Abstract
Visualizing cell behavior and effector function on a single cell level has been crucial for understanding key aspects of mammalian biology. Due to their small size, large number and rapid recruitment into thrombi, there is a lack of data on fate and behavior of individual platelets in thrombosis and hemostasis. Here we report the use of platelet lineage restricted multi-color reporter mouse strains to delineate platelet function on a single cell level. We show that genetic labeling allows for single platelet and megakaryocyte (MK) tracking and morphological analysis in vivo and in vitro, while not affecting lineage functions. Using Cre-driven Confetti expression, we provide insights into temporal gene expression patterns as well as spatial clustering of MK in the bone marrow. In the vasculature, shape analysis of activated platelets recruited to thrombi identifies ubiquitous filopodia formation with no evidence of lamellipodia formation. Single cell tracking in complex thrombi reveals prominent myosin-dependent motility of platelets and highlights thrombus formation as a highly dynamic process amenable to modification and intervention of the acto-myosin cytoskeleton. Platelet function assays combining flow cytrometry, as well as in vivo, ex vivo and in vitro imaging show unaltered platelet functions of multicolor reporter mice compared to wild-type controls. In conclusion, platelet lineage multicolor reporter mice prove useful in furthering our understanding of platelet and MK biology on a single cell level.
Introduction
Platelets are anucleate cells of 1-2 µm diameter and are the second most abundant blood cell type. They are rapidly recruited upon vascular injury, undergo an adhesion and activation cascade and subsequently form a hemostatic plug. This process is highly coordinated and tightly interconnected with the coagulation system.1 In atherosclerosis, erosion or rupture of plaques leads to unwanted platelet recruitment and thrombus formation, causing vessel occlusion and ischemia.2
In vivo imaging of murine thrombosis models has considerably advanced our understanding of this process.3 For example, recent work has contributed to understanding thrombus architecture, defining layers of differently activated platelets in growing thrombi.4 Fluorescent tracking of key components like tissue factor, procoagulant surface and thrombin generation have shed light on thrombus formation in detail.4-6 Nevertheless, tracking and analysis of individual platelets has remained elusive because of their small size, large number and rapid re-cruitment.7
Platelets originate from megakaryocytes (MK) in the bone marrow, spleen and lung.8,9 Maturation of MK into platelet-producing cells occurs through differentiation from hematopoietic stem cells (HSC) to multipotent progenitor, common myeloid progenitor, MK-erythroid progenitor, and MK progenitor.10 Upon final differentiation, MK progenitors undergo DNA replication without cell division, a process termed endomitosis.11 This process leads to accumulation of DNA content of 4n, 8n, 16n, 32n, 64n, and even up to 128n in a single polylobulated nucleus.11,12 This process results in functional gene amplification most likely necessary for a ramp-up in protein synthesis necessary for platelet production.13 Genetic targeting after/during endomitosis can lead to stochastic recombination events in each set of chromosomes, leading to distinct genotypes within one cell.
In a final step, MK start to produce proplatelets in a shear-dependent manner by protrusion of cytoplasmic extensions into the blood flow. Despite their large size, MK turnover, behavior and positioning in the bone marrow niche are not well understood, warranting in vivo and in vitro imaging approaches.14-16
Multicolor reporter mouse strains have proven to be very useful in understanding cellular dynamics and cell fate in a wide range of cell types in vivo.17-21 The Brainbow imaging technique, originally developed to visualize and distinguish individual neurons and their fine axonal processes in the brain, allowed for the first-time multicolor tracking of individual cells.22 Recently, a Cre-re-porter mouse, termed R26R-Confetti was developed to achieve tissue-specific Brainbow expression.23
Here we report the use of a platelet lineage restricted R26R-Confetti multicolor reporter mouse strain which allows for single platelet and MK tracking and morphological analysis in vivo and in vitro, while not affecting lineage functions.
Methods
Mouse strains
PF4-Cre24 and Rosa26-Confetti23 were purchased from The Jackson Laboratory and maintained and cross-bred at our animal facility (stock no: 008535 and 017492). vWF-Cre mice were a gift of W. Aird.25 Mhy9 fl/fl mice were a gift from Dr. Gachet.26 All strains were backcrossed to and maintained on C57BL/6J-background. If not otherwise stated, animals of same sex and age were randomly assigned to experimental groups, and PF4-Cre negative control animals were used. We used Rosa26Confetti/+ animals expressing Cre for Online Supplementary Figure S1. The other experiments were performed with homozygous Rosa26Confetti/Confetti mice expressing Cre. Cre negative animals were used as controls. All procedures performed on mice were approved by the local legislation on the protection of animals (Regierung von Oberbayern, Munich).
Confocal intravital microscopy of the mesentery
After induction of narcosis, animals received 15 µg of X488 antibody (Emfret) and 80 µg of Fibrinogen-Alex-aFluor594 conjugate to visualize platelets and fibrin(ogen), respectively. Confocal microscopy of the mesentery was performed as previously described.27 Briefly, the skin and peritoneum were opened along the midline, and 200 µL of warm phosphate-buffered saline was pipetted into the abdominal cavity. Next, the proximal bowel was carefully exteriorized onto a glass coverslip and the mesenteric vessels was exposed. Wet tissue paper was used to stabilize the bowel and prevent artifacts from bowel movements. A 1x1 mm piece of filter paper was saturated with 10% ferric chloride solution and was carefully placed in direct contact with a mesenteric vein for 3 minutes (min). Next, the ensuing thrombus formation was imaged on an inverted Zeiss LSM 880 in AiryScan Fast Mode (20x/0.8 obj., 990 ms/frame, laser power: 0.96%).
Bone marrow whole mount stains
Bone marrow from PF4cre-Confetti animals was harvested and processed as described previously. For whole mounts, bones were snap frozen after paraformaldehyde (PFA) fixation (4%, 60 min) and sucrose treatment (30%, overnight). Bones were then cut longitudinally with a cryotome to expose marrow, and were stained with CD41-AF647, (1:100), CD144-AF647 (1:100), FarRed Nuclear Stain (1:1000) and were imaged with a Zeiss LSM 880 confocal microscope (20x/0.8 obj.) in AiryScan mode.
