Abstract
Introduction
Despite the disproportional impact of SLE on historically marginalised communities, the individual and sociocultural factors underlying these health disparities remain elusive. We report the design and methods for a study aimed at identifying epigenetic biomarkers associated with racism and resiliency that affect gene function and thereby influence SLE in a health disparity population.
Methods and analysis
The Social Factors, Epigenomics and Lupus in African American Women (SELA) Study is a cross-sectional, case–control study. A total of 600 self-reported African American women will be invited to participate. All participants will respond to questionnaires that capture detailed sociodemographic and medical history, validated measures of racial discrimination, social support, as well as disease activity and damage for cases. Participants who wish will receive their genetic ancestry estimates and be involved in research. Blood samples are required to provide peripheral blood mononuclear cell counts, DNA and RNA. The primary goals of SELA are to identify variation in DNA methylation (DNAm) associated with self-reported exposure to racial discrimination and social support, to evaluate whether social DNAm sites affect gene expression, to identify the synergistic effects of social factors on DNAm changes on SLE and to develop a social factors-DNAm predictive model for disease outcomes. This study is conducted in cooperation with the Sea Island Families Project Citizen Advisory Committee.
Discussion and dissemination
SELA will respond to the pressing need to clarify the interplay and regulatory mechanism by which various positive and negative social exposures influence SLE. Results will be published and shared with patients and the community. Knowledge of the biological impact of social exposures on SLE, as informed by the results of this study, can be leveraged by advocacy efforts to develop psychosocial interventions that prevent or mitigate risk exposures, and services or interventions that promote positive exposures. Implementation of such interventions is paramount to the closure of the health disparities gap.
Keywords: Systemic Lupus Erythematosus; Epidemiology; Lupus Erythematosus, Systemic; Polymorphism, Genetic
What is already known about this topic?
Although multiple social and individual factors affect SLE outcomes and contribute to its disparities, the biological mechanisms by which adverse and protective factors synergistically modulate disease outcomes are not currently well understood.
What does this study add?
A socioecological model of SLE outcomes that integrates multiple social, demographic, behavioural, genomic, epigenomic and transcriptomic factors to understand the effects of positive and negative social environments on SLE through epigenomic changes.
The identification of epigenetic biomarkers by which risk and resiliency factors affect gene expression and thereby influence SLE in African American women.
How this study might affect research, practice or policy?
Epigenetic biomarkers might be used prospectively as biomarkers of previous social exposures to identify individuals at risk of SLE or worse outcomes.
Knowledge of the effects of social exposures on biological changes can be leveraged by advocacy efforts to develop psychosocial interventions that mitigate risk exposures, and services or interventions that promote positive exposures, which are paramount to the closure of the health disparities gap.
Introduction
Health disparities in SLE are well established. As recently reviewed, women are 8–10 times more likely than men to develop SLE; relative to European Americans, African Americans are three to four times more likely to develop SLE, suffer from remarkably higher disease severity and death rates, and are more likely to suffer from multiple comorbidities such as depression, cardiovascular disease, diabetes, and worse health-related quality of life.1 SLE is among the leading causes of death in young women (highest for African American and Hispanic women),2 3 underscoring its impact as an important public health issue.
Despite the disproportionate impact of SLE on minority racial and ethnic communities, the causes for these health disparities remain elusive. The causal mechanisms underlying SLE risk and outcomes among and within ethnic groups are complex, involving biological, sociocultural, physical and other environmental exposures. Differences in disease risk allele frequency in populations might underlie some of the health disparities, as multiple genetic risk factors for SLE vary among populations.4 Additionally, multiple social stressors (eg, poverty, low household income, unemployment, perceived stress, racial discrimination) negatively affect SLE outcomes, while protective factors (eg, social support, healthy lifestyles) can help improve SLE outcomes.1 However, the mechanisms by which these adverse and protective social factors synergistically modulate disease outcomes are not currently well understood.
Most SLE research to date has focused on biological mechanisms independent of the effects of social exposures, and health disparities research has focused primarily on the influence of socioeconomic determinants on outcomes without considering the biological mechanisms involved. This has resulted in a knowledge gap regarding the interactions among individual and social factors that contribute to disparities in SLE outcomes. We propose a socioecological model of SLE outcomes that emphasises the importance of integrating sociocultural and individual determinants to understand and address health disparities in SLE.1
Despite the influence that social or environmental experiences have on SLE in African American women, it is not known how these experiences influence disease outcomes. We postulate that in African American women, exposure to adverse and protective social contexts is associated with epigenomic changes that in turn are associated with disease outcomes. We specifically hypothesise that social support compensates for the detrimental, independent effect of racism on SLE, considering other sociodemographic and behavioural characteristics, through epigenetic and gene regulatory mechanisms. We will investigate the role of DNA methylation (DNAm) in mediating the effects of social exposures on SLE.
