Abstract
Cancer cells secrete pronociceptive mediators that sensitize adjacent sensory neurons and cause pain. Identification and characterization of these mediators could pinpoint novel targets for cancer pain treatment. In this study, we identified candidate genes in cancer cell lines that encode for secreted or cell surface proteins that may drive nociception. To undertake this work, we used an acute cancer pain mouse model, transcriptomic analysis of publicly available human tumor-derived cell line data, and a literature review. Cancer cell line supernatants were assigned a phenotype based on evoked nociceptive behavior in an acute cancer pain mouse model. We compared gene expression data from nociceptive and nonnociceptive cell lines. Our analyses revealed differentially expressed genes and pathways; many of the identified genes were not previously associated with cancer pain signaling. Epidermal growth factor receptor (EGFR) and disintegrin metalloprotease domain 17 (ADAM17) were identified as potential targets among the differentially expressed genes. We found that the nociceptive cell lines contained significantly more ADAM17 protein in the cell culture supernatant compared to nonnociceptive cell lines. Cytoplasmic EGFR was present in almost all (>90%) tongue primary afferent neurons in mice. Monoclonal antibody against EGFR, cetuximab, inhibited cell line supernatant-induced nociceptive behavior in an acute oral cancer pain mouse model. We infer from these data that ADAM17-EGFR signaling is involved in cancer mediator-induced nociception. The differentially expressed genes and their secreted protein products may serve as candidate therapeutic targets for oral cancer pain and warrant further evaluation.
Keywords: Cancer, Pain, Nociception, EGFR, ADAM17, Gene expression
1. Introduction
Cancer pain is the most severe symptom in a variety of cancers and predicts poor prognosis.56 Most forms of cancer pain are hypothesized to arise from cancer-secreted mediators that sensitize and activate primary afferent neurons innervating the cancer microenvironment.73 We previously demonstrated that oral cancer cell culture supernatant induces nociceptive behavior in mice in the absence of tumor burden or illness associated with malignancy.38,72,86 Algogenic mediators released by tumor cells include endothelin-1 (ET-1),35,61 nerve growth factor,84,88 interleukins,1,18,76 and tumor necrosis factor alpha.72 However, cancer pain therapies that target these mediators exhibit limited efficacy.5,11,80 Identification and characterization of novel cancer-secreted mediators could provide targets for cancer pain treatment. We seek to identify candidate genes that encode for the secreted and cell surface proteins that drive cancer pain.
Here, we develop a bioinformatic pipeline to couple preclinical characterization (nociceptive vs non-nociceptive) of human nontumorigenic and cancer cell lines in mice with transcriptomic analysis of the cell lines using publicly available data and a literature review of the resultant proteome. Human global gene expression profiling is often used to identify novel cancer therapy targets9,16,34; however, performance of gene expression profiling for cancer pain biomarker discovery has not been reported. The Gene Expression Omnibus (GEO) database, a growing National Center for Biotechnology Information (NCBI) repository for gene expression data generated from human and nonhuman tissues and cell lines,3 is underutilized as a resource to evaluate new hypotheses related to cancer pain. We use the GEO database to pursue studies of cancer pain by accessing deposited genomic data sets of cell lines previously characterized as nociceptive or nonnociceptive using an acute supernatant cancer pain mouse model.72
Our analyses comparing nociceptive cell lines to nonnociceptive cell lines reveal protein-encoding gene sets (pathways); many of the identified genes were not previously associated with cancer pain signaling. The phosphatidylinositol 3′-kinase (PI3K)-Akt signaling pathway is the most differentially expressed gene (DEG) set and includes EGFR and disintegrin metalloprotease domain 17 (ADAM17); these 2 molecules play a role in cancer progression.4,53,57,74 At least 10% of all cell surface proteins are proteolytically cleaved and release soluble proteins21,30; many EGFR ligands are substrates for ADAM17 cleavage.6 However, the role of ADAM17-EGFR signaling in cancer pain remains unknown. We hypothesize that EGFR ligands undergo ADAM17-mediated proteolytic cleavage from the plasma membrane of oral cancer cells, to activate the trigeminal primary afferent sensory neurons in the cancer microenvironment. To begin to investigate whether ADAM17-EGFR signaling contributes to oral cancer pain, we used an oral cancer pain model created by injecting supernatant from human cancer cell lines into the tongue. The dolognawmeter assay was used to quantify a behavioral index (gnawing) of orofacial nociception.15 The data demonstrate that factors secreted by oral cancers cell lines (ie, ADAM17) have the capability to induce pain, EGFR is expressed on trigeminal neurons, and EGFR inhibitors effectively block this pain.
2. Materials and methods
2.1. Cell culture and supernatant collection
Supernatant from 5 cancer cell lines (oral squamous cell carcinoma [HSC-3,55 SCC-4,65 SCC-966], skin melanoma [SkMel-2820], or pancreatic carcinoma [PANC-142]) and 3 nontumorigenic cell lines (normal primary oral keratinocyte [NOK], immortalized skin keratinocyte [HaCaT8], and oral dysplastic keratinocyte [DOK12]) was used to produce the acute cancer pain models. Cells were maintained and culture supernatant was collected as previously described.72,84,85 All cell lines were cultured in 10 cm cell culture dishes at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium (Gibco, Waltham, MA) supplemented with 10% fetal bovine serum and penicillin/streptomycin (50 U/mL). When cells reached 70% to 80% confluency (1.5 × 106 cells), the culture medium was changed to serum-free phenol-free Dulbecco’s modified Eagle’s medium (3 mL total volume), and incubated for 48 hours. Culture supernatant was collected, centrifuged at 300×g for 4 minutes to remove cell debris, and frozen for storage at −20°C. All cell lines used for supernatant collection were successfully authenticated by ATCC using short-tandem repeat profiling.
2.2. Acute cancer pain model
2.2.1. Animals
Adult (10-12 weeks, 20-30 g) male C57BL/6 mice (stock #000664; Jackson Labs, Bar Harbor, ME) were used for all experiments. Mice were housed in a temperature-controlled room on a 12:12-hour light:dark cycle (07:00-19:00 hours light), with unrestricted access to food and water. Researchers were trained under the Animal Welfare Assurance Program. All procedures were approved by the New York University Institutional Animal Care and Use Committee and performed in accordance with National Institutes of Health guidelines for the use of laboratory animals in research. All mice used for the acute cancer pain model (Methods section 2.2.3) were euthanized 12 hours after supernatant injection, after the conclusion of the orofacial pain behavior assay, dolognawmeter, to ensure the experience of pain was minimized.
2.2.2. Orofacial behavior
A behavioral index of orofacial nociception was quantified in a dolognawmeter,15 a device and assay designed to quantify gnawing activity. Each mouse was placed into a dolognawmeter confinement tube; escape from the tube is obstructed by 2 dowels in series in front of the mouse. The mouse voluntarily gnaws through both dowels to escape the device. Each obstructing dowel is connected to a digital timer that automatically records the duration required for the mouse to sever each dowel. The outcome variable is the gnaw-time for the second dowel and is a validated index of orofacial nociception in mice with oral cancer.15 To acclimatize the mice and improve consistency in gnawing behavior, all mice were trained for 5 to 7 sessions in the dolognawmeter. Training is accomplished by placing a mouse in the device and allowing it to gnaw through the obstructing dowels in the same manner as the subsequent experimental gnawing trials. For acute oral cancer pain models, a baseline gnaw-time (mean of the final 3 training sessions) was established for each mouse. The investigator was blinded to the treatment groups.
2.2.3. Acute cancer pain model
We generated acute oral cancer pain models by injecting cell line supernatant into the tongue as previously described.72 Mice received 50-μL injections (under isoflurane general anesthesia) of cell line supernatant over a 5-second period, into the left lateral tongue on 1 day or 3 consecutive days depending on the experiment. Nociceptive orofacial behavior measurements were recorded in awake mice 1 hour after the final supernatant injection.
2.3. Proteomics literature review
The NCBI PubMed search engine (https://www.ncbi.nlm.nih.gov/pubmed/) was used to identify published studies that performed proteomic or secretomic analyses on any of the 3 oral squamous cell carcinoma (SCC) cell lines HSC-3,55 SCC-4,65 SCC-966 or on the pancreatic cell line PANC-1.42 We used the keywords: “HSC-3,” “SCC-4,” “SCC-9,” “PANC-1” AND “proteomic” or “secretomic” or “proteome” or “secretome.” The search results were further evaluated based on the following inclusion criteria: (1) the study included “untreated” cell lines; (2) the study included a minimum of 2 samples per group; and (3) the study reported relative protein expression. Candidate proteins were cataloged for each eligible study, compiled, and reviewed as a group by a subset of the coauthors (N.N.S., Z.C., and B.E.A.). The PMID of eligible articles included in the analysis are shown in Supplementary Table S1 (available at http://links.lww.com/PAIN/B47). The protein list was further refined using GeneCards77 and Uniprot78 to identify proteins that are secreted or plasma membrane bound in the functional cell.
2.4. Gene expression analyses
2.4.1. Publicly available cell line gene expression data
Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and Array Express (https://www.ebi.ac.uk/arrayexpress/) were queried for available gene expression data sets generated using the cell lines evaluated in the acute oral cancer pain model. Gene expression data sets generated using the Affymetrix U133 Plus 2.0 microarray platform were selected for analysis because the largest number of data sets was available in this array format (n = 55). Eligibility criteria for cell line data sets are tabulated in Supplementary Table S2 (available at http://links.lww.com/PAIN/B47) and included: (1) data sets generated with one of the following human-derived cell lines: NOK, HaCaT, DOK, HSC-3, SCC-4, SCC-9, SkMel-28, PANC-1; (2) an untreated comparator was used (eg, the control sample for a given experiment); (3) cell line authentication was used in the original study; and (4) cell lines originated from males. After the GEO search, no additional unique entries were identified from the Array Express database. Raw GeneChip probe results files (.CEL) were extracted through the R statistical environment using the Bioconductor package GEOquery.
