Abstract
Hepatocellular carcinoma (HCC) is a common malignant tumour with high rates of morbidity and mortality worldwide. Therefore, it is of great significance to find new molecular markers for HCC diagnosis and treatment. G6PD is known to be dysregulated in a variety of tumours. In addition, the ceRNA network plays a crucial role in the occurrence and development of HCC. However, the mechanism by which the ceRNA network regulates G6PD in HCC remains unclear. We used TCGA-LIHC data to analyse the possibility of using G6PD as an independent prognostic marker. Univariate Cox proportional hazards regression, multivariate Cox proportional hazards regression, and receiver operating characteristic curve analysis were used to analyse the influence of G6PD overexpression on the prognosis of HCC patients. We also analysed the biological function of G6PD, its effect on the immune microenvironment, and drug sensitivity. Finally, we constructed a ceRNA network of lncRNAs/miR-122-5p/G6PD to explore the regulatory mechanism of G6PD. G6PD was highly expressed in HCC, was related to pathological stage and poor prognosis, and could be used as an independent prognostic indicator of HCC. The expression of G6PD was closely related to the immune microenvironment of HCC. In addition, the expression of G6PD in HCC could be regulated by the ceRNA network. Therefore, G6PD can be used as an immunotherapy target to improve the survival and prognosis of HCC patients, and the ceRNA regulatory network of G6PD has potential diagnostic and therapeutic value for HCC.
Keywords: biomarkers, ceRNA, G6PD, HCC, immunomodulation, miR-122-5p
Introduction
Primary liver cancer is the third leading cause of cancer-related deaths worldwide and the second leading cause of cancer-related deaths in China. The number of new liver cancer cases ranks sixth among all tumour types. Hepatocellular carcinoma (HCC) is the most common form, accounting for approximately 85% of all primary liver cancers [1]. The long-term survival rate of patients with HCC is generally low, and recent data from the National Cancer Center show that the 5-year relative survival rate for HCC in China is only 12.1% [2,3]. HCC occurrence and development involve the regulation of multiple genes and different factors; for example, chronic hepatitis B infection, aflatoxin-containing food consumption, alcohol abuse, smoking, and Type 2 diabetes might increase the risk of HCC [4]. Currently, the diagnosis of HCC mainly depends on imaging and serological indicators. Imaging techniques tend to have a higher diagnostic efficacy in advanced stages of HCC [5]. Alpha-fetoprotein is a recognised serum marker of HCC. However, it is not used as a diagnostic test owing to its inadequate sensitivity and specificity [6]. Given the high incidence, high mortality, and poor prognosis of HCC, it is urgent to further study the pathogenesis of this disease at the molecular level to identify new targets for diagnosis and treatment.
The glucose-6-phosphate dehydrogenase (G6PD) gene located on chromosome Xq28 encodes the G6PD protein, a rate-limiting enzyme in the pentose phosphate pathway (PPP). The main biological function of the PPP is to produce reduced nicotinamide adenine dinucleotide phosphate (NADPH) and ribo-5-phosphate. The PPP is a key metabolic pathway and plays an important role in tumour cell proliferation, invasion, and metastasis [7,8]. Research has been mostly focused on the study of diseases, such as haemolysis and jaundice, caused by deletion or mutations of G6PD [9,10]. However, it was recently observed that G6PD can regulate cell transformation, proliferation, apoptosis, and angiogenesis in tumour tissues [11–13]. Increased G6PD expression, at both the mRNA and protein levels, in different tumour tissues increases PPP activity and promotes tumour cell growth and survival [14–16]. Therefore, to explore the biological function of G6PD in HCC, we aimed to test it as a new molecular marker for HCC diagnosis and treatment.
