Abstract
NADPH-dependent alkylaldehyde reducing enzyme, which was greatly induced by n-hexadecane, from Acinetobacter sp. strain M-1 was purified and characterized. The purified enzyme had molecular masses of 40 kDa as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and 160 kDa as determined by gel filtration chromatography. The enzyme, which was shown to be highly thermostable, was most active toward n-heptanal and could use n-alkylaldehydes ranging from C2 to C14 and several substituted benzaldehydes, including the industrially important compounds cinnamyl aldehyde and anisaldehyde, as substrates. The alrA gene coding for this enzyme was cloned, and its nucleotide sequence was determined. The deduced amino acid sequence encoded by the alrA gene exhibited homology to the amino acid sequences of zinc-containing alcohol dehydrogenases from various sources. The gene could be highly expressed in Escherichia coli, and the product was purified to homogeneity by simpler procedures from the recombinant than from the original host. Our results show that this enzyme can be used for industrial bioconversion of useful alcohols and aldehydes.
The enzymatic aspect of alkane degradation in microorganisms has attracted much interest for the development of conversion processes for petrochemicals, as well as for oil-contaminated environments. Not only do the intermediate compounds in the alkane degradation pathway have great potential as components of industrial products, but the enzymes in the pathway also can be used as catalysts for many bioconversion reactions, as in the case of the alkane hydroxylase complex from Pseudomonas oleovorans (10).
We have been studying the alkane degradation pathway in Acinetobacter sp. strain M-1. In the course of our study, we found a novel NADPH-dependent aldehyde-reducing activity, namely, NADP-dependent alcohol dehydrogenase activity, which is induced in cells grown on hexadecane (7). Alcohol dehydrogenases have also been reported in some other n-alkane-degrading Acinetobacter strains. Constitutive NADP-dependent alcohol dehydrogenases have been purified from Acinetobacter calcoaceticus NCIB8250 (12) and A. calcoaceticus strain HO1-N (11), but the physiological role of these enzymes has not been determined. Unstable, NAD-dependent alcohol dehydrogenase activity was detected in A. calcoaceticus strain HO1-N (8), and the enzyme exhibited activity with hexadecanol; however, the level of activity was very low. Acinetobacter sp. strain M-1, which utilizes long-chain alkanes with chain lengths ranging from C13 to C44, also showed negligible NAD+- and NADP+-dependent alcohol dehydrogenase activities. However, we found metabolically significant levels of the reverse activities, which were strongly induced by n-hexadecane. In this report, we describe purification and characterization of the NADPH-dependent aldehyde-reducing enzyme. Since this enzyme exhibited both thermostability and a high level of activity with broad substrate specificity, it should have great potential for industrial utilization. We also describe high-level production of this enzyme with a recombinant Escherichia coli strain.
MATERIALS AND METHODS
Chemicals and enzymes.
n-Dodecanal, n-tridecanal, n-tetradecanal, 2-decanone, 3-octanone, and 2-methylundecanal were purchased from Aldrich Chemical Co., Inc. (Milwaukee, Wis.). 2,4-Hexadienal and trans-2-decenal were obtained from Tokyo Kasei Organic Chemicals (Tokyo, Japan). Alkylaldehydes with carbon chain lengths of 2 to 10, benzaldehyde, phenylaldehyde, and alkylalcohols were obtained from Nacalai Tesque (Osaka, Japan). Undecanal, substituted benzaldehydes, p-anisaldehyde, and trans-cinnamaldehyde were obtained from Wako Pure Chemical Ind. Ltd. (Osaka, Japan). Most of the aldehydes and ketones were highly purified products (>90% pure); the exceptions were tetradecanal (80% pure), trans-2-decenal (10% in ethanol), 2,4-hexadienal (80%), o-methyl benzaldehyde (80%), and phenylaldehyde (50% in diethyl phthalate). All of the substrates were used without further purification. Plysurf A210G was obtained from Daiichi Kogyo Seiyaku (Tokyo, Japan). Superdex 200, Q-Sepharose, and Phenyl-Sepharose were products of Amersham Pharmacia Biotech (Uppsala, Sweden). DEAE-Toyopearl and Butyl-Toyopearl were obtained from Tosoh Co., Ltd. (Tokyo, Japan). Dye Matrex Red A was obtained from Amicon Inc. (Beverly, Mass.). Restriction enzymes, alkaline phosphatase (calf intestine), T4 DNA ligase, and Ex Taq DNA polymerase were products of Takara Shuzo Co., Ltd. (Kyoto, Japan). A dye deoxy terminator cycle sequencing kit was purchased from Applied Biosystems Inc. (Foster City, Calif.), and [α-32P]dCTP was obtained from Amersham Corp. (Arlington Heights, Ill.). NAD+-dependent alcohol dehydrogenases from baker's yeast and horse liver were products of Sigma Chemical Co. (St. Louis, Mo.).