2.5. Enzyme-linked immunosorbent assay
The ADAM17 protein concentration in cell line supernatant was measured with an ELISA (MyBioSource, Inc; San Diego, CA). Supernatant was collected as described above from 3 separate cultures within the same cell passage and treated with protease inhibitor cocktail (Pierce, Thermo Fisher Scientific, Waltham, MA). Protein concentrations were determined using a Pierce bicinchoninic acid assay (Thermo Fisher Scientific). ELISA was performed per the manufacturer’s instructions; each sample was run in triplicate. The optical densities of the standards and samples were read at 450 nm using a Model 680 Microplate Reader (Bio-Rad Laboratories, Inc, Hercules, CA).
2.6. Immunohistochemistical staining for epidermal growth factor receptor in trigeminal from the mouse models
At least 10 days before tissue harvest, the retrograde tracer 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI; Invitrogen, Carlsbad, CA) was injected peripherally into the anterior lateral aspect of the tongue to label tongue afferents. The tracer was dissolved at 170 mg/mL in dimethylsufoxide (DMSO), diluted 1:10 in sterile saline, and injected bilaterally using a 30-g needle for a total volume of 5 to 7 μL per tongue under isoflurane (Abbott Laboratories, North Chicago, IL) anesthesia. Mice were euthanized with an overdose of inhaled isoflurane and perfused transcardially with 4% paraformaldehyde (PFA, Sigma Aldrich, St. Louis, MO). Trigeminal (TG) were dissected, postfixed for 1 hour in PFA, and cryoprotected in 30% sucrose in 0.1 M phosphate buffer at 4°C. TG were embedded in Tissue-Tek OCT compound (Sakura Finetek, Torrance, CA), sectioned (14 μm), and mounted on Superfrost Plus slides (Fisher Scientific Company, Pittsburgh, PA). After several washes, sections were incubated in primary antibody in PBS containing 1% bovine serum albumin overnight at room temperature. The primary antibody used was rabbit anti-EGFR (1:500; Abcam, Cambridge, United Kingdom). Slides were thoroughly washed in PBS and incubated in goat anti-rabbit secondary antibodies conjugated to cyanine 2 (Jackson ImmunoResearch, West Grove, PA) at 1:250 for 2.5 hours, thoroughly washed, and coverslipped with UltraCruz Aqueous Mounting Medium with nuclear stain, DAPI (Santa Cruz Biotechnology, Dallas, TX). Sections were photographed using NIS Elements software and a Nikon Eclipse Ti microscope.
2.7. RNA sequencing
Total RNA isolation of each cell line sample was achieved with a Qiagen AllPrep DNA/RNA Micro Kit (Qiagen, Inc, Valencia, CA). For each cell line tested, 3 cell pellets containing 5 × 105 to 1 × 106 cells from different cell culture passages were used to extract RNA at a concentration of 20 to 50 ng/μL. Quality control (QC) was done using fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) (accessed March 7, 2019) and summarized with multiqc.17 All the RNA samples passed QC. HISAT233 was used to align the raw data to Ensembl 38 version 95, followed by SAMtools41 to convert BAM files into SAM files. Transcript assembly and quantification was done using StringTie60 and transcript counts generated by prepde.py, a python script given from the StringTie manual. Differential transcript analysis was done using DeSeq2.46 Transcripts counts of less than 10 were excluded as were transcripts with an average of less than 10 counts across the sequenced samples. Correction for batch effect was done using the Bioconductor package sva. The function svaseq was used to remove unknown batch effects.39 Annotation of the transcripts was done using Ensembl 38 version 95. The genes identified as being differentially expressed using publicly available microarray data were then examined in the RNAseq-based gene expression data set. A P-value of less than 0.05 was the cutoff for declaring a transcript’s expression to differ between the comparison groups.
2.8. Bioinformatic and statistical analysis
2.8.1. Statistical analyses of acute cancer pain model experiments
Nociceptive orofacial behavior was measured using a dolognawmeter. Each mouse was compared to its own baseline gnaw-time (mean of 3 training trials before pretreatment trial). Data are analyzed as a two-way analysis of variance (ANOVA) with repeated measures (2 time points) to determine an interaction between time and treatment. Holm–Sidak test was used for post hoc analysis to control for multiple comparisons. Data are presented as a percent change from baseline gnaw-time ± SEM. Statistical significance was set at P < 0.05. Prism 8.3.0 (Graphpad Software LLC) was used for statistical analysis and figure generation.
2.8.2. Statistical analyses of protein expression
Single protein ELISA data were analyzed as the target protein concentration normalized to total protein concentration in each sample, as measured by bicinchoninic acid assay. Each sample was run in triplicate and averaged together. One-way ANOVA was used to evaluate the difference in protein concentration between groups. The protein concentration was below the level of detection in all 3 samples for NOK, DOK, and HaCaT; therefore, the concentration was marked as 0 pg/mg. Homogeneity of variance testing was not run due to repeated values in 3 of 8 samples. Holm–Sidak multiple comparisons test was used to compare all cell lines to nontumorigenic immortalized keratinocyte cell line (HaCaT). HaCaT was selected as the control because it is nonoral, immortalized, and did not produce any orofacial behavior response using the dolognawmeter assay.
2.8.3. Human oral cancer gene expression analysis
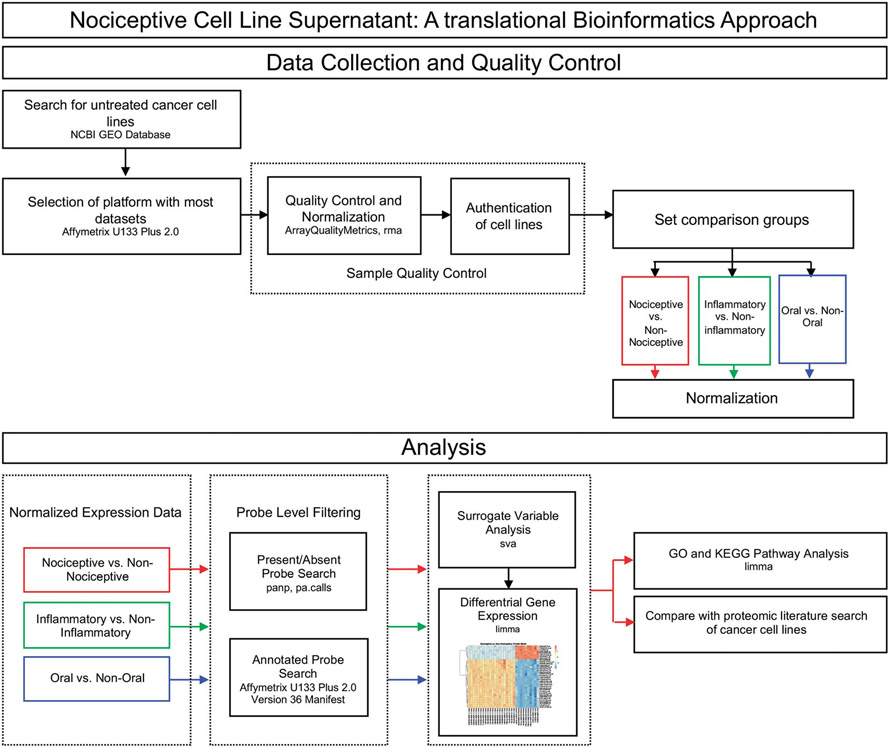
Quality control was performed on all cell lines (n = 55) using the Bioconductor package arrayQualityMetrics.29 Samples that failed any one of arrayQualityMetrics’s 3 outlier detection methods were removed from the study; 3 samples failed, leaving 52 cell lines. We determined the authentication status (eg, verification of cell line purity by STR profiling) of each identified data set. If the authentication status was not reported in the associated publication, the group that contributed the data set was contacted to authenticate the status of the cell lines used for the respective gene expression analyses. Thirty cell line samples met all inclusion criteria and passed QC (Supplementary Table S2, available at http://links.lww.com/PAIN/B47). The experimental workflow is depicted in Figure 1. Differential gene expression analyses were performed with the Bioconductor package limma,67 which fits a linear model to find DEGs. Oligo10 was used for background correction, quantile normalization, and log2 transformation. Differential gene expression was assessed between nociceptive (PANC-1, SCC-9, SCC-4, and HSC-3) and nonnociceptive (NOK, DOK, HaCaT, and SkMel-28) cell lines. Normalized expression data were further filtered using the Bioconductor package panp,82 which retains probes that pass the cutoff for signal above background (P < 0.05), while filtering out absent and marginal probes, resulting in 19,847 probes retained for further evaluation. Then, probes with ambiguous annotation (eg, no annotation or more than one gene associated with the probe) from the Affymetrix U133 Plus 2.0 Version 36 manifest were excluded, leaving 18,541 probes. Correction for batch effects was performed using the Leek surrogate variable analysis method with the Bioconductor package sva.40 Surrogate variable estimation was performed using control probes; control probes were then excluded before differential gene expression analysis of the remaining 18,488 probes was performed.
Figure 1.
Study design for bioinformatics and statistical analyses. The flow chart illustrates the QC filtering and analysis pipeline using publicly available cancer cell line gene expression data from the NCBI GEO database. Cell line samples from the Affymetrix U133 Plus 2.0 platform were subjected to sample QC assessment. Probe level filtering was applied independently to the normalized expression data of the 3 comparison groups (Nociceptive, Inflammatory, Oral) before differential gene expression analysis was performed. Differentially expressed genes (significance threshold set at P < 0.05) for the nociceptive trait (red line) were then subjected to pathway analysis and a search for candidate proteins of interest identified from the literature. GEO, Gene Expression Omnibus.
