Abstract
Background
Despite advances in B7 homolog 3 protein (B7-H3) based immunotherapy, the development of drug resistance remains a major clinical concern. The heterogeneity and emerging loss of B7-H3 expression are the main causes of drug resistance and treatment failure in targeted therapies, which reveals an urgent need to elucidate the mechanism underlying the regulation of B7-H3 expression. In this study, we identified and explored the crucial role of the transcription factor SPT20 homolog (SP20H) in B7-H3 expression and tumor progression.
Methods
Here, we performed CRISPR/Cas9-based genome scale loss-of-function screening to identify regulators of B7-H3 in human ovarian cancer cells. Signaling pathways altered by SP20H knockout were revealed by RNA sequencing. The regulatory role and mechanism of SP20H in B7-H3 expression were validated using loss-of-function and gain-of-function assays in vitro. The effects of inhibiting SP20H on tumor growth and efficacy of anti-B7-H3 treatment were evaluated in tumor-bearing mice.
Results
We identified SUPT20H (SP20H) as negative and eIF4E as positive regulators of B7-H3 expression in various cancer cells. Furthermore, we provided evidence that either SP20H loss or TNF-α stimulation in tumor cells constitutively activates p38 MAPK-eIF4E signaling, thereby upregulating B7-H3 expression. Loss of SP20H upregulated B7-H3 expression both in vitro and in vivo. Additionally, deletion of SP20H significantly suppressed tumor growth and increased immune cells infiltration in tumor microenvironment. More importantly, antibody–drug conjugates targeting B7-H3 exhibited superior antitumor performance against SP20H-deficient tumors relative to control groups.
Conclusions
Activation of p38 MAPK-eIF4E signaling serves as a key event in the transcription initiation and B7-H3 protein expression in tumor cells. Genetically targeting SP20H upregulates target antigen expression and sensitizes tumors to anti-B7-H3 treatment. Collectively, our findings provide new insight into the mechanisms underlying B7-H3 expression and introduce a potential synergistic target for existing antibody-based targeted therapy against B7-H3.
Keywords: immunotherapy, antigenic modulation, tumor biomarkers
WHAT IS ALREADY KNOWN ON THIS TOPIC
As a famous pan-cancer antigen and immune checkpoint, B7-H3 is a very attractive target for cancer therapeutics.
WHAT THIS STUDY ADDS
For the first time, we applied pooled CRISPR screening to investigate the molecular mechanisms that support B7-H3 expression and identified SP20H as a potential synergistic target in anti-B7-H3 treatment.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE, OR POLICY
This work uncovered the critical molecular mechanism underlying B7-H3 expression and provided a basis for development of future therapeutic strategies targeting B7-H3.
Background
B7 homolog 3 protein (B7-H3, also known as CD276), a member of the immunoglobulin family, is characterized as a pan-cancer antigen that is aberrantly overexpressed in various types of cancer. It was initially characterized as a T cell stimulating protein, which binds to an unknown counter-receptor on activated T cells and increases the production of IFN-γ during T cell activation,1 while an increasing number of recent studies have reported its negative regulatory function in both human and mouse T cell immune responses by suppressing T cells activation.2 3
Aberrant protein expression of B7-H3 is detected in many types of malignancies, while limited protein level is detected in normal human tissues. Whereas transcripts of B7-H3 ubiquitously express in human nonlymphoid and lymphoid organs, these indicate that strict post-transcriptional regulation happens.4 5 Highly expressed B7-H3 can promote tumor progression through immunological and non-immunological mechanisms. Accumulated evidence demonstrates that high levels of B7-H3 affect tumor progression by activating the JAK/STAT3,6 7 PI3K/Akt/mTOR,8 9 and TLR4/NF-κB pathways10 and increasing the expression of IL8, MMP-2/9 and STAT3.11 12 Moreover, B7-H3 is also reported to reprogram cancer glucose metabolism by regulating HK2 and HIFα levels.13 14 Collectively, abnormal expression of B7-H3 correlates with poor prognosis, increased tumor grade and metastasis, resistance to therapy, and decreased overall survival.15 16
The immunosuppressive role of B7-H3 in the tumor microenvironment makes it an appealing immuno-therapy target in cancer therapy. A large number of preclinical assessments of the efficacy and specificity of B7-H3-targeted antibody monotherapy and combination therapies have obtained encouraging outcomes in different cancer types, including hepatocellular carcinomas,17 colon carcinoma,18 ovarian cancer,19 central nervous system cancer,20 and renal cell and bladder carcinoma.21 Clinical trials testing the efficacy of B7-H3-targeted therapies involved monoclonal antibodies (mAbs) (NCT02982941), CAR-T cells (NCT04483778; NCT04185038), antibody-conjugated drugs (NCT03729596), and synergistic therapies (NCT04634825; NCT04637503). Among them, enoblituzumab (MGA271; anti-B7-H3 antibody) and radioiodinated anti-B7-H3 murine mAb I-omburtamab have been reported to be well tolerated with minimal toxicities in phase I studies and feasible for further combinatory therapies.22 23
However, despite significant advances in B7-H3-targeted immunotherapies, the mechanisms underlying the regulation of B7-H3 expression in cancer cells are not fully understood. Existing studies have reported discrepancies in the mechanisms underlying the induction and maintenance of B7-H3 expression within immune cells and tumor cells. Scholars such as Chapoval et al have confirmed that inflammatory cytokines such as GM-CSF and IFN-γ, LPS, and the phorbol myristate acetate+ionomycin combination can induce the expression of B7-H3 on dendritic cells and monocytes.1 24 In tumor cells, limited studies have reported that B7-H3 expression can be upregulated by ILT4 via PI3K/AKT/mTOR activation in non-small cell lung cancer (NSCLC) cells,25 and BRD4 regulates B7-H3 expression at the transcriptional level in PDAC cells.26 In addition, microRNA-29/187/143 are reported to directly interact with the 3′-untranslated regions of B7-H3 mRNAs, further suppressing their protein translation.27–29
Improving the understanding of the mechanistic basis of B7-H3 expression is still needed to develop novel strategies that enhance or act in concert with B7-H3-targeted therapy. Hence, we performed genome-wide CRISPR knockout screening to identify regulators of B7-H3 in human tumor cells and seek alternative approaches to augment antitumor efficacy.
Methods
Antibodies and reagents
The following antibodies were used: anti-B7-H3 (Proteintech #66 481–1-Ig); antiphospho-p38 MAPK (Thr180/Tyr182) (CST #4511); anti-p38 (CST #9212); antiphospho-eIF4E (S209) (HUABIO #ET1608-66); anti-eIF4E (Proteintech #66 655–1-Ig); anti-HER2 (Proteintech #18 299–1-AP); anti-α-tubulin (Beyotime Biotechnology #AF5012); anti-β-actin (Proteintech #6009–1-Ig); anti-PDL1 (CSB-MA878942A1 m); anti-EGFR (ZENBIO# 201012); HRP conjugated goat antirabbit IgG goat polyclonal antibody (HUABIO #HA1001); and HRP-conjugated goat antimouse IgG goat polyclonal antibody. Antibodies for flow cytometry, including PerCP-Cy5.5-antimouse CD45, FITC-antimouse CD11B, APC-antimouse B7-H3, APC-Cy7-antimouse CD3, FITC-antimouse CD4, BV510-antimouse CD8, APC-antimouse F4-80, BV421-antimouse CD86, PE-antimouse CD206, and PE-antihuman B7-H3, were all purchased from Biolegend.
Cells were treated with the following reagents: SB203580 (Beyotime Biotechnology# S1863); recombinant human TNF-alpha protein (Sinobiological# 10602-HNAE); and lipopolysaccharide (LPS) (Biosharp#BS904).
Cell culture
Human HEK293T, SK-OV-3, and HeLa cell lines were all cultured in DMEM, mouse 4T1 and human A375, and DU145 cell lines were cultured in RPMI 1640 media (Gibco), and both were supplemented with 10% FBS (Gemini), 100 µg/mL penicillin and 100 U/mL streptomycin (HyClone). Experiments were all started when cells reached log phase at 37°C and 5% CO2. For p38 MAPK pathway activation, vehicle control, or eIF4E, MNK1-knockout cells were treated with (0–500 ng/mL) recombinant human TNF-alpha protein for 16 hours. For p38 MAPK pathway inhibition, 125 ng/mL TNF-α pretreated or SP20H-knockout cells were treated with 25 µM SB203580 for 2 hours before detection. For H2O2 treatment, DU145 cells were incubated with 0, 50, 150, 250, 500, and 800 µM H2O2 for 2 hours, respectively. For LPS stimulation, a series of concentrations of LPS (0~500 ng/mL) were added for 16 hours at 37°C. All cellular experiments were repeated at least three times.