Static thrombus formation
Static thrombi were generated by activating platelet-rich plasma (PRP) with thrombin (0.1 U) for 5 min, dilution of these thrombi 1:10 and assessment of platelets was done with a LSM 880 confocal microscope in AiryScan mode.
Histology
For mesentery, lung and cremaster muscle histology, organs were harvested and kept in 1% PFA for 1 hour, before transfer in 20% sucrose overnight. After embedding in TissueTek OCT mounting medium (Sakura), organs were snap frozen and cut using a cryotome (10 µm slices). Staining was performed using antibodies as indicated.
Flow chamber experiments
As previously described,28 heparinized whole blood was perfused over a collagen coated surface at arterial shear rate (1,000/s) for 5 min. Formed thrombi were assessed on a LSM 880 confocal microscope in AiryScan mode (20x/0.8 obj., 2x zoom for overviews, and 5x zoom for detailed views, laser power: 0.96%).
Data analysis
Data analysis was realized using FIJI (ImageJ) and ZenBlack (Zeiss). 4D in vivo data was stabilized with the “ImageStabilizer” plugin and dimensions were reduced by maximum intensity projection (MIP). Cells were tracked with “Manual tracking” FIJI plugins. For secondary analysis, IBIDI Chemotaxis tool was used. For form analysis, cells were manually outlined, masked and converted to binary images. Solidity also known as convexity, was computed as the proportion of the pixels in the convex hull that are also in the object (Area/ConvexArea). Aspect ratio as a maker of polarization was computed by major_axis divided by minor_axis. Number of filopodia were quantified manually. For quantification, FIJI shape descriptors were used. Data were post-processed in Excel (Microsoft), Prism (GraphPad) and Illustrator (Adobe).
Statistical analysis and reproducibility
Data are shown as means ± standard error of the mean (SEM). Statistical parameters, including the exact value of replicates (‘‘n’’) for individual experiments can be found within the figure legends. In order to evaluate statistical differences between groups, t-tests and analysis of variance (ANOVA) were performed. A P value of <0.05 was considered statistically significant. Analyses were performed with Prism (GraphPad Software) and Excel (Microsoft).
Results
A multicolor platelet lineage reporter mouse
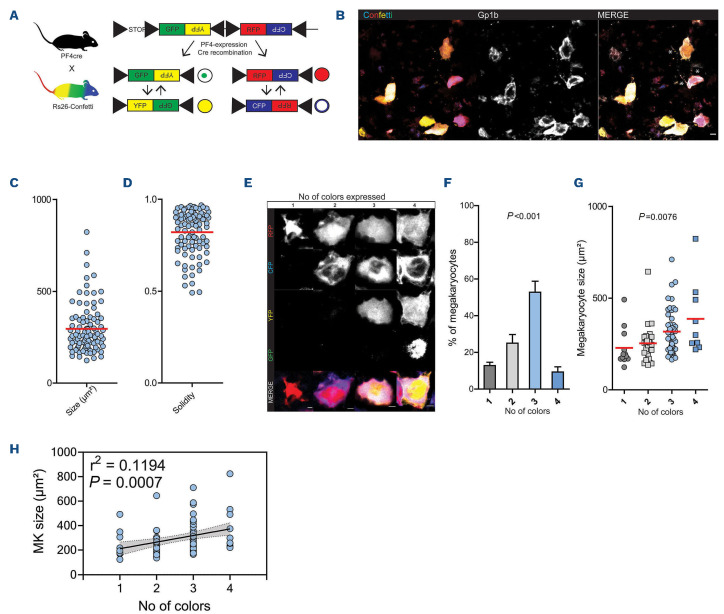
Livet et al. introduced the Cre-recombinase dependent Brainbow 2.1 construct (loxP-STOP-loxP-GFP-PFY-PxolloxP-RFP-PFC-Pxol) to allow genetic fluorescent labeling of neuronal networks.22 The construct consists of two color tandems, yellow fluorescent protein (YFP) and inverted green fluorescent protein (GFP) and red green fluorescent protein (RFP) and inverted cyan green fluorescent protein (CFP), creating YFP-, GFP-, RFP- or CFP-positive populations (Figure 1A). GFP is expressed in the nucleus, CFP is membrane bound, whereas YFP and RFP show cytosolic expression. Flanking loxP sites enable Cre driven recombination, leading to stochastically driven removal and/or inversion of tandem constructs. Repeated/ongoing expression of the Cre recombinase can lead to expression of the anti-sense fluorescent protein by inversion of the tandem after initial recombination. Snippert et al. recently developed a Cre-reporter mouse, termed R26R-Confetti-stop-flox, allowing tissue specific Brainbow 2.1 expression under the control of a Cre-Deleter.23 We crossed R26R-Confetti-stop-flox mice to a platelet factor 4 (PF4)-Cre transgenic mice to limit recombination events to the platelet/MK lineage (Figure 1A).24
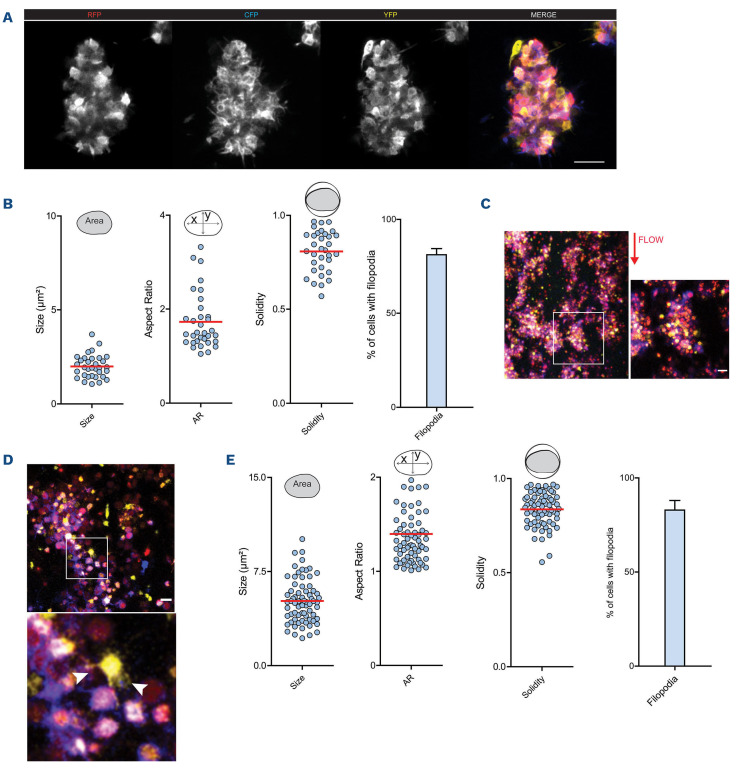
Confocal microscopy of bone marrow whole mounts revealed large, brightly labeled cells that were positive for platelet/MK marker Gp1b, confirming that these cells were indeed MK (Figure 1B; Online Supplementary Figure S1A). Also, we detected proplatelet formation in a subpopulation of Confetti-expressing cells indicating intact thrombopoiesis (Figure 1B). Multicolor expression enabled precise size and shape analyses of labeled MK (Fig-
