Abstract
Pseudomonas putida KT2440 uses proline as the sole C and N source. Utilization of this amino acid involves its uptake, which is mediated by the PutP protein, and its conversion into glutamate, mediated by the PutA protein. Sequence analysis revealed that the putA and putP genes are transcribed divergently. Expression from the putP and putA genes was analyzed at the mRNA level in different host backgrounds in the absence and presence of proline. Expression from the put promoters was induced by proline. The transcription initiation points of the putP and putA genes were precisely mapped via primer extension, and sequence analysis of the upstream DNA region showed well-separated promoters for these two genes. The PutA protein acts as a repressor of put gene expression in P. putida because expression from the put promoters is constitutive in a host background with a knockout putA gene. This regulatory activity is independent of the catabolic activity of PutA, because we show that a point mutation (Glu896→Lys) that prevents catalytic activity allowed the protein to retain its regulatory activity. Expression from the put promoters in the presence of proline in a putA-proficient background requires a positive regulatory protein, still unidentified, whose expression seems to be ς54 dependent because the put genes were not expressed in a ς54-deficient background. Expression of the putA and putP genes was equally high in the presence of proline in ς38- and ihf-deficient P. putida backgrounds.
Pseudomonas putida mt-2 is a saprophytic soil bacterium able to use m-methylbenzoate as the sole C source (34). This strain and its DNA restriction-deficient mutant called KT2440 have been shown to be aggressive root colonizers and are considered rhizosphere microorganisms (23).
Recent studies have focused on the possible role of amino acids as alternative carbon substrates that can support the growth of microorganisms in the rhizosphere of plants (9, 15, 29, 35). All of the 20 amino acids present in the proteins can be detected in plant exudates. Our group and others have shown that proline is one of the most abundant amino acids in corn root exudates (31, 32); therefore, this amino acid could be an important energy source for bacteria during the first stages of colonization of plant roots.
We have found that P. putida KT2440 can use proline as the sole C and N source, and we have recently cloned the genes of P. putida involved in proline utilization (named put for proline utilization) (32). In enteric bacteria and P. putida (2, 17, 19, 32), two genes were found to be essential for proline metabolism: the putP gene, whose gene product is involved in the uptake of proline to the cytoplasm of the cell, and the putA gene product, a multifunctional protein that not only catalyzes the formation of glutamate from proline via pyrroline-5′-carboxylic acid but is also involved in control of expression of the put genes (24–27). The putA gene has also been identified in Rhodobacter and Agrobacterium species and members of the family Rhizobiaceae (8, 11, 12, 14).
Using the transcriptional fusions of the putA and putP promoters to ′lacZ, it was shown that the putA and putP genes are regulated at the transcriptional level in P. putida, with proline acting as an inducer, since β-galactosidase levels from the putA and putP gene promoters increased by about 20- and 4-fold, respectively, in liquid culture medium in the presence of proline (32). However, the promoter regions of these genes and their pattern of expression are unknown. Using the put promoter fusions to ′lacZ, it was shown that in a putA mutant background, high levels of expression from these genes occurred, suggesting that the P. putida PutA protein acts as a repressor of putA and putP gene expression, as also described for enteric bacteria (27) and Rhodobacter capsulatus (14). In enteric bacteria, in addition to the putA gene, two other host factors, integration host factor (IHF) and ς54, are involved in control of expression of the put genes (4, 26, 27). rpoN and ihfA mutants of P. putida KT2440 deficient in the synthesis of ς54 and IHF, respectively, are available; however, the patterns of expression of the put genes in these backgrounds are unknown.
In this study we analyzed expression from the put promoters at the mRNA level in different P. putida backgrounds in the absence and presence of proline, and we describe a plausible model for the control of expression of the proline utilization genes in P. putida. We also report that the regulatory activity of the P. putida PutA protein is independent of the catabolic activities of this multifunctional protein.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The P. putida strains used in this study are shown in Table 1. P. putida KT2440-Pro21 is a spontaneous mutant unable to use proline as the sole C and N source. It has a point mutation that causes Glu-896 to be replaced by Lys in the translated protein (our unpublished results).
TABLE 1.
