of the greater sensitivity of the Bradford method, which allows a more acute determination of protein concentration in crude extract compared to A280 or A254 measurements.
Surprisingly, analysis of the structure of the purified bacteriocin produced by B. coagulans I4 and the sequence deduced from the gene show a strong similarity with that of pediocins AcH and PA-1. The only difference is a threonine residue at position 41 instead of asparagine (N41T). This single modification in the structure is confirmed by the difference observed between the molecular masses of purified coagulin and AcH, estimated by mass spectrometry to be 4,612 and 4,624 Da, respectively. Furthermore, this difference in measured (4,612 Da) and estimated (4,616 Da) molecular masses may suggest the existence of two disulfide bonds linking the four cysteine residues on the sequence of coagulin. The N41T modification occurs at the C-terminal part of the peptide, which is less conserved than the N-terminal region within the family of pediocin-like bacteriocins (22). Recently, pediocin AcH substitution mutants with altered bactericidal activity have been described (36). Such single-residue mutants obtained in the C-terminal I25-H42 region (I26T, M31T, A34D, G37E, G37R, and H42L) displayed a modified hydrophobic moment. The authors suggested that this domain, when folded into a putative α helix, becomes amphiphatic and would mediate the adsorption to the bacterial membrane. Modification of its hydrophobic moment would change the angle of contact at which pediocin adsorbs to the membrane interface. This modification was observed for all mutants, except for one case (N41K), which showed a 10-fold-reduced activity, although the substitution did not change the hydrophobic moment of the sequence. Interestingly, the discovery of coagulin shows that an N41T variant exists naturally and that this modification does not result in a significant change in average hydrophobicity and hydrophobic moment of the I25-H42 C-terminal part (Table 2). As suggested (36), the formation of the C-24–C-44 disulphide bond probably interferes with the formation of an α helix. A theoretical loss of specific activity of coagulin compared to pediocin AcH and the N41K variant could not be calculated. Such studies would be interesting to elucidate the influence of such a mutation on activity and host spectrum. Preliminary studies indicate that coagulin is not active on Lactobacillus plantarum and Staphylococcus aureus, contrary to pediocin AcH. Such results seem to be contradictory to those obtained by Miller et al. (36), who obtained a single mutant, A34D, whose mutation may have a species-specific effect.
TABLE 2.
Average hydrophobicities and hydrophobic momentsa of the I25-H42 C-terminal sequences of various bacteriocins
| Bacteriocin | Hydrophobicity | Hydrophobic moment (μδ) |
|---|---|---|
| Pediocin AcH | 0.22 | 0.30 |
| Substitution mutant N41Kb | 0.18 | 0.31 |
| Coagulin (N41T) | 0.24 | 0.30 |
Values were calculated by the method of Eisenberg et al. (12); hydrophobic moments are a measure of the amphipathicity of a sequence.
Pediocin AcH substitution mutant as previously described (36).
Pediocins have been isolated from different P. acidilactici strains—PAC1.0 (32), LB 42-923 (3, 37), SJ-1 (48), M (14)—and from Pediococcus pentosaceus strains FBB61 (44) and L7230 (7). These strains were isolated from fermented sausages or cucumbers. Recently, a pediocin-like antilisterial bacteriocin was purified from an L. plantarum strain named WHE 92, which was isolated from munster cheese (15). To our knowledge, the present study provides the first evidence for pediocin production by a non-LAB bacterium. Interestingly, available data concerning the host spectra of these bacteriocins reveal that pediocins PA-1–AcH (3, 22), SJ-1 (48), AcM (14), and A (44) have been found to be active against L. plantarum and that pediocin AcM is active against B. coagulans (14).