Confeti expression in polyploid megakaryocytes generates unique color paterns and reveals late expression of platelet factor 4 in the megakaryocyte lineage
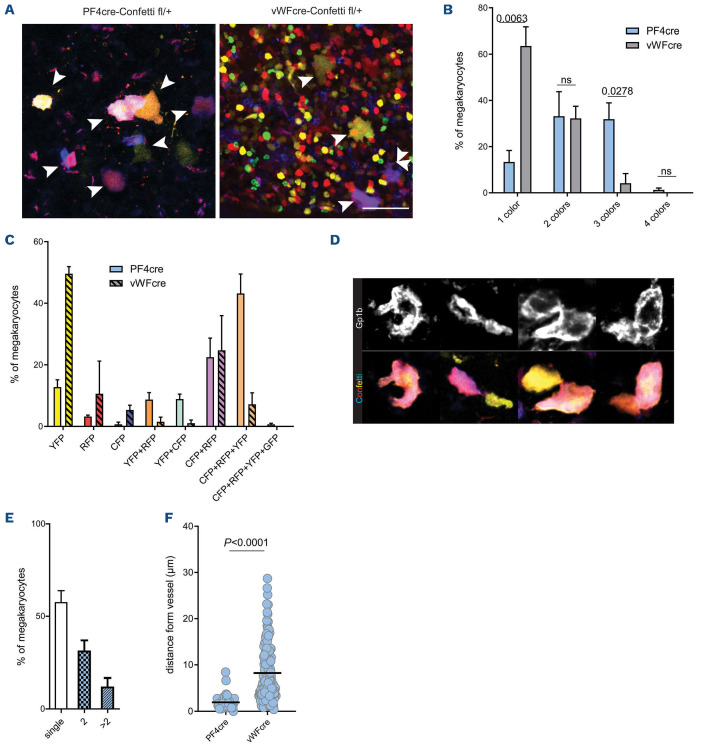
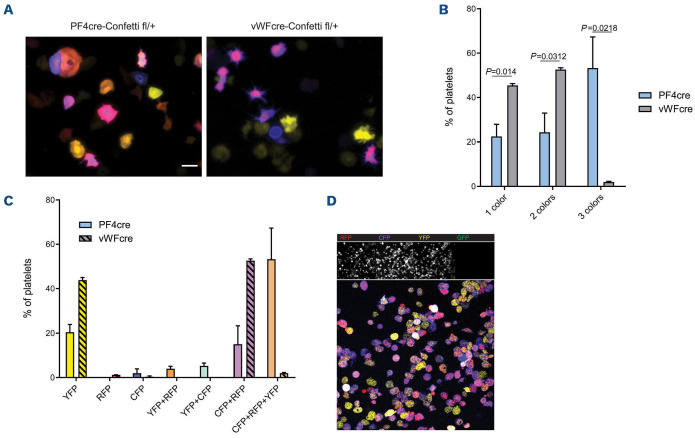
In contrast to previously reported Confetti-expression patterns in other cell types showing a single XFP expressed per cell, individual MK expressed one up to all four possible XFP (Figure 1E).19,23 The majority of MK expressed two to three XFP (Figure 1F). MK are unique as they undergo endomitosis during their maturation. Therefore, genetic targeting after endomitosis can theoretically lead to multiple genotypes in one cell. This could explain the recombination of multiple Confetti-constructs in PF4crepositive MK. Indeed, Confetti-positive cells contained multilobulated nuclei and correlation analysis of color expression to MK size revealed that color expression diversity increased with cell size, a parameter that strongly correlates with ploidy and DNA content (Online Supplementary Figure S1B; Figure 1G, H).30 In order to confirm this observation, we generated von Willebrand factor (vWF)-Cre; R26RConfetti/+ mice. vWF is expressed early in the MK lineage, at the level of platelet-biased stem cells, and should therefore lead to recombination prior to endomitosis - and thus also prior to polyploidization.31 Indeed, in contrast to PF4-Cre; R26RConfetti/+ mice, vWF-Cre; R26RConfetti/+ MK expressed either one color (mainly YFP) or two colors (RFP and CFP) (Figure 2A-C). The simultaneous coexpression of RFP and CFP is possible since their two open reading frames are positioned in a head-to-tail tandem dimer and are therefore able to permanently flip during megakaryopoiesis due to a persistent Cre recombinase expression under the control of the PF4 or vWF promoter.22 We confirmed this finding in peripheral blood platelets of these mice (Figure 3A-C). Together, our data provide new insights into the expression pattern of the PF4-Cre transgene in the MK lineage: robust expression in MK seems to occur rather late in MK development, with important implications regarding the use of PF4-Cre in studying MK biology. As reported previously by others, we also detected PF4-Cre-driven expression of XFP in cells beyond the MK lineage for example in the mesenterial fat and cremaster muscle tissue (Online Supplementary Figure S1C, D).32 These cells appear to be mainly of macrophage origin as evidenced by their morphology.33
Analysis of the megakaryocyte niche in Confeti mice reveals clusters of mature megakaryocytes
The MK bone marrow niche is still only partially understood, and analysis/differentiation of single or clustered MK is hampered by their unconventional shape in different states of thrombopoiesis, forming long protrusions or extending pro-platelets. Gp1b staining revealed multiple MK that could not be clearly defined as single or clustered cells (Figure 2D). The Confetti signal enabled separation of single and clustered MK, and unexpectedly revealed that almost 50% of MK were found in clusters of two or more cells (Figure 2D, E). Large, mature MK (PF4+) were found in close vicinity to CD144+ blood vessels (Figure 2F). In contrast, small cells expressing vWF-Cre-driven Confetti (a mixed cell population containing both immature MK progenitors and non-MK cells) localized further away from vessels, highlighting the preferential localization of mature MK near vessels (Figure 2F).34 In conclusion, multicolor reporter mice may provide a useful tool to investigate clonality and expansion of the MK within the bone marrow niche.