DNAm can respond to multiple environmental stimuli, such as exercise, diet, smoking or pollutants. In addition, altered DNAm patterns are associated with a broad range of age-related diseases, including cancer, cardiovascular disease and Alzheimer disease.5 DNAm variation is also associated with psychosocial factors, including socioeconomic status and general perceived stress.6–8 Additionally, although DNAm varies between populations, and this variation is partially explained by their distinct genetic ancestry, environmental factors not captured by genetic ancestry are significant contributors to variation in DNAm, underscoring the notion that an interaction between social, genetic and epigenetic factors underlies the health disparity in SLE.9–16
Epigenetic age acceleration, that is, the difference between chronological age and epigenetic age (estimated by subsets of DNA-methylated sites associated with chronological age) is associated with multiple disorders including blood pressure, cancer and osteoarthritis.5 Epigenetic age acceleration also has been linked to social factors including economic hardship,17 lifetime stress,18 education19 and adversity (education, income, neighbourhood disadvantage and discrimination).20 Early childhood exposure to abuse, financial hardship or neighbourhood disadvantage is associated with epigenetic age acceleration,21 and even maternal preconception adverse childhood experiences have been associated with epigenetic ageing in their offspring.22 Perceived racial discrimination is associated with accelerated epigenetic ageing among African Americans.23 Other biological markers for age acceleration, such as telomere length, also suggest there is accelerated biological ageing associated with experiencing racial discrimination.24–26 Although epigenetic age acceleration has not been reported in SLE, accelerated telomere shortening has been previously noted in rheumatic diseases,27 including with SLE within our local study population.28
The goal of this study is to identify epigenetic biomarkers associated with positive and negative social factors that affect gene expression and therefore influence SLE in African American women. To understand the effects of positive and negative social environments on SLE through epigenomic changes, we will test a conceptual model that integrates multiple social, demographic, behavioural, genomic, epigenomic and transcriptomic factors. The research will identify the epigenetic biomarkers by which risk and resiliency factors affect gene function and thereby influence SLE in African American women.
Methods and analysis
Social Factors, Epigenomics and Lupus in African American Women Study overview
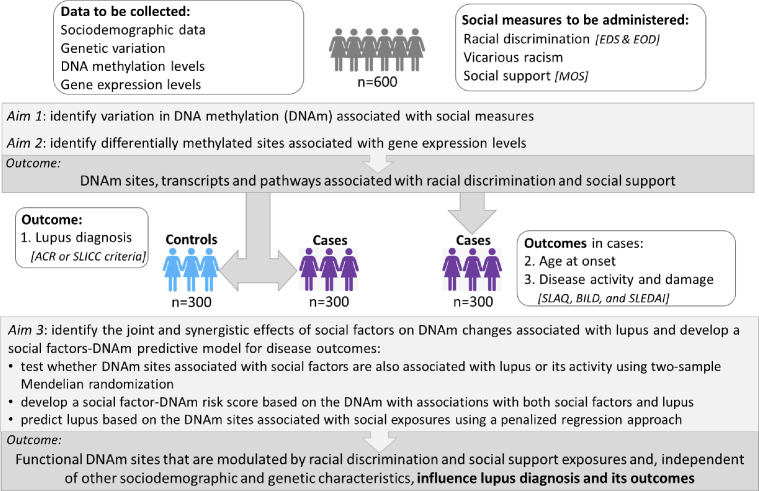
Social Factors, Epigenomics and Lupus in African American Women (SELA) is an observational study whose goal is to evaluate the effects of racial discrimination and social support on SLE outcomes through changes in DNAm and gene expression (figure 1). The study has three major aims: (aim 1) to identify variation in DNAm associated with self-reported (a) exposure to racial discrimination, (b) exposure to social support, and (c) assess whether these exposures and SLE are associated with epigenetic age acceleration; (aim 2) to assess whether social DNAm sites affect gene expression; (aim 3) to identify the synergistic effects of social factors on DNAm changes on SLE and develop a social factors-DNAm predictive model for disease outcomes.
Figure 1.
Overall SELA Study description. The goal of this study is to identify functional DNA methylation (DNAm) and/or mediated effects of social factors with effects on the likelihood of lupus and its outcomes. The primary outcomes are noted in the dark shaded boxes. ACR, American College of Rheumatology; BILD, Brief Index of Lupus Damage; EDS, Everyday Discrimination Scale; EOD, Experiences of Discrimination; MOS, Medical Outcomes Study; SELA, Social Factors, Epigenomics and Lupus in African American Women; SLAQ, Systemic Lupus Activity Questionnaire; SLEDAI, Systemic Lupus Erythematosus Disease Activity Index; SLICC, Systemic Lupus Erythematosus International Collaborating Clinics.
SELA involves three sites. The Medical University of South Carolina (MUSC) is the coordinating and recruitment centre. Wake Forest School of Medicine will generate the molecular data and lead the statistical and bioinformatics analyses of the genotypic, DNAm and RNA-seq data. Emory University will lead the racial discrimination aspects of this project. A total of 600 participants will be enrolled, 300 cases and 300 controls. Recruitment began in January 2022 and will be ongoing for 3 years. Data collection will finish during the third year, and final analyses of the primary outcomes of this study will take place during the fourth year. These primary outcomes (figure 1) include the identification of DNAm sites and gene transcripts associated with exposure to racial discrimination and exposure to social support, and the identification of causal relationships between DNAm sites and SLE or activity.
Study population
SLE disproportionately affects African Americans and women, so it is important to study these health disparity populations. In addition to SLE, social stressors like racial discrimination also place a disproportionate burden on African American women.29–31 Since the SLE disparity is already established in African American women, this study focuses on this disparity group, minimising confounding effects due to ethnicity and gender. Healthy adult controls (age matched) will be invited to volunteer to respond to questionnaires and donate blood.
Eligibility criteria for patients with SLE and controls are shown in tables 1 and 2. Patients will be primarily recruited from a longitudinal registry of patients for research studies at the MUSC.
Table 1.
Eligibility criteria for cases
| Inclusion criteria | Exclusion criteria |
|
|
PI, Principal Investigator.
Table 2.
Eligibility criteria for controls
| Inclusion criteria | Exclusion criteria |
|
|
PI, Principal Investigator.