Production of pediocin by L. plantarum WHE 92 has been reported to involve genes that are located on an 11-kb plasmid. This size is different from those of the plasmids involved in pediocin production in P. acidilactici (37). These authors envisaged an intergeneric conjugal transfer of a plasmid harboring the pediocin determinants, followed by a modification event (15). Analysis of the 5-kb fragment harboring the coa locus revealed extremely high conservation of these determinants, with only six mismatches out of 3.532-kb, which may support the idea of a similar genetic transfer by conjugation between Pediococcus and Bacillus spp. Nevertheless, the following observations remain intriguing: (i) the operon is located between two regions (2.545 and 1.885-kb) with no homology to plasmid pSMB 74 DNA, (ii) the G+C content of the coa operon and the upstream 2.545-kb sequence (39%) is lower than that reported for the chromosome of B. coagulans (45 to 47%), and (iii) the promoter identified in P. acidilactici is absent in pI4. It is tempting to speculate that these elements may be relevant to an alternative situation, involving the extremely precise excision of the Pediococcus operon sequence devoided of its promoter, and its rearrangement downstream of a promoter on a different resident plasmid. Although the mobilizing capacity of pI4 remains untested, the presence of a Mob-RSA module downstream of coa could explain the following step, i.e., plasmid transmission between bacteria. The interaction of the oriT-RSA site and Mob protein are well known to contribute to the dissemination of small antibiotic plasmids among gram-positive bacteria (53). Hence, the suggested precise recombination, then combined to a conjugative mobilization and/or interplasmidic recombination event, would have enabled the transfer of the pediocin determinants to other gram-positive bacteria, such as B. coagulans.
Complete nucleotide sequence of pI4 and detailed functional analysis will be necessary to understand the organization of this plasmid and to enable some conclusions about its evolution. In particular, the availability of this sequence should help to determine whether the bacteriocin determinants are contemporary additions or not and to understand the mechanism that led to their inheritance by B. coagulans I4. Recent comparative studies of the sequences of the S. aureus antimicrobial multiresistance plasmid pSK41 (16) and the Lactococcus lactis bacteriocin production plasmid pMRC01 (11) suggested that insertion sequence-mediated cointegrate formation may play an important role in the evolution of these plasmids, allowing bacteria to collect and subsequently disseminate advantageous traits. To date, little information is available on plasmids replicating in B. coagulans, except for the report of a cryptic 1.6-kb plasmid called pBC1 (9). This rolling-circle type plasmid (RCR) was classified in the pC194/pUB110 family (19, 27). RCR plasmids are characterized by the modular organization of functional regions and known to be highly recombinogenic, a feature that may favor their intra- and intergeneric dissemination (27).
ACKNOWLEDGMENTS
We acknowledge the assistance of J. C. Truffert (PE Applied Biosystems) for mass spectral analysis and Kathryn Mayo for critical reading of the manuscript.
REFERENCES
- 1.Aymerich T, Holo H, Havarstein L S, Hugas M, Garriga M, Nes I F. Biochemical and genetic characterization of enterococcin A from Enterococcus faecium, a new anti-listerial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol. 1996;62:1676–1682. doi: 10.1128/aem.62.5.1676-1682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhugaloo-Vial P, Dousset X, Métivier A, Sorokine O, Anglade P, Boyaval P, Marion D. Purification and amino acid sequences of piscicocins V1a and V1b, two class IIa bacteriocins secreted by Carnobacterium piscicola V1 that display significantly different levels of specific inhibitory activity. Appl Environ Microbiol. 1996;62:4410–4416. doi: 10.1128/aem.62.12.4410-4416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhunia A, Johnson M C, Ray B. Purification, characterization and antimicrobial spectrum of a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol. 1988;65:261–268. doi: 10.1111/j.1365-2672.1988.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Ann Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bradley D E. Ultrastructure of bacteriophages and bacteriocins. Bacteriol Rev. 1967;31:230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chikindas M L, Garcia-Garcera M J, Driessen A J M, Ledeboer A M, Nissen-Meyer J, Nes I F, Abee T, Konings W N, Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC 1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol. 1993;59:3577–3584. doi: 10.1128/aem.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daeschel M A, Klaenhammer T R. Association of a 13.6-megadalton plasmid in Pediococcus pentosaceus with bacteriocin activity. Appl Environ Microbiol. 1985;50:1538–1541. doi: 10.1128/aem.50.6.1538-1541.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Borjac H, Lajudie J. Mise en évidence de facteurs antagonistes du type des bactériocines chez Bacillus thuriengensis. Ann Microbiol (Paris) Ser B. 1974;125:529–537. [PubMed] [Google Scholar]