Exclusion of functional effects on the megakaryocyte/platelet lineage in PFR4-Cre-Confeti mice
Confocal imaging of isolated platelets from PF4-Cre-Rs26-Confetti mice revealed multicolor expression in YFP, RFP and CFP, but no nGFP labeling, as this fluorescent protein is restricted to the nucleus (Figure 3D). Next, we excluded effects of this construct on the function of the platelet/MK lineage as this would affect generalization of findings drawn from using this mouse line. Platelet counts, mean platelet volume and hemoglobin content in peripheral blood did not show significant changes between Cre+ and Cre- animals (Online Supplementary Figure S2A). Flow cytometric analysis of isolated platelets confirmed strong fluorescence in PE and FITC channels in Cre+ mice, which was absent in Cre- mice (Online Supplementary Figure S2B), while morphology was unaltered (Online Supplementary Figure S2C). Flow cytometric assessment of quiescent platelets showed no alterations in size, surface expression of Gp1b and GpIIbIIIa, activation status (P-selectin expression) or fibrinogen binding in response to activation (Online Supplementary Figure S2D). Upon stimulation with ADP, collagen or thrombin, markers of activation did not differ significantly between Cre+ animals and Cre- littermates (Online Supplementary Figure S2E-G). Efficiency of platelet activation was independent of XFP-expression as gating on XFP (YFP+, RFP+, and YFP+RFP+) revealed no difference in fibrinogen binding upon thrombin stimulation (Online Supplementary Figure S2H).
Figure 1.
Rs26-Confetti as a tool to study megakaryocytes in vivo. (A) Rs26-Confetti gene product and Cre-dependent recombination; (B) representative endogenous Confetti fluorescence (YFP, RFP, GFP and CFP) and Gp1b (AlexaFluor647) staining of a bone marrow whole mount. *Indicates pro-platelet formation; (C and D) size and shape analysis of megakaryocytes (MK) in bone marrow whole mounts, n=93 cells in 8 bones; (E) representative MK expressing 1-4 colors; (F) percentage of MK expressing 1,2,3 or 4 colors, n=8 bones, ANOVA across color expression groups; (G to H) MK size related to color expression (1, 2, 3, 4 colors), n=93 cells in 8 bones; (G) ANOVA across color expression groups; (H) correlation analysis using linear regression. YFP: yellow fluorescent protein; GFP: green fluorescent protein; RFP: red green fluorescent protein; CFP: cyan green fluorescent protein.
Next, we assessed hemostatic function in reporter and littermate mice. Time to initial hemostasis and total bleeding time were unaltered in tail bleeding assays (Online Supplementary Figure S3A). In vitro assessment of clot retraction revealed a similar degree of retraction (Online Supplementary Figure S3B).
A ferric-chloride induced thrombosis model of the carotid artery revealed similar thrombus formation kinetics in Cre+ and Cre- animals (Online Supplementary Figure S3C; Online Supplementary Video S1). Simultaneous visualization of XFP (RFP and YFP) and intravenously (i.v.) injected plateletlabeling antibody (Far Red; Dylight 649) revealed the same platelet recruitment pattern indicating that the endogenous fluorescence signal in Cre+ mice, which was absent in Cre- mice allowed reliable tracking of thrombus formation in vivo (Online Supplementary Figure S3D). In addition, similar amounts of isolated platelets were recruited to collagen and fibrinogen coated surfaces and showed comparable spreading and lamellipodium formation (Online Supplementary Figure S3E-H).
Figure 2.
Rs26-Confetti expression in the platelet lineage reveals megakaryocyte clustering in the bone marrow. (A-C) Analysis of megakaryocytes (MK) in bone marrow whole mounts from von Willebrand factor (vWF)-Cre and PF4-Cre dependent Rs26-Confetti reporter mice. (A) Representative micrographs showing YFP, CFP, RFP and GFP overlay, arrows: MK, scale bar: 50 µm. Of note, vWF is expressed early during megkaryopoiesis in pluripotent platelet biased hematopoietic stem cells, leading to vWF-Credriven expression of Confetti in a large proportion of small, multi-lineage bone marrow cells in addition to large MK. (B) Quantification of color expression n=3 mice per group, t-test; (C) detailed analysis of color expression. (D) Confetti fluorescence (YFP, RFP, GFP and CFP) and Gp1b (DyLight647) staining of representative single/grouped cells. Image was cropped from Figure 1B and shown at higher magnification. Compare Figure 1B and E percentage of single and grouped MK, n=10 bones, ANOVA. (F) Mean distance of Confetti positive cells in the bone marrow of PF4-cre (n=50) and vWF-cre (n=157) dependent Rs26-Confetti reporter mice from CD144+ blood vessels. Analyzed in n=3 bones from individual mice per group, t-test. YFP: yellow fluorescent protein; GFP: green fluorescent protein; RFP: red green fluorescent protein; CFP: cyan green fluorescent protein.
Figure 3.