2.8.4. Pathway analysis
To evaluate for enrichment of DEGs among pathways, pathway analysis was conducted using 2 complementary and overlapping annotations: gene ontology (GO2) and Kyoto Encyclopedia of Genes and Genomes (KEGG28). Pathway analysis using KEGG and GO annotations was performed using the kegga and goana commands in limma, respectively, with nonsignificant DEGs specified as the “background universe.” The moderated t test statistic was calculated for each term; the nominal significance threshold was used to declare a pathway to be differentially perturbed (P < 0.05). Given the overlap in KEGG- and GO-based pathway analyses, the latter was listed in Supplemental Table S3 (available at http://links.lww.com/PAIN/B47). The Bioconductor package pathview45 was used to generate additional images of significant KEGG pathways of interest.
3. Results
3.1. Cancer cell line-secreted mediators evoke nociceptive orofacial behavior
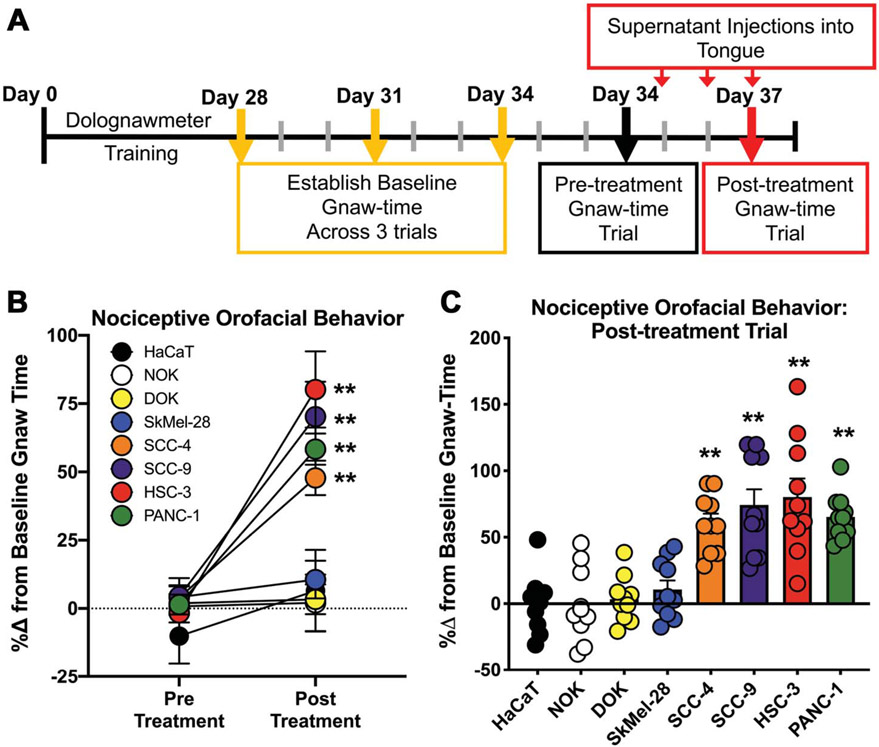
We previously reported that the oral SCC cell line, HSC-3, releases high concentrations of algogenic proteins that evoke nociceptive behavior when injected into the mouse tongue.71,72,86 Supernatant from nontumorigenic keratinocyte cell lines did not elicit nociceptive behavior.72 We sought to determine whether nociceptive behavior secondary to injection with cancer-secreted proteins was unique to oral SCC supernatant in male mice. We limited our investigations to human cancer cell lines generated from male patients and tested nociceptive behavior in male mice to avoid the confounding effects associated with hormone signaling. We measured nociceptive behavior in male mice after injection of cell culture supernatant from 1 of the following 5 cancer cell lines: oral cancer cell lines (HSC-3, SCC-9, and SCC-4); melanoma skin cancer cell line (SkMel-28), and pancreatic cancer cell line (PANC-1) (Table 1). We also used clinically nonpainful dysplastic oral keratinocyte cell line, DOK, and non-tumorigenic oral and skin keratinocyte cell lines, NOK and HaCaT, respectively. As indicated in the timeline in Figure 2A, mice were trained to complete an orofacial gnawing assay, dolognawmeter, for 4 weeks. After baseline gnaw-times were calculated (mean of the final 3 trials in each respective mouse), mice were randomized into 10 mice per group and injected with cell line supernatant into the anterior lateral tongue for 3 consecutive days. Three consecutive injections of supernatant were used to mimic a persistent exposure to mediators released by the tumor, without the impact of tumor burden or illness associated with carcinogenesis. Functional allodynia, as indicated by an increase in gnaw-time, was measured 1 hour after the final injection of cell line supernatant (Fig. 2A). Data are depicted as a percent change from baseline gnaw-time and were analyzed as a two-way repeated-measures ANOVA to determine an interaction between time and treatment (two-way ANOVA, F(7,70) = 6.931). Post hoc analysis revealed a significant increase in gnaw-time in response to cell line supernatant from HSC-3 (P < 0.0001), SCC-9 (P < 0.0001), SCC-4 (P = 0.007), and PANC-1 (P = 0.0002) compared to respective baseline gnaw-times. SkMel-28 (P = 0.999) and the control cell lines DOK (P > 0.999), NOK (P > 0.999), and HaCaT (P = 0.780) did not elicit nociceptive behavior (Fig. 2B). Data in Figure 2C are presented to demonstrate biological variability in response to supernatant injection.
Table 1.
Differential nociceptive behavior and designated behavior phenotype in response to cell line culture supernatant.
| Cell type | Cell line | Source | %Δ in gnaw-time | Behavior phenotype |
|---|---|---|---|---|
| Skin keratinocytes (immortalized) | HaCaT | AddexBio | 6.5 ± 14.9 | Nonnociceptive |
| Normal oral keratinocytes (primary) | NOK | Lifeline Cell Tech | 2.1 ± 10.4 | Nonnociceptive |
| Dysplastic oral keratinocytes | DOK | Sigma | 3.3 ± 7.7 | Nonnociceptive |
| Melanoma | SkMel-28 | ATCC | 12.9 ± 8.1 | Nonnociceptive |
| Oral squamous cell carcinoma | HSC-3 | JCRB | 80.2 ± 13.9 | Nociceptive |
| Oral squamous cell carcinoma | SCC-4 | ATCC | 70.2 ± 13.6 | Nociceptive |
| Oral squamous cell carcinoma | SCC-9 | ATCC | 50.7 ± 6.7 | Nociceptive |
| Pancreatic carcinoma | PANC-1 | ATCC | 55.0 ± 6.2 | Nociceptive |
NOK, normal primary oral keratinocyte; DOK, oral dysplastic keratinocyte; SCC, squamous cell carcinoma.
Figure 2.
Cancer cell line supernatant evokes orofacial nociceptive behavior in male mice. (A) Experimental timeline for the cell line supernatant model of oral cancer pain. Adult C57BL/6 male mice were trained in a dolognawmeter over 4 weeks or until a consistent baseline was achieved. Three additional baseline trials were completed and then mice underwent 3 consecutive injections of cell line supernatant followed by assessment in the dolognawmeter (Postinjection Gnaw-Time Measurement). There was no change in gnaw-time between groups; therefore, the mean of the 3 baseline trials for each group were calculated and analyzed as a percent change. (B) Pooled data for Pre-Treatment and Post-treatment trials as well as (C) individual values for each mouse in the Post-Treatment Trial are represented as a percent change from the average baseline gnaw-time (two-way ANOVA, ** P < 0.01). ANOVA, analysis of variance.
3.2. Proteomics literature identified secreted proteins previously reported in nociceptive cancer cell lines
We categorized each cell line as either nociceptive or nonnociceptive depending on the effect of the respective cell line supernatant (injected into the tongue) on the outcome variable in a dolognawmeter (Table 1). We infer that specific cancer-secreted proteins cause nociceptive behavior and thus we sought to create a summary of proteins secreted by the nociceptive cell lines. We executed a literature search using the NCBI PubMed search engine accessing primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics. The search included articles published from January 1, 1980, to June 1, 2018, that used the identified 4 nociceptive cell lines (HSC-3, SCC-4, SCC-9, and PANC-1) and search terms related to measurement of proteomic profiles through techniques such as gel electrophoresis and mass spectrometry (ie, proteome, proteomic, secretome, and secretomic). We identified 36 peer-reviewed published articles with accessible proteomic and/or secretomic data sets and identified 1370 overexpressed proteins among the nociceptive cancer cell lines; the data set was refined to 698 proteins identified as secreted or membrane-bound proteins (Supplementary Table S1, available at http://links.lww.com/PAIN/B47).
3.3. Transcriptome analyses revealed that nociceptive and nonnociceptive cell lines exhibit distinct gene expression profiles
We subsequently used the GEO database, a gene expression repository, to perform transcriptome analysis of publicly available data sets generated using the cell lines evaluated in the nociceptive supernatant assay. Available gene expression data sets using the Affymetrix U133 Plus 2.0 microarray platform were downloaded from NCBI GEO; this array format was selected because it had the largest number of data sets available. Eligibility criteria for cell line data sets are tabulated in Supplementary Table S2 (available at http://links.lww.com/PAIN/B47). We compared 20 data sets from nociceptive cell lines (HSC-3, SCC-4, SCC-9, and PANC-1) to 10 data sets from nonnociceptive cell lines (DOK, NOK, HaCaT, and SkMel-28). We identified 2225 differentially expressed probes mapping to 1760 DEGs (moderated t test, all P < 0.05) associated with the nociceptive phenotype (Supplementary Table S4, available at http://links.lww.com/PAIN/B47).