Library virus production and determination of virus titration
The Human CRISPR Knockout Pooled Library (Brunello) was purchased from Addgene (Pooled Library #73179). Library viruses were produced by cotransfecting pooled library plasmids with packaging plasmids (psPAX2 and pMD2. G) into HEK293T cells in a 4:3:2 ratio using PEI reagent. Fresh medium was replaced 12 hours after incubation. Lentiviral supernatants were harvested 48 and 72 hours post-transfection, and cell debris was removed by centrifugation (400× g, 300 min) and filtration (0.45 µm). Supernatants were then concentrated and purified by centrifugation at 20 000× g and 4°C for 2 hours. The pellet was resuspended in serum-free medium, aliquoted, and stored at −80°C until use.
B7-H3-positive SK-OV-3 cells were transduced by a serial dilution of concentrated virus with 8 µg/mL polybrene (Sigma-Aldrich) at 30~40% confluence in 24-well plates for 8 hours before fresh medium replacement. Cells were subjected to 3 µg/mL puromycin selection 48 hours after transfection for 3 days, and the dilution ratio that gave a 30~40% cell survival rate (corresponding to a multiplicity of infection (MOI) of 0.3~0.5) was selected as the optimal large-scale transduction condition.
CRISPR knockout (KO) library screening and next-generation sequencing (NGS) analysis
A total of 1.5×108 cells were initially infected with pooled CRISPR knockout virus to achieve 400-fold sgRNA coverage. Genomic DNA of 1×107 resistant cells collected on day 0 after puromycin withdrawal was served as a baseline control, and an equal count of cells collected before sorting was used as unsorted control. Sorted samples were obtained by gating on 1% cells with extremely high and low expression of B7-H3 and followed by FACS after 7 days of incubation. Overall, a total of ~3×108 infected cells were sorted, ~2×106 B7-H3low cells and ~3×106 B7-H3high cells were collected, respectively.
For sgRNA amplicon sequencing, sorted cells were directly subjected to genomic DNA extraction using QuickExtrac Solution (Epicentre, QE09050), and genomic regions containing sgRNA-targeted sites were amplified by two rounds of PCR. First round PCR amplified in 25 reactions with the following primers: outer F: 5′-aatggactatcatatgcttaccgtaacttgaaagta-3′; outer R: 5′-aacgttcacggcgactactgcacttatatacggttctc-3′; second round PCR amplified using 2 µL products of first round PCR as templates with the following primers: P5: 5′-ttgtggaaaggacgaaacaccg-3′; P7: 5′- ccaattcccactcctttcaagacct-3′. Four groups were set: day 0, unsorted control, sorted B7-H3high, and sorted B7-H3low, giving approximately 7 million reads in each sample. Purified PCR products were pooled into libraries and subjected to NGS with a dual-end read of 150 bp. Read mapping, normalization, quality control (QC), hit identification and functional analysis of our screen data were performed using the MAGeCKFlute R package.30
Gene knockdown and overexpression
The designed sgRNAs were cloned into the lentiCRISPRv2-puro vector (Addgene plasmid #98290) using the BsmBI site, while all shRNAs were cloned into the EcoRI-digested and PacI-digested pLKO.3G vector (Addgene plasmid #14748) individually. Synthesized SUPT20H and FAM118A cDNA were cloned into a modified pLenti CMV GFP Puro vector (Addgene#17448) for fusion with C-terminal or N-terminal GFP. The detailed sequence information is listed in the online supplemental information. Stable gene knockout or overexpression cells were generated by lentivirus infection and puromycin selection as previously described, while pLKO.3G-transduced cells were identified by GFP+. To control for sgRNA-specific effects, two sgRNAs or shRNAs were used for each targeted gene. Additional sequence information is provided in online supplemental table S1.
jitc-2022-004875supp001.pdf (1.4MB, pdf)
RNA sequencing (RNA-seq) and analysis
For transcriptome sequencing (RNA-seq), all of our libraries were sequenced in a paired-end 150-base (PE150) strategy on an Illumina NovaSeq 6000 system by Guangzhou Huayin Health Medical Group Co, Ltd (Guangzhou, China). Control (Cas9 alone), SP20H-knockout, and F118A-knockout cells collected 3 days and 7 days postselection were lysed in TRIzol (Invitrogen, 15506026). RNA-seq libraries were generated from triplicate samples per condition. The expression level of mRNA was calculated using RSEM (RNA-Seq by Expectation Maximization) (V.1.3.1) and normalized to fragments per kilobase per million (FPKM) reads.31 The differentially expressed mRNAs were screened using the edgeR package (https://bioconductor.org/packages/release/bioc/html/edgeR.html) in R.32 |log2Foldchange|≥1 and p value <0.05 were considered significant. Gene Ontology (GO) enrichment analysis, KEGG pathway enrichment analysis, and gene set enrichment analysis (GSEA) were completed using the cluster profiler package in R.33 Unless otherwise noted, all data analyses were performed in R, and the data were visualized using TBtools and the online tools available at https://hiplot.com.cn.
Flow cytometry analysis
Cultured cells were digested with 0.25% trypsin-EDTA into single cells. For tumor tissues obtained from sacrificed mice, single-cell suspensions were prepared on a gentleMACS Octo Dissociator (Miltenyi Biotec) using gentleMACS C tubes with the m_impTumor_01 program and filtered through 70 µm cell strainers. After blocking with 5% BSA in PBS, cell surface markers such as B7-H3, CD45, CD11B, CD3, CD4, CD8, F4_80, and CD86 were stained first with antibodies diluted at 1:200 for 40 min on ice, accompanied by Live/Dead staining using Fixable Viability Stain 700 (BD Biosciences) for 15 min. Stained cells were fixed with 4% paraformaldehyde (PFA) for 15 min and permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature, followed by intracellular CD206 staining for 30 min. Labeled cells were washed once with PBS and resuspended in 500 µL PBS for further analysis. All FACS data were collected on BD FACSymphony A5 and analyzed using FlowJo software.
Western blot analysis
Treated or transfected cells were scraped off the plates and collected by centrifugation at 4°C. Cell pellets were lysed in RIPA buffer with protease/phosphatase inhibitors (Beyotime Biotechnology, P0013B) on ice. After centrifugation, the supernatant protein content was determined using a BCA Protein Assay Kit (Beyotime Biotechnology, P0010). Equal amounts of total protein were loaded, subjected to 10% SDS-PAGE, and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked in 5% non-fat milk for 1 hour at room temperature and then incubated with specific primary antibodies at 4 °C overnight. After washing in TBS/T (TBS with 0.1% Tween-20), the membranes were exposed to secondary antibodies for 1 hour. The blots were stripped and reprobed for total protein after phosphorylated protein detection. All antibodies were diluted as recommended ratios according to the manufacturer’s instructions. Signals were visualized by the enhanced chemiluminescence method using Super ECL Plus substrate (Uelandy, S6009 M) and imaged on a Dako REALTM EnVision Detection System.
Preparation and characterization of antibody–drug conjugates
To prepare a mouse mAb against human B7-H3 protein, hybridoma cells expressing anti-B7-H3 antibody were transferred into Balbc mice, and ascites fluid was harvested after 9~14 days. The mAbs were further purified from ascites fluid by protein G affinity purification. Conjugation of mAbs and DM1-MCC was conjugated at a molar ratio of 1:9 in reaction buffer (50 mM potassium phosphate, 50 mM sodium chloride, 2 mM EDTA, pH 7.2) and then left at 25°C with stirring (220 rpm) overnight. The drug-to-antibody ratios were determined based on the absorbance of conjugates at 280 nm and 252 nm, calculated as previously described.34 The DAR of each batch was kept in the range of 2.2~3.0. The integrity and purity of conjugated and unconjugated antibodies were confirmed by SDS-PAGE and Coomassie staining.
For the in vitro cytotoxicity assay of αB7-H3-DM1, 1.5×103 control or edited cancer cells per well were seeded in 96-well plates for 24 hours and subjected to treatment with serially diluted MCC-DM1 conjugated or free antibodies targeting B7-H3. Triplicate experiments were performed at each concentration. Follow-up was performed with 72 hours of continuous monitoring using the IncuCyte live-cell imaging system (Essen BioScience).