All control participants will be recruited via approved advertisements in print and electronic format, including social media platforms, through word of mouth from patients with SLE or from our longitudinal registry of volunteers for research studies. Information about the research study will be provided at educational events for the public. Controls will be asked questions about potential symptoms of autoimmune disease as part of the Connective Tissue Disease Screening Questionnaire, which has been validated in African American populations.32 33 Controls who present multiple signs or symptoms of autoimmune disease or are on current steroid or immunosuppressive therapy will be provided resources for healthcare evaluation and intervention and excluded from analyses. Although the anticipated number of individuals on current steroid or immunosuppressive therapy is small, a sensitivity analysis will inform the exclusion of cases on steroids or immunosuppression for other conditions than SLE. Controls will be self-reported African American women without a history of SLE, systemic sclerosis or other connective tissue diseases that are matched based on patients’ age (±5 years).
Participant enrolment and interview
For study enrolment, 300 self-reported African American women with SLE and 300 age-matched African American female controls will be recruited. The informed consent document is sent to potential participants prior to the screening visit, either by mail or email, for their review as requested. Currently, written informed consent of the participant is obtained at the screening visit. We are seeking approval to allow the participants who prefer to complete the informed consent electronically prior to the visit following MUSC-approved platforms (ie, Doxy.me, REDCap). Once approved, and if preferred by the participant, she can receive questionnaires electronically through the REDCap eSurvey system for completion prior to the visit. Any questions the participant might have will be answered by the study coordinator, physician or investigator as appropriate, and she will be specifically informed that consent to donate a specimen will in no way obligate her to do so at a future date, nor affect her care in any way. The consent form will allow the participants to consent to future research use and submission of their data to publicly accessible data repositories, hence allowing the sharing of individual, de-identified data. The participants will be informed that their global genetic ancestry estimates will be generated as part of this study and will agree or decline to receive their genetic ancestry composition results after the data are analysed. If they agree to participate, the study coordinator asks the participant to sign the combined informed consent with Health Insurance Portability and Accountability Act document. No study procedures are performed prior to obtaining written informed consent. Once the participants give written informed consent, participants will be asked to complete questionnaires and donate biospecimens (see below).
Questionnaires
The questionnaires will capture sociodemographic and behavioural information (eg, education, income, occupation, marital status, health insurance, educational attainment, smoking, alcohol use, physical activity, chronic illnesses). Gender identity will be asked (eg, cis-woman, trans-woman). Since different individuals might prefer more specific terms when defining their race, participants will be asked to describe how they prefer to self-report (eg, black Caribbean, Afro-Latino). They will also be asked if they consider themselves Sea Islanders, and their parents’ place of birth.
The questionnaires include two validated measures of racial discrimination, one measure of social support and one measure of depression (table 3).30 34–37 Our primary measure of racial discrimination will be the validated Experiences of Discrimination measure.34 We will augment this measure with the validated Everyday Discrimination Scale.37 We will also perform the same analyses using the measure of vicarious racism stress we previously used; this measure has high internal consistency and reliability (Cronbach α=0.83).30 All participants will be asked to respond to healthcare utilisation and lost productivity questionnaire,38–40 and to a brief debriefing questionnaire to identify potential distress from responding to the questionnaires. If the participant has a positive screen, we have compiled a list of mental health resources that the study coordinator will be able to provide to the participant. All participants will also receive a short questionnaire to assess their interest in being included in this study’s progress and involved in our research.
Table 3.
Questionnaires used to assess racial discrimination, vicarious racism stress, social support, depression and SLE disease activity
| Racial discrimination | Experiences of Discrimination34 |
| Everyday Discrimination Scale37 | |
| Vicarious racism stress | Vicarious racism stress questionnaire from BeWell Study30 |
| Social support | Medical Outcomes Study-Social Support Survey35 |
| Depression | Patient Health Questionnaire-836 |
| SLE disease activity | Systemic Lupus Activity Questionnaire39 |
| SLE disease damage | Brief Index of Lupus Damage38 |
In addition, all patients with SLE will also complete validated questionnaires that assess disease activity (Systemic Lupus Activity Questionnaire (SLAQ))39 and disease damage (Brief Index of Lupus Damage).38 The study rheumatologist will evaluate and assess the patients using the Systemic Lupus Erythematosus Disease Activity Index 2000 (SLEDAI-2K).41
Biospecimens
Peripheral blood mononuclear cells (PBMCs), plasma and serum will be isolated from 30 mL of blood collected from participants who agree to participate in this research study. In order to get a current assessment of disease activity (SLEDAI), urine will also be requested from new patients with SLE and MUSC patients without recent (within 6 months) visits.
Cell subset composition will be assessed in order to generate reference matrices for the deconvolution of the PBMC methylomic and transcriptomic data. The main PBMC populations (T cells, B cells, natural killer cells and monocytes) from each participant will be counted using analytical flow cytometry. Genetic material (DNA and RNA) will be isolated from the PBMCs.
Data generation and analyses
Using genomic DNA or RNA isolated from PBMCs, the Infinium Global Diversity Array (Illumina) will be used for genotyping; the MethylationEPIC BeadChip (Illumina) will be used to assess DNA methylation levels and RNA-seq will be used to measure transcript levels. These assays will be performed at Wake Forest School of Medicine. For genome-wide single nucleotide polymorphism (SNP) genotyping, variants with missing position, missing allele, allele mismatch, call rates ≤95%, departure from Hardy-Weinberg equilibrium (HWE) (p<10-6) and minor allele frequency <0.01 will be removed. Samples will be excluded if showing low call rates (≤95%), gender discordance, DNA contamination or if duplicated. Genetic variants that pass quality control filters will be imputed to the African Genome Resources Panel. These genotypic data will be used for inference of genetic ancestry and identification of population stratification. The genetic ancestry estimates will be shared with individual participants if requested during the consent process.