- 9.De Rossi E, Milano A, Brigidi P, Bini F, Riccardi G. Structural organization of pBCl, a cryptic plasmid from Bacillus coagulans. J Bacteriol. 1992;174:638–642. doi: 10.1128/jb.174.2.638-642.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vos W M, Kuipers O P, Van der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 11.Dougherty B A, Hill C, Weidman J F, Richardson D R, Venter J C, Ross R P. Sequence analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRCO1 from Lactococcus lactis DPC3147. Mol Microbiol. 1998;29:1029–1038. doi: 10.1046/j.1365-2958.1998.00988.x. [DOI] [PubMed] [Google Scholar]
- 12.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 13.Eisjink V M, Skeie M, Middelhoven P H, Brurberg M B, Nes I F. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl Environ Microbiol. 1998;64:3275–3281. doi: 10.1128/aem.64.9.3275-3281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elegado F B, Kim W J, Kwon D Y. Rapid purification, partial characterization, and antimicrobial spectrum of the bacteriocin, pediocin AcM from Pediococcus acidilactici M. Int J Food Microbiol. 1997;37:1–11. doi: 10.1016/s0168-1605(97)00037-8. [DOI] [PubMed] [Google Scholar]
- 15.Ennahar S, Aoude-Werner D, Sorokine O, Van Dorsselaer A, Bringel F, Hubert J C, Hasselmann C. Production of pediocin AcH by Lactobacillus plantarum WHE 92 isolated from cheese. Appl Environ Microbiol. 1996;62:4381–4387. doi: 10.1128/aem.62.12.4381-4387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firth N, Berg T, Skurray R A. Evolution of conjugative plasmids from gram-positive bacteria. Mol Microbiol. 1998;31:1598–1600. [PubMed] [Google Scholar]
- 17.Gennaro M L, Kornblum J, Novick R P. A site-specific recombination function in Staphylococcus aureus plasmids. J Bacteriol. 1987;169:2601–2610. doi: 10.1128/jb.169.6.2601-2610.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez C, Kunka B. Plasmid-associated bacteriocin production and sucrose fermentation in Pediococcus acidilactici. Appl Environ Microbiol. 1987;53:2534–2538. doi: 10.1128/aem.53.10.2534-2538.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruss A, Gruss S D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989;53:231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman L M, Espinosa M. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J Mol Biol. 1997;266:688–702. doi: 10.1006/jmbi.1996.0824. [DOI] [PubMed] [Google Scholar]
- 21.Héchard Y, Derijard B, Letellier F, Cenatiempo Y. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. J Gen Microbiol. 1992;138:2725–2731. doi: 10.1099/00221287-138-12-2725. [DOI] [PubMed] [Google Scholar]
- 22.Henderson J, Chopko A L, Van Wassenaar P D. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 23.Hyronimus B, Le Marrec C, Urdaci M C. Coagulin, a bacteriocin-like inhibitory substance produced by Bacillus coagulans I4. J Appl Microbiol. 1998;85:42–50. doi: 10.1046/j.1365-2672.1998.00466.x. [DOI] [PubMed] [Google Scholar]
- 24.Ivanovics G. Bacteriocins and bacteriocin-like substances. Bacteriol Rev. 1962;26:108–118. [PMC free article] [PubMed] [Google Scholar]
- 25.Jack R W, Tagg J R, Ray B. Bacteriocins of Gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen E F, Hirschmann D J. Subtilin, an antibacterial substance of Bacillus subtilis. Culturing conditions and properties. Arch Biochem. 1944;4:297–309. [Google Scholar]
- 27.Khan S. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klaenhammer T R. Bacteriocins of lactic acid bacteria. Biochimie. 1988;70:337–349. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- 29.Klaenhammer T R. Genetics of bacteriocins by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 30.Klein C, Kaletta C, Schnell N, Entian K. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol. 1992;58:132–142. doi: 10.1128/aem.58.1.132-142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 32.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression, and nucleotide sequence of genes involved in the production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mead D A, Skorupa E S, Kemper B. Single stranded DNA SP6 promoter plasmid for engineering mutant RNAs and proteins; synthesis of a ‘stretched’ preproparathyroid hormone. Nucleic Acids Res. 1985;13:1103–1118. doi: 10.1093/nar/13.