Rs26-Confetti expression in platelets. (A-C) Analysis of peripheral blood platelets of von Wllebrand factor (vWF)-Cre and PF4-Cre dependent Rs26-Confetti reporter mice. (A) Representative micrographs of spread platelets showing YFP, CFP, RFP overlay; (B) quantification of color expression n=3 mice per group, t-tests; (C) detailed analysis of color expression. (D) Fluorescence expression in PF4-Cre-Rs26-Confetti platelets. Scale bars= 5 µm. YFP: yellow fluorescent protein; GFP: green fluorescent protein; RFP: red green fluorescent protein; CFP: cyan green fluorescent protein.
Analysis of multicolor platelets in reporter mice
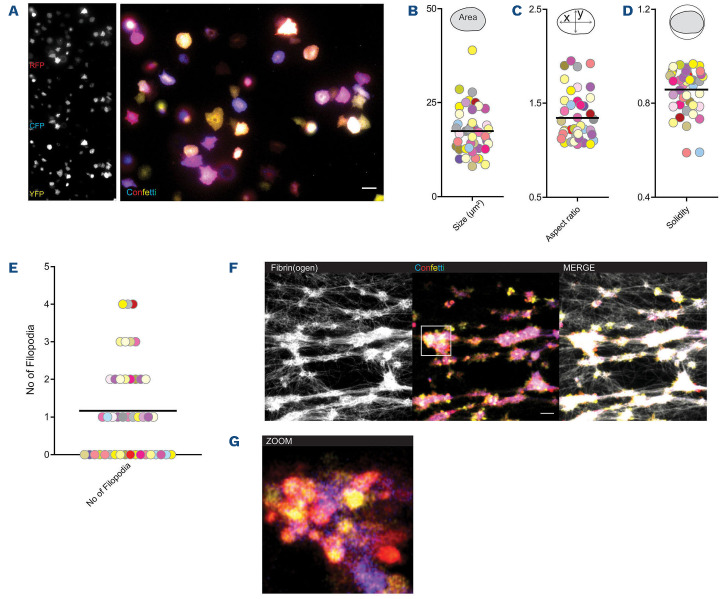
Reporter platelets spreading on fibrinogen revealed homogenous distribution of fluorescence signal in individual cells (Figure 4A). As expected from our data on fluorescence expression in MK, platelets revealed a range of colors depending on (i) the expression of one to three XFP and (ii) intensity of expression of each XFP (Figure 4A). Mapping color and size, aspect ratio (marker of polarization) and solidity (marker of irregular shape) and number of filopodia did not reveal overt clustering of cells dependent on color (Figure 4B-E). Similarly, analysis based on expressed color (YFP, RFP and CFP) did not reveal differences in size or shape or migration behavior (Online Supplementary Figure S4A-C).
Morphology analysis of platelets within thrombi shows prominent filopodia formation
Thrombosis and hemostasis trigger rapid recruitment of activated platelets in large numbers and therefore complicate single cell tracking. In order to test if multi-color labeling by R26R-Confetti allows segmentation of individual platelets within the cell-packed environment of a thrombus we generated fibrin-rich platelet thrombi under static conditions in vitro. Confocal microscopy revealed reorganization of the fibrin network by platelets, leading to tight platelet-fibrin aggregates (Figure 4F). Stochastic expression of XFP in R26R-Confetti platelets still allowed identification of individual platelets in dense aggregates (Figure 4G).
Next, we generated static microthrombi from isolated Confetti platelets (Figure 5A). Confocal analysis enabled us to discriminate single platelets in these tightly packed aggregates, as well as protrusions formed by individual cells classified as filopodia (Online Supplementary Video S2). Recruited platelets did not form discernible sheet-like lamellipodia (Figure 5A). In addition, the morphology of the individual cells could be quantified and shape descriptors of aggregated platelets were determined. Over 80% of recruited cells formed filopodia of various lengths (Figure 5B). Flow chamber-based assays are an important tool to study thrombus formation in vitro. By using heparinized blood from R26R-Confetti reporter mice, we were able to visualize multicolor aggregate formation under blood shear conditions, and higher magnification revealed single platelet morphology (Figure 5C, D). Similar to static thrombi generated from isolated platelets, the majority of recruited platelets formed filopodia, but no lamellipodia (Figure 5D, E).
Figure 4.
Rs26-Confetti expression in activated platelets in vitro. (A-E) Spreading of Rs26-Confetti platelets on fibrinogen. (A) Representative micrograph; (B) size; (C), aspect ratio and (D) solidity in relation to color expression, n=48 cells; (E) filopodia formation. (F, G) In vitro generated fibrin clots using Rs26-Confetti platelet-rich plasma, (G) revealing cell aggregates.
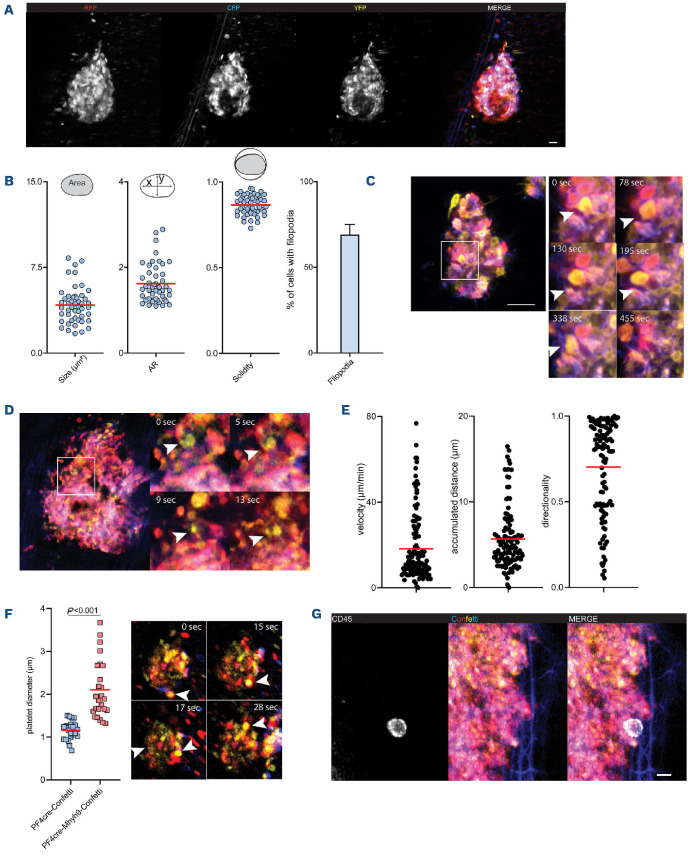
In vitro approaches are limited when studying complex biological events. Therefore, we assessed the applicability of platelet R26R-Confetti expression in vivo. In order to study thrombus formation, we utilized a ferric-chloride induced mesenteric thrombosis model. Similar to our in vitro findings, RFP, YFP and CFP fluorescence expression allowed for intravital discrimination of individual cells in growing thrombi (Figure 6A; Online Supplementary Video S3). Moreover, analysis of single platelet morphology revealed formation of filopodia of the majority of recruited individual platelets (Figure 6B).