Immunofluorescence and immunohistochemistry analysis
For indirect immunofluorescence staining, cells were seeded in 8-well chamber slides. GFP N-terminal-tagged or C-terminal-tagged SUPT20H-overexpressing or FAM118A-overexpressing cells were fixed prior to staining for surface antigens. We labeled surface B7-H3 protein using our homemade mouse antihuman B7-H3 antibody diluted at 1:500 for 1 hour at room temperature. The cells were then washed in PBS, followed by Alexa Fluor 594-conjugated secondary antibody (Huabio #HA1112) staining for 45 min, and nuclei were stained with DAPI for 2~3 min. Confocal imaging was performed on an LSM 880 microscope (Carl Zeiss).
For immunohistochemistry staining, tumor tissues were fixed in 4% PFA, embedded in paraffin and sectioned at 3 µm. Dewaxed and rehydrated sections were boiled in 10 mM sodium citrate buffer (pH 6.0) for heat-induced epitope retrieval. After cooling, the slides were blocked in 2% BSA in TBST and then incubated with primary antibodies diluted in 1% BSA in TBST at 4 °C overnight. The endogenous peroxidase blocking, HRP secondary antibody incubation, and DAB substrate-chromogen reaction were performed using the Polymer HRP (mouse/rabbit) IHC kit (ZSGB-BIO, PV-6000 and ZLI-9019) according to the manual. After counterstaining of nuclei with hematoxylin (Beyotime, C0107), the slides were dehydrated and sealed. Images were acquired on a Pannoramic MIDI (3DHISTECH Ltd).
In vivo experiments
All experiments were performed on female mice at 7–8 weeks. A total of 2×106 SK-OV-3 (control, SP20H-KO, or B7-H3-KO) cells and 2×105 4T1 (control or SP20H-KO) cells were subcutaneously injected into the right flanks of NOD/SCID and BALB/c mice, respectively. Tumor sizes were measured by calipers every 3~5 days. Tumors were measured at the indicated times and volume was calculated as volume (mm3) = (width [mm])2 × (length (mm))2. For in vivo monitoring of tumor growth, control and SP20H-knockout SK-OV-3 cells were transduced with lentivirus stably expressing luciferase-GFP (SK-OV-3-eGFP.Luc). A total of 2×106 sorted GFP-positive cells were inoculated into NOD-SCID mice and followed by two injections of 10 mg/kg αB7-H3-DM1 at day 15 and day 20, respectively. Bioluminescence imaging and analysis were performed on an IVIS imaging system. Mice were all sacrificed to collect tumors when the control tumor volume reached 1500 mm3.
Statistics
All statistical analyses were performed using Prism (GraphPad, V.8.2.1) or Excel (Microsoft). Statistical analysis was performed using Student’s t-test for single comparisons and two-way analyses of variance followed by Dunnett’s for multiple comparisons. Data are shown as the means±SD unless otherwise indicated, and p values are indicated by *p<0.05, **p<0.01, and ***p<0.001, ****p<0.0001.
Results
A pooled genome-wide CRISPR screen to identify regulators of B7-H3
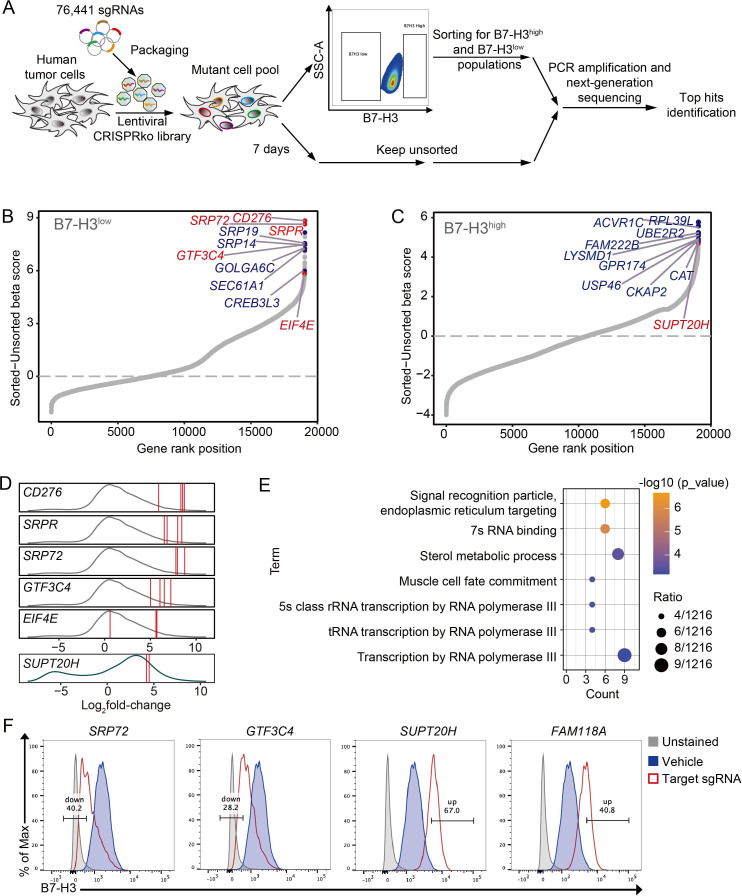
Here, we employed SK-OV-3, a B7-H3 endogenously high expressed cell line, to perform an unbiased genome-wide CRISPR/Cas9 loss-of-function screen for B7-H3 regulators. The experimental workflow was illustrated in figure 1A. Four thousand five hundred and eighty-two and 1540 candidates (of 19 060 mapped genes) were upregulated in B7-H3high and B7-H3low population, respectively. Candidates enriched in sorted B7-H3high and B7-H3low populations were ranked, and top hits were plotted in figure 1B, C, respectively. At least two sgRNAs per gene, with log2FC>1, were highly enriched (figure 1D). GO annotation of 41 genes enriched in the B7-H3low population shows significant enrichment for signal recognition particle (SRP)-dependent ER translocation and RNA transcription processes (figure 1E). No significantly enriched GO terms were found in B7-H3high group. Top hits were validated with their most frequently enriched sgRNA in vitro, and surface B7-H3 expression levels of each sgRNA-edited SK-OV-3 cells were analyzed individually. Results showed that targeting both positive hits (SRP72 and GTF3C4) and negative hits (SUP20H and FAM118A) have varying degrees of regulatory effects on B7-H3 expression (figure 1F). These results were also confirmed in another human melanoma cell line (online supplemental figure S1A). Bulk PCR and Sanger sequencing confirmed genomic edits in each sgRNA construct transduced clones (online supplemental figure S1B). Thus, these results together confirmed the validity of our screen setup and its capacity to identify gene expression regulators.
Figure 1.
A genome-wide CRISPR–Cas9 screen identifies regulators of B7-H3 in human cancer cells. (A) Schematic representation of the in vitro loss-of-function screen for regulators of B7-H3 using the human CRISPR Brunello lentiviral pooled library in cancer cells. (B and C) Candidates enriched in B7-H3high and B7-H3low populations were plotted (measured by β-score), respectively. Y-axis indicates the distribution of normalized enrichment scores (sorted vs unsorted) of each gene enrichment. Top-ranked genes were marked in orange, and genes highlighted in red were selected for validation. The plots were generated using the R package ggplot2. (D) Density plots show the distribution of the log2-fold change of individual sgRNAs counts (sorted vs unsorted) enriched in B7-H3high (green curve) and B7-H3low (gray curve) populations. sgRNAs targeting selected genes were denoted by a red line. (E) Gene ontology (GO) analysis of genes enriched in B7-H3low population. (F) Flow cytometry analysis of surface B7-H3 expression on SK-OV-3 cells expressing Cas9 alone (vehicle) or sgRNA targeting CD276, GTF3C4, SUPT20H, or FAM118A. Experiments were repeated three times with similar results.