P. putida strains used in this study
| Strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| KT2440 | Prototroph, Cmr | 10 |
| KT2442 | Rifr derivative of KT2440 | 10 |
| KT2440-IHF3 | ifhA::ΩKm Kmr Rifr | 20 |
| KT2440-rpoN | rpoN::Tn5 Kmr Rifr | 16 |
| MAD2 | KT2440 Δpst | 6 |
| S14D2 | putA::mini-Tn5-luxAB Kmr Rifr | 32 |
| KT2442-Pro21 | Point mutation at putA so that the strain does not use proline | Our laboratory |
Cmr, Kmr, and Rifr, resistance to chloramphenicol, kanamycin, and rifampin, respectively.
Bacterial cells were usually grown on M9 minimal medium with succinate (20 mM) and/or proline (20 mM) as the sole C source (1). When proline (20 mM) was used as the sole C and N source, M9 depleted of ammonium, called M8, was used. When necessary, kanamycin and rifampin were added to final concentrations of 25 and 10 μg/ml, respectively.
Nucleic acid techniques.
The 5′ mRNA start of the transcript that originated from the put promoter was determined by the method of Marqués et al. (22). The primer used to analyze expression from putA was 5′-CACCACTTCCTGCTCGGGGCGG-3′, and the primer used to determine putP expression was 5′-GGCGATCCAGGCCTCGGACAGGCCCG-3′. These oligonucleotides were 5′ end labeled with [γ-32P]ATP and polynucleotide kinase, and about 105 cpm of the labeled primers was annealed to 20 to 30 μg of total RNA prepared from the different P. putida strains grown under different conditions. cDNA was synthesized by using avian myeloblastosis virus reverse transcriptase as previously described (22). The products of reverse transcription were analyzed in urea-polyacrylamide sequencing gels. Gels were exposed, during the time required, to Amersham RPN-8 films for autoradiography.
Nucleotide sequence accession number. The DNA sequence of the intergenic region between the putA and putP genes can be retrieved from GenBank under accession number AF153207.
RESULTS AND DISCUSSION
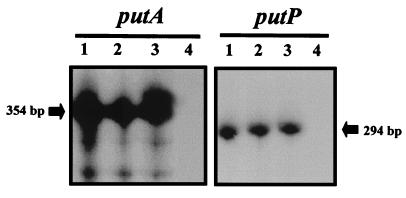
We have found that as in members of the family Enterobacteriaceae, in P. putida the put genes are transcribed divergently, and we have located the intergenic region at about 400 bp (Fig. 1). Therefore, this region should contain all the elements necessary to control expression from the putA and putP genes. To study the transcription of the put genes, we have grown P. putida cells on M9 minimal medium with proline as the sole C source (ammonium as the N source) or N source (succinic acid as the C source) or with proline as the sole C and N source (1). As a control, we have grown P. putida cells on M9 minimal medium with ammonium as the N source and succinate as the sole C source. mRNA from cells growing exponentially with these nutrients was isolated, and both the presence and amount of the put mRNAs were analyzed by primer extension. The results obtained are shown in Fig. 2. It was found that expression of the put genes was induced because no mRNAs were detected in cells grown on M9 minimal medium with succinate, whereas in the presence of proline, regardless of its utilization as C, N, or C and N source, both genes were expressed. In addition, the absolute expression levels were similar in the proline concentration range of 200 μM to 20 mM.
FIG. 1.
DNA sequence of the intergenic region between the putA and putP genes. The ATG start codon of the genes is boxed; the transcription initiation point of each gene is marked by an asterisk, and the −10 and −35 regions of each promoter are underlined.
FIG. 2.
Expression of the putA and putP genes of P. putida KT2440 under different growth conditions. mRNA was prepared as described previously (20). Cells were grown in different media as follows. Lane 1, M8 minimal medium with 20 mM proline; lane 2, M9 minimal medium with 20 mM proline; lane 3, M9 with 20 mM proline and succinate; lane 4, M9 with succinate. Primer extension analysis was done as described in Materials and Methods with a primer complementary to putA or putP mRNA.