Microorganisms, culture conditions, and vectors.
Acinetobacter sp. strain M-1 was grown on medium containing hexadecane (1.0%, wt/vol) and glycerol (0.5%), as reported previously (7). E. coli JM109 was used for gene cloning and expression and was usually grown on 2 × YT medium (pH 7.0) containing Bacto Yeast Extract (10/liter), Bacto Tryptone (16 g/liter), and NaCl (5 g/liter) in the presence of ampicillin (10 μg/ml) when necessary (Difco). pT7Blue (Novagen, Madison, Wis.) was used for subcloning PCR products. pBluescript II SK+ (Toyobo, Osaka, Japan) and pUC118 (Takara Shuzo Co., Ltd.) were used as cloning and expression vectors, respectively.
Enzyme assay.
NADP+-dependent alcohol dehydrogenase was assayed in a reaction mixture (1.0 ml) containing 50 μmol of Tris-Cl buffer (pH 9.5), 1.3 μmol of NADP+, 1.0 μmol of n-heptanol, and 0.01% Plysurf A210G. Since the activity of the reverse reaction (NADPH-dependent aldehyde reduction) was much greater than that of the forward reaction, the activity of the enzyme was routinely measured by determining NADPH oxidation in a reaction mixture (1.0 ml) containing 50 μmol of Tris-Cl buffer (pH 7.0), 0.13 μmol of NADPH, 1.0 μmol of n-heptanal, and 0.01% Plysurf A210G. Each reaction mixture was sonicated (150 W for 1 min) and then allowed to equilibrate for 1 min at 30°C, and then an appropriate quantity of enzyme was added to initiate the reaction. The activities of the forward and reverse reactions were assayed by measuring the increase and decrease in absorbance at 340 nm, respectively, with a Shimadzu spectrophotometer (UV-160) with a 1-cm-light-path cuvette. As a reference, a reaction mixture without a substrate was used. One unit of enzyme activity was defined as the amount of enzyme that catalyzed the reduction and oxidation of 1.0 μmol of NADP+ and 1.0 μmol of NADPH, respectively, per min at 30°C. When the activity of NAD+-dependent alcohol dehydrogenase was assayed, the NADP+ and NADPH in the above mixtures were replaced by NAD+ and NADH, respectively.
Analysis.
Protein was measured with a Bio-Rad protein assay kit (Japan Bio-Rad Laboratories, Tokyo, Japan) by using bovine serum albumin as the standard (1). The relative molecular mass of the native enzyme was measured by gel filtration by using a fast protein liquid chromatography system (Amersham Pharmacia Biotech) and a Superdex 200 pg column equilibrated with 50 mM sodium phosphate buffer (pH. 8.0) containing 100 mM KCl. The standard protein markers used were obtained from Oriental Yeast Co. Ltd. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (3), and Bio-Rad standard proteins (Low Range) were used for molecular mass measurement. Nondenaturing PAGE (10% gel) was performed at 4°C as described by Fox et al. (2). After electrophoresis, the gels were either stained for proteins with Coomassie brilliant blue R-250 or used for activity staining. Each gel was incubated in the dark at 30°C for 2 h in a reaction mixture containing 50 mM Tris-Cl (pH 8.8), 55 mM nitroblue tetrazolium, 550 mM phenazine methosulfate, 1 mM NADP+, and 2 mM n-heptanol. The purified enzyme was digested with Protease V8 (Sigma Chemical Co.), and the resulting peptides (or the purified enzyme) were separated by SDS-PAGE and then electroblotted onto a polyvinylidene difluoride membrane (PsqPVDF; Millipore Corp., Bedford, Mass.) at 14 V overnight by using a transfer buffer containing 75 mM Tris base and 580 mM glycine in 20% (vol/vol) methanol. The N-terminal amino acid sequence was determined by Edman's method with a Perkin-Elmer protein sequencer (model 476A).