SP20H negatively regulates B7-H3 expression in cancer cells
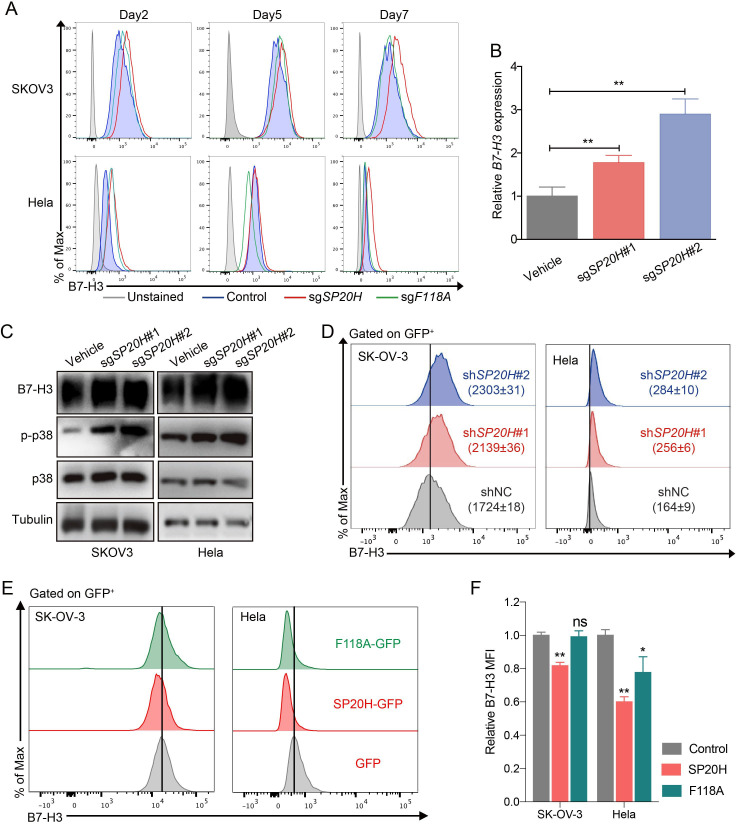
Given that we aimed to search for synergistic therapeutic targets in B7-H3-targeting therapies, we were particularly interested in validation of negative hits. Indeed, we found that highly scored SUP20H (SP20H) had two targeting sgRNAs enriched, whereas FAM118A (F118A) was lowly scored, with only one targeting sgRNA strongly enriched (data not shown). Differential regulatory effects of SP20H (67.0%) and F118A (40.8%) on B7-H3 expression were confirmed in figure 1F. We next investigated the effects of SP20H and F118A knockout on SK-OV-3 cells and another B7-H3 moderately expressed cell line-Hela. Through continuous monitoring of B7-H3 expression on SP20H and F118A knockout cells by flow cytometry, we observed stable upregulation of B7-H3 in both SP20H-KO SK-OV-3 and Hela cells but not in F118A-KO cells (figure 2A). We speculated that the regulation of B7-H3 expression mediated by F118A is time and cell type dependent. Therefore, specific attention was dedicated to SP20H in subsequent studies.
Figure 2.
SP20H acts as a negative regulator of B7-H3. (A) The expressions of B7-H3 in control and gene knockout (SP20H or F118A) SK-OV-3 and Hela cells at different timepoints post infection were analyzed. (B) The relative expressions of B7-H3 mRNA relative to β-actin mRNA in Control (Cas9 alone) and SP20H-KO SK-OV-3 cells are shown. Error bars show mean±SD (n=3; **p<0.01). (C) Western blot analyses of SP20H-knockout cancer cells for total B7-H3 and phospho-p38 protein expression. (D) Cells expressing shRNA against SP20H, or control scramble shRNA (shNC) were identified for B7-H3 expression analysis, which was performed on gated GFP+ cells. Values in histogram plots showed mean MFI ±SD of triplicates. (E) Histograms show flow cytometric analysis of cell surface expression of B7-H3 in SP20H or F118A-overexpressed SK-OV-3 and Hela cells, compared with empty vector control (GFP). (F) Relative B7-H3 MFI of gene overexpressed cells were shown in bar graph. Error bars show mean±SD (*p<0.05; **p<0.01). All p values were calculated by paired two-tailed Student’s t-test. Experiments were repeated three times with similar results.
We further identified upregulation of B7-H3 in SP20H-knockout cells at both mRNA and protein levels by qPCR and western blot (figure 2B, C). Particularly, the level of phospho-p38 had significant increased, which demonstrated the potential regulatory role of SP20H on p38 activity. Similar results obtained in SP20H knockdown cells, which were transduced with control or SP20H-targeted pLKO.3-shRNA constructs (figure 2D). Immunofluorescence staining and flow cytometry analysis were performed in cells infected with lentiviruses that direct the expression of GFP or GFP-fusion protein containing full-length SP20H and F118A. Confocal imaging showed that F118A was mainly located in the intermediate filaments in the cytosol and that B7-H3 was located in the cell membrane, whereas the C-terminal GFP-fused SP20H protein was only found as a punctate cytosolic pool largely excluded from the nucleus with limited expression levels, and N-terminal GFP-fused SP20H was highly expressed in the nucleus (online supplemental figure S2), which are in line with previous studies.35 In addition, overexpression of SP20H exhibited varying degrees of B7-H3 inhibition (figure 2E, F). These results together demonstrated the regulatory role of SP20H on B7-H3 expression.
TNF-p38 MAPK pathway activation induces upregulation of B7-H3
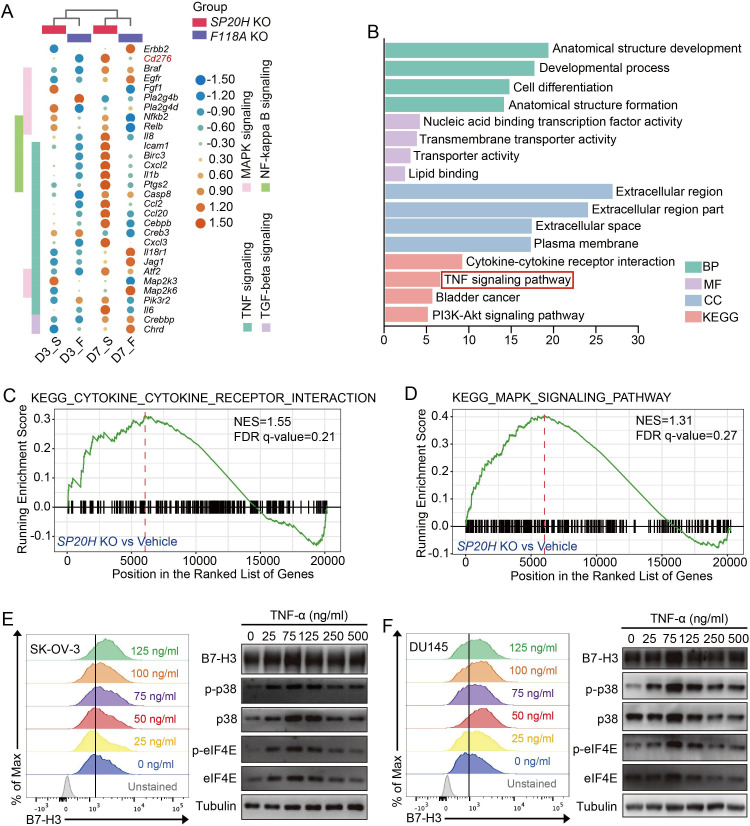
To reveal the possible mechanisms that drive B7-H3 upregulation, we performed RNA sequencing (RNA-seq) of short-term (3 days) and long-term (7 days) cultured SP20H-KO and F118A-KO ovarian cells, respectively. Analysis of RNA-seq data revealed a large discrepancy between long-term and short-term cultured gene-edited cells, especially multiple proinflammatory pathways, such as the TNF/NF-κB/MAPK pathways, and several key inflammatory mediators (eg, IL-1β, IL-6, CXCL2 and CCL2) were significantly upregulated (figure 3A). GO and KEGG pathway enrichment analysis results of differentially expressed genes (DEGs) in long-term cultured SP20H-depleted cells were plotted and revealed an enrichment of the TNF pathway (figure 3B). The GSEA plot illustrated the enrichment of DEGs in the KEGG MAPK pathway and cytokine–cytokine receptor interaction, with FDR q-value <0.25 (figure 3C, D). Previous publications have reported that MAP kinases act both upstream and downstream of TNF-α/TNFR1 signaling.36 In addition, we found that eukaryotic translation initiation factor 4E (eIF4E), which is known as a downstream effector of p38 MAPK signaling that mediates translational regulation, was identified as a positive regulator of B7-H3 in figure 1D. Combining with the previous observed upregulation of phoshhospho-p38 in SP20H-KO cells, we speculated that the TNF-p38 MAPK-eIF4E pathway is responsible for B7-H3 induction.
Figure 3.