Some long non-coding RNAs (lncRNAs), circular RNAs, pseudogenes, messenger RNAs (mRNAs), and other noncoding RNAs (ncRNAs) have microRNA (miRNA)-response elements (MREs), which regulate the expression of miRNA-target genes by competitively binding with miRNA [17]. Molecules in the regulatory network that perform this function are called ceRNAs. It has been recently found that the regulatory mechanism underlying the ceRNA network plays a crucial role in HCC occurrence and development [18]. The ceRNA network in HCC is involved in various biological processes, such as cancer cell growth, epithelial–mesenchymal transformation, invasion, and metastasis [19]. LncRNAs overexpressed in the ceRNA network are also involved in different biological processes in HCC, such as cell cycle progression, autophagy, and apoptosis [20]. MiRNAs are small and evolutionarily conserving ncRNAs that regulate gene expression by degrading mRNA or binding to the 3′-UTR region of the target mRNA to inhibit its translation [21]. Abnormal expression of miRNA can affect the growth and migration of cells and affect the occurrence of cancer.
MiR-122-5p is highly expressed in normal liver cells and not expressed or less expressed in other cell types. MiR-122-5p is specifically expressed in the liver to maintain liver homeostasis and metabolism and is also a new marker of liver injury [22,23]. MiR-122-5p has been shown to be a cancer suppressor molecule that inhibits the growth, metastasis, and infiltration of cancer cells in HCC [24,25]. In the ceRNA network, lncRNAs prevent the combination of miRNA and its target gene by binding to the target miRNA, thus upregulating the expression of the mRNA. However, the mechanism underlying G6PD regulation by the ceRNA network in HCC has not yet been clarified. Therefore, studying the ceRNA network centred on G6PD regulation could reveal the molecular mechanism underlying HCC occurrence and development.
To study the mechanism underlying the effects of G6PD in HCC, we analysed microarray data of human HCC samples from The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/) [26]. We used R to determine the expression levels of G6PD in tumour and normal tissues, and based on the information obtained, we analysed the correlation between G6PD and the prognosis of patients with HCC as well as the biological role of G6PD. Finally, we constructed a ceRNA network of lncRNAs/miR-122-5p/G6PD, providing a new strategy for elucidating HCC pathogenesis.
Methods
Data collection and processing
On October 7, 2021, information about 424 individuals (including 374 patients with HCC and 50 normal controls) was obtained from TCGA database, including mRNA sequencing (seq), miRNAseq, and survival data. RNA expression profiles were annotated using the Ensembl gene ID. The mean of the expression levels of duplicate genes was considered the expression level of the gene. LncRNAs, miRNAs, and mRNAs in low abundance were eliminated (low abundance was defined as the mean of gene expression being >0). The study met the research requirements of TCGA and did not require ethics approval.
Identification of differentially expressed genes (DEGs)
We used the DESeq2 package from Bioconductor in R (Version 4.1.0) to analyse the difference between the mRNA matrix files of HCC and normal samples [27]. Genes with |log2 fold-change (FC)| > 1.5 and P-adj < 0.01 were considered differentially expressed. After the identification of differentially expressed genes (DEGs), we used the pheatmap package to visualise the expression of the DEGs, after which the ggplot2 package was used to visualise the volcano plot of the up-regulated and down-regulated genes [28]. Genes with P-adj < 0.01, but log2 FC > 1.5 or log2 FC < -1.5 were defined as up-regulated and down-regulated, respectively.
Pan-cancer analysis
Pan-oncogenic transcriptome data were obtained from the University of California Santa Cruz (UCSC) database (http://xena.ucsc.edu/) for 33 different cancers [29]. The differential expression of G6PD in these tumours was analysed using the limma package in R. Tumour types with fewer than five normal samples in each tumour were removed, and the expression levels of G6PD in the remaining tumour tissues were analysed.
Expression of G6PD in HCC
HCC data in TCGA were used to analyse the differential expression of G6PD in normal tissues and HCC tumour tissues. Furthermore, we used an immunohistochemical (IHC) database (Human Protein Atlas, HPA, https://www.proteinatlas.org/) to determine the expression level of G6PD protein in cancer and normal tissues [30]. The HPA database uses the protein analysis method of IHC from normal and tumour tissues to determine protein levels. Based on the clinical data and gene expression profile of HCC in TCGA, we analysed the relationship between the expression level of G6PD and clinic-pathological features.