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijer W J J, Wisman G B A, Terpstra P, Thorsted P B, Thomas C M, Holsappel S, Venema G, Bron S. Rolling-circle plasmids from B. subtilis: complete nucleotide sequences and analyses of genes of pTA1015, pTA1040, pTA1050 and pTA1060, and comparisons with related plasmids from Gram-positive bacteria. FEMS Microbiol Rev. 1998;21:337–368. doi: 10.1111/j.1574-6976.1998.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 35.Métivier A, Pilet M F, Dousset X, Sorokine O, Anglade P, Zagorec M, Piard J C, Marion D, Cenatiempo Y, Frémaux C. Divercin V41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergens V41: primary structure and genomic organization. Microbiology. 1998;144:2837–2844. doi: 10.1099/00221287-144-10-2837. [DOI] [PubMed] [Google Scholar]
- 36.Miller K W, Schamber R, Osmanagaoglu O, Ray B. Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl Environ Microbiol. 1998;64:1997–2005. doi: 10.1128/aem.64.6.1997-2005.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motlagh A M, Bhunia A K, Szostek F, Hansen T R, Johnson M G, Ray B. Nucleotide and amino acid sequence of pap-gene (pediocin AcH production) in Pediococcus acidilactici H. Lett Appl Microbiol. 1992;15:45–48. doi: 10.1111/j.1472-765x.1992.tb00721.x. [DOI] [PubMed] [Google Scholar]
- 38.Motlagh A M, Bukhtiyarova M, Ray B. Complete nucleotide sequence of pSMB74, a plasmid encoding production of pediocin AcH in Pediococcus acidilactici. Lett Appl Microbiol. 1994;18:305–312. doi: 10.1111/j.1472-765x.1994.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 39.Naclerio G, Ricca E, Sacco M, De Felice M. Antimicrobial activity of a newly identified bacteriocin of Bacillus cereus. Appl Environ Microbiol. 1993;59:4313–4316. doi: 10.1128/aem.59.12.4313-4316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura L K, Blumenstock I, Claus D. Taxonomic studies of Bacillus coagulans Hammer 1915 with a proposal for Bacillus smithii sp. nov. Int J Syst Bacteriol. 1998;38:63–73. [Google Scholar]
- 41.Nes I F, Diep D B, Håvarstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 42.Novotny J F, Perry J J. Characterization of bacteriocins from two strains of Bacillus thermoleovorans, a thermophilic hydrocarbon-utilizing species. Appl Environ Microbiol. 1992;58:2393–2396. doi: 10.1128/aem.58.8.2393-2396.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paik H D, Bae S S, Park S H, Pan J P. Identification and partial characterization of tochicin, a bacteriocin produced by Bacillus thuringiensis subsp. tochigiensis. J Ind Microbiol Biotechnol. 1997;19:294–298. doi: 10.1038/sj.jim.2900462. [DOI] [PubMed] [Google Scholar]
- 44.Piva A, Headon D R. Pediocin A, a bacteriocin produced by Pediococcus pentosaceus FBB61. Microbiology. 1994;140:697–702. doi: 10.1099/00221287-140-4-697. [DOI] [PubMed] [Google Scholar]
- 45.Quadri L E N, Sailer M, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17 B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schved F, Lalazar Y H, Juven B. Purification, partial characterization and plasmid-linkage of pediocin SJ-1, a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol. 1993;74:67–77. doi: 10.1111/j.1365-2672.1993.tb02998.x. [DOI] [PubMed] [Google Scholar]
- 49.Shafia F. Thermocins of B. stearothermophilus. J Bacteriol. 1966;92:524–525. doi: 10.1128/jb.92.2.524-525.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stiles M E. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:331–345. doi: 10.1007/BF00395940. [DOI] [PubMed] [Google Scholar]
- 51.Tagg J R, McGiven A R. Assay system for bacteriocins. Appl Microbiol. 1971;21:943–948. doi: 10.1128/am.21.5.943-943.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson J D, Higgins D J, Gibson T J. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalities and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Lelie D, Bron S, Venema G, Oskam L. Similarity of minus origins of replication and flanking open reading frames of plasmids pUB110, pTB913 and pMV158. Nucleic Acids Res. 1989;18:7283–7294. doi: 10.1093/nar/17.18.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng G, Slavik M F. Isolation, partial purification and characterization of a bacteriocin produced by a newly isolated Bacillus subtilis strain. Lett Appl Microbiol. 1999;28:363–367. doi: 10.1046/j.1365-2672.1999.00545.x. [DOI] [PubMed] [Google Scholar]
- 55.Zheng G, Yan L Z, Vederas J C, Zuber P. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J Bacteriol. 1999;181:7346–7355. doi: 10.1128/jb.181.23.7346-7355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]