In six independent assays, the level of cDNA resulting from extension with the putA primer was found to be 3.2- to 5.7-fold higher than the levels obtained for the putP promoter, which suggests that the putA promoter is stronger than the putP promoter. This induction ratio was independent of the concentration of proline used for induction within the range between 200 μM and 20 mM. In addition, it should be mentioned that glutamate, the first stable metabolite of proline metabolism, is neither an inducer of the put genes nor a repressor in cultures growing with proline and glutamate (not shown). This contrasts with the complex control of proline utilization in higher microorganisms such as Aspergillus nidulans (9).
The primer extension analysis shown in Fig. 2 allowed us to determine the main transcription start site corresponding to nucleotide 140 for putA and to nucleotide 400 for the putP genes (Fig. 1). Analysis of the DNA sequence of the region upstream from the start of each transcript was carried out. In both cases, sequences that resembled those recognized by RNA polymerase with ς70 were found (Fig. 1).
P. putida PutA protein is involved in putA and putP gene expression, and its regulatory role is independent of its catalytic activity.
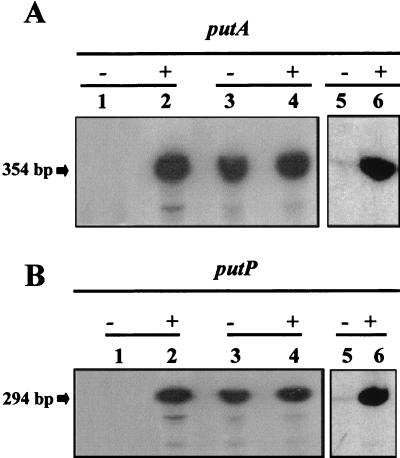
In Escherichia coli and Salmonella enterica serovar Typhimurium, it has been suggested that not only does the putA gene product have two enzymatic activities but that it also regulates expression of the put promoters (27). Its role has been suggested to be that of a repressor protein that binds to the put region, hindering the access of the RNA polymerase (27). We previously generated a P. putida putA null mutant carrying the insertion of a mini-Tn5 at the 5′ end of the putA gene (32). We tested the expression from the putA and putP promoters in this isogenic PutA-deficient background. We found that transcription from the putA and putP promoters was constitutive (Fig. 3). Since the mini-Tn5 insertion can exert polar effects on downstream genes, we cannot exclude the possibility that the constitutivity of put genes in this background is the result of the lack of a yet unidentified regulator located downstream of putA. To this end, we analyzed in detail the 3′ region of putA. A hairpin (5′-AAGGAGAGCCTCGGCTCTCCTT-3′) that could destabilize RNA polymerase was found 22 bp downstream from the stop codon. In addition, no open reading frames were found within the contiguous 600 bp. This strongly suggests that PutA is involved as a repressor of expression of the put promoters in P. putida.
FIG. 3.
Expression from the putA and putP promoters in P. putida KT2442, the PutA-deficient derivative S14D2, and the PutA Glu896→Lys mutant. Cells were grown in the absence (−) or presence (+) of proline. The strains were P. putida KT2442 (lanes 1 and 2), P. putida S14D2 (lanes 3 and 4), and P. putida KT2442-Pro21 (lanes 5 and 6). Other conditions are as described in the legend for Fig. 2.
P. putida KT2442-Pro21 is a mutant unable to use proline as the sole C or N source. We have identified that in this mutant the PutA protein lacks the ability to mediate proline-to-glutamate conversion due to a single point mutation that resulted in the single amino acid change Glu896→Lys (S. Vílchez, unpublished results). We have analyzed the pattern of expression from the put promoters in cells growing with succinate and ammonium in the absence and presence of proline. The results obtained are shown in Fig. 3, where it can be observed that expression from the put promoters followed the same pattern as in the wild-type strain. This indicates that the regulatory role of PutA is independent of its catabolic activities and makes PutA a peculiar protein in the sense that this 1,315-amino-acid protein has two catabolic activities and a gene regulatory function.
Involvement of different sigma factors in the control of expression of the put genes.