Survival analysis
Kaplan–Meier (K-M) survival analysis was used to analyse survival data from the HCC dataset [31]. The Survminer package in R was used to visualise the relationship between gene expression and overall survival (OS) of patients with HCC. The prognosis and diagnostic values of G6PD in HCC were evaluated by K-M survival analysis combined with receiver operating characteristic curve (ROC) analysis. The predictive value of G6PD, univariate Cox proportional hazards regression (UCR), and multivariate Cox proportional hazards regression (MCR) analysis were performed to understand whether G6PD could be used as an independent prognostic marker. Statistical significance was set at P<0.05.
Analysis of the biological function of G6PD in HCC
Different analysed HCCs were grouped according to G6PD expression levels, and gene set enrichment analysis (GSEA) was conducted to determine the biological processes affected by G6PD and its role in HCC development [32]. The screening conditions were nominal P<0.05, |normalize enrichment score (NES)| > 1, and false discovery rate < 0.25. R software was then used to extract the expression levels of immunomarker genes in HCC tissues, and the correlation between G6PD expression and tumour immune cell infiltration was analysed. In total, 28 immunomarker genes were extracted from eight types of immune cells as follows: B cells, CD8+ T cells, CD4+ T cells, natural killer (NK) cells, M1 and M2 macrophages, neutrophils, and dendritic cells [33–35]. Finally, the correlation between G6PD expression and immune checkpoint molecules (PDCD1, CD274 and CTLA-4) was studied to explore its relationship with immune checkpoint molecular blocking therapy.
Building the ceRNA network
A lncRNA-miRNA-G6PD network was constructed for G6PD. The miRNA-mRNA module of the ENCORI online database (https://starbase.sysu.edu.cn) was used to identify the miRNAs combination of G6PD [36]. In the module, we set the conditions as follows: genome = human, target = G6PD, programNum ≥ 2. Next, the miRDB database (http://mirdb.org/mirdb/) is used to predict the target miRNAs that regulate G6PD [37]. Finally, the target miRNA was matched with the miRNA data in TCGA-LIHC, and the miRNA with expression level of 0 was removed. We conducted co-expression analysis (with the criteria of |cor| > 0.2 and P<0.001) of target miRNA and G6PD to screen out more reliable miRNA. Further, we identified the interaction of miRNA and G6PD 3′-UTR by bioinformatic analysis using Targetscan (https://www.targetscan.org/vert_72/) [38].
Next, lncRNAs bound to miRNAs were predicted by the ENCORI under the following conditions: genome = human, miRNA = miR-122-5p, target = all. We conducted co-expression analysis of target lncRNAs and miR-122-5p, lncRNAs and G6PD to screen out more reliable lncRNAs. Finally, Cytoscape software was used to construct the ceRNA network regulating the G6PD experssion [39].
Drug susceptibility prediction
To evaluate the value of novel biomarkers in the clinical treatment of HCC, we used R to analyse the sensitivity of therapeutic drugs from two aspects of chemotherapy drugs and targeted inhibitors. The pRRophetic package was used to calculate the half-maximum inhibitory concentration (IC50) to predict the degree of drug response in the high G6PD and low G6PD groups [40].
Results
G6PD is overexpressed in HCC and is a potential HCC biomarker
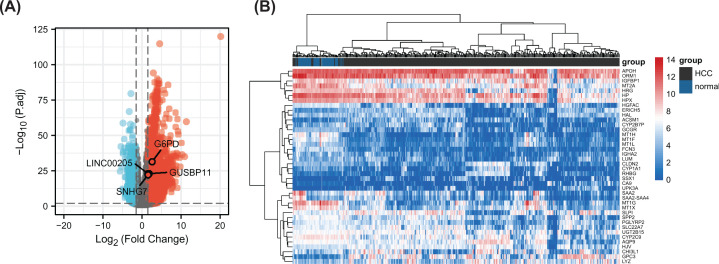
To analyse G6PD expression levels in HCC, RNAseq data from 374 HCC tissues and 50 normal tissues were obtained from TCGA. Followed the criteria of gene |log2FC| > 1.5 and P-adj <0.01, we identified 4,800 DEGs in tumour tissues (n=374) when compared with the gene expression in normal tissues (n=50); these included 3,971 up-regulated and 829 down-regulated genes (Figure 1A). G6PD was identified as a DEG in HCC, with a log2 FC of 2.598596. The top 40 DEGs were visualised using heatmaps (Figure 1B). These observations suggested that G6PD overexpression is associated with HCC.