For genome-wide DNAm from PBMCs, data will be analysed with the R package Chip Analysis Methylation Pipeline (ChAMP)42; probes will be excluded if the bead count <3, the CpG site assayed includes a common SNP or the detection p>0.05. The data will be normalised using beta-mixture quantile normalisation method. To adjust for potential batch effects, we will use ChAMP42 to employ ComBat, which uses Empirical Bayes’ methods to correct for technical variation. For genome-wide RNA-seq from PBMCs, Illumina’s TruSeq Stranded Total RNA kit will be used for rRNA reduction followed by stranded preparation and sequencing at 40M reads per sample at PE50. After FastQC is used for stringent quality control, reads will be aligned to the gene annotations of UCSC hg19 using STAR.43 FeatureCounts44 will be used to generate count data for each sample. These data will be examined for batch and time effects and corrected.
The methylation and gene expression data will be deconvoluted in order to adjust for cell type heterogeneity.45 46 Currently, reference data sets do not match the age, gender, clinical characteristics or ancestry of the individuals under study.47 Of relevance, chronic and acute stressors might also alter blood cell composition, further underscoring the importance of this confounding source of variability when conducting epigenomic and transcriptomic studies using heterogeneous cell mixtures. We will improve the statistical deconvolution by incorporating cell counts from our samples.
To identify DNAm sites associated with life course racial discrimination, we will compute a linear regression model with levels of racial discrimination as a predictor of the methylation level at each CpG site, controlling for age, smoking, white blood cell proportions, SLE status, medication use, and Principal Components or admixture proportions. Similar regression analysis will be computed to test for an association between DNAm at individual CpG sites and levels of social support. Power analyses were calculated using the power evaluation tool pwrEWAS48 available on GitHub. To detect differences up to 5% and 10% in CpG-specific methylation across 2500 CpGs between groups (1:1 case:control ratio) with at least 80% power, about 280 and 125 total subjects are needed, respectively. For a total sample size of 280 subjects, half are needed who report the social exposure outcome (ie, racial discrimination or social support). Hence, even of only 24% of all participants report racial discrimination or social support, this study is well powered to detect differences up to 5% in DNAm.
To identify differentially expressed genes associated with (1) racial discrimination and (2) social support, we will use DESeq249 and edgeR50 adjusting for age, smoking, estimated white blood cell proportions, SLE status and medication use as covariates. Power analyses were calculated using the Bioconductor RNASeqPower package.51 Using 300 samples per group and α=0.0001 (which based on our data is a good approximation to false discovery rate (FDR)=0.05), we are well powered to detect differential expression of fold change ≥1.28.
We will also test whether multiple measures of epigenetic age acceleration (eg, DNAm PhenoAge52) are associated with exposure to racial discrimination, exposure to social support, SLE diagnosis, SLE age of onset and disease activity (as measured by the SLAQ39 and SLEDAI41 scores).
In order to understand the mechanisms and functional consequences of socially induced epigenetic changes, we will identify associations between methylation levels and gene expression levels, that is, expression quantitative trait methylation (eQTM) loci. We will compute linear regression models with the methylation level of each CpG site as a predictor of transcript expression for any gene within 250 kb of the CpG site, adjusting for age, cell proportions and Principal Components. It is unclear how many differentially methylated regions associated with social exposures are likely to be detected. However, if we assume that we will detect 200 differentially methylated regions, with an average of three genes within 250 kb from the top associated CpG site and therefore a Bonferroni-adjusted type 1 error rate of α=0.00008 (0.05/600), then in the entire sample (n=600) the study has 80% power to detect eQTMs that explain 3.7% of the variance in log2 expression. Assuming that 40% (n=240) to 60% (n=360) of all participants report racial discrimination (or social support), then the study has 80% power to detect eQTMs that explain 9.0% and 6.1% of the variance in log2 expression in that subset of individuals, respectively.
Given the effects of genetic variation on DNAm and gene expression, as secondary analyses, we will also identify associations between genetic variation and methylation levels (cis-methylation quantitative trait loci (meQTL)) and associations between genetic variation and gene expression levels (cis-expression quantitative trait loci (eQTL)). Joint effects of racial discrimination and social support on DNAm will be modelled through a DNAm regression model that includes social support, racial discrimination and their interaction, controlling for age, smoking, white blood cell proportions, SLE status, medication use, and principal components or admixture proportions. We will use two-sample Mendelian randomisation to identify CpG sites that may mediate the effect of social exposures (racial discrimination and social support) on SLE, including admixture estimates as covariates. We will test if biological pathway-driven DNAm risk scores (eg, weighted averages of identified DNAm, where weights are the effect size) correlate with SLE or SLE activity (as measured by the SLAQ39 and SLEDAI41 scores). Finally, we will use machine learning methods (ie, penalised regression) to develop a social factor-DNAm predictive model for SLE and SLE activity. The power to detect DNAm sites associated with social exposures and with gene expression changes using the two-sample Mendelian randomisation analysis was evaluated using the approach described by Burgess.53 Given a sample of 300 cases and 300 controls and assuming R2=0.1, we have 80% power to detect OR ≥2.1 using unadjusted significance level α=0.05 or an OR ≥2.75 using a Bonferroni-adjusted significance of α=0.0025 for 20 comparisons.