3.4. Proteomic literature and database search refined potential therapeutic targets
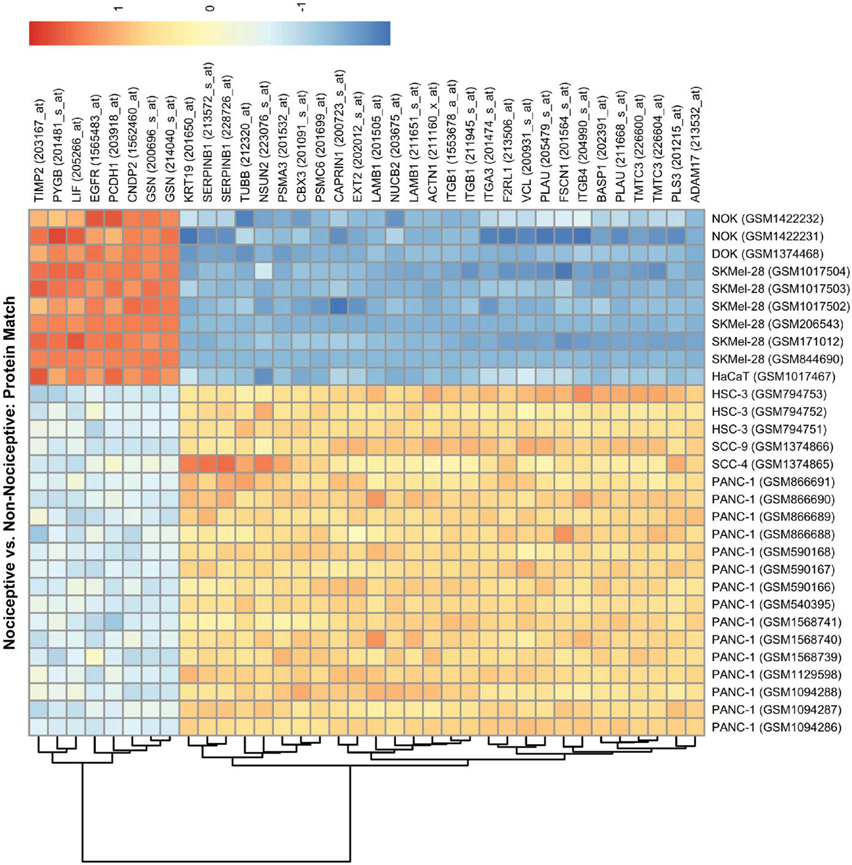
To prioritize DEGs that are membrane-bound or secreted, we cross-referenced genes encoding for the proteins identified in the proteomic literature search with the DEGs identified from the nociceptive vs nonnociceptive comparison. There is recognized incongruence between gene expression and resulting protein products22,43,81; we considered genes that exhibit a significant increase or decrease in expression. We identified 30 DEGs in nociceptive cell lines and potentially involved in supernatant-evoked nociception; 7 genes had significantly decreased expression and 23 genes had significantly increased expression (Table 2). In Figure 3, a heatmap visually represents the relative expression of the 30 protein-encoding genes across the 30 cell line data sets. In addition, we cross-referenced the 30 protein-encoding genes with 3 pain gene databases (ie, the Pain Genes Database,37 the Human Pain Genetics Database,52 and the Pain Research Forum14). These 3 pain gene databases have been derived from studies primarily focused on neuronal and/or neuropathic mechanisms (not cancer pain) and are not comprehensive. We found that 45 of the 1760 DEGs and 1 of the 30 protein-encoding genes (F2RL1) were cited in at least one of the pain gene databases. We infer from these results that our experimental paradigm is capable of identifying genes and pathways relevant to cancer pain. We then explored the drug–gene product interaction database DGIdb for targets of known drugs for these 30 protein-encoding gene targets to identify new compounds that can be repurposed or redesigned to target pain (Supplementary Table S5, available at http://links.lww.com/PAIN/B47). Epidermal growth factor receptor was the most commonly cited target in the DGIdb; antagonists of EGFR signaling (eg, cetuximab and gefitinib) are currently available to treat cancer.26,36,54.
Table 2.
Differentially expressed genes for nociception also identified in a proteomic literature review.
| Gene symbol | Protein name | Probe ID 1 | logFC 1 | Probe ID 2 | logFC 2 | Articles (PMID) |
|---|---|---|---|---|---|---|
| ACTN1 | Alpha-actinin-1 | 211160_x_at | 0.78 | — | — | 24495306; 26850699 |
| ADAM17 | ADAM17 | 213532_at | 1.24 | — | — | 23090842 |
| BASP1 | Brain acid soluble protein 1 | 202391_at | 5.49 | — | — | 26850699 |
| CAPRIN1 | Caprin-1 | 200723_s_at | 0.99 | — | — | 24495306 |
| CBX3 | Chromobox protein homolog 3 | 201091_s_at | 1.44 | — | — | 24211406 |
| CNDP2 | Camosine dipeptidase 2 | 1562460_at | −1.05 | — | — | 26850699 |
| EGFR | Epidermal growth factor receptor | 1565483_at | −1.67 | — | — | 22902387; 23090842 |
| EXT2 | Exostosin-2 | 202012_s_at | 0.88 | — | — | 23090842 |
| F2RL1 | Proteinase-activated receptor 2 | 213506_at | 3.49 | — | — | 23090842 |
| FSCN1 | Fascin | 201564_s_at | 2.37 | — | — | 24495306 |
| GSN | Isoform 1 of gelsolin | 214040_s_at | −4.48 | 200696_s_at | −3.25 | 22905270 |
| ITGA3 | Integrin α3 | 201474_s_at | 3.05 | — | — | 23090842 |
| ITGB1 | Fibronectin receptor (integrin β1) | 1553678_a_at | 2.48 | 211945_s_at | 1.37 | 16215274 |
| ITGB4 | Integrin β4 | 204990_s_at | 3.21 | — | — | 23090842 |
| KRT19 | Cytokeratin 19 | 201650_at | 6.99 | — | — | 23463621 |
| LAMB1 | Laminin, beta 1 | 211651_s_at | 2.19 | 201505_at | 1.79 | 25484078 |
| LIF | Leukemia inhibitory factor | 205266_at | −1.91 | — | — | 25484078 |
| NSUN2 | tRNA (cytosine(34)-C(5))-methyltransferase | 223076_s_at | 1.25 | — | — | 24211406 |
| NUCB2 | Nucleobindin-2 | 203675_at | 2.23 | — | — | 24495306 |
| PCDH1 | Protocadherin-1 | 203918_at | −1.77 | — | — | 24211406 |
| PLAU | Urokinase-type plasminogen activator | 211668_s_at | 4.74 | 205479_s_at | 3.80 | 26850699 |
| PLS3 | Plastin-3 | 201215_at | 1.04 | — | — | 26850699 |
| PSMA3 | Proteasome subunit alpha type-3 | 201532_at | 0.95 | — | — | 23090842 |
| PSMC6 | 26S protease regulatory subunit S10B | 201699_at | 1.36 | — | — | 23090842; 24495306 |
| PYGB | Glycogen phosphorylase | 201481_s_at | −2.29 | — | — | 26850699 |
| SERPINB1 | Plasminogen activator inhibitor 1 | 213572_s_at | 1.74 | 228726_at | 2.20 | 26850699 |
| TIMP2 | Metalloproteinase inhibitor 2 | 203167_at | −1.75 | — | — | 25484078; 26850699 |
| TMTC3 | Transmembrane and TPR repeat-containing protein 3 | 226600_at | 2.36 | 226604_at | 2.81 | 23090842 |
| TUBB | Tubulin beta chain | 212320_at | 0.87 | — | — | 23090842 |
| VCL | Vinculin | 200931_s_at | 1.51 | — | — | 16215274 |
Genes identified in the proteomic literature review were also differentially expressed in the nociceptive cell line analysis and corresponding Affymetrix U133 Plus 2.0 probe IDs. The log fold-change (FC) values for each significant gene-probe (P < 0.05 is estimated as significant using the moderated ttest employed in limma) are listed. The PMID is the PubMed publication identifier.
EGFR, epidermal growth factor receptor.
Figure 3.
Heatmap of genes that are differentially expressed in nociceptive trait and also identified in the proteomic literature review. The heatmap depicts normalized gene expression for DEGs (significance at P < 0.05) for the nociceptive trait comparison group that were also identified in the proteomic literature review. Each row represents one of 30 DEGs, some with multiple probes, among the 30 cell line samples used in the nonnociceptive and nociceptive cell line comparison. Each cell line sample (ie, each column) includes the unique accession number assigned in GEO (eg, GSM1094286 for the leftmost PANC-1 sample). GEO, Gene Expression Omnibus.