Figure 1. Volcano plot and heatmap maps of DEGs.
(A) Volcano plot, red for up-regulated genes, blue for down-regulated genes. (B) Heatmap, heatmap of the top 40 DEGs. Red and blue show differential gene expression in grouped samples. Red indicates that the expression value is high; blue indicates that the expression value is low.
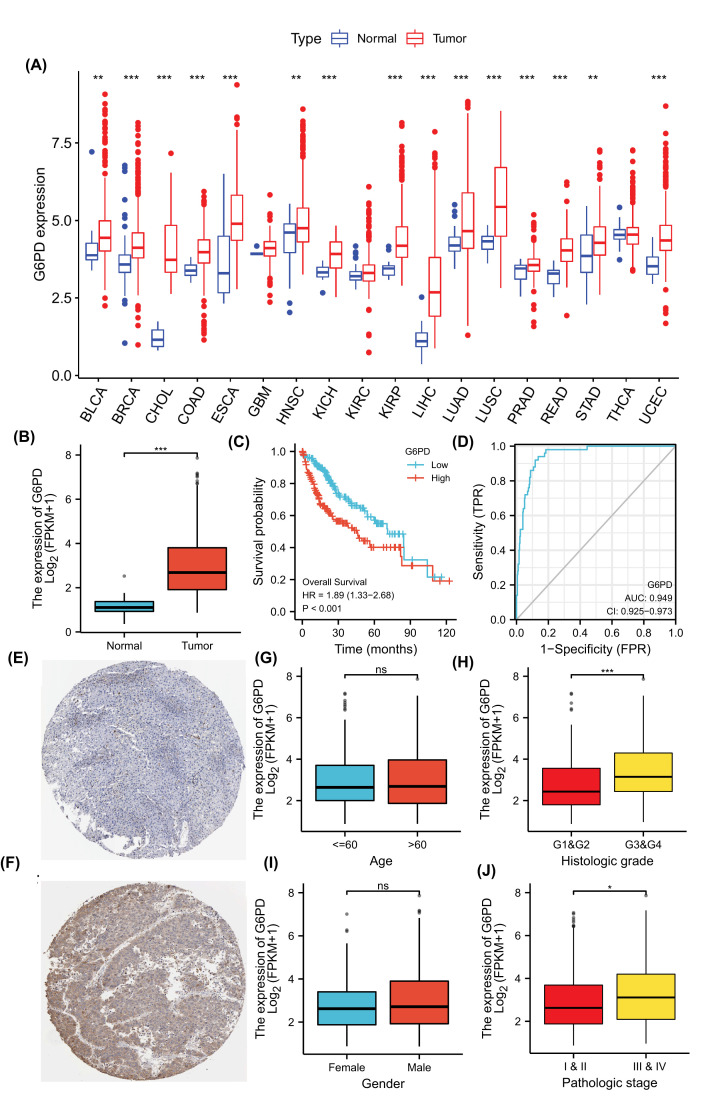
To understand the variation in G6PD expression in different tissue-derived cancers, we analysed G6PD mRNA expression levels in tissues from 33 different cancers and their respective normal tissues using RNAseq data from the UCSC database. Pan-cancer analysis showed that G6PD was significantly overexpressed in different tumour tissues, including HCC (Figure 2A,B).
Figure 2. Expression and prognostic value of G6PD.