Purification of the enzyme.
Purification was performed at 4°C. Acinetobacter sp. strain M-1 was grown on the medium for 24 h, harvested by centrifugation at 6,500 × g for 20 min, washed with 0.85% NaCl, and then kept at −20°C until it was used. Cells (260 g, wet weight) were suspended in 50 mM potassium phosphate buffer (pH 8.0) (buffer A; total volume, 700 ml), disrupted with a model 200M Insonator (Kubota, Osaka, Japan) at 150 W for 60 min at 4°C, and then centrifuged at 18,000 × g for 20 min. In order to remove the membrane fraction, the supernatant was centrifuged at 110,000 × g for 60 min. The clear supernatant (soluble fraction) was used for purification of the enzyme. Ammonium sulfate was added to the soluble fraction at a concentration of 1.2 M, and then the supernatant was mixed with Butyl-Toyopearl gel (300 ml) preequilibrated with buffer A containing 1.2 M ammonium sulfate. After gentle stirring for 30 min, the gel was washed with buffer A by using a Buchner funnel with appropriate filter paper. Ammonium sulfate was added to the eluted fraction again at a concentration of 1.2 M, and then the supernatant was put on a Phenyl-Sepharose column (3.0 by 14 cm) that was preequilibrated with buffer A containing 1.2 M ammonium sulfate. Elution was performed with a linear gradient containing decreasing ammonium sulfate concentrations (1.2 to 0 M) and increasing ethylene glycol concentrations (0 to 20%) in buffer A. The active fractions were collected and precipitated by the addition of ammonium sulfate at concentrations up to 3 M, and then the precipitate was dialyzed against buffer A. The concentrated enzyme was put on a Dye Matrex Red A column (1.5 by 5.0 cm) that was preequilibrated with buffer A. After the column was washed with buffer A, the enzyme was eluted with buffer A containing 2 mM NADPH and then concentrated by ultrafiltration with a Diaflow membrane (YM30; Amicon Inc.). The concentrated sample was chromatographed on a Superdex 200-pg column (1.6 by 60 cm) equilibrated with buffer A containing 100 mM KCl. The active fractions were concentrated, desalted with YM30, and then applied to a Q-Sepharose column (1.8 by 10 cm) that was preequilibrated with buffer A. The enzyme was eluted with a linear KCl gradient (0 to 500 mM), concentrated with YM30, and then stored at 0°C until it was used.
The recombinant enzyme was purified from E. coli JM109 (pUCalrA) cells (see below). Forty-two grams (wet weight) of cells was suspended in 100 ml of buffer A. The cell extract, which was prepared by sonication, was chromatographed on a Butyl-Toyopearl column by using the elution conditions described above for the Phenyl-Toyopearl step. The enzyme was concentrated with ammonium sulfate (3 M) and then dialyzed against buffer A. The enzyme solution was heated at 65°C for 15 min and then centrifuged to remove the inactivated protein. The purified enzyme preparation was stored at 0°C until it was used.
Cloning of the alcohol dehydrogenase-encoding gene.
Upstream and downstream primers were designed on the basis of the N terminus and an internal amino acid sequence, respectively, to PCR amplify the DNA fragment coding for the alcohol dehydrogenase gene from Acinetobacter sp. strain M-1 chromosomal DNA. The sequences of the primers used were as follows: N-terminal primer A-N, 5′-GC(A/G/T/C)ATGCA(A/G)GC(A/T/C)GA(A/G)CA-3′; and internal primer A-I1, 5′-TGGTC(A/G/T/C)GC(A/G/T)CCCAT(A/G/T/C)GC(T/C)TT-3′. Chromosomal DNA extracted from Acinetobacter sp. strain M-1 by the method of Marmur (4) was used as a template for amplification. The PCR mixtures (25 μl) contained 0.25 μg of chromosomal DNA, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 100 pmol of each primer, 2.5 μl of 10× Ex Taq buffer, and 0.75 U of Ex Taq DNA polymerase. Each reaction was performed with a Perkin-Elmer Cetus thermal cycler (Takara Shuzo Co.). The initial template denaturation step consisted of 3 min at 95°C. The PCR profile consisted of 25 cycles of 1 min at 58°C, 1 min at 72°C, and 1 min at 95°C. The PCR product was electrophoresed on a 0.7% low-melting-temperature agarose gel, extracted from the gel with SUPREC-01 (Takara Shuzo), ligated into the pT7Blue vector, and then introduced into E. coli JM109 cells.