3.5. Altered pathways identified in nociceptive cell lines
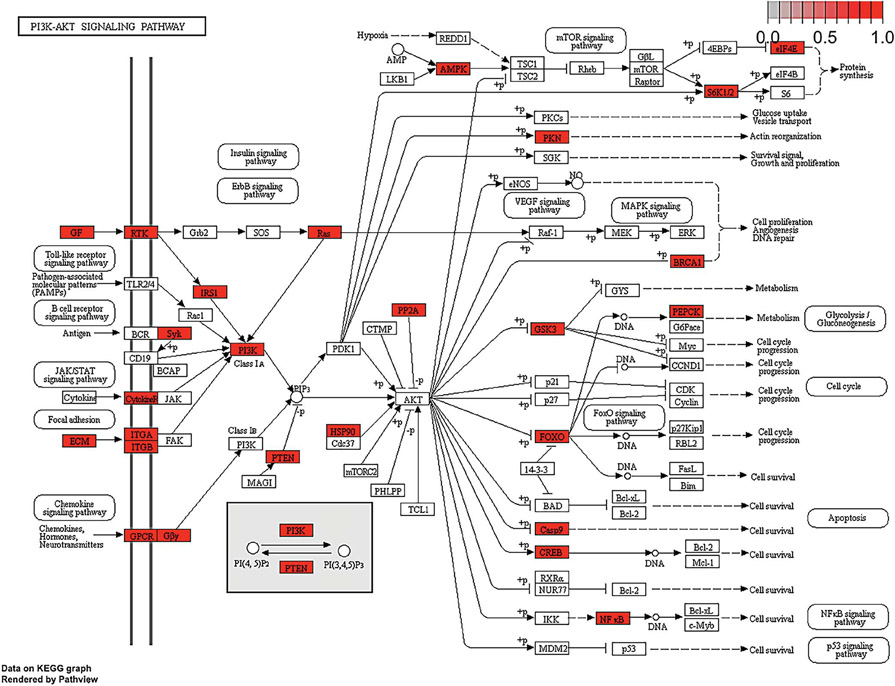
We used pathway analysis to evaluate for the enrichment of DEGs in pathways that could drive cancer supernatant-induced nociception. This approach allows for additional prioritization of proteins identified as druggable targets. We used the full list of DEGs (n = 1760) to explore beyond what is known from the proteomic literature review (n = 698 proteins). Thirteen significant KEGG pathways were identified (Student t-test, P < 0.05) (Table 3). Pathway analysis conducted using GO term annotations identified similar pain signaling pathways (Supplemental Table S3, available at http://links.lww.com/PAIN/B47). The PI3K-Akt signaling pathway (hsa04151, P = 0.008) contained the greatest number of differential expressed genes in relation to the nociceptive trait. Protein products of DEGs found in the PI3K-Akt signaling pathway are highlighted in Figure 4. The signaling components in this pathway may serve as cancer pain drug targets; EGFR protein, a receptor tyrosine kinase, is present in the PI3K-Akt signaling pathway (Fig. 4). This pathway also includes mammalian target of rapamycin signaling, which has been implicated in neuropathic and injury-induced pain.50,51,62
Table 3.
The results of KEGG pathway analysis for the DEGs in the nociceptive trait comparison.
| KEGG pathway ID | Pathway | N | DE | P |
|---|---|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | 199 | 45 | 0.00799 |
| hsa04360 | Axon guidance | 118 | 37 | 2.06E-05 |
| hsa04510 | Focal adhesion | 137 | 32 | 0.01443 |
| hsa04931 | Insulin resistance | 79 | 25 | 0.00037 |
| hsa04380 | Osteoclast differentiation | 76 | 20 | 0.01366 |
| hsa04625 | C-type lectin receptor signaling pathway | 72 | 19 | 0.01551 |
| hsa04670 | Leukocyte transendothelial migration | 72 | 18 | 0.03074 |
| hsa01524 | Platinum drug resistance | 60 | 16 | 0.02277 |
| hsa04660 | T cell receptor signaling pathway | 64 | 16 | 0.04013 |
| hsa04146 | Peroxisome | 65 | 16 | 0.04565 |
| hsa05100 | Bacterial invasion of epithelial cells | 65 | 16 | 0.04565 |
| hsa03430 | Mismatch repair | 21 | 7 | 0.03841 |
| hsa04710 | Circadian rhythm | 22 | 7 | 0.04884 |
The “N” column denotes the total number of genes in each pathway. The “DE” column denotes the number of genes in the pathway identified as significantly expressed in the nociceptive trait. The “P.DE” column lists the P-value statistic of the genes found in each pathway. Significant (P.DE < 0.05) pathways are listed.
KEGG, Kyoto Encyclopedia of Genes and Genome.
Figure 4.
KEGG’s PI3K-Akt pathway and differentially expressed results from the nociceptive trait. Pathway analysis of the DEGs in the cell lines that generated nociceptive behavior compared to the cell lines that did not generate nociception produced 13 KEGG pathways (P < 0.05). Significant pathways included those involved in PI3K-Akt (hsa04151; P = 0.008). Protein products of DEGs found in the pathways are highlighted red. KEGG, Kyoto Encyclopedia of Genes and Genome.
3.6. A role of ADAM17 in epidermal growth factor receptor-mediated cancer pain
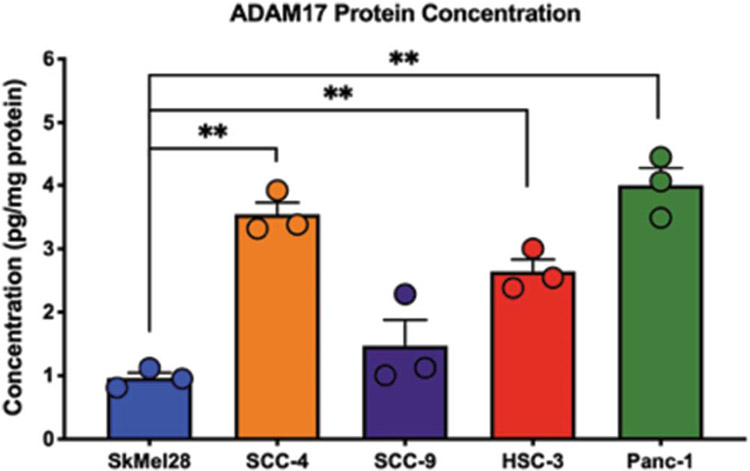
The primary goal of our analysis is to identify potential therapeutic targets for the treatment of cancer pain. Using the bioinformatic pipeline described above, we identified 30 potential therapeutic targets that may be involved in cancer supernatant-evoked nociception; EGFR was differentially expressed and classified as a druggable target. Ligands that bind to and activate EGFR include epithelial growth factor (EGF), transforming growth factor alpha (TGFα), heparin-binding EGF-like growth factor, amphiregulin, betacellulin, and epiregulin.13 TGFA was found to be a significant DEG associated with the cancer-induced nociception (logFC = 1.78, P = 0.0370, Supplemental Table S2, available at http://links.lww.com/PAIN/B47). However, the subsequent genes for known ligands of EGFR were not detected by the Affy 2.0 array, did not pass our QC filters, or were not identified as a significant DEG. To confirm the predicted profile of EGFR signaling in nociceptive cell lines, we analyzed total gene expression by performing RNA sequencing (RNAseq) in all 8 cell lines of interest. We found that none of the known EGFR ligands were significantly differentially expressed (Table 4). However, ADAM17, a gene that encodes the shedding protease responsible for the cleavage and systemic activation of EGFR ligands,6 was identified as one of the 30 DEGs (Table 2) and confirmed to be upregulated in the nociceptive cell lines using RNAseq (logFC = 10.45, P = 0.015) (Table 4). We validated the ADAM17 protein profile with an ELISA assay. Nociceptive cell line supernatant contained significantly more ADAM17 protein compared to nonnociceptive cell line NOK supernatant (oneway ANOVA; F(7,8) = 401.5; HSC-3 P < 0.0001; SCC-4 P = <0.0001; SCC-9 P = 0.001; PANC-1 P < 0.0001) (Fig. 5). However, despite the nonnociceptive phenotype, SkMel-28 supernatant also contained increased ADAM17 protein (P = 0.004). ADAM17 protein concentration was below the level of detection in HaCaT and DOK cell line supernatant (Fig. 5).
Table 4.
Differentially expressed EGFR-associated genes for nociception.
| Gene symbol | Protein name | Ensembl gene ID | logFC | P |
|---|---|---|---|---|
| AREG | Amphiregulin | ENSG00000109321 | 1.47 | 0.55747 |
| BTC | Betacellulin | ENSG00000174808 | 0.73 | 0.85470 |
| EGF | Epidermal growth factor | ENSG00000138798 | 0.17 | 0.98282 |
| EREG | Epiregulin | ENSG00000124882 | 0.28 | 0.96207 |
| HBEGF | Heparin-binding EGF-like growth factor | ENSG00000113070 | 1.11 | 0.98345 |
| TGFA | Transforming growth factor-alpha | ENSG00000163235 | −0.07 | 0.11805 |
| EPGN | Epigen | ENSG00000182585 | −0.68 | 0.91514 |
| ADAM17 | A disintegrin and metallopeptidase domain 17 | ENSG00000151694 | 10.45 | 0.01515 |
The log fold-change (FC) values for each significant gene (P < 0.05 is estimated as significant using the Wald test statistic) are listed.
EGF, epithelial growth factor; EGFR, epidermal growth factor receptor.
Figure 5.
ADAM17 expression in oral cancer cell lines. ADAM17 protein was measured in cell line culture supernatant. ADAM17 protein concentration was significantly higher in SkMel-28 HSC-3, SCC-4, SCC-9, and PANC-1 compared to NOK. Data are normalized to total protein in the supernatant. **P < 0.01 by one-way ANOVA. ANOVA, analysis of variance; NOK, normal primary oral keratinocyte; SCC, squamous cell carcinoma.