Single cell tracking of individual cells in complex thrombi reveals myosin dependent motility in vivo and in vitro
For a long time, activated recruited platelets were considered stationary cells. Recently, platelets were shown to actively migrate at sites of vascular injury.7,35 In addition, myosin IIa-dependent pulling forces trigger collective platelet motion in a process termed clot retraction.26 We utilized the Confetti model to track individual platelets in static microthrombi over time using time lapse microscopy and identified single motile platelets (Figure 6C; Online Supplementary Video S2).
Similarly, single cell tracking in vivo showed relocation dynamics of individual platelets in complex thrombi (Figure 6D). Particularly in the consolidation phase of thrombi, collective motility of recruited platelets was observed (Online Supplementary Video S3). We tracked >100 platelets in a single thrombus, which on the one hand revealed platelet repositioning in all parts of the thrombus. On the other hand, the observed motility patterns were highly heterogenous, as accumulated distance, velocity and directionality varied between cells (Figure 6E).
Figure 5.
Rs26-Confetti enables studying of platelet dynamics on a single cell level in vitro. (A, B) In vitro generated static platelet thrombi. (A) Representative micrograph and (B) shape descriptors and filopodia of single platelets; n=33 cells in two thrombi. (C-E) Flow chamber experiments with arterial shear rate (1,000/s) over collagen; (C, D) representative micrographs and (E) shape descriptors and filopodia of single platelets; n=33 cells in two thrombi; n=71 cells from three micrographs.
Finally, we bred PF4cre; R26RConfetti mice to Mhy9-floxed mice to generate a multicolor reporter mouse of myosin heavy chain deficient platelets.26 Mhy9-disorders are characterized by increased platelet size and reduced number – macrothrombocytopenia. In vitro, Mhy9-deficient platelets spread normally with no apparent morphological difference from control platelets (Online Supplementary Figure S4D). By intravital microscopy of injured mesenteric vessels in PF4cre; R26RConfetti; Myh9flox mice we were able to perform size measurements of individual platelets in vivo confirming enlarged Mhy9-/- platelets (Figure 6F). Single cell tracking within thrombi revealed reduced velocity and accumulated distances of Myh9-deficient platelets compared to control mice (Online Supplementary Figure S4E).26 The associated lack of collective contractile behavior led to increased embolization and less consolidation of the thrombus as the clot was not retracted (Online Supplementary Video S4; Figure 6F).
Figure 6.
Rs26-Confetti enables studying of platelet dynamics on a single cell level in vivo. (A, B) In vivo FeCl3-dependent thrombus formation in the mesentery. (A) Representative micrograph and (B) shape descriptors and filopodia of single platelets; n=48 cells in two thrombi. (C) Tracking of individual platelets over time (arrows) in static in vitro thrombi. Micrograph shows higher magnification of a region of interest cropped from (A). (D, E) Tracking of thrombus recruited platelets in vivo; (D) exemplary micrograph of individual motile platelet (arrow) and (E) tracking of n=118 individual platelets in single thrombus allows for computation of single cell velocity, covered distance and directionality. (F, left) Size differences of control and Mhy9-deficient platelets on the Rs26-Confetti background are evident in vivo and (F, right) differences of individual platelet behavior in Mhy9-deficient animals in Fe Cl3-dependent thrombus formation in the mesentery. Arrow marks exemplary platelet moving through thrombus with flow, unable to retract. (G) Visualization of CD45+ leukocytes in Rs26-Confetti thrombi. Scale bar =10 mm.
Thrombus formation is a complex process involving the concerted action of additional cell types, for example neutrophils and monocytes. It is therefore important to be able to label additional cell populations in deployed fluorescence reporter strains, and to exclude passive platelet motility patterns depending on leukocyte migration.35 We injected CD45 antibody conjugated to a Far Red emitter (Alexa Fluor 647) to stain circulating leukocytes. Excitation/emission spectra revealed no overlap with Confetti signal and enabled side-by-side visualization of leukocytes and platelets in multicolor thrombi (Figure 6G; Online Supplementary Figure S4F).
Discussion
Here, we describe in detail a novel mouse model that can be used to assess single cell dynamics of the platelet/MK lineage. We previously utilized this model to study single cell behavior mainly under inflammatory conditions in vivo, which helped to reveal the platelet migratory phenotype.7 We further extended our understanding of the employed mouse model, and ruled out a major effect of Confetti fluorescence expression/ Cre recombination on platelet and MK biology. In addition to tracking of individual cells, this model allows to discern cell shape in vivo and in vitro, which is important for understanding platelet effector functions.35 We demonstrate the versatility of this model by showing that it also allows for additional staining of populations/antigens using far red emitters.
We substantiate the relevance of the reporter model for a better understanding of the biology of MK and platelets by revealing previously unkown aspects of this lineage. First, we identified pairs and clusters of MK in the bone marrow. These clusters may indicate clonal expansion of MK precursors within the MK-specific bone marrow niche, where MK precursors proliferate before differentiating and forming platelets.31,34 Additional work will be required to resolve megakaryopoiesis spatially and temporally in greater detail.