Acinetobacter sp. strain M-1 chromosomal DNA was digested with various restriction enzymes. The digests were electrophoresed on a 0.7% agarose gel and then transferred to a Biodyne nylon membrane (Pall Bio Support Corp., East Hills, N.Y.). Hybridization was performed with the random primed 32P-labeled PCR product as a probe under the highly stringent conditions recommended by Southern (9). The probe hybridized to a 6.0-kb HindIII fragment. The HindIII-digested DNA fragments corresponding to this size were ligated into pBluescript II SK+ (pAlr1) and then transformed into E. coli JM109. Colonies that formed on the master plates were transferred to a Biodyne nylon membrane. After lysis of the E. coli cells and binding of the liberated DNA to the nylon membrane, the resulting blot was used for colony hybridization under the conditions used for genomic Southern hybridization.
Nucleotide sequencing.
The cloned HindIII fragment was digested with a variety of restriction enzymes to obtain convenient DNA fragments for subcloning into pBluescript II SK+ (pAlr1), and then DNA sequencing was performed by the dideoxy chain termination method using a DNA sequencer (Applied Biosystems model 373A). The sequencing reaction was performed as described in the manuals supplied with Taq dye terminator and Taq dye primer cycle sequencing kits (Applied Biosystems). Sequence data were analyzed with the BLAST program (GenBank, EMBL, and SWISSPROT databases).
Expression of the alcohol dehydrogenase gene in E. coli.
A PCR was performed by using pAlr1 (see below as the template and the synthesized oligonucleotide primers 5′-GGAATTCCAAGGAGGTTTTTATATGAGCAATCATCAAATTGG-3′ and GGAATTCCTTAGTCGAAGTCTGCTTTGA-3′, each of which contained an EcoRI site (underlined in the sequence) and a Shine-Dalgarno sequence (italicized in the sequence). The amplification reaction was performed as described above. The PCR product was digested with EcoRI, separated on an agarose gel, ligated into the EcoRI site of pUC118, and then introduced into E. coli JM109 cells. Transformants were selected on 2× YT agar plates containing ampicillin (10 μg/ml), IPTG, (isopropyl-β-d-thiogalactopyranoside) (10 μg/ml), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (0.05 mM). The DNA sequence of the PCR product was confirmed. The recombinant plasmid (pUCalrA) was recovered from the positive clone.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper has been deposited in the DDBJ/EMBL/GenBank nucleotide sequence database under accession number AB047854.
RESULTS
Pyridine nucleotide-dependent alcohol dehydrogenase activities in the soluble fraction of Acinetobacter sp. strain M-1.
Significant levels of NADH- and NADPH-dependent aldehyde-reducing activities (0.1 to 0.2 and 0.3 to 0.5 U·mg−1, respectively) were found in the soluble fraction of hexadecane-grown strain M-1, and the activities were strongly induced by n-hexadecane (2.5- and 12-fold, respectively). The NADH-dependent activity was too unstable for purification, although some of the activity could be restored by the addition of 1 mM Cu2+ to the assay mixture. We purified the NADPH-dependent enzyme that was significantly thermostable as described below. On the other hand, only negligible NAD+- and NADP+-dependent alcohol dehydrogenase activities were detected in the soluble fraction of Acinetobacter sp. strain M-1 cells grown on n-hexadecane.
Purification of the NADPH-dependent aldehyde-reducing enzyme.