TNF pathway activation mediated p38 MAPK-eIF4E axis signaling and B7-H3 upregulation. (A) RNA-seq heatmap represents the differential gene expressions in SP20H or F118A knockout cells incubated for various periods of time. The gene expression value in heatmap was normalized by log2-fold change in each row. (B) Bar plot shows the most significant enriched GO terms and KEGG pathways in D7_SP20H-knockout cells. (C and D) GSEA plots of indicated signature genes enriched in long-term SP20H depleted cells (D7_S). (E–F) SK-OV-3 and DU145 cells treated or untreated with different concentrations of TNF-α were analyzed for surface B7-H3 protein expression by flow cytometric analysis and detected for total protein expression of B7-H3, phospho-p38, and phospho-eIF4E by immunoblotting. At least two independent experiments were performed with similar results; representative data are shown. D3/D7: day3, day7; F, F118A; GO, Gene Ontology; GSEA, gene set enrichment analysis; S, SP20H.
To test our idea, we treated SK-OV-3 and DU145 cells with different concentrations (0, 25, 75, 125, 250, and 500 ng/mL) of TNF-α for 16 hours, followed by flow cytometry and western blot analysis. Consistent upregulation of cell-surface and total B7-H3 protein expression was observed in a concentration-dependent manner in these two cell lines (figure 3E, F). Meanwhile, elevated levels of phospho-p38, phospho-eIF4E and B7-H3 expression were detected in TNF-α-stimulated tumor cells (figure 3E, F, right panels), whereas a series concentration of LPS and hydrogen peroxide (H2O2) stimulation both induced no detectable changes in B7-H3 expression level on TLR4 highly expressed DU145 cells (online supplemental figure S3A, B, respectively). Therefore, we demonstrated that TNF-p38 MAPK pathway activation participates in the regulation of B7-H3 expression in tumor cells but not oxidative stress or LPS-TLR4 signaling.
EIF4E acts as a crucial p38 downstream effector responsible for B7-H3 translation
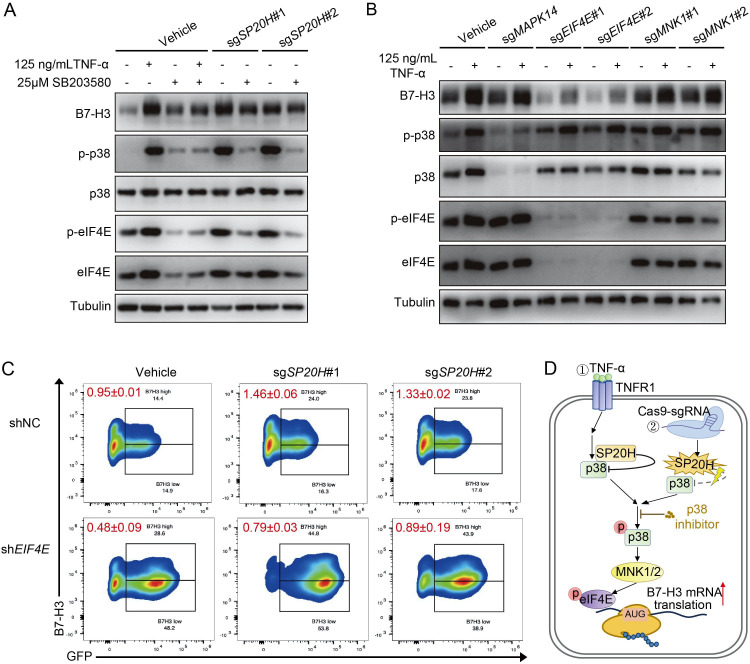
As we know, SP20H is also known as p38-interacting protein. So we speculated that SP20H deficiency activated TNF downstream p38 MAPK-EIF4E axis signaling, which was responsible for further induction of B7-H3. To prove our hypothesis, we added 25 µmol/L SB203580 (a p38-selective inhibitor) to SP20H-knockout cells and TNF-α pretreated WT cells, and the protein expression levels were measured 2 hours after treatment. Western blot analysis showed that inhibition of p38 MAPK activation successfully blocked both TNF-α stimulation and SP20H deficiency-induced upregulation of B7-H3 expression by suppressing phosphorylation of p38 MAPK and EIF4E (figure 4A).
Figure 4.
Activation of eIF4E is responsible for both SP20H knockout and TNF-α stimulation induced B7-H3 translation. (A) Total protein expression of B7-H3, phospho-p38, and phospho-eIF4E in SB203580 treated SP20H knockout or TNF-α pretreated SK-OV-3 cells. (B) Changes of p38 MAPK-eIF4E signaling and B7-H3 expression in SK-OV-3 cells transduced with the indicated sgRNAs under TNF-α stimulation. (C) The expression levels of B7-H3 after interfering with eIF4E in the SP20H knockout SK-OV-3 cells. The values represent the mean ratios of B7-H3high/B7-H3low population of triplicate measurements±SD. (D) Schematic illustration of two ways to induce B7-H3 expression via the p38 MAPK- eIF4E signaling.
To further verify the essential role of eIF4E in p38 MAPK activation-induced B7-H3 expression, we examined the protein levels of the B7-H3, phospho-p38, and phospho-eIF4E in TNF-α stimulated MAPK14-KO, eIF4E-KO, and MNK1-KO SK-OV-3 cells. A significant reduction of B7-H3 was detected in eIF4E-knockout cells, but no difference was observed on p38 phosphorylation; moreover, no clear effects were detected in MAPK14-KO and MNK1-KO cells on both B7-H3 expression and downstream eIF4E phosphorylation, which was considered as consequences of incomplete inhibition of upstream signaling pathways that acting through p38-eIF4E axis (figure 4B). Similarly, decreased ratios of the B7-H3high/B7-H3low population in both control and SP20H-depleted SK-OV-3 cells were detected after delivering of unique shRNA targeting eIF4E, compared with the scramble shRNA control (figure 4C). Therefore, our data confirmed that B7-H3 expression can be upregulated by two ways: TNF-α stimulation and SP20H knockout (figure 4D). Together, these results supported our initial hypothesis that loss of SP20H induces B7-H3 upregulation by activating the p38 MAPK-EIF4E signaling axis.
SP20H loss strongly suppresses tumor progression in both immune-compromised and immune-competent mouse models
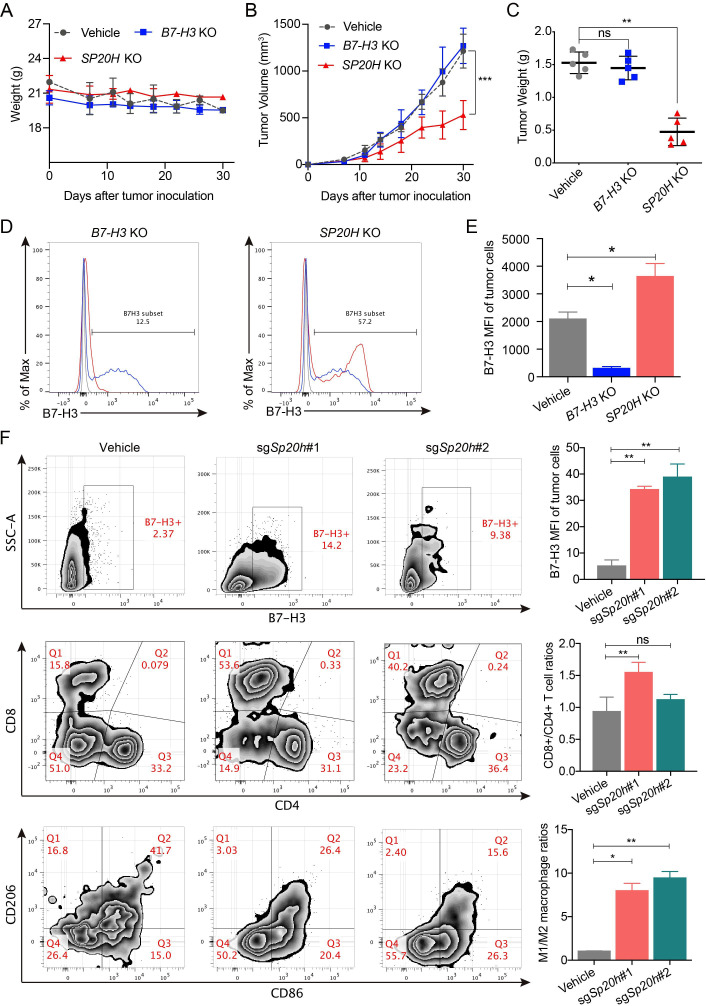
We next determined whether SP20H deficiency affects tumor progression in two mouse models under different immune backgrounds. To evaluate the effect of B7-H3 expression on tumor growth in vivo, the established sgB7-H3, sgSP20H, and control SK-OV-3 cells were injected subcutaneously into NOD-SCID mice. Body weight and tumor volume were monitored until resection. No significant body weight changes were observed in the control and gene knockout groups (figure 5A). SP20H-deficient tumors exhibited superior growth control compared with vehicle tumors, while B7-H3 loss had no apparent effect (figure 5B, C). Moreover, significantly increased expression levels of B7-H3 were detected in SP20H-deficient tumors (figure 5D, E), while its impact on other tumor antigens (eg, HER2, PD-L1, and EGFR) is not apparent (online supplemental figure S4A). These findings suggest that SP20H negatively regulates B7-H3 expression both in vitro and in vivo.