Conceptual model
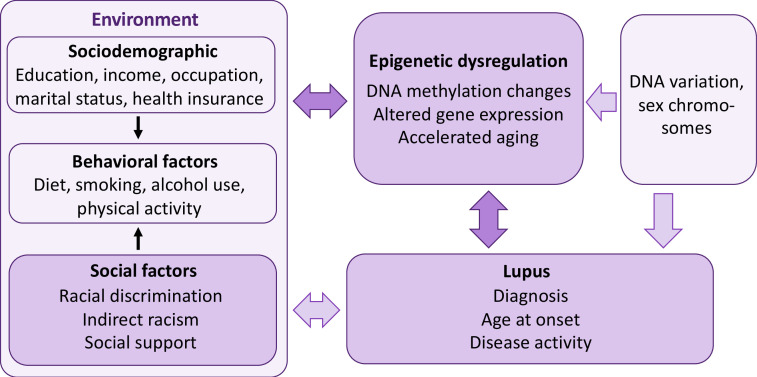
Our conceptual model (figure 2) is based on the Biopsychosocial Model, which asserts that African Americans who perceive certain circumstances as racist experience physiological stress responses that can be modulated by adverse or protective sociodemographic (eg, socioeconomic status) and psychological characteristics (eg, depression), behavioural factors (eg, smoking, alcohol use) and coping responses (eg, ability to mobilise social support) to such experiences.54 This conceptual model for African American women integrates social determinants of health with biological factors. Although not a comprehensive model of SLE risk factors, it models how interdependencies among genetic background, environmental exposures and epigenetic signatures may contribute to increased risk of SLE in African American women. The model (figure 2) depicts how genetic factors (which include ancestry-related variation in allele frequencies and sex) can increase the risk of SLE directly, or through variation in DNA methylation and gene expression. Our analyses will account for this genetic variation by adjusting for genetic ancestry and sex in the model. Since genetic ancestry partly accounts for variation in DNAm, it is critical to account for genetic variation among the participants.9–16 The model also shows how multiple environmental factors, including social exposures, can directly affect or indirectly affect, through epigenetic dysregulation, risk for SLE. Lupus can also directly affect the response to environmental exposures, causing epigenetic changes. These reverse causation and confounding factors will be considered during analyses.
Figure 2.
Study description. We posit that social support compensates for the detrimental, independent effect of racism on lupus outcomes, taking into account other sociodemographic and behavioural characteristics, through epigenetic and gene regulatory mechanisms. We will: (1) identify DNA methylation (DNAm) variation associated with exposure to racial discrimination and to social support; (2) assess whether these DNAm sites influence gene expression, taking genetic variation into account; (3) identify the joint and synergistic effects of social factors on DNAm changes associated with lupus outcomes; and (4) develop a social factors-DNAm predictive model for disease outcomes.
Community engagement and patient involvement
Community engagement is essential to advance understanding of and eliminate racial and ethnic disparities impacting patients with SLE, particularly since economic and social determinants of health, such as poverty, discrimination and community-level social stressors account for over 40% of the modifiable contributors to healthy outcomes for a population.55 Our SLE genetics projects involve African American community members from rural South Carolina and are conducted in cooperation with the Sea Island Families Project (SIFP) Citizen Advisory Committee (CAC).56 Interdisciplinary research teams from the MUSC developed community-engaged research projects between academic researchers and Gullah African Americans residing in rural South Carolina, leading to the formation of SIFP-CAC.56 This 28-year partnership meets quarterly for sharing of research results and providing guidance and recommendations to new research. The proposed community-engaged study was approved by and will be conducted in cooperation with the SIFP-CAC.
During the study visit, participants will be asked about their willingness to partner with us in research studies trying to understand the causes of autoimmune diseases like SLE. Specifically, they will be asked the following three questions: (1) ‘Would you like to partner with us, participating with suggestions for and feedback on research studies during annual or biannual meetings?’; (2) ‘If you are not willing or able to commit to a partnership, are there any research questions or feedback that you have about issues that are important to you, regarding the research we do?’; and (3) ‘Would you like to be included in annual newsletters reporting the progress of this research study?’ Participants who respond affirmatively will be involved as they choose.
Discussion
There is increasing awareness and need for integrative mechanistic studies that examine the dynamic interplay of multiple factors across the life course, in order to better understand and address the drivers of health disparities and inform the development of effective interventions. This SELA Study was developed in response to PAR-19-372 (Social Epigenomics Research Focused on Minority Health and Health Disparities), whose purpose is to support epigenomic investigations focused on identifying and characterising the mechanisms by which social context and experiences, both positive and negative, affect gene function and thereby influence health trajectories or modify disease risk in health disparity populations.
The conceptual framework for this study is based on the socioecological model that emphasises the importance of integrating societal, community, interpersonal and individual determinants to understand and address health disparities in SLE.1 Social determinants of health span the socioeconomic (employment, income, housing and food security), community (family and social support), neighbourhood and physical environment (access to food and housing, crime and violence, safety, transportation, air and water quality), and the healthcare system (access, quality). Individual determinants include genetic (sex chromosomes, DNA, epigenetic and gene expression variation) and behavioural factors (diet, smoking, alcohol use, physical and mental health). Since exposures and experiences vary across individuals from different populations, locations and cultures, it is critical to study population differences in SLE health disparities within the sociocultural context. This is further underscored by both the paucity of disadvantaged communities in research and the genetic and cultural heterogeneity of racial and ethnic groups.