The enzyme was purified 1,300-fold from the soluble fraction (Table 1). The specific activity of the purified enzyme was 560 U·mg of protein−1, and the yield was 3.0%. The purified preparation gave a single band on both SDS-PAGE and native PAGE gels. When Q-Sepharose column chromatography was used, the activity was divided into two peaks. The first activity peak was purified further. The enzyme in the second peak, whose total activity was about 60% of the activity in the first peak, gave a single activity band on a nondenatured PAGE gel, and the activity was at the same position as the other activity. Since the N-terminal amino acid sequences of the two enzymes were identical and only one gene for the enzyme was found in the chromosomal DNA, as mentioned below, we concluded that the separation into two peaks was an artifact of chromatography and that the two enzymes were the products of one gene.
TABLE 1.
Purification of alcohol dehydrogenase from Acinetobacter sp. strain M-1
| Purification step | Total activity (U) | Total protein (mg) | Sp act (U · mg−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Soluble fraction | 820 | 1,900 | 0.43 | 1 | 100 |
| Butyl-Toyopearl | 630 | 940 | 0.67 | 1.6 | 77 |
| Phenyl-Sepharose | 400 | 84 | 4.8 | 11 | 49 |
| Dye Matrex Red A | 350 | 4.9 | 72 | 170 | 43 |
| Superdex 200 | 130 | 0.26 | 490 | 1,200 | 16 |
| Q-Sepharose | 24 | 0.043 | 560 | 1,300 | 3.0 |
Molecular mass, subunit structure, and amino acid sequence.
The relative molecular mass of the purified enzyme was estimated to be 40 kDa by SDS-PAGE and 160 kDa by gel filtration. Only one N-terminal amino acid sequence, SNHQIRAYAAMQAGEQVVYQFDAGELKKHQ-, was found when Edman degradation was used. Judging from the results, the enzyme is tetrameric. The amino acid sequence of an internal peptide fragment was -LKAMGADHVVNSRDAQAIKA-.
General properties of the purified enzyme.
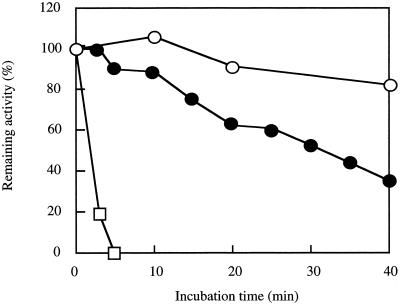
The enzyme was thermostable and retained 90% of its activity after incubation for 40 min at 70°C (Fig. 1). The enzyme assay was linear for 1 min at 60°C, and the rate observed at 60°C was 1.8 times higher than that observed at 30°C.
FIG. 1.
Heat stability of the enzyme. A purified enzyme solution (0.02 mg·ml−1) in buffer A containing 0.1 M KCl was incubated at various temperatures, and then the remaining activity was assayed under the standard conditions. Symbols: ○, 70°C; ●, 75°C; □, 80°C.
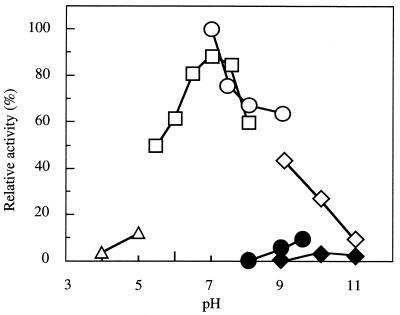
Maximum activities were found at pH 7.0 for n-heptanal reduction and at pH 9.5 for n-heptanol oxidation; the latter activity was 10% of the reduction activity (Fig. 2). The enzyme was stable at pH values ranging from 8.0 (Tris-Cl) to 9.0 (glycine-NaOH).
FIG. 2.
Effects of pH on enzyme activities. The aldehyde reduction activity (open symbols) with heptanal and the alcohol dehydrogenation activity (solid symbols) with heptanol were assayed at various pHs. In the case of alcohol dehydrogenation activity, NADP+ was used as the cofactor. Symbols: triangles, acetate buffer; squares, sodium phosphate buffer; circles, Tris-Cl buffer; diamonds, glycine-NaOH buffer.
Various reagents and metal ions were added to the reaction mixture. The enzyme was susceptible to sulfhydryl reagents, including 0.1 mM p-mercuribenzoate (96% inhibition), 1 mM N-ethylmaleimide (49%), and 1 mM iodoacetamide (33%). Chelating agents (2,2′-dipyridyl and EDTA) or serine esterase inhibitors (diisopropylfluorophosphate and phenylmethanesulfonyl fluoride) did not inhibit the enzyme at concentrations of 1 mM. The enzyme was inhibited by the following metal ions at concentrations of 1 mM; Ag+ (100% inhibition), Zn2+ (100%), Hg2+ (100%), Ca2+ (26%), Mn2+ (29%), Co2+ (91%), and Ni2+ (84%). These results suggest that the sulfhydryl group of the enzyme is important for activity.