PF4 is expressed in mature MK where it has been shown to act as negative regulator of megakarypoesis and hematopoiesis.15,36 Thus, late PF4 expression possibly serves as a paracrine negative feedback loop to control MK differentiation. Expression of multiple XFP following PF4-Cre driven recombination of the Confetti construct supports the notion that PF4 is mainly expressed after initiatiation of MK endomitosis in mature MK. Therefore, it might serve as a useful late genetic marker for studying megakaryopoesis, however, with the drawback of potenial off-target effects in monocytic cells.32,33 In contrast, using the vWF promotor for specific and robust Cre expression in early MK progenitors, might prove useful in studying the contribution of cytoskeletal proteins or transcription factors in early megakaryopoiesis.
Despite great progress in recent years, it remains difficult to assess individual platelet behavior in vivo.37 Especially data on platelet cytoskeletal rearrangements during activation and recruitment are almost exclusively derived from in vitro studies. For example, platelet lamellipodia formation, termed “spreading”, is a key element of in vitro assessment of platelet function.38 However, there is no evidence that platelets form lamellipodia in hemostatic plugs in vivo.39 Nesbitt et al. have demonstrated that platelets might not undergo dramatic shape changes in thrombosis.40 Recently, using mouse lines in which lamellipodia formation is disrupted, we identified lamellipodia as a key cytoskeletal feature of immune responsive platelets with a neglible role in classical thrombosis and hemostasis.35 Along these lines, we here consolidate these data by excluding lamellipodia formation but showing prominent filopodia formation of activated platelets in three different thrombosis models in vivo and in vitro. These needle-like protrusions might serve as anchor points as well as sensors registering the microenviroment.41 In addition, they might increase platelet surface area and redistribute adhesion receptors,42 affecting the platelets propensity to activate. This fits very well with data showing normal hemostasis and thrombosis in mice unable to form lamellipodia.35,39,43,44 As interference with the cytoskeleton might prove an interesting pharmacological target, our data highlight the need to further investigate the regulation of filopodia formation in platelets.
Regarding platelet motility within complex thrombi, our experiments show ubiquitous and global repositioning of platelets in thrombus formation. Single cell tracking in our model revealed a collective motion of platelets towards the thrombus core largely depending on force generation by myosinIIa, as genetic ablation of Mhy9 drastically alters clot formation and reduces platelet re-location, confiming previous reports.26 In addition to consolidation of the clot, the centripedal, collective motion of platelets may generate forces required for the mechanical extrusion of procoagulant platelets to reinforce thrombin generation and fibrin formation at the thrombus surface as recently proposed by others.45 Thus, multicolor tracking of platelets highlights thrombus formation as a highly dynamic process amenable to modification and intervention of the acto-myosin cytoskeleton. Combination of platelet-specific multicolor reporter mouse lines with genetic/pharmacological targeting and labeling of neutrophils or other immune cells might help to better understand clot initiation, consolidation and lysis.
Future studies could take advantage of this model to understand single platelet and MK dynamics in a wide range of disease models and conditions.
Supplementary Material
Acknowledgments
The authors thank Anna Titova, Sebastian Helmer, Nicole Blount and Beate Jantz for technical assistance.
Funding Statement
Funding: This study was supported by the Deutsche Forschungsgemeinschaft (DFG) SFB 914 (to SM [B02 and Z01]), the DFG SFB 1123 (to SM [B06]), the DFG FOR 2033 (to SM), the German Center for Cardiovascular Research (DZHK) (Clinician Scientist Programme), MHA 1.4VD (to SM), Postdoc Start-up Grant, 81X3600213 (to FG), 81X3600222 (to LN), the FP7 program (project 260309, PRESTIGE [to SM]). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 83344, ERC-2018-ADG “IMMUNOTHROMBOSIS” [to SM] and the Marie Skłodowska Curie Individual Fellowship (EU project 747687, LamelliActin [to FG]).
References
- 1.Versteeg HH, Heemskerk JWM, Levi M, Reitsma PH. New fundamentals in hemostasis. Physiol Rev. 2013;93(1):327-358. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP. Arterial thrombosis--insidious, unpredictable and deadly. Nat Med. 2011;17(11):1423-1436. [DOI] [PubMed] [Google Scholar]
- 3.Furie B, Furie BC. Mechanisms of thrombus formation. N Engl J Med. 2008;359(9):938-949. [DOI] [PubMed] [Google Scholar]
- 4.Stalker TJT, Wu E, Wannemacher J, et al. Hierarchical organization in the hemostatic response and its relationship to the platelet-signaling network. Blood. 2013;121(10):1875-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stalker TJ, Welsh JD, Brass LF. Shaping the platelet response to vascular injury. Curr Opin Hematol. 2014;21(5):410-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong PC, Hoefer T, Knowles RB, et al. Newly formed reticulated platelets undermine pharmacokinetically short-lived antiplatelet therapies. Arterioscler Thromb Vasc Biol. 2017;37(5):949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaertner F, Ahmad Z, Rosenberger G, et al. Migrating platelets are mechano-scavengers that collect and bundle bacteria. Cell. 2017;171(6):1368-1382.e23. [DOI] [PubMed] [Google Scholar]
- 8.Lefrançais E, Ortiz-Muñoz G, Caudrillier A, et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017;544(7648):105-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junt T, Schulze H, Chen Z, et al. Dynamic visualization of thrombopoiesis within bone marrow. Science. 2007;317(5845):1767-1770. [DOI] [PubMed] [Google Scholar]
- 10.Haas S, Hansson J, Klimmeck D, et al. Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell- like megakaryocyte progenitors.. Cell Stem Cell. 2015;17(4):422-434. [DOI] [PubMed] [Google Scholar]