3.7. Epidermal growth factor receptor/ADAM17 signaling contributes to oral cancer pain
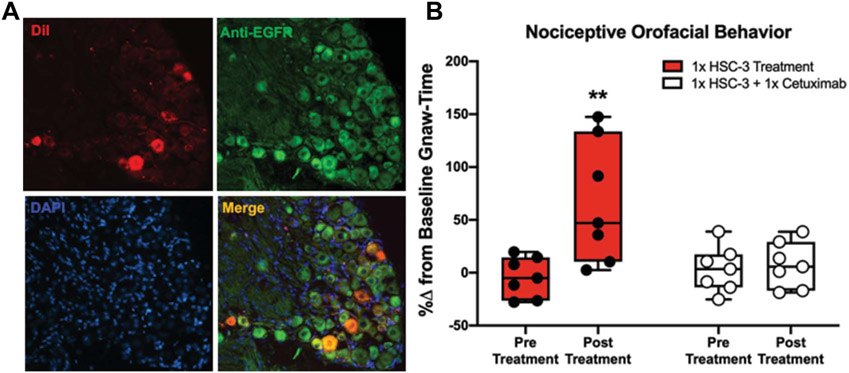
ADAM17 function in EGFR biology has been shown to play a role in cancer progression.70,83 Epidermal growth factor receptor is also present in dorsal root ganglia neurons in humans25 and mice48,59; EGFR expression in sensory TG neurons innervating the tongue is unknown. Using immunohistochemistry, we examined the expression of EGFR in trigeminal tongue afferent neurons. We used TG sections from retrogradely labeled mice to locate EGFR-like immunoreactivity in primary afferent neurons innervating the tongue. Cytoplasmic EGFR was present in almost all (>90%) retrograde labeled (DiI+) neurons (Fig. 6A). To reveal the effect of ADAM17-EGFR signaling on cancer-induced nociception, we administered systemic cetuximab, a chimeric monoclonal antibody that extracellularly inhibits EGFR, in a cancer pain mouse model using a single injection of HSC-3 cell line supernatant (Fig. 2A).68 This cell line was selected because HSC-3 supernatant produced the most robust nociceptive behavior as measured by the dolognawmeter assay (Figs. 2B and C). Mice were trained to complete an orofacial gnawing assay, dolognawmeter, for 4 weeks. After baseline gnaw-times were calculated (mean of the 3 trials in each respective mouse), mice randomized into 7 mice per group and completed the dolognawmeter assay to generate the pretreatment timepoint. Two days later, mice were injected with HSC-3 cell line supernatant into the tongue. After 1 hour, mice were injected with cetuximab (1 mg/mouse, intraperitoneal) and the dolognawmeter assay was initiated. Injection of HSC-3 with the vehicle (saline) was also performed for comparison. The pretreatment and posttreatment time points were calculated and analyzed as the percent change from the baseline gnaw-time for each mouse. There was a significant interaction between time and treatment (two-way ANOVA, F(1, 12) = 9.194, P = 0.010). HSC-3 supernatant injection yielded an increase in gnaw-time compared to baseline (71.1 ± 15.6%, P = 0.001) (Fig. 6B). Inhibition of EGFR signaling using cetuximab blocked the supernatant-induced nociceptive behavior. HSC-3 supernatant in the presence of cetuximab produced no significant change in gnaw-time compared to baseline (4.28 ± 15.6%, P = 0.955).
Figure 6.
Inhibition of EGFR signaling in oral cancer pain. (A) Representative anti-EGFR immunoreactivity (Green) in TG sections (12 μm) from a mouse with retrograde labeling from the tongue (DiI, Red). DAPI was used to identify nuclei (Blue). More than 90% of all DiI-labeled neurons overlapped (Merge, Yellow) with EGFR immunoreactivity (×20 magnification). (B) Orofacial nociceptive behavior measured in mice 1 hour after HSC-3 supernatant injection into the tongue immediately followed by saline (Solid bar) or cetuximab treatment (Striped bar). Data were analyzed as a percent change from baseline gnawing behavior. Two-way ANOVA, **P < 0.01. ANOVA, analysis of variance; EGFR, epidermal growth factor receptor.
4. Discussion
The question addressed by this study was whether a bioinformatic pipeline coupling preclinical nociceptive characterization of human cell line supernatant with transcriptomic analysis of publicly available data can identify candidate protein-encoding genes which drive cancer pain. The main finding is that ADAM17 is overexpressed in nociception-producing cancer cells. The many molecules shed by ADAM17 are ligands for EGFR89; we found that EGFR inhibitor, cetuximab, inhibits oral cancer supernatant-induced nociception. Our experimental paradigm identified nociceptive mediators secreted by cancer cell lines using an unbiased gene set enrichment analysis. We used publicly available gene expression data by applying contemporary QC and surrogate variable analyses not available when the data sets were generated. We found that EGFR was expressed at a higher level in nonnociceptive cell lines; however, the rationale for the additional investigation into EGFR came from cross-referencing the differential gene expression results with additional databases (ie, drug–gene product interaction database); others have demonstrated that EGFR is overexpressed in oral tumor tissue,44 which may be composed of cancer cells, immune infiltrate, nerve terminals, and stromal cells.
Epidermal growth factor receptor is a current therapeutic target for cancer treatment.48 The downstream effects of EGFR are mediated through the PI3K-Akt signaling pathway, which is known to regulate pain27,87; we identified this pathway to have the most perturbed DEGs associated with the nociceptive trait. Systemic administration of EGFR inhibitors for treating cancer pain has not been studied in patients. Kersten et al.32 found an analgesic effect of cetuximab in patients with cancer-related neuropathic pain; however, the study did not control for the source of the cancer-related neuropathic pain (ie, tumor burden and surgical resection). Small molecule EGFR inhibitors that target the tyrosine kinase site of the EGFR receptor, including gefitinib and lapatinib, are effective analgesics in mice for inflammatory and neuropathic pain.48 The prophylactic effect of cetuximab against chemotherapy-induced peripheral neuropathy has been demonstrated in 2 large randomized trials, with the neuropathy-inducing chemotherapeutic oxaliplatin.49,79 One case report demonstrated marked pain relief after infusion of cetuximab in a 68-year-old male patient with metastatic rectal cancer; the analgesic effect lasted for 10 to 12 days; the drug was readministered approximately every 12 days for three and a half years.31 The mechanism of action for EGFR-induced cancer pain is not yet known. We found that EGFR is expressed on trigeminal neurons innervating the tongue and have previously reported that the EGFR ligand, TGFα, protein concentration is significantly elevated in HSC-3 supernatant.72 We found that cetuximab inhibited nociceptive behavior in response to a single HSC-3 supernatant injection suggesting direct cancer-neuron communication underlying oral cancer-evoked pain. We have previously demonstrated prolonged gnawing time in response to both a single injection38,86 and 3 consecutive injections71,72; the latter evokes a robust immune response composed of both innate and adaptive immune cells.72 Epidermal growth factor receptor is expressed on many different immune cell types and its expression is importance for immune function.47,69 Additional studies are needed using tumorbearing mouse models to understand the impact of the immune system in cetuximab-induced inhibition of cancer pain.
ADAM17 is responsible for the cleavage and activation of all EGFR ligands; overexpression of ADAM17 in SCC-9 cells increases heparin-binding EGF-like growth factor shedding from the membrane.75 Endothelin-1, an algogenic peptide overexpressed and secreted by several cancers,58,61,64 activates ADAM17 to release EGFR ligands from cancer cells.24 Our DEG analysis revealed significant overexpression of TGFA in nociceptive cancer cell lines; however, this finding was not supported by RNAseq analysis. Alternatively, we found that nociceptive cell line supernatant contained more ADAM17 protein than nonnociceptive cell line supernatant with the exception of SkMel-28. However, the consequence of increased ADAM17-mediated shedding activity in the cancer microenvironment is unknown. We previously determined that TGFα protein is significantly increased in supernatant from HSC-3, SCC-9, and SkMel-28 cell lines compared to supernatant from HaCaT.72 Previous studies have found that epiregulin is the primary endogenous activator of EGFR-related hyperalgesia in mice; late-phase formalin-induced nociception was exacerbated in a dose-dependent manner after intrathecal injection of epiregulin.48 Mice with reduced ADAM17 activity were hyposensitive to noxious stimulation, showing elevated mechanical thresholds as well as impaired heat and cold sensitivity.63 Although the ADAM17-mediated shedding activity extends beyond EGFR ligands, the search for specific inhibitors that target only a subset of the shedding events performed by ADAM17 is ongoing.57 Additional study is warranted to understand how elevated levels of ADAM17 in painful cancers contribute to disease progression and pain.
There are 3 notable limitations to our current study. First, more pancreatic cancer cell line data sets were included in the DEG analysis than other cancer cell line data sets. This proportional imbalance could have produced enrichment of genes that encode for mediators more common to pancreatic cancer than to other cancers. However, in a post hoc analysis comparing the expression of the nociceptive DEGs in PANC1 (n = 15) vs that in HSC-3, SCC-4, and SCC-9 (n = 5), only 1 probe for KRT19 met the Bonferroni-adjusted moderated t test statistic (P < 0.00139), suggesting that it is unlikely that the identified genes are driven primarily by pancreatic cancer (Supplemental Table S6, available at http://links.lww.com/PAIN/B47). The second limitation is that no commercially available gene expression microarray can interrogate all known genes; the Affymetrix 2.0 array does not feature all the currently annotated genes. In addition, a subset of genes did not pass our QC assessment. In an attempt to mitigate this limitation, we validated our list of protein-encoding genes with RNAseq in the cell lines of interest. We found that 19 of the 30 protein-encoding genes were significantly overexpressed or underexpressed. This result suggests that the bioinformatic pipeline using publicly available data identified several DEGs related to nociception; however, additional gene expression analyses such as RNA sequencing at greater sequencing depth in an independent sample is warranted. The final limitation of our study is the inclusion of only one sex for genomic data and preclinical mouse models. Research on sex differences in pain has increased substantially in recent years.19 Several preclinical studies demonstrate that hormonal regulation of cancer7,23 may affect downstream nociceptive signaling. To control possible hormonal confounders, we used only cancer cell lines derived from men and measured nociceptive responses in male mice. This approach limited the number of cell lines available for phenotypic characterization and precluded the use of clinically nonpainful cancers such as breast cancer. Further exploration in female-derived cell lines is warranted.