Substrate specificity.
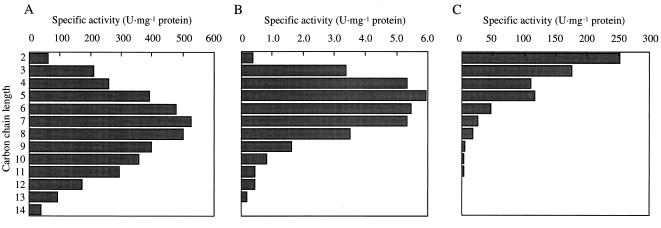
As shown in Table 2, the enzyme catalyzed reduction of a broad range of n-alkylaldehydes (C2 to C14), and n-heptanal was the most suitable substrate tested. The enzyme was also active toward benzaldehyde, benzaldehydes with fluoro and methyl substitutions, p-anisaldehyde, and trans-cinnamyl aldehyde. Ketones and aldoses were hardly reduced by the enzyme. The reduction activities of the enzyme toward several n-alkylaldehydes were compared with those of NAD-dependent alcohol dehydrogenases from baker's yeast and horse liver under the same conditions (Fig. 3). The enzyme from Acinetobacter sp. strain M-1 exhibited notably higher activities with medium-chain alkylaldehydes, and this substrate specificity was evidently different from those of other enzymes. NADPH could not be replaced by NADH (at concentrations up to 5 mM) for aldehyde reduction. The apparent Kms were 16 μM for NADPH and 36 μM for NADP+ when n-heptanal and n-heptanol, respectively, were used as the substrates.
TABLE 2.
Substrate specificity of the enzyme
| Substrate | Concn (mM) | Relative activity (%)a |
|---|---|---|
| Acetaldehyde | 100 | 5.7 |
| Propanal | 100 | 40 |
| Butanal | 50 | 49 |
| Pentanal | 20 | 74 |
| Hexanal | 10 | 90 |
| Heptanal | 1 | 100 |
| Octanal | 1 | 95 |
| Nonanal | 1 | 76 |
| Decanal | 1 | 68 |
| Undecanal | 1 | 56 |
| Dodecanal | 1 | 33 |
| Tridecanal | 1 | 18 |
| Tetradecanal | 1 | 7.2 |
| 2-Methylundecanal | 1 | 7.7 |
| 2-Decanone | 1 | 5.0 |
| 3-Heptanone | 1 | 6.7 |
| 3-Octanone | 1 | 5.1 |
| trans-2-Decenal | 1 | 12 |
| 2,4-Hexadienal | 1 | 16 |
| Vitamin A aldehyde | 1 | 5.2 |
| Benzaldehyde | 1 | 55 |
| o-Fluorobenzaldehyde | 1 | 57 |
| m-Fluorobenzaldehyde | 1 | 65 |
| p-Fluorobenzaldehyde | 1 | 68 |
| o-Methylbenzaldehyde | 1 | 3.8 |
| m-Methylbenzaldehyde | 1 | 27 |
| p-Methylbenzaldehyde | 1 | 40 |
| Phenylacetaldehyde | 1 | 6.5 |
| p-Anisaldehyde | 1 | 63 |
| trans-Cinnamaldehyde | 1 | 21 |
| d-Glyceraldehyde | 10 | <1 |
| d-Ribose | 10 | <1 |
| d-Glucose | 10 | <1 |
| d-Mannose | 10 | <1 |
| d-Galactose | 10 | <1 |
The activity with heptanal (specific activity, 560 U · mg−1) was defined as 100%. Reactions were carried out under the standard conditions, except that the substrates listed were used.
FIG. 3.