(A) Pan-cancer analysis, G6PD is expressed in different human tumours. (B) G6PD is expressed differently in HCC tumour tissues and normal tissues. (C) Kaplan–Meier survival curves were used to analyze the prognostic value of G6PD in HCC. (D) ROC analysis of the G6PD signatures in HCC. (E) Immunohistochemical images. The protein expression of G6PD in normal liver tissue was derived from HPA. (F) Immunohistochemical images. The protein expression of G6PD in HCC was derived from HPA. (G–J) The expression of G6PD in age, gender, histologic grade and pathological stage, respectively. ns: no sense; *: P<0.05; **: P<0.01; ***: P<0.001.
Survival analysis showed that G6PD expression was correlated with the prognosis of patients with HCC, as patients with high G6PD expression had worse prognosis than those with low G6PD expression (Figure 2C). ROC analysis was used to further verify the predictive value of G6PD as a molecular target for HCC diagnosis and treatment. The area under the curve value for G6PD was 0.949 (95%CI: 0.925–0.973) (Figure 2D), indicating that the G6PD expression level could distinguish normal tissues from HCC tissues and can be used as a potential HCC biomarker. Furthermore, the result of ROC analysis showed that G6PD had good specificity (0.864) and sensitivity (0.940) as a marker of HCC. UCR analysis showed that pathological stage, T stage, M stage, and G6PD expression were correlated with OS. MCR analysis showed that only G6PD expression was correlated with OS. UCR and MCR analyses (Table 1) showed that the G6PD expression level could serve as an independent prognostic marker for patients with HCC (both P<0.05).
Table 1. Univariate and multivariate analyses of factors associated with survival of patients with HCC in TCGA database.
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |
| Age, years (≤60 vs. >60) | 1.205 (0.850–1.708) | 0.295 | 1.217 (0.757–1.956) | 0.417 |
| Gender (Woman vs. Man) | 0.793 (0.557–1.130) | 0.200 | 0.908 (0.553–1.491) | 0.703 |
| Grade (G1&G2 vs. G3&G4) | 1.091 (0.761–1.564) | 0.636 | 1.098 (0.682–1.768) | 0.699 |
| Stage (I& II vs. III& IV) | 2.504 (1.727–3.631) | <0.001 | 0.188 (0.010–3.548) | 0.265 |
| T stage (T1&T2 vs. T3&T4) | 2.598 (1.826–3.697) | <0.001 | 16.950 (0.925–310.459) | 0.056 |
| N stage (N0 vs. N1) | 2.029 (0.497–8.281) | 0.324 | 6.060 (0.778–47.176) | 0.085 |
| M stage (M0 vs. M1) | 4.077 (1.281–12.973) | 0.017 | 1.840 (0.529–6.399) | 0.338 |
| G6PD expression (Low vs. High) | 1.889 (1.331–2.682) | <0.001 | 2.165 (1.335–3.510) | 0.002 |
The expression of G6PD in HCC was further confirmed by immunohistochemistry, and the protein expression level of G6PD was analysed by the HPA. The results showed that G6PD was mainly expressed in the cytoplasm, and the protein expression of G6PD in HCC tissues was significantly higher than that in normal tissues (Figure 2E,F). Moreover, high expression of G6PD in HCC Li-7 cell lines was observed (Table 2).
Table 2. The expression level of hsa-miR-122-5p in Li-7 cell line was among the top 10 predicted targets.
| Target rank | Target score | miRNA name | Gene symbol | Li-7 expression |
|---|---|---|---|---|
| 1 | 86 | hsa-miR-122-5p | G6PD | 310 |
| 2 | 82 | hsa-miR-122-5p | PKM | 305 |
| 3 | 56 | hsa-miR-122-5p | CLIC1 | 294 |
| 4 | 51 | hsa-miR-122-5p | CANX | 270 |
| 5 | 51 | hsa-miR-122-5p | NONO | 142 |
| 6 | 69 | hsa-miR-122-5p | ALDOA | 116 |
| 7 | 85 | hsa-miR-122-5p | CCNG1 | 80 |
| 8 | 97 | hsa-miR-122-5p | HNRNPU | 78 |
| 9 | 80 | hsa-miR-122-5p | TUBA1C | 64 |
| 10 | 73 | hsa-miR-122-5p | LRP10 | 64 |
Notes: A predicted target with prediction score > 80 is most likely to be real. Targets with high or moderate expression are more likely to be relevant in Li-7 (High expression > 20; moderate expression 5-20; low expression 1-5).