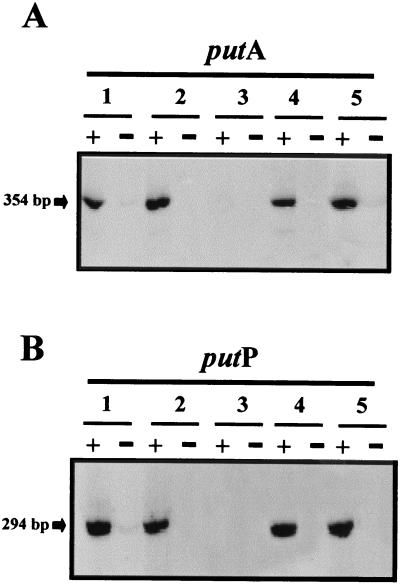
Since the analysis of the upstream sequences of the put promoters revealed −10 and −35 regions similar to those recognized by ς70 and because some promoters can be transcribed in vivo by ς70 or ς38 according to the growth phase (13, 30), we tested expression from the put promoters in cells in the early stationary phase for both the wild type and the isogenic ς38 mutant (25). Proline (2 mM) was added to cells in the stationary phase, mRNA was isolated 30 min later, and the level of expression from the putA and putP promoters was analyzed. It was found that expression from these promoters was similar in both backgrounds (Fig. 4).
FIG. 4.
Expression from the putA and putP gene promoters in different host backgrounds. P. putida cells were grown in the presence (+) or absence (−) of proline. The strains used were the wild type (lane 1), IHF-deficient mutant (lane 2), ς54-deficient mutant (lane 3), ς38-deficient mutant (lane 4), and ptsN-deficient mutant (lane 5). Other conditions are as described in the legend for Fig. 2.
In enterobacteria, several proteins have been proposed to be involved in transcription from the put promoters: Nac, ς54, and IHF (18, 27). We had previously generated P. putida knockout mutations in the rpoN gene encoding ς54 (14) and in the ihfA gene (20). To date, the nac gene has not been identified in pseudomonads. In the IHF- and RpoN-deficient isogenic backgrounds, we assayed the expression from the put promoters in cells growing in the presence of 2 mM proline, and we compared the results with those obtained for the wild-type cells growing under similar conditions. Culture samples were taken for mRNA analysis after 120 min of incubation. The results obtained are shown in Fig. 4, where it can be observed that in the IHF-deficient background, the expression from the put promoters was similar to that from these promoters in the wild-type background. However, in the ς54-deficient background, there was no expression in the presence of proline.
Cases et al. (6) recently reported that in P. putida KT2440, the pts genes lie downstream of rpoN. The pts genes are part of an operon with rpoN. Their study has suggested a regulatory role for these genes in processes related to the use of different C sources by P. putida. Since the ς54-deficient P. putida strain carries a Tn5 insertion within the rpoN gene and because the insertion exerted a polar effect on downstream genes, we examined expression from the put promoters in P. putida MAD2 (Table 1) which is ς54 proficient and Pts deficient (Fig. 4). We have found that in the MAD2 strain, the pattern of expression from the put promoters in the presence of proline was similar to that found in the wild-type strain. This result leads to the suggestion that the lack of expression of the put promoters in the P. putida rpoN mutant is due to the lack of ς54 rather than to other proteins of the rpoN-pts operon.
The ς54 promoter recognition sequence includes short elements at nucleotides −12 (GC) and −24 (GC) with extensive conservation between the two (5, 33). Such ς54 promoter sequences have not been found upstream from the transcription initiation start points of the put genes in Pseudomonas. Therefore, we ascribed the lack of expression from the put promoters to the lack of a regulator involved in the control of the put promoters whose expression is ς54 dependent. In Klebsiella aerogenes, the Nac protein is involved in control of a number of promoters subject to nitrogen regulation (hutUH, gdh, putA, and ureA) whose transcription is mediated by RNA polymerase with ς70. No expression from these promoters occurred in a ς54-deficient background. The reason for this is that the nac system is under the control of the ntr system and the nac gene expression is dependent on ς54 (3, 18, 24). Therefore, Nac represents a form of nitrogen regulation that is not independent of the Ntr system in enterobacteria. To date, neither the nac nor the ntr system has been reported in P. putida. As a hypothesis, we propose a model for proline utilization in Pseudomonas in which an analog of NtrC activates ς54-dependent expression of an analog of Nac. The Nac protein, thus produced, displaces PutA from the put promoter and allows ς70-dependent expression of put genes. We propose that the main role of this Nac-like activator in Pseudomonas is to overcome PutA repression, since in a PutA-deficient background, expression from put is proline independent.