Figure 5.
SP20H inactivation represses tumor growth in vivo and increases populations of proinflammation T cells and macrophages. (A–B) Body weight changes and subcutaneous tumor growth curve of SK-OV-3 vehicle, SP20H-KO, or B7-H3-KO cells in NOD-SCID mice were plotted (n=5; ***p<0.001). (C) Tumors were excised and weighed 30 days after tumor challenge (n=5; **p<0.01; ns, no significant). (D) Analysis of surface B7-H3 expressions on B7-H3-deficient or SP20H-deficient tumor cells from SK-OV-3 tumor-bearing NOD-SCID mice by flow cytometry. (E) Statistical analysis of corresponding MFI of B7-H3 in (D); *p<0.05. (F) Flow cytometry analysis of surface B7-H3 expression of tumor cells(upper), frequencies of tumor-infiltrating T cells (middle) and macrophages (lower) in control or sgSp20h-4T1 tumor-bearing BALB/c mice. Tumor cells were gated on CD45− CD11B− live cells; the CD45+ CD3+ live cells were divided into CD4+ and CD8+ T cells; the CD45+ CD11B+ F4_80+ live cells were divided into M1-like (CD86+ CD206−) and M2-like (CD86− CD206+) macrophages. Data shown are representative results. The p values were determined by a two-tailed paired Student’s t-test (n=3; *p<0.05; **p<0.01; ns, no significant).
To further determine the effect of SP20H depletion on tumor immune microenvironment, we expanded our studies by subcutaneous injection of either parental or Sp20h-deficient 4T1 murine mammary carcinoma cells into immunocompetent BALB/c mice. Encouragingly, consistent tumor growth inhibitions were observed in Sp20h-deficient tumors compared with control tumors (online supplemental figure S4B, C). Moreover, ~6-fold increase of tumor-infiltrating CD8+ T cells and ~3-fold decrease of M2 tumor-infiltrating macrophages were detected in Sp20h-deficient tumors compared with control groups. These data suggested that loss of SP20H increase the infiltration of immune cells and dramatically change the immune status of tumor microenvironment (figure 5F). Representative gating strategies to identify different cell types from tumor tissues are presented in online supplemental figure S5. These findings suggest a consistent regulatory role of SP20H on B7-H3 expression in vivo and the tumor growth suppression effect of SP20H depletion.
SP20H inactivation cooperates with anti-B7-H3 to eliminate tumors in vivo
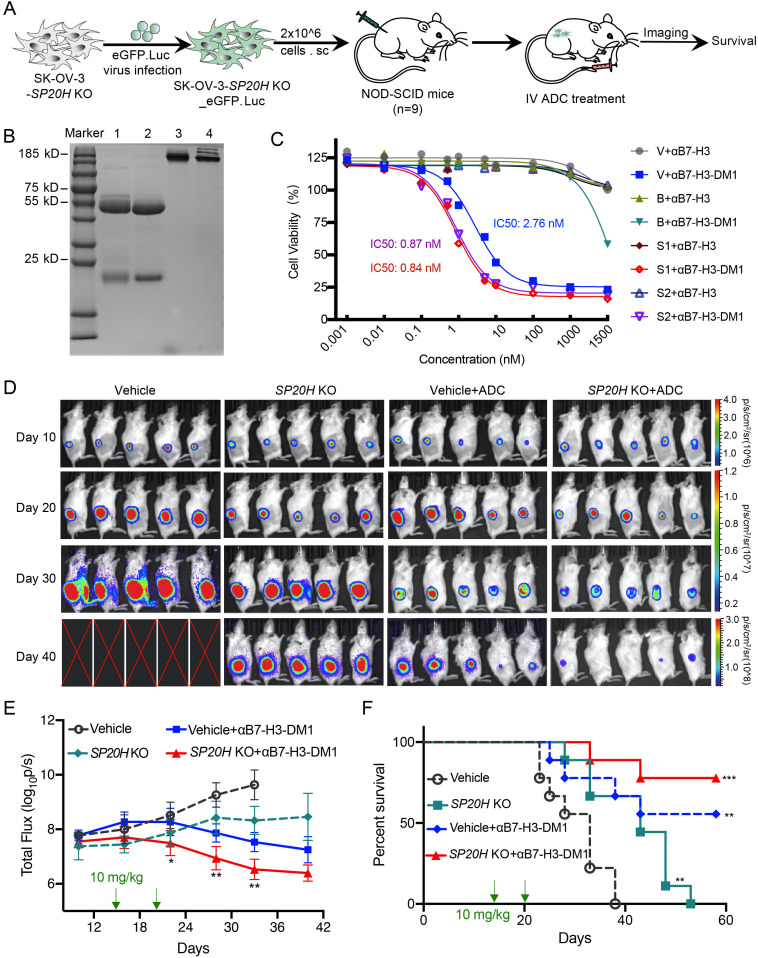
To investigate whether SP20H deficiency has any cooperative effect on anti-B7-H3 therapy in vivo, we performed subcutaneous injection of luciferase-GFP-labeled control SP20H-knockout SK-OV-3 cells in NOD-SCID mice and monitored tumor development by bioluminescence imaging, as shown in figure 6A. Prior to this, emtansine (MCC-DM1) conjugated mAbs against human B7-H3 protein were prepared as described in a previous report.34 The anti-B7-H3 mAbs used for conjugation reaction were prepared by our lab.37 The integrity and purity of the anti-B7-H3 and MCC-DM1 conjugates (αB7-H3-DM1) were verified by SDS-PAGE, and there was no significant molecular weight difference between the conjugated and unconjugated antibodies under both reducing and nonreducing conditions (figure 6B). For the antigen-specific in vitro killing assay, we tracked cell killing of αB7-H3-DM1 conjugates on GFP-labeled tumor cells using the IncuCyte imaging system, and images after 36 hours of treatment were captured (online supplemental figure S6). The 72-hour cell growth inhibition rates of αB7-H3-DM1 conjugates were calculated and the cytotoxicity of conjugates showed a B7-H3 expression-dependent manner. IC50 values in control (B7-H3++), SP20H-KO (B7-H3+++), and B7-H3-KO (B7-H3−) SK-OV-3 cells were 2.8, 0.9, and >1200 nM, respectively (figure 6C).
Figure 6.
Loss of SP20H augments antitumor efficacy of B7-H3 antibody–drug conjugates in vivo. (A) Schematic illustration of the treatment procedure of αB7-H3-DM1 on control or SP20H-knockout tumor-bearing NOD-SCID mice. (B) DM1-MCC conjugated and unconjugated anti-hB7-H3 constructs were resolved by SDS-PAGE and stained with Coomassie blue. Lane 1: reduced αB7-H3; lane 2: reduced αB7-H3-DM1; lane 3: non-reduced αB7-H3; lane 4: non-reduced αB7-H3-DM1; (C) Dose–response curves of DM1-MCC conjugated or unconjugated anti-hB7-H3 antibody (αB7-H3, αB7-H3-DM1) against SK-OV-3 cells with normal (V, Vehicle), negative (B, sgB7-H3), or high (S1, sgSP20H#1; S2, sgSP20H#2) expression of B7-H3 were shown. (D) In vivo bioluminescence imaging of NOD-SCID mice inoculated with ovarian tumors. Five mice were randomly selected from each group (n=9) for IVIS imaging. Symbol X indicates mice that either died or underwent euthanasia. Experiments were repeated twice with similar results and representative images are shown. (E) Quantification of bioluminescence intensity is shown. Statistical analysis was performed by two-way analysis of variance. (F) Survival curves were estimated using the Kaplan-Meier analysis with a log-rank test (n=9). *p<0.05; **p<0.01; ***p<0.001.