In summary, we identified a role for ADAM17 and EGFR signaling based on a preclinical animal model and an unbiased transcriptomic analysis of gene expression data from cancer lines. A better understanding of the cancer-secreted mediators that control cross-talk between the cancer and the peripheral nervous system will ultimately lead to more effective cancer pain management. The DEGs and their secreted protein products warrant further evaluation because they may serve as therapeutic targets for cancer pain.
Supplementary Material
Acknowledgements
F32 DE027269 (NNS), American Association of Dental Student Research Fellowship (ZRC), NIH NIDCR R03 DE027777 (YY), NIH NIDCR R01DE026806 (BLS).
Footnotes
Conflict of interest statement
The authors have no conflicts of interest to declare.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.painjournalonline.com).
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B47.
References
- [1].Al-Mazidi S, Farhat K, Nedjadi T, Chaudhary A, Zin Al-Abdin O, Rabah D, Al-Zoghaibi M, Djouhri L. Association of interleukin-6 and other cytokines with self-reported pain in prostate cancer patients receiving chemotherapy. Pain Med 2018;19:1058–66. [DOI] [PubMed] [Google Scholar]
- [2].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology: The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 2013;41:D991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bates T, Kennedy M, Diajil A, Goodson M, Thomson P, Doran E, Farrimond H, Thavaraj S, Sloan P, Kist R, Robinson M. Changes in epidermal growth factor receptor gene copy number during oral carcinogenesis. Cancer Epidemiol Biomarkers Prev 2016;25:927–35. [DOI] [PubMed] [Google Scholar]
- [5].Battistini B, Berthiaume N, Kelland NF, Webb DJ, Kohan DE. Profile of past and current clinical trials involving endothelin receptor antagonists: the novel “-sentan” class of drug. Exp Biol Med (Maywood) 2006;231: 653–95. [PubMed] [Google Scholar]
- [6].Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol 2005;6:32–43. [DOI] [PubMed] [Google Scholar]
- [7].Bohra A, Bhateja S. Carcinogenesis and sex hormones: a review. Endocrinol Metab Synd 2015;4:156. [Google Scholar]
- [8].Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 1988;106:761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Campos-Parra AD, Padua-Bracho A, Pedroza-Torres A, Figueroa-Gonzalez G, Fernandez-Retana J, Millan-Catalan O, Peralta-Zaragoza O, Cantu de Leon D, Herrera LA, Perez-Plasencia C. Comprehensive transcriptome analysis identifies pathways with therapeutic potential in locally advanced cervical cancer. Gynecol Oncol 2016;143:406–13. [DOI] [PubMed] [Google Scholar]
- [10].Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010;26:2363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chang DS, Hsu E, Hottinger DG, Cohen SP. Anti-nerve growth factor in pain management: current evidence. J Pain Res 2016;9:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chang SE, Foster S, Betts D, Marnock WE. DOK, a cell line established from human dysplastic oral mucosa, shows a partially transformed non-malignant phenotype. Int J Cancer 1992;52:896–902. [DOI] [PubMed] [Google Scholar]
- [13].Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006;7:505–16. [DOI] [PubMed] [Google Scholar]
- [14].Das S, McCaffrey PG, Talkington MW, Andrews NA, Corlosquet S, Ivinson AJ, Clark T. Pain Research Forum: application of scientific social media frameworks in neuroscience. Front Neuroinform 2014; 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dolan JC, Lam DK, Achdjian SH, Schmidt BL. The dolognawmeter: a novel instrument and assay to quantify nociception in rodent models of orofacial pain. J Neurosci Methods 2010;187:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Durand S, Trillet K, Uguen A, Saint-Pierre A, Le Jossic-Corcos C, Corcos L. A transcriptome-based protein network that identifies new therapeutic targets in colorectal cancer. BMC Genomics 2017;18:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fang D, Kong LY, Cai J, Li S, Liu XD, Han JS, Xing GG. Interleukin-6-mediated functional upregulation of TRPV1 receptors in dorsal root ganglion neurons through the activation of JAK/PI3K signaling pathway: roles in the development of bone cancer pain in a rat model. PAIN 2015; 156:1124–44. [DOI] [PubMed] [Google Scholar]
- [19].Fillingim RB, King CD, Ribeiro-Dasilva MC, Rahim-Williams B, Riley JL III. Sex, gender, and pain: a review of recent clinical and experimental findings. J Pain 2009;10:447–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst 1977;59:221–6. [DOI] [PubMed] [Google Scholar]
- [21].Fong KP, Barry C, Tran AN, Traxler EA, Wannemacher KM, Tang HY, Speicher KD, Blair IA, Speicher DW, Grosser T, Brass LF. Deciphering the human platelet sheddome. Blood 2011;117:e15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 2003;4:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Grimm M, Biegner T, Teriete P, Hoefert S, Krimmel M, Munz A, Reinert S. Estrogen and Progesterone hormone receptor expression in oral cavity cancer. Med Oral Patol Oral Cir Bucal 2016;21:e554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hinsley EE, Hunt S, Hunter KD, Whawell SA, Lambert DW. Endothelin-1 stimulates motility of head and neck squamous carcinoma cells by promoting stromal-epithelial interactions. Int J Cancer 2012;130:40–7. [DOI] [PubMed] [Google Scholar]
- [25].Huerta JJ, Diaz-Trelles R, Naves FJ, Llamosas MM, Del Valle ME, Vega JA. Epidermal growth factor receptor in adult human dorsal root ganglia. Anat Embryol (Berl) 1996;194:253–7. [DOI] [PubMed] [Google Scholar]
- [26].Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, Tebbutt NC, van Hazel G, Wierzbicki R, Langer C, Moore MJ. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357:2040–8. [DOI] [PubMed] [Google Scholar]
- [27].Kaganoi J, Watanabe G, Okabe M, Nagatani S, Kawabe A, Shimada Y, Imamura M, Sakai Y. STAT1 activation-induced apoptosis of esophageal squamous cell carcinoma cells in vivo. Ann Surg Oncol 2007;14: 1405–15. [DOI] [PubMed] [Google Scholar]
- [28].Kanehisa M The KEGG database. Novartis Found Symp 2002;247: 91–101; discussion 101-103, 119-128, 244-152. [PubMed] [Google Scholar]
- [29].Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 2009;25:415–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kawahara R, Lima RN, Domingues RR, Pauletti BA, Meirelles GV, Assis M, Figueira AC, Paes Leme AF. Deciphering the role of the ADAM17-dependent secretome in cell signaling. J Proteome Res 2014;13: 2080–93. [DOI] [PubMed] [Google Scholar]
- [31].Kersten C, Cameron MG. Cetuximab alleviates neuropathic pain despite tumour progression. BMJ Case Rep 2012;2012: bcr1220115374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kersten C, Cameron MG, Laird B, Mjaland S. Epidermal growth factor receptor-inhibition (EGFR-I) in the treatment of neuropathic pain. Br J Anaesth 2015;115:761–7. [DOI] [PubMed] [Google Scholar]
- [33].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ko JJ, Grewal JK, Ng T, Lavoie JM, Thibodeau ML, Shen Y, Mungall AJ, Taylor G, Schrader KA, Jones SJM, Kollmannsberger C, Laskin J, Marra MA. Whole-genome and transcriptome profiling of a metastatic thyroid-like follicular renal cell carcinoma. Cold Spring Harb Mol Case Stud 2018; 4:a003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kopetz ES, Nelson JB, Carducci MA. Endothelin-1 as a target for therapeutic intervention in prostate cancer. Invest New Drugs 2002;20: 173–82. [DOI] [PubMed] [Google Scholar]
- [36].Kris MG, Natale RB, Herbst RS, Lynch TJ Jr, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA 2003;290:2149–58. [DOI] [PubMed] [Google Scholar]
- [37].Lacroix-Fralish ML, Ledoux JB, Mogil JS. The Pain Genes Database: an interactive web browser of pain-related transgenic knockout studies. PAIN 2007;131:3.e1–4. [DOI] [PubMed] [Google Scholar]
- [38].Lam DK, Dang D, Zhang J, Dolan JC, Schmidt BL. Novel animal models of acute and chronic cancer pain: a pivotal role for PAR2. J Neurosci 2012;32:14178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res 2014;42:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome project data processing S. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lieber M, Mazzetta J, Nelson-Rees W, Kaplan M, Todaro G. Establishment of a continuous tumor-cell line (panc-1) from a human carcinoma of the exocrine pancreas. Int J Cancer 1975;15:741–7. [DOI] [PubMed] [Google Scholar]
- [43].Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016;165:535–50. [DOI] [PubMed] [Google Scholar]
- [44].Lo WY, Wang HJ, Chiu CW, Chen SF. miR-27b-regulated TCTP as a novel plasma biomarker for oral cancer: from quantitative proteomics to post-transcriptional study. J Proteomics 2012;77:154–66. [DOI] [PubMed] [Google Scholar]