ACKNOWLEDGMENTS
This work was supported in part by grants from GX-Biosystems España and the European Commission (BIO4-CT98-0283).
We thank I. Cases and S. Marqués for kindly providing strains.
REFERENCES
- 1.Abril M A, Michán C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen S W, Sentis Willis A, Maloy S R. DNA sequence of the putA gene from Salmonella typhimurium: a bifunctional membrane-associated dehydrogenase that binds DNA. Nucleic Acids Res. 1993;21:1676–1686. doi: 10.1093/nar/21.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender R A, Snyder P M, Bueno R, Quinto M, Magasanik B. Nitrogen regulation system of Klebsiella aerogenes: the nac gene. J Bacteriol. 1983;156:444–446. doi: 10.1128/jb.156.1.444-446.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown E D, Wood J M. Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J Biol Chem. 1993;268:8972–8979. [PubMed] [Google Scholar]
- 5.Buck M, Gallegos M-T, Studholme D J, Guo Y, Gralla J D. The bacterial enhancer-dependent ς54 (ςN) transcription factor. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cases I, Pérez-Martín J, de Lorenzo V. The IIANtr (PtsN) protein of Pseudomonas putida mediated C-source inhibition of the ς54-dependent Pu promoter of the TOL plasmid. J Biol Chem. 1999;274:15562–15568. doi: 10.1074/jbc.274.22.15562. [DOI] [PubMed] [Google Scholar]
- 7.Chen C S, Yoshida A. Enzymatic properties of the proline encoded by newly cloned human alcohol dehydrogenase ADH6 gene. Biochem Biophys Res Commun. 1991;181:743–747. doi: 10.1016/0006-291x(91)91253-9. [DOI] [PubMed] [Google Scholar]
- 8.Cho K, Fuqua C, Winans S C. Transcriptional regulation and locations of Agrobacterium tumefaciens genes required for complete catabolism of octopine. J Bacteriol. 1997;179:1–8. doi: 10.1128/jb.179.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cubero B, Gómez D, Scazzocchio C. Metabolite repression and inducer exclusion in the proline utilization gene cluster of Aspergillus. J Bacteriol. 2000;182:233–235. doi: 10.1128/jb.182.1.233-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin F G H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiménez-Zurdo J I, van Dillewijn P, Soto M J, de Felipe M R, Olivares J, Toro N. Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol Plant-Microbe Interact. 1995;8:492–498. doi: 10.1094/mpmi-8-0492. [DOI] [PubMed] [Google Scholar]
- 12.Jiménez-Zurdo J I, García-Rodríguez F M, Toro N. The Rhizobium meliloti putA gene: its role in the establishment of the symbiotic interaction with alfalfa. Mol Microbiol. 1997;23:85–93. doi: 10.1046/j.1365-2958.1997.1861555.x. [DOI] [PubMed] [Google Scholar]
- 13.Jishage M, Iwata A, Veda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keuntje B, Masepohl B, Klipp W. Expression of the putA gene encoding proline dehydrogenase from Rhodobacter capsulatus is independent of NtrC regulation but requires an Lrp-like activator protein. J Bacteriol. 1995;177:6432–6439. doi: 10.1128/jb.177.22.6432-6439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohl D H, Schubert K R, Carter M B, Hagedam C H, Shearer G. Proline metabolism in N2-fixing root nodules: energy transfer and regulation of purine synthesis. Proc Natl Acad Sci USA. 1988;85:2036–2040. doi: 10.1073/pnas.85.7.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köhler T, Harayama S, Ramos J-L, Timmis K N. Involvement of Pseudomonas putida RpoN ς factor in regulation of various metabolic functions. J Bacteriol. 1989;171:4326–4333. doi: 10.1128/jb.171.8.4326-4333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling M, Allen S W, Wood J M. Sequence analysis identifies the proline dehydrogenase and Δ1-pyrroline-5-carboxylate dehydrogenase domains of the multifunctional Escherichia coli PutA protein. J Mol Biol. 1994;243:950–956. doi: 10.1006/jmbi.1994.1696. [DOI] [PubMed] [Google Scholar]