A tolerability experiment preverified a well-tolerated dose of 30 mg/kg conjugated and unconjugated B7-H3 antibody via intravenous injection. Tumor-bearing mice received systemic injections of αB7-H3-DM1 (10 mg/kg) when control tumors grew to an average size of 300 mm3. Tumor growth was monitored by bioluminescence imaging, and emissions were quantified by the average total flux of each group of mice (figure 6D, E). Significantly reduced tumor burden and prolonged overall survival were observed in αB7-H3-DM1treated groups. Notably, five (out of nine) mice in the αB7-H3-DM1 treated vehicle group and seven (out of nine) mice in the αB7-H3-DM1treated SP20H-KO group survived over 58 days (figure 6E, F). Taken together, these data suggest SP20H inhibition exerts synergistic antitumor effects with B7-H3 targeted antitumor therapies.
Discussion
In our previous study, we found that heterogeneous expression and loss of tumor antigen are the major cause of treatment failure and cancer recurrence.38 39 There is thus a great need to gain a comprehensive understanding of the intrinsic tumor antigen expression mechanism and develop novel strategies to improve therapeutic efficacy. To this end, we employed and validated unbiased high-throughput CRISPR knockout library screening for B7-H3 regulators. This effort identified the TNF-p38 MAPK pathway as the major signaling process supporting B7-H3 expression in tumor cells and revealed a novel role of SP20H in B7-H3 upregulation, acting together with its downstream effector eIF4E. Moreover, SP20H-deficient tumors exhibit significant inhibition of tumor growth and extended overall survival in either immunodeficient or immunocompetent backgrounds, with increasing ratios of CD8+/CD4+ T cells and decreased M2 macrophage infiltration in SP20H-deficient tumors in the context of an immunocompetent. More importantly, SP20H depletion-induced eIF4E-dependent translation of B7-H3 augments the antitumor activity of the antibody–drug conjugate in vivo. In summary, our findings confirm a new signaling cascade responsible for the upregulation of B7-H3 in tumor cells and suggest new strategies to enhance immunotherapy efficacy.
SP20H, also named p38IP, was first identified in a yeast two-hybrid screening using a p38α bait (YL and JH, unpublished data, 1998; NCBI accession #AF093250). Further investigation demonstrated that it is required for p38MAPK activation in vivo and critical for gastrulation.40 SP20H has been reported to be involved in multiple biological processes, such as autophagy,35 ER stress‐responsive gene expression,41 the cell cycle,42 and monocyte–macrophage differentiation.43 P38 mitogen-activated protein (MAP) kinases are a class of MAPKs that regulate many biological processes, including inflammation, the cell cycle, cell death, development, cell differentiation, senescence and tumorigenesis.44 As a signal transduction nexus, the p38 MAPK pathway can be activated by various extracellular stimuli, such as UV light, heat, osmotic shock, inflammatory cytokines, and growth factors.44 45 Unfortunately, multiple commercial antibodies against SP20H (Abcam#ab122517, Proteintech#24 419–1-AP, and Bioworld#BS8867) were employed in our experiments but failed to identify changes of SP20H protein expression in our edited cells. Thus, the depletion of SP20H in this study were confirmed by real-time PCR and genomic DNA sequencing; future work is needed to develop SP20H-specific antibodies.
To date, few studies have elucidated the role of p38 MAPK and SP20H in tumorigenesis and development. Here, we demonstrate that B7-H3 expression on tumor cells is regulated by p38 signaling. A previous study identified the role of SP20H as a negative regulator controlling T cell receptor and LPS-activated NF-κB and p38 signaling in Jurkat T cells.46 In line with this, activation of the TNF-p38 MAPK pathway is found in SP20H-knockout tumor cells, which is responsible for further induction of B7-H3 expression. Perhaps even more importantly, human eukaryotic translation initiation factor 4E (eIF4E), which interacts with eIF4A and eIF4G to assemble an active eIF4F complex that assumes cap-dependent translation, was characterized as a positive regulator of B7-H3 expression in our study.47 48 As a convergent signaling point, eIF4E activity is regulated in two manners: PI3K/AKT/mTOR-mediated phosphorylation of 4E-BP liberates eIF4E, and ERK- or p38MAPK-MNK1/2-mediated phosphorylation of eIF4E.49–51 Our findings demonstrate that constitutive activation of the p38-eIF4E signaling axis is responsible for both TNF-α stimulation and SP20H depletion-induced B7-H3 expression, thus providing additional evidence that eIF4E facilitates abnormal gene expression.
In vivo investigations revealed consistent upregulation of B7-H3 expression and potent growth inhibition of SP20H-deficient tumors in immunodeficient mice. Moreover, we observed different distributions of B7-H3 and HER2 antigens in ovarian tumor tissues, which indicates the probability of HER2 as an actionable target in ovarian cancers in multitarget collaborative antitumor therapies (online supplemental figure S4A). In addition, the increasing infiltration of proinflammatory macrophages and T cells in SP20H-deficient tumors indicates its immunomodulatory functions in the tumor microenvironment. The synergistic cytotoxic activity of αB7-H3-DM1 observed in SP20H-deficient tumors shows the promise of SP20H as a combinatory target in antitumor therapy. Currently, the main hurdle to this strategy is the lack of small molecule inhibitors targeting SP20H, thus providing direction for future research. Alternatively, knockdown of SP20H by RNA interference using nanoparticles or degradation by proteolysis-targeting chimera (PROTAC) technology may be useful methods for further in vivo assays.
Collectively, our findings fill in the knowledge gaps regarding the mechanisms underlying B7-H3 expression in cancer cells and provide new insight into the development of combinatory therapeutic strategy targeting B7-H3. In the future, more investigations are needed to translate these findings to clinical practice.
Acknowledgments
We are grateful to the Lab of Infectious Diseases and Vaccine and Research Core Facility of West China Hospital of Sichuan University for providing experiment platform and technical assistances. We would thank to thank Yating Hu (Institute of Molecular Medicine, College of Future Technology, Peking University) for providing the bioinformatic technical support.
Footnotes
SZ and YW contributed equally.
Contributors: AT, XZ, and LZ cosupervised and directed this work. SZ and YW designed and performed the experiment. CN, GG, SZ and NY visualized and interpreted data. MM, HL, ZW, KZ, MZ, and HY helped conduct the experiments. XT, ZZ, CH, and GW contributed to the writing and editing of the manuscript. TC was responsible for all FACs-experiments. AT is responsible for the overall content as the guarantor.
Funding: This work was supported by National Natural Science Foundation of China (82073404, 82102898, 82002648 & 81773188); The 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYJC18007); CAMS Innovation Fund for Medical Sciences (2021-I2M-C&T-B-096); Major Subject of the Science and Technology Department of Sichuan Province (2019YFS0110); Chengdu Science and Technology Project-Technical innovation research and development project (2021-YF05-00920-SN).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Medical Ethics Committee of Hospital of West China Hospital of Sichuan University Biomedical Ethics Committee (20210535A) and was performed according to the Institutional Guidelines.