Innovative aspects of this study include the focus on culturally distinct Gullah and non-Gullah African American women, the community partnership, and the integrative analysis of multiple individual and social factors, including risk and protective social effects. This integrative, multiomic research that integrates social and genomic data requires complementary expertise from multiple health centres partnered together to provide expertise in minority health and health disparities, social epidemiology, SLE, genomics, epigenomics, transcriptomics and statistical methods.
Allowing the participants to further describe how they self-identify beyond the generic ‘black or African American’ racial category or expand on their gender identities beyond the dichotomous male/female categories allows for more inclusive participation. Indeed, close to 2% of all individuals are born without being clearly sexed,57 and according to the 2020 US Census Bureau, ‘Other’ is now the second most common racial group in the USA, and the third most common in South Carolina together with ‘Two or More Races’.58 59 We expect that the increased inclusivity and more granular data might help interpret the results of this study. The option to receive their genetic ancestry estimates has been welcomed by the participants so far.
A potential limitation of this study is the lack of previous research linking specific experiences or behaviours to epigenetic changes in SLE. However, mounting evidence across several traits suggests that epigenetic mechanisms may provide a causal link between social adversity and health disparity.60 Another limitation is the current lack of published genome-wide association studies (GWAS) of SLE in patients with African ancestry, thus current polygenic risk score data for African Americans are lacking. If GWAS data in African Americans become available during this study, we will integrate the SLE African American polygenic risk scores as potential confounders in our models.
This study will identify epigenetic biomarkers by which risk and resiliency factors affect gene function and thereby influence SLE in African American women—the most vulnerable and susceptible group to this prototypical autoimmune disease without any safe and effective treatments. The epigenetic biomarkers identified in this study can be used prospectively as biomarkers of previous social exposures to identify individuals at risk of SLE or worse outcomes. Knowledge of the effects of social exposures on biological changes can be leveraged by advocacy efforts to develop psychosocial interventions that prevent or mitigate risk exposures, and services or interventions that promote positive exposures. Implementation of these novel treatments and preventative interventions, as supported by the results of this study, is paramount to the closure of the health disparities gap. Due to the shared aetiological mechanisms, the implications of this research extend across autoimmune diseases and beyond, as an overarching paradigm of the mechanisms for how social stressors physiologically affect human health.
Footnotes
Twitter: @PSRamos_PhD
Contributors: ELV wrote the original draft preparation. CDL and BJW contributed to methodology. QQ, LHM, LQK, IDM, SLB and LAU contributed to investigation. LAU, DLK and PSR contributed to supervision. PSR contributed to project administration. CDL, BJW, TDH, GAH, SSL, EMW, DLK and PSR contributed to the conceptualisation of this study and funding acquisition. All authors reviewed, edited and agreed on the final version of the manuscript.
Funding: This work is supported by the US National Institute on Minority Health and Health Disparities of the National Institutes of Health (NIH) under Award Number R01 MD015395 (CDL, BJW, TDH, GAH, LQK, IDM, SSL, EMW, DLK, PSR), by the US National Institute of Arthritis and Musculoskeletal and Skin Diseases under Award Numbers T32 AR050958 (ELV), P30 AR072582 (BJW, SLB, LAU, EMW, DLK, PSR) and K24 R068406 (DLK), and by the National Center for Chronic Disease Prevention and Health Promotion (NCCDPHP) under Award Number U01 DP006488 (SSL).
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
Data sharing not applicable as no data sets generated and/or analysed for this study.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
This study was approved by the Institutional Review Board at the Medical University of South Carolina (Pro# 112945). Informed consent will be obtained from all participants. All research included in this manuscript conforms with the Declaration of Helsinki. Progress and results will be presented at national or international conferences, to colleagues at seminars and talks, to the community at patient education events and the study participants through annual newsletters. Results will be published in peer-reviewed journals in a timely fashion. The investigators will abide by the recent 'Updated Guidance on the Reporting of Race and Ethnicity in Medical and Science Journals' published in JAMA (Flanagin et al, 2021; https://pubmed.ncbi.nlm.nih.gov/34402850/). All final peer-reviewed manuscripts that arise from this proposal will be submitted to the digital archive PubMed Central. Research data that document, support and validate research findings will be made available at the same time the main findings from the final research data set have been accepted for publication. The investigators will work to facilitate any request made for data produced under this proposal upon publication of data, using standard, university-approved material/data transfer agreements. The genotyping data will be deposited in the National Institutes of Health (NIH) database of Genotypes and Phenotypes (dbGaP), and the DNA methylation and RNA-seq data will be deposited for public access in NCBI’s Gene Expression Omnibus (GEO) database. No identifying information will accompany these data. According to the Extramural Institutional Certification of the genotypic data submitted to dbGaP, these data are considered to have particular ‘sensitivities’ related to potential for group harm and will only to be made available through controlled access. In addition to the genomic data, relevant associated data (eg, phenotype, study protocols) will be concomitantly submitted.