- 18.Macaluso A, Best E A, Bender R A. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J Bacteriol. 1990;172:7249–7255. doi: 10.1128/jb.172.12.7249-7255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maloy S R. The proline utilization operon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1513–1519. [Google Scholar]
- 20.Marqués S, Gallegos M-T, Manzanera M, Holtel A, Timmis K N, Ramos J L. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J Bacteriol. 1998;180:2889–2894. doi: 10.1128/jb.180.11.2889-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marqués S, Gallegos M T, Ramos J L. Role of ςS in transcription from the positively controlled Pm promoter of the TOL plasmid of Pseudomonas putida. Mol Microbiol. 1995;18:851–857. doi: 10.1111/j.1365-2958.1995.18050851.x. [DOI] [PubMed] [Google Scholar]
- 22.Marqués S, Ramos J L, Timmis K N. Analysis of the mRNA structure of the Pseudomonas putida TOL meta fission pathway operon around the transcription initiation point, the xylTE and the xylFJ regions. Biochim Biophys Acta. 1993;1216:227–236. doi: 10.1016/0167-4781(93)90149-8. [DOI] [PubMed] [Google Scholar]
- 23.Molina L, Ramos C, Duque E, Ronchel M C, García J M, Wyke L, Ramos J L. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol Biochem. 2000;32:315–321. [Google Scholar]
- 24.Muro-Pastor A M, Ostrowsky P, Maloy S. Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J Bacteriol. 1997;179:2788–2791. doi: 10.1128/jb.179.8.2788-2791.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieuwkoop A J, Baldauf S A, Hudspeth M E S, Bender R A. Bidirectional promoter in the hut(P) region of the histidine utilization (hut) operons from Klebsiella aerogenes. J Bacteriol. 1988;170:2240–2246. doi: 10.1128/jb.170.5.2240-2246.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Brien K, Deno G, Ostrovsky de Spicer P, Gadner J F, Maloy S. Integration host factor facilitates repression of the put operon in Salmonella typhimurium. Gene. 1992;118:13–19. doi: 10.1016/0378-1119(92)90243-i. [DOI] [PubMed] [Google Scholar]
- 27.Ostrovsky de Spicer P, O'Brien K, Maloy S. Regulation of proline utilization in Salmonella typhimurium: a membrane-associated dehydrogenase binds DNA in vitro. J Bacteriol. 1991;173:211–219. doi: 10.1128/jb.173.1.211-219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos-González M I, Molin S. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J Bacteriol. 1998;180:3421–3431. doi: 10.1128/jb.180.13.3421-3431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soto M J, Jiménez-Zurdo J I, van Dillewijn P, Toro N. Sinorhizobium meliloti putA gene regulation: a new model within the family Rhizobiaceae. J Bacteriol. 2000;182:1935–1941. doi: 10.1128/jb.182.7.1935-1941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka K, Tanayanagi Y, Fujita N, Ishihama A, Takahashi H. Heterogeneity of the principal ς factor in Escherichia coli: the rpoS gene product, ς38, is second principal ς factor of RNA polymerase in stationary-phase. Proc Natl Acad Sci USA. 1993;90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vancura V. Plant metabolites in soil. In: Kunc F, Vancura V, editors. Soil microbial associations: control of structures and functions. Amsterdam, The Netherlands: Elsevier; 1988. pp. 156–168. [Google Scholar]
- 32.Vílchez S, Molina L, Ramos C, Ramos J L. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes. J Bacteriol. 2000;182:91–99. doi: 10.1128/jb.182.1.91-99.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Gralla J D. Multiple in vivo roles for the −12-region elements of ς54 promoters. J Bacteriol. 1998;180:5626–5631. doi: 10.1128/jb.180.21.5626-5631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y-X, Shearer G, Kohl D H. Proline fed to intact soybean plants influences acetylene reducing activity, and content and metabolism of proline in bacteroids. Plant Physiol. 1992;98:1020–1028. doi: 10.1104/pp.98.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]