- 11.Machlus KR, Italiano JE. The incredible journey: from megakaryocyte development to platelet formation. J Cell Biol. 2013;201(6):785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmet J, Ravid K. Polyploidy: occurrence in nature, mechanisms, and significance for the megakaryocyte-platelet system. Exp Hematol. 2000;28(1):3-16. [DOI] [PubMed] [Google Scholar]
- 13.Machlus KR, Italiano JE. 2 - Megakaryocyte development and platelet formation. In: Michelson AD, editor. Platelets (Fourth Edition). Academic Press; 2019. p. 25-46. [Google Scholar]
- 14.Italiano JE. Megakaryopoiesis and Platelet Biogenesis. Molecular and Cellular Biology of Platelet Formation. Springer; 2016. p. 3-22. [Google Scholar]
- 15.Bruns I, Lucas D, Pinho S, et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat Med. 2014;20(11):1315-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Urtz N, Gaertner F, et al. Sphingosine kinase 2 (Sphk2) regulates platelet biogenesis by providing intracellular sphingosine 1-phosphate (S1P). Blood. 2013;122(5):791-802. [DOI] [PubMed] [Google Scholar]
- 17.Abe T, Fujimori T. Reporter mouse lines for fluorescence imaging. Dev Growth Diff. 2013;55(4):390-405. [DOI] [PubMed] [Google Scholar]
- 18.Chappell J, Harman JL, Narasimhan VM, et al. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ Res. 2016;119(12):1313-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tas JMJ, Mesin L, Pasqual G, et al. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351(6277):1048-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng T, Tian Y, Boogerd CJ, et al. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013;500(7464):589-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148(1-2):33-45. [DOI] [PubMed] [Google Scholar]
- 22.Livet J, Weissman TA, Kang H, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56-62. [DOI] [PubMed] [Google Scholar]
- 23.Snippert HJ, Van Der Flier LG, Sato T, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143(1):134-144. [DOI] [PubMed] [Google Scholar]
- 24.Tiedt R, Schomber T, Hao-Shen H, Skoda RC. Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood. 2007;109(4):1503-1506. [DOI] [PubMed] [Google Scholar]
- 25.Yuan L, Chan GC, Beeler D, et al. A role of stochastic phenotype switching in generating mosaic endothelial cell heterogeneity. Nat Commun. 2016;7:10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leon C, Eckly A, Hechler B, et al. Megakaryocyte-restricted MYH9 inactivation dramatically affects hemostasis while preserving platelet aggregation and secretion. Blood. 2007;110(9):3183-3191. [DOI] [PubMed] [Google Scholar]
- 27.Samson AL, Alwis I, Maclean JA, et al. Endogenous fibrinolysis facilitates clot retraction in vivo. Blood. 2017;130(23):2453-2462. [DOI] [PubMed] [Google Scholar]
- 28.Petzold T, Thienel M, Konrad I, et al. Oral thrombin inhibitor aggravates platelet adhesion and aggregation during arterial thrombosis. Sci Transl Medi. 2016;8(367):367ra168. [DOI] [PubMed] [Google Scholar]
- 29.Levine RF, Hazzard KC, Lamberg JD. The significance of megakaryocyte size. Blood. 1982;60(5):1122-1131. [PubMed] [Google Scholar]
- 30.Tomer A. Human marrow megakaryocyte differentiation: multiparameter correlative analysis identifies von Willebrand factor as a sensi 2004;104(9):2722-2727. [DOI] [PubMed] [Google Scholar]
- 31.Sanjuan-Pla A, Macaulay IC, Jensen CT, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502(7470):232-236. [DOI] [PubMed] [Google Scholar]
- 32.Nagy Z, Vögtle T, Geer MJ, et al. The Gp1ba-Cre transgenic mouse: a new model to delineate platelet and leukocyte functions. Blood. 2019;133(4):331-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pertuy F, Aguilar A, Strassel C, et al. Broader expression of the mouse platelet factor 4-cre transgene beyond the megakaryocyte lineage. J Thromb Haemost. 2015;13(1):115-125. [DOI] [PubMed] [Google Scholar]
- 34.Stegner D, Judith MM, Angay O, et al. Thrombopoiesis is spatially regulated by the bone marrow vasculature. Nat Commun. 2017;8(1):1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicolai L, Schiefelbein K, Lipsky S, et al. Vascular surveillance by haptotactic blood platelets in inflammation and infection. Nat Commun. 2020;11(1):5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert MP, Wang Y, Bdeir KH, Nguyen Y, Kowalska MA, Poncz M. Platelet factor 4 regulates megakaryopoiesis through low-density lipoprotein receptor-related protein 1 (LRP1) on megakaryocytes. Blood. 2009;114(11):2290-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson SP, Nesbitt WS, Westein E. Dynamics of platelet thrombus formation. J Thromb Haemost. 2009;7(Suppl 1):S17-20. [DOI] [PubMed] [Google Scholar]
- 38.Aslan JE, Itakura A, Gertz JM, McCarty OJ. Platelet shape change and spreading. Methods Mol Biol. 2012;788:91-100. [DOI] [PubMed] [Google Scholar]
- 39.Schurr Y, Sperr A, Volz J, et al. Platelet lamellipodium formation is not required for thrombus formation and stability. Blood. 2019;134(25):2318-2329. [DOI] [PubMed] [Google Scholar]
- 40.Nesbitt WS, Westein E, Tovar-Lopez FJ, et al. A shear gradientdependent platelet aggregation mechanism drives thrombus formation. Nat Med. 2009;15(6):665-673. [DOI] [PubMed] [Google Scholar]
- 41.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9(6):446-454. [DOI] [PubMed] [Google Scholar]
- 42.Galbraith CG, Yamada KM, Galbraith JA. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 2007;315(5814):992-995. [DOI] [PubMed] [Google Scholar]
- 43.Paul DS, Casari C, Wu C, et al. Deletion of the Arp2/3 complex in megakaryocytes leads to microthrombocytopenia in mice. Blood Adv. 2017;1(18):1398-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stenberg PE, Barrie RJ, Pestina TI, et al. Prolonged bleeding time with defective platelet filopodia formation in the Wistar Furth rat. Blood. 1998;91(5):1599-1608. [PubMed] [Google Scholar]
- 45.Nechipurenko DY, Receveur N, Yakimenko AO, et al. Clot contraction drives the translocation of procoagulant platelets to thrombus surface. Arterioscler Thromb Vasc Biol. 2019;39(1):37-47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.