References
- 1.Ramos PS. Integrating genetic and social factors to understand health disparities in lupus. Curr Opin Rheumatol 2021;33:598–604. 10.1097/BOR.0000000000000840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yen EY, Singh RR. Brief report: Lupus-An unrecognized leading cause of death in young females: a population-based study using nationwide death certificates, 2000-2015. Arthritis Rheumatol 2018;70:1251–5. 10.1002/art.40512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim SS, Helmick CG, Bao G, et al. Racial Disparities in Mortality Associated with Systemic Lupus Erythematosus - Fulton and DeKalb Counties, Georgia, 2002-2016. MMWR Morb Mortal Wkly Rep 2019;68:419–22. 10.15585/mmwr.mm6818a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanata CM, Blazer A, Criswell LA. The contribution of genetics and epigenetics to our understanding of health disparities in rheumatic diseases. Rheum Dis Clin North Am 2021;47:65–81. 10.1016/j.rdc.2020.09.005 [DOI] [PubMed] [Google Scholar]
- 5.Seale K, Horvath S, Teschendorff A, et al. Making sense of the ageing methylome. Nat Rev Genet 2022. 10.1038/s41576-022-00477-6. [Epub ahead of print: 02 May 2022]. [DOI] [PubMed] [Google Scholar]
- 6.Lam LL, Emberly E, Fraser HB, et al. Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci U S A 2012;109 Suppl 2:17253–60. 10.1073/pnas.1121249109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borghol N, Suderman M, McArdle W, et al. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol 2012;41:62–74. 10.1093/ije/dyr147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vidal AC, Benjamin Neelon SE, Liu Y, et al. Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans. Genet Epigenet 2014;6:GEG.S18067–44. 10.4137/GEG.S18067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barfield RT, Almli LM, Kilaru V, et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol 2014;38:231–41. 10.1002/gepi.21789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galanter JM, Gignoux CR, Oh SS, et al. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. Elife 2017;6. 10.7554/eLife.20532. [Epub ahead of print: 03 01 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husquin LT, Rotival M, Fagny M, et al. Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol 2018;19:222. 10.1186/s13059-018-1601-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopalan S, Carja O, Fagny M, et al. Trends in DNA methylation with age replicate across diverse human populations. Genetics 2017;206:1659–74. 10.1534/genetics.116.195594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fagny M, Patin E, MacIsaac JL, et al. The epigenomic landscape of African rainforest hunter-gatherers and farmers. Nat Commun 2015;6:10047. 10.1038/ncomms10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heyn H, Moran S, Hernando-Herraez I, et al. DNA methylation contributes to natural human variation. Genome Res 2013;23:1363–72. 10.1101/gr.154187.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods 2013;10:949–55. 10.1038/nmeth.2632 [DOI] [PubMed] [Google Scholar]
- 16.Quach H, Rotival M, Pothlichet J, et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 2016;167:643–56. 10.1016/j.cell.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simons RL, Lei MK, Beach SRH, et al. Economic hardship and biological weathering: the epigenetics of aging in a U.S. sample of black women. Soc Sci Med 2016;150:192–200. 10.1016/j.socscimed.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zannas AS, Arloth J, Carrillo-Roa T, et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol 2015;16:266. 10.1186/s13059-015-0828-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiorito G, Polidoro S, Dugué P-A, et al. Social adversity and epigenetic aging: a multi-cohort study on socioeconomic differences in peripheral blood DNA methylation. Sci Rep 2017;7:16266. 10.1038/s41598-017-16391-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons RL, Lei M-K, Klopack E, et al. The effects of social adversity, discrimination, and health risk behaviors on the accelerated aging of African Americans: further support for the weathering hypothesis. Soc Sci Med 2021;282:113169. 10.1016/j.socscimed.2020.113169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marini S, Davis KA, Soare TW, et al. Adversity exposure during sensitive periods predicts accelerated epigenetic aging in children. Psychoneuroendocrinology 2020;113:104484. 10.1016/j.psyneuen.2019.104484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nwanaji-Enwerem JC, Van Der Laan L, Kogut K, et al. Maternal adverse childhood experiences before pregnancy are associated with epigenetic aging changes in their children. Aging 2021;13:25653–69. 10.18632/aging.203776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brody GH, Miller GE, Yu T, et al. Supportive Family Environments Ameliorate the Link Between Racial Discrimination and Epigenetic Aging: A Replication Across Two Longitudinal Cohorts. Psychol Sci 2016;27:530–41. 10.1177/0956797615626703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee DB, Kim ES, Neblett EW. The link between discrimination and telomere length in African American adults. Health Psychol 2017;36:458–67. 10.1037/hea0000450 [DOI] [PubMed] [Google Scholar]
- 25.Chae DH, Epel ES, Nuru-Jeter AM, et al. Discrimination, mental health, and leukocyte telomere length among African American men. Psychoneuroendocrinology 2016;63:10–16. 10.1016/j.psyneuen.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chae DH, Wang Y, Martz CD, et al. Racial discrimination and telomere shortening among African Americans: the coronary artery risk development in young adults (cardia) study. Health Psychol 2020;39:209–19. 10.1037/hea0000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dehbi AZA, Radstake TRDJ, Broen JCA. Accelerated telomere shortening in rheumatic diseases: cause or consequence? Expert Rev Clin Immunol 2013;9:1193–204. 10.1586/1744666X.2013.850031 [DOI] [PubMed] [Google Scholar]