References
- 1.Chapoval AI, Ni J, Lau JS, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol 2001;2:269–74. 10.1038/85339 [DOI] [PubMed] [Google Scholar]
- 2.Suh W-K, Gajewska BU, Okada H, et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat Immunol 2003;4:899–906. 10.1038/ni967 [DOI] [PubMed] [Google Scholar]
- 3.Prasad DVR, Nguyen T, Li Z, et al. Murine B7-H3 is a negative regulator of T cells. J Immunol 2004;173:2500–6. 10.4049/jimmunol.173.4.2500 [DOI] [PubMed] [Google Scholar]
- 4.Hofmeyer KA, Ray A, Zang X. The contrasting role of B7-H3. Proc Natl Acad Sci U S A 2008;105:10277–8. 10.1073/pnas.0805458105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loos M, Hedderich DM, Friess H, et al. B7-H3 and its role in antitumor immunity. Clin Dev Immunol 2010;2010:1–7. 10.1155/2010/683875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong C, Tao B, Chen Y, et al. B7-H3 regulates glioma growth and cell invasion through a JAK2/STAT3/Slug-Dependent signaling pathway. Onco Targets Ther 2020;13:2215–24. 10.2147/OTT.S237841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, Tekle C, Chen Y-W, et al. B7-H3 silencing increases paclitaxel sensitivity by abrogating JAK2/STAT3 phosphorylation. Mol Cancer Ther 2011;10:960–71. 10.1158/1535-7163.MCT-11-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L, Zhao Y. B7-H3 induces ovarian cancer drugs resistance through an PI3K/AKT/BCL-2 signaling pathway. Cancer Manag Res 2019;11:10205–14. 10.2147/CMAR.S222224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flem-Karlsen K, Tekle C, Andersson Y, et al. Immunoregulatory protein B7-H3 promotes growth and decreases sensitivity to therapy in metastatic melanoma cells. Pigment Cell Melanoma Res 2017;30:467–76. 10.1111/pcmr.12599 [DOI] [PubMed] [Google Scholar]
- 10.Xie C, Liu D, Chen Q, et al. Soluble B7-H3 promotes the invasion and metastasis of pancreatic carcinoma cells through the TLR4/NF-κB pathway. Sci Rep 2016;6:27528. 10.1038/srep27528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tekle C, Nygren MK, Chen Y-W, et al. B7-H3 contributes to the metastatic capacity of melanoma cells by modulation of known metastasis-associated genes. Int J Cancer 2012;130:2282–90. 10.1002/ijc.26238 [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Zhang Q, Chen W, et al. B7-H3 is overexpressed in patients suffering osteosarcoma and associated with tumor aggressiveness and metastasis. PLoS One 2013;8:e70689. 10.1371/journal.pone.0070689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi T, Ma Y, Cao L, et al. B7-H3 promotes aerobic glycolysis and chemoresistance in colorectal cancer cells by regulating HK2. Cell Death Dis 2019;10:308. 10.1038/s41419-019-1549-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim S, Liu H, Madeira da Silva L, et al. Immunoregulatory protein B7-H3 reprograms glucose metabolism in cancer cells by ROS-mediated stabilization of HIF1α. Cancer Res 2016;76:2231–42. 10.1158/0008-5472.CAN-15-1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picarda E, Ohaegbulam KC, Zang X. Molecular pathways: targeting B7-H3 (CD276) for human cancer immunotherapy. Clin Cancer Res 2016;22:3425–31. 10.1158/1078-0432.CCR-15-2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inamura K, Yokouchi Y, Kobayashi M, et al. Tumor B7-H3 (CD276) expression and smoking history in relation to lung adenocarcinoma prognosis. Lung Cancer 2017;103:44–51. 10.1016/j.lungcan.2016.11.013 [DOI] [PubMed] [Google Scholar]
- 17.Ma L, Luo L, Qiao H, et al. Complete eradication of hepatocellular carcinomas by combined vasostatin gene therapy and B7H3-mediated immunotherapy. J Hepatol 2007;46:98–106. 10.1016/j.jhep.2006.07.031 [DOI] [PubMed] [Google Scholar]
- 18.Seaman S, Zhu Z, Saha S, et al. Eradication of tumors through simultaneous ablation of CD276/B7-H3-Positive tumor cells and tumor vasculature. Cancer Cell 2017;31:501–15. 10.1016/j.ccell.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du H, Hirabayashi K, Ahn S, et al. Antitumor responses in the absence of toxicity in solid tumors by targeting B7-H3 via chimeric antigen receptor T cells. Cancer Cell 2019;35:221–37. 10.1016/j.ccell.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theruvath J, Sotillo E, Mount CW, et al. Locoregionally administered B7-H3-targeted CAR T cells for treatment of atypical teratoid/rhabdoid tumors. Nat Med 2020;26:712–9. 10.1038/s41591-020-0821-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo D, Alderson RF, Chen FZ, et al. Development of an Fc-enhanced anti-B7-H3 monoclonal antibody with potent antitumor activity. Clin Cancer Res 2012;18:3834–45. 10.1158/1078-0432.CCR-12-0715 [DOI] [PubMed] [Google Scholar]
- 22.Powderly J, Cote G, Flaherty K, et al. Interim results of an ongoing phase I, dose escalation study of MGA271 (Fc-optimized humanized anti-B7-H3 monoclonal antibody) in patients with refractory B7-H3-expressing neoplasms or neoplasms whose vasculature expresses B7-H3. J Immunother Cancer 2015;3:O8. 10.1186/2051-1426-3-S2-O8 [DOI] [Google Scholar]
- 23.Modak S, Zanzonico P, Grkovski M, et al. B7H3-directed intraperitoneal radioimmunotherapy with radioiodinated Omburtamab for desmoplastic small round cell tumor and other peritoneal tumors: results of a phase I study. J Clin Oncol 2020;38:4283–91. 10.1200/JCO.20.01974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Dong Q, Xu Y, et al. B7-H3: another molecule marker for Mo-DCs? Cell Mol Immunol 2005;2:307–11. [PubMed] [Google Scholar]
- 25.Zhang P, Yu S, Li H, et al. ILT4 drives B7-H3 expression via PI3K/AKT/mTOR signalling and ILT4/B7-H3 co-expression correlates with poor prognosis in non-small cell lung cancer. FEBS Lett 2015;589:2248–56. 10.1016/j.febslet.2015.06.037 [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Meng Z, Xie C, et al. B7-H3 is regulated by BRD4 and promotes TLR4 expression in pancreatic ductal adenocarcinoma. Int J Biochem Cell Biol 2019;108:84–91. 10.1016/j.biocel.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 27.Xu H, Cheung IY, Guo H-F, et al. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res 2009;69:6275–81. 10.1158/0008-5472.CAN-08-4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, Lei T, Xu C, et al. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem Biophys Res Commun 2013;438:439–44. 10.1016/j.bbrc.2013.07.095 [DOI] [PubMed] [Google Scholar]
- 29.Zhou X, Mao Y, Zhu J, et al. TGF-β1 promotes colorectal cancer immune escape by elevating B7-H3 and B7-H4 via the miR-155/miR-143 axis. Oncotarget 2016;7:67196–211. 10.18632/oncotarget.11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Wang M, Zhang W, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc 2019;14:756–80. 10.1038/s41596-018-0113-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Wang Y, Wu Y, et al. Therapeutic potential of an anti-HER2 single chain antibody-DM1 conjugates for the treatment of HER2-positive cancer. Signal Transduct Target Ther 2017;2:17015. 10.1038/sigtrans.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. Embo J 2010;29:27–40. 10.1038/emboj.2009.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabio G, Davis RJ. TNF and MAP kinase signalling pathways. Semin Immunol 2014;26:237–45. 10.1016/j.smim.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Jiang C, Liu Z, et al. B7-H3-targeted CAR-T cells exhibit potent antitumor effects on hematologic and solid tumors. Mol Ther Oncolytics 2020;17:180–9. 10.1016/j.omto.2020.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X, Wang Y, Huang J, et al. Administration of B7-H3 targeted chimeric antigen receptor-T cells induce regression of glioblastoma. Signal Transduct Target Ther 2021;6:125. 10.1038/s41392-021-00505-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang X, Liu F, Liu Z, et al. Bioactivity and safety of B7-H3-targeted chimeric antigen receptor T cells against anaplastic meningioma. Clin Transl Immunology 2020;9:e1137. 10.1002/cti2.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zohn IE, Li Y, Skolnik EY, et al. P38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 2006;125:957–69. 10.1016/j.cell.2006.03.048 [DOI] [PubMed] [Google Scholar]
- 41.Nagy Z, Riss A, Romier C, et al. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol Cell Biol 2009;29:1649–60. 10.1128/MCB.01076-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Xiao W, Wang X-D, et al. The p38-interacting protein (p38IP) regulates G2/M progression by promoting α-tubulin acetylation via inhibiting ubiquitination-induced degradation of the acetyltransferase GCN5. J Biol Chem 2013;288:36648–61. 10.1074/jbc.M113.486910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X, Wang Q-L, Li Y-F, et al. A novel miR-200b-3p/p38IP pair regulates monocyte/macrophage differentiation. Cell Discov 2016;2:15043. 10.1038/celldisc.2015.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res 2005;15:11–18. 10.1038/sj.cr.7290257 [DOI] [PubMed] [Google Scholar]
- 45.Han J, Lee JD, Bibbs L, et al. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994;265:808–11. 10.1126/science.7914033 [DOI] [PubMed] [Google Scholar]
- 46.Wang X-D, Zhao C-S, Wang Q-L, et al. The p38-interacting protein p38IP suppresses TCR and LPS signaling by targeting TAK1. EMBO Rep 2020;21:e48035. 10.15252/embr.201948035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009;136:731–45. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Preiss T, W Hentze M. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 2003;25:1201–11. 10.1002/bies.10362 [DOI] [PubMed] [Google Scholar]
- 49.Waskiewicz AJ, Flynn A, Proud CG, et al. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. Embo J 1997;16:1909–20. 10.1093/emboj/16.8.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waskiewicz AJ, Johnson JC, Penn B, et al. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol 1999;19:1871–80. 10.1128/MCB.19.3.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun S-Y, Rosenberg LM, Wang X, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res 2005;65:7052–8. 10.1158/0008-5472.CAN-05-0917 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004875supp001.pdf (1.4MB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.