- [45].Lou WJ, Brouwer C. Pathview: an R/bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013;29:1830–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].MacDonald F, Zaiss DMW. The immune system’s contribution to the clinical efficacy of EGFR antagonist treatment. Front Pharmacol 2017;8: 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Martin LJ, Smith SB, Khoutorsky A, Magnussen CA, Samoshkin A, Sorge RE, Cho C, Yosefpour N, Sivaselvachandran S, Tohyama S, Cole T, Khuong TM, Mir E, Gibson DG, Wieskopf JS, Sotocinal SG, Austin JS, Meloto CB, Gitt JH, Gkogkas C, Sonenberg N, Greenspan JD, Fillingim RB, Ohrbach R, Slade GD, Knott C, Dubner R, Nackley AG, Ribeiro-da-Silva A, Neely GG, Maixner W, Zaykin DV, Mogil JS, Diatchenko L. Epiregulin and EGFR interactions are involved in pain processing. J Clin Invest 2017;127:3353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL, Kay E, Mitchell JK, Madi A, Jasani B, James MD, Bridgewater J, Kennedy MJ, Claes B, Lambrechts D, Kaplan R, Cheadle JP; Investigators MCT. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet 2011;377:2103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Megat S, Price TJ. Therapeutic opportunities for pain medicines via targeting of specific translation signaling mechanisms. Neurobiol Pain 2018;4:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Melemedjian OK, Asiedu MN, Tillu DV, Sanoja R, Yan J, Lark A, Khoutorsky A, Johnson J, Peebles KA, Lepow T, Sonenberg N, Dussor G, Price TJ. Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol Pain 2011;7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Meloto CB, Benavides R, Lichtenwalter RN, Wen X, Tugarinov N, Zorina-Lichtenwalter K, Chabot-Dore AJ, Piltonen MH, Cattaneo S, Verma V, Klares R III, Khoury S, Parisien M, Diatchenko L. Human pain genetics database: a resource dedicated to human pain genetics research. PAIN 2018;159:749–63. [DOI] [PubMed] [Google Scholar]
- [53].Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 2010;277:301–8. [DOI] [PubMed] [Google Scholar]
- [54].Modjtahedi H, Essapen S. Epidermal growth factor receptor inhibitors in cancer treatment: advances, challenges and opportunities. Anticancer Drugs 2009;20:851–5. [DOI] [PubMed] [Google Scholar]
- [55].Momose F, Araida T, Negishi A, Ichijo H, Shioda S, Sasaki S. Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J Oral Pathol Med 1989;18: 391–5. [DOI] [PubMed] [Google Scholar]
- [56].Montazeri A Quality of life data as prognostic indicators of survival in cancer patients: an overview of the literature from 1982 to 2008. Health Qual Life Outcomes 2009;7:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Moss ML, Minond D. Recent advances in ADAM17 research: a promising target for cancer and inflammation. Mediators Inflamm 2017;2017: 9673537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nelson JB, Nguyen SH, Wu-Wong JR, Opgenorth TJ, Dixon DB, Chung LW, Inoue N. New bone formation in an osteoblastic tumor model is increased by endothelin-1 overexpression and decreased by endothelin A receptor blockade. Urology 1999;53:1063–9. [DOI] [PubMed] [Google Scholar]
- [59].Neto E, Alves CJ, Leitao L, Sousa DM, Alencastre IS, Conceicao F, Lamghari M. Axonal outgrowth, neuropeptides expression and receptors tyrosine kinase phosphorylation in 3D organotypic cultures of adult dorsal root ganglia. PLoS One 2017;12:e0181612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pickering V, Jay Gupta R, Quang P, Jordan RC, Schmidt BL. Effect of peripheral endothelin-1 concentration on carcinoma-induced pain in mice. Eur J Pain 2008;12:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Price TJ, Dussor G. AMPK: an emerging target for modification of injury-induced pain plasticity. Neurosci Lett 2013;557(pt A):9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Quarta S, Mitric M, Kalpachidou T, Mair N, Schiefermeier-Mach N, Andratsch M, Qi Y, Langeslag M, Malsch P, Rose-John S, Kress M. Impaired mechanical, heat, and cold nociception in a murine model of genetic TACE/ADAM17 knockdown. FASEB J 2019;33:4418–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ratna A, Das SK. Endothelin: ominous player in breast cancer. J Cancer Clin Trials 2016;1:e102. [PMC free article] [PubMed] [Google Scholar]
- [65].Rheinwald JG, Beckett MA. Defective terminal differentiation in culture as a consistent and selectable character of malignant human keratinocytes. Cell 1980;22(2 pt 2):629–32. [DOI] [PubMed] [Google Scholar]
- [66].Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res 1981;41:1657–63. [PubMed] [Google Scholar]
- [67].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Russell JS, Colevas AD. The use of epidermal growth factor receptor monoclonal antibodies in squamous cell carcinoma of the head and neck. Chemother Res Pract 2012;2012:761518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sasada T, Azuma K, Ohtake J, Fujimoto Y. Immune responses to epidermal growth factor receptor (EGFR) and their application for cancer treatment. Front Pharmacol 2016;7:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sasaki T, Hiroki K, Yamashita Y. The role of epidermal growth factor receptor in cancer metastasis and microenvironment. Biomed Res Int 2013;2013:546318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Scheff NN, Bhattacharya A, Dowse E, Dang RX, Dolan JC, Wang S, Kim H, Albertson DG, Schmidt BL. Neutrophil-mediated endogenous analgesia contributes to sex differences in oral cancer pain. Front Integr Neurosci 2018;12:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Scheff NN, Ye Y, Bhattacharya A, MacRae J, Hickman DN, Sharma AK, Dolan JC, Schmidt BL. Tumor necrosis factor alpha secreted from oral squamous cell carcinoma contributes to cancer pain and associated inflammation. PAIN 2017;158:2396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Schmidt BL. The neurobiology of cancer pain. Neuroscientist 2014;20: 546–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shen H, Li L, Zhou S, Yu D, Yang S, Chen X, Wang D, Zhong S, Zhao J, Tang J. The role of ADAM17 in tumorigenesis and progression of breast cancer. Tumour Biol 2016;37:15359–370. [DOI] [PubMed] [Google Scholar]
- [75].Simabuco FM, Kawahara R, Yokoo S, Granato DC, Miguel L, Agostini M, Aragao AZ, Domingues RR, Flores IL, Macedo CC, Della Coletta R, Graner E, Paes Leme AF. ADAM17 mediates OSCC development in an orthotopic murine model. Mol Cancer 2014;13:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Singh PK, Chandra G, Bogra J, Gupta R, Kumar V, Hussain SR, Jain A, Mahdi AA, Ahmad MK. Association of genetic polymorphism in the interleukin-8 gene with risk of oral cancer and its correlation with pain. Biochem Genet 2016;54:95–106. [DOI] [PubMed] [Google Scholar]
- [77].Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016;54:1.30.31–33. [DOI] [PubMed] [Google Scholar]
- [78].The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017;45:D158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Tveit KM, Guren T, Glimelius B, Pfeiffer P, Sorbye H, Pyrhonen S, Sigurdsson F, Kure E, Ikdahl T, Skovlund E, Fokstuen T, Hansen F, Hofsli E, Birkemeyer E, Johnsson A, Starkhammar H, Yilmaz MK, Keldsen N, Erdal AB, Dajani O, Dahl O, Christoffersen T. Phase III trial of cetuximab with continuous or intermittent fluorouracil, leucovorin, and oxaliplatin (Nordic FLOX) versus FLOX alone in first-line treatment of metastatic colorectal cancer: the NORDIC-VII study. J Clin Oncol 2012;30:1755–62. [DOI] [PubMed] [Google Scholar]
- [80].Vendrell I, Macedo D, Alho I, Dionisio MR, Costa L. Treatment of cancer pain by targeting cytokines. Mediators Inflamm 2015;2015:984570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012;13:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Warren P,Taylor D, Martini PGV, JAckson J, Bienkowska J. Panp—a new method of gene detection on Oligonucleotide Expression Arrays. 2007 IEEE 7th International Symposium on BioInformatics and BioEngineering, 6-8 July 2007. 2007:108–115. [Google Scholar]
- [83].Witters L, Scherle P, Friedman S, Fridman J, Caulder E, Newton R, Lipton A. Synergistic inhibition with a dual epidermal growth factor receptor/HER-2/neu tyrosine kinase inhibitor and a disintegrin and metalloprotease inhibitor. Cancer Res 2008;68:7083–9. [DOI] [PubMed] [Google Scholar]
- [84].Ye Y, Dang D, Zhang J, Viet CT, Lam DK, Dolan JC, Gibbs JL, Schmidt BL. Nerve growth factor links oral cancer progression, pain, and cachexia. Mol Cancer Ther 2011;10:1667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ye Y, Ono K, Bernabe DG, Viet CT, Pickering V, Dolan JC, Hardt M, Ford AP, Schmidt BL. Adenosine triphosphate drives head and neck cancer pain through P2X2/3 heterotrimers. Acta Neuropathol Commun 2014;2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ye Y, Scheff NN, Bernabe D, Salvo E, Ono K, Liu C, Veeramachaneni R, Viet CT, Viet DT, Dolan JC, Schmidt BL. Anti-cancer and analgesic effects of resolvin D2 in oral squamous cell carcinoma. Neuropharmacology 2018;139:182–93. [DOI] [PubMed] [Google Scholar]
- [87].Zhang Y, Zheng L, Zhang J, Dai B, Wang N, Chen Y, He L. Antitumor activity of taspine by modulating the EGFR signaling pathway of Erk1/2 and Akt in vitro and in vivo. Planta Med 2011;77:1774–81. [DOI] [PubMed] [Google Scholar]
- [88].Zhu Z, Friess H, diMola FF, Zimmermann A, Graber HU, Korc M, Buchler MW. Nerve growth factor expression correlates with perineural invasion and pain in human pancreatic cancer. J Clin Oncol 1999;17:2419–28. [DOI] [PubMed] [Google Scholar]
- [89].Zunke F, Rose-John S. The shedding protease ADAM17: physiology and pathophysiology. Biochim Biophys Acta Mol Cell Res 2017;1864(11 pt B): 2059–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.