- 28.Hoffecker BM, Raffield LM, Kamen DL, et al. Systemic lupus erythematosus and vitamin D deficiency are associated with shorter telomere length among African Americans: a case-control study. PLoS One 2013;8:e63725. 10.1371/journal.pone.0063725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chae DH, Martz CD, Fuller-Rowell TE, et al. Racial discrimination, disease activity, and organ damage: the black women's experiences living with lupus (BeWELL) study. Am J Epidemiol 2019;188:1434–43. 10.1093/aje/kwz105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martz CD, Allen AM, Fuller-Rowell TE, et al. Vicarious racism stress and disease activity: the black women's experiences living with lupus (BeWELL) study. J Racial Ethn Health Disparities 2019;6:1044–51. 10.1007/s40615-019-00606-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spears EC, Allen AM, Chung KW, et al. Anticipatory racism stress, smoking and disease activity: the black women's experiences living with lupus (BeWELL) study. J Behav Med 2021;44:760–71. 10.1007/s10865-021-00235-9 [DOI] [PubMed] [Google Scholar]
- 32.Karlson EW, Sanchez-Guerrero J, Wright EA, et al. A connective tissue disease screening questionnaire for population studies. Ann Epidemiol 1995;5:297–302. 10.1016/1047-2797(94)00096-C [DOI] [PubMed] [Google Scholar]
- 33.Karlson EW, Costenbader KH, McAlindon TE, et al. High sensitivity, specificity and predictive value of the connective tissue disease screening questionnaire among urban African-American women. Lupus 2005;14:832–6. 10.1191/0961203305lu2227oa [DOI] [PubMed] [Google Scholar]
- 34.Krieger N, Smith K, Naishadham D, et al. Experiences of discrimination: validity and reliability of a self-report measure for population health research on racism and health. Soc Sci Med 2005;61:1576–96. 10.1016/j.socscimed.2005.03.006 [DOI] [PubMed] [Google Scholar]
- 35.Sherbourne CD, Stewart AL. The mos social support survey. Soc Sci Med 1991;32:705–14. 10.1016/0277-9536(91)90150-B [DOI] [PubMed] [Google Scholar]
- 36.Kroenke K, Spitzer RL, Williams JB. The PHQ-9: validity of a brief depression severity measure. J Gen Intern Med 2001;16:606–13. 10.1046/j.1525-1497.2001.016009606.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams DR, Yan Yu, Jackson JS, et al. Racial differences in physical and mental health: socio-economic status, stress and discrimination. J Health Psychol 1997;2:335–51. 10.1177/135910539700200305 [DOI] [PubMed] [Google Scholar]
- 38.Yazdany J, Trupin L, Gansky SA, et al. Brief index of lupus damage: a patient-reported measure of damage in systemic lupus erythematosus. Arthritis Care Res 2011;63:1170–7. 10.1002/acr.20503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlson EW, Daltroy LH, Rivest C, et al. Validation of a systemic lupus activity questionnaire (SLAQ) for population studies. Lupus 2003;12:280–6. 10.1191/0961203303lu332oa [DOI] [PubMed] [Google Scholar]
- 40.Katz P, Trupin L, Rush S, et al. Longitudinal validation of the brief index of lupus damage. Arthritis Care Res 2014;66:1057–62. 10.1002/acr.22268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bombardier C, Gladman DD, Urowitz MB, et al. Derivation of the sledai. A disease activity index for lupus patients. Arthritis & Rheumatism 1992;35:630–40. 10.1002/art.1780350606 [DOI] [PubMed] [Google Scholar]
- 42.Morris TJ, Butcher LM, Feber A, et al. Champ: 450k CHIP analysis methylation pipeline. Bioinformatics 2014;30:428–30. 10.1093/bioinformatics/btt684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, Davis CA, Schlesinger F, et al. Star: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W. featureCounts: an efficient General purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 45.Houseman EA, Kim S, Kelsey KT, et al. Dna methylation in whole blood: uses and challenges. Curr Environ Health Rep 2015;2:145–54. 10.1007/s40572-015-0050-3 [DOI] [PubMed] [Google Scholar]
- 46.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 2014;15:R31. 10.1186/gb-2014-15-2-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahmani E, Schweiger R, Shenhav L, et al. BayesCCE: a Bayesian framework for estimating cell-type composition from DNA methylation without the need for methylation reference. Genome Biol 2018;19:141. 10.1186/s13059-018-1513-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graw S, Henn R, Thompson JA, et al. pwrEWAS: a user-friendly tool for comprehensive power estimation for epigenome wide association studies (EWAS). BMC Bioinformatics 2019;20:218. 10.1186/s12859-019-2804-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hart SN, Therneau TM, Zhang Y, et al. Calculating sample size estimates for RNA sequencing data. J Comput Biol 2013;20:970–8. 10.1089/cmb.2012.0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018;10:573–91. 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol 2014;43:922–9. 10.1093/ije/dyu005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark R, Anderson NB, Clark VR. Racism as a stressor for African Americans. A biopsychosocial model. Am Psychol 1999;54:805–16. [DOI] [PubMed] [Google Scholar]
- 55.Hood CM, Gennuso KP, Swain GR, et al. County health rankings: relationships between determinant factors and health outcomes. Am J Prev Med 2016;50:129–35. 10.1016/j.amepre.2015.08.024 [DOI] [PubMed] [Google Scholar]
- 56.Spruill IJ, Leite RS, Fernandes JK, et al. Successes, challenges and lessons learned: Community-engaged research with South Carolina's Gullah population. Gateways 2013;6. 10.5130/ijcre.v6i1.2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blackless M, Charuvastra A, Derryck A, et al. How sexually dimorphic are we? review and synthesis. Am J Hum Biol 2000;12:151–66. [DOI] [PubMed] [Google Scholar]
- 58.Jones N, Marks R, Ramirez R. 2020 census illuminates racial and ethnic composition of the country, 2021. Available: https://www.census.gov/library/stories/2021/08/improved-race-ethnicity-measures-reveal-united-states-population-much-more-multiracial.html
- 59.Staff . South Carolina gained almost half a million people last decade: United States census bureau, 2021. Available: https://www.census.gov/library/stories/state-by-state/south-carolina-population-change-between-census-decade.html
- 60.Peschken CA. Health disparities in systemic lupus erythematosus. Rheum Dis Clin North Am 2020;46:673–83. 10.1016/j.rdc.2020.07.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable as no data sets generated and/or analysed for this study.