Comparison of the substrate specificity of alcohol dehydrogenase from Acinetobacter sp. strain M-1 with the substrate specificities of horse liver alcohol dehydrogenase and baker's yeast alcohol dehydrogenase. The purified enzyme (A), NAD+-dependent horse liver alcohol dehydrogenase (B), and NADP+-dependent yeast alcohol dehydrogenase (C) were compared by comparing their substrate specificities and specific activities with alkylaldehydes having various chain lengths. The substrate concentrations used were the same as those shown in Table 2. Other assay conditions are described in Materials and Methods, except that NADH was used as the cofactor.
Cloning and sequence of the alcohol dehydrogenase gene.
An approximately 600-bp DNA fragment was amplified by PCR by using Acinetobacter sp. strain M-1 chromosomal DNA as the template and primers A-N and A-I1. During colony hybridization selection, one positive clone was isolated from an HindIII gene library. This clone, pAlr1, had a 6.0-kb insert and was introduced into E. coli JM109 cells.
Nucleotide and deduced amino acid sequences.
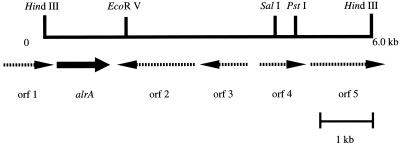
Determination of the entire nucleotide sequence of the 6.0-kb HindIII insert in pAlr1 revealed four complete and two partial open reading frames (ORFs) (Fig. 4). The ORF for alrA consists of 1,023 bp, and the deduced amino acid sequence includes 340 amino acid residues with a theoretical molecular mass of 36,379 Da. This value is close to the molecular mass of the purified enzyme from Acinetobacter sp. strain M-1 determined by SDS-PAGE, 40 kDa. The N-terminal and internal amino acid sequences of the purified enzyme were found in the deduced amino acid sequence. From these results and those of the gene expression study (see below), we concluded that this ORF encoded the alcohol dehydrogenase gene. The deduced amino acid sequences of other ORFs were homologous to those of the following proteins, as judged by a database search performed with the BLAST and FASTA programs: orf1, DNA-methyladenine glycosidase of Haemophilus influenzae (54% identity and 73% similarity); orf2, l-tyrosine:2-oxoglutarate aminotransferase of E. coli (31% identity and 51% similarity); orf3, hypothetical transcriptional regulator of E. coli (43% identity and 66% similarity); orf4, probable aldehyde reductase of E. coli (67% identity and 80% similarity); and orf5, putative transmembrane efflux protein of Streptomyces coelicolor (39% identity and 53% similarity).
FIG. 4.
Restriction map of the genomic HindIII fragment in the pAlrA1 plasmid carrying the alrA gene. The arrows indicate the orientations of the ORFs.
Expression of alrA in E. coli.
The specific activity of a cell extract of E. coli JM109(pUCalrA) grown on Luria-Bertani medium with induction by IPTG was 190 U · mg of protein−1, and this value was 620-fold higher than that of the original host strain. The enzyme was purified from the transformant cells to apparent homogeneity by SDS-PAGE with a much simpler procedure involving only two steps (hydrophobic interaction chromatography and heat treatment), and the yield was higher (19%) than the yield obtained with the original strain (Table 3). The N-terminal sequence (SNHQIRAYAAMQAGE-), molecular mass (40 kDa as determined by SDS-PAGE and 160 kDa as determined by gel filtration), substrate specificity, and heat stability of the purified recombinant enzyme were very similar to those of the purified enzyme from the original strain.
TABLE 3.
Purification of recombinant alcohol dehydrogenase from E. coli JM109(pUCalrA)a
| Purification step | Total activity (U) | Sp act (U · mg−1) | Total protein (mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Cell extract | 250,000 | 190 | 1,400 | 1 | 100 |
| Ultracentrifugation supernatant | 270,000 | 170 | 1,600 | 0.91 | 110 |
| Butyl-Toyopearl | 97,000 | 410 | 230 | 2.2 | 39 |
| Heat precipitation | 47,000 | 1,700 | 30 | 9.1 | 19 |
Heat precipitation was perfomed at 65°C for 15 min, and the enzyme was recovered from the centrifugation supernatant. Other purification procedures and the enzyme activity assay were the same as those used for the original strain.