To further determine the potential value of G6PD in clinical work, we analysed the influence of G6PD expression in TCGA-LIHC samples on clinical data. The results showed that the expression of G6PD was independent of age and gender (Figure 2G,H). The increased expression of G6PD was significantly correlated with histologic grade (Figure 2I). Moreover, in pathological stages, the expression of G6PD in stages III and IV was higher than that in stages I and II (Figure 2J). The expression of G6PD was elevated in the early pathological stage of HCC and increased with the progression of the disease.
Our analysis showed that G6PD is overexpressed in different cancer tissues, including HCC, and can be used as an HCC biomarker.
G6PD overexpression correlates with alteration of different signalling pathways
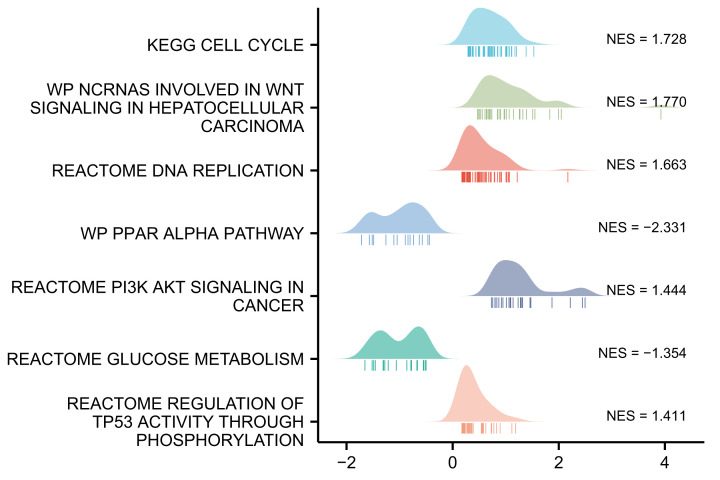
The mRNA expression matrix of patients with HCC was obtained from TCGA, and samples were grouped according to the G6PD expression level. GSEA was conducted using a molecular characteristic database to further understand the biological processes, signalling pathways, and roles of G6PD in HCC. GSEA showed that high G6PD mRNA expression correlated with cell cycle progression (P=0.001045, NES = 1.728131), DNA replication (P=0.003119, NES = 1.662617), and involvement of non-coding RNA in the WNT pathway in HCC (P=0.001087, NES = 1.7703589). Additionally, we observed a correlation between G6PD expression levels and the PI3K-AKT signalling pathway in cancer (P=0.021368, NES = 1.443601) and the regulation of P53 activity via phosphorylation (P=0.036756, NES = 1.411444). In contrast, low G6PD mRNA expression correlated with the PPARα pathway (P=0.004386, NES = −2.330786) and glucose metabolism (P=0.024390, NES = −1.353922) (Figure 3). These observations suggest an association between G6PD overexpression and alterations in a variety of signalling pathways.
Figure 3. A merged enrichment plot from gene set enrichment analysis including normalised enrichment score and gene sets.
When G6PD was highly expressed, the significantly enriched signalling pathways were cell cycle, DNA replication, ncRNAs in WNT signalling in hepatocellular carcinoma, PI3K-AKT signalling in cancer and regulation of P53 activity through phosphorylation.
G6PD overexpression correlates with the up-regulation of immune cell markers and immune checkpoint genes in HCC
To explore the relationship between G6PD expression and different immune infiltrating cells, the expression levels of different immune cell markers in HCC tumours were extracted using R software and correlated with G6PD expression levels. B cells, CD8+T cells, CD4+T cells, NK cells, M1 macrophages, M2 macrophages, neutrophils, and dendritic cells were positively correlated with G6PD expression in HCC tumours (P<0.05, cor > 0.1; Table 3). These results suggest that G6PD expression in HCC is closely related to immune cells in the tumour immune microenvironment.