DISCUSSION
Pyridine nucleotide-dependent alcohol dehydrogenases have been found in the cytosolic fractions of several strains of alkane-utilizing Acinetobacter spp., and the physiological roles of these enzymes have not been elucidated, because the activities were very low and the enzymes seemed to be constitutive. In this study, we found that such enzymes were greatly induced by n-hexadecane when the activities were measured by determining NAD(P)H-dependent aldehyde reduction. This means that each enzyme not only has a housekeeping function in metabolism, as described for strain HO1-N (11), but also plays a specific role in n-alkane metabolism. However, the enzyme is assumed not to participate in the main pathway of alkane oxidation to fatty acids in strain M-1 for the following reasons: (i) since the integral membrane terminal alkane hydroxylase is evidently essential for n-alkane oxidation in Acinetobacter spp. (5), alcohol oxidation is most probably catalyzed by a membrane-bound enzyme; (ii) the cytosolic enzymes were active toward medium-chain alcohols (or aldehydes), although strain M-1 can grow on longer-chain n-alkanes; and (iii) the alcohol oxidation activity was much lower than the aldehyde reduction activity.
Acinetobacter spp. are known to accumulate intracellular wax esters as cell reserves when they are grown on n-alkanes. A recent publication reported that the wax esters are synthesized from an acyl coenzyme A and an alkyl alcohol by acyl coenzyme A:alcohol transacylase and that the alcohol moiety is formed from acyl coenzyme A through two reduction steps with acyl coenzyme A reductase and aldehyde reductase (6). The cytosolic NAD(P)-dependent alcohol dehydrogenase characterized in this study may be the latter enzyme for the following reasons: (i) the enzyme was induced more by n-alkanes than the NAD-dependent enzyme was and (ii) NADPH-dependent reduction is reasonable for synthesis of wax esters, judging from the fact that NADPH provides the reducing equivalent.
Apart from its physiological significance, some features of the enzyme, such as its thermostability, broad substrate specificity, and high levels of activity toward medium-chain aldehydes, are attractive for application to enzymatic conversion of aldehydes. Among the substrates tested, for example, cinnamyl aldehyde and cinnamyl alcohol are used in the flavor and perfume industries, anisaldehyde is used for perfume and toilet soaps, and decylalcohol is used in the manufacture of plasticizers. The high-level production system for this enzyme established in this study may be useful for industrial application of this enzyme as a biocatalyst in the future.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Fox M G A, Dickinson M, Ratledge C. Long-chain alcohol and aldehyde dehydrogenase activities in Acinetobacter calcoaceticus strain HO1-N. J Gen Microbiol. 1992;138:1963–1972. doi: 10.1099/00221287-138-9-1963. [DOI] [PubMed] [Google Scholar]
- 3.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 4.Marmur J A. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 5.Ratajczak A, Geissdörfer W, Hillen W. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl Environ Microbiol. 1998;64:1175–1179. doi: 10.1128/aem.64.4.1175-1179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiser S, Somerville C. Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J Bacteriol. 1997;179:2969–2975. doi: 10.1128/jb.179.9.2969-2975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai Y, Maeng J H, Kubota S, Tani A, Tani Y, Kato N. A non-conventional dissimilation pathway for long chain n-alkanes in Acinetobacter sp. M-1 that starts with a dioxygenase reaction. J Ferment Bioeng. 1996;81:286–291. [Google Scholar]
- 8.Singer M E, Finnerty W R. Alcohol dehydrogenases in Acinetobacter sp. strain HO1-N: role in hexadecane and hexadecanol metabolism. J Bacteriol. 1985;164:1017–1024. doi: 10.1128/jb.164.3.1017-1024.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 10.van Beilen J B, Kingma J, Witholt B. Substrate specificity of the alkane hydroxylase system of Pseudomonas oleovorans GPo1. Enzyme Microb Technol. 1994;16:904–911. [Google Scholar]
- 11.Wales M R, Fewson C A. Constitutive NADP-dependent alcohol dehydrogenase of Acinetobacter sp. strain HO1-N. Curr Microbiol. 1994;29:273–277. [Google Scholar]
- 12.Wales M R, Fewson C A. NADP-dependent alcohol dehydrogenases in bacteria and yeast: purification and partial characterization of the enzymes from Acinetobacter calcoaceticus and Saccharomyces cerevisiae. Microbiology. 1994;140:173–183. doi: 10.1099/13500872-140-1-173. [DOI] [PubMed] [Google Scholar]