Table 3. Correlation analysis between G6PD and markers of immune cells.
| Immune cell | Gene | cor | P value |
|---|---|---|---|
| B cell | CD19 | 0.282931 | <0.001 |
| B cell | CD79A | 0.191088 | 0.000201 |
| CD8+ T cell | CD8A | 0.241276 | 0.000003 |
| CD8+ T cell | CD8B | 0.264401 | <0.001 |
| CD4+ T cell | CD4 | 0.179514 | 0.000495 |
| M1 macrophage | NOS2 | -0.098838 | 0.056170 |
| M1 macrophage | IRF5 | 0.342395 | <0.001 |
| M1 macrophage | PTGS2 | 0.132085 | 0.010556 |
| M2 macrophage | CD163 | 0.193447 | 0.000172 |
| M2 macrophage | VSIG4 | 0.276690 | <0.001 |
| M2 macrophage | MS4A4A | 0.246326 | 0.000002 |
| Neutrophil | CEACAM8 | 0.091118 | 0.078423 |
| Neutrophil | ITGAM | 0.482054 | <0.001 |
| Neutrophil | CCR7 | 0.099191 | 0.055319 |
| Dendritic cell | HLA-DPB1 | 0.354284 | <0.001 |
| Dendritic cell | HLA-DQB1 | 0.275923 | <0.001 |
| Dendritic cell | HLA-DRA | 0.318123 | <0.001 |
| Dendritic cell | HLA-DPA1 | 0.279320 | <0.001 |
| Dendritic cell | CD1C | 0.151630 | 0.003287 |
| Dendritic cell | NRP1 | 0.262827 | <0.001 |
| Dendritic cell | ITGAX | 0.436201 | <0.001 |
| Natural killer cell | KIR2DL1 | 0.003966 | 0.939062 |
| Natural killer cell | KIR2DL3 | 0.076028 | 0.142236 |
| Natural killer cell | KIR2DL4 | 0.233855 | 0.000005 |
| Natural killer cell | KIR3DL1 | -0.025447 | 0.623740 |
| Natural killer cell | KIR3DL2 | 0.130731 | 0.011386 |
| Natural killer cell | KIR3DL3 | 0.072229 | 0.163319 |
| Natural killer cell | KIR2DS4 | -0.005967 | 0.908433 |
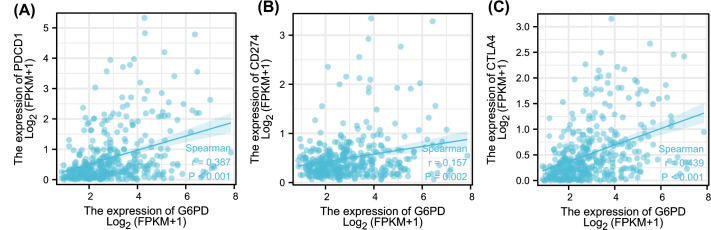
To further explore the role of G6PD in the tumour immune microenvironment, we also analysed the association between G6PD and immune checkpoint molecules (PDCD1, CD274 and CTLA4). The levels of G6PD and PDCD1 (r=0.387, P<0.001), CD274 (r=0.157, P=0.002), and CTLA4 (r=0.439, P<0.001) were positively correlated in HCC (Figure 4), suggesting that G6PD in HCC affects the efficacy of immunotherapy.
Figure 4. Scatter plot of correlation between G6PD and immune checkpoint.
(A) The expression level of G6PD was positively correlated with PDCD1 (r=0.387, P<0.001). (B) The expression level of G6PD was positively correlated with CD274 (r = 0.157, P=0.002). (C) The expression level of G6PD was positively correlated with CTLA4 (r=0.439, P<0.001).
Correlation between G6PD expression level and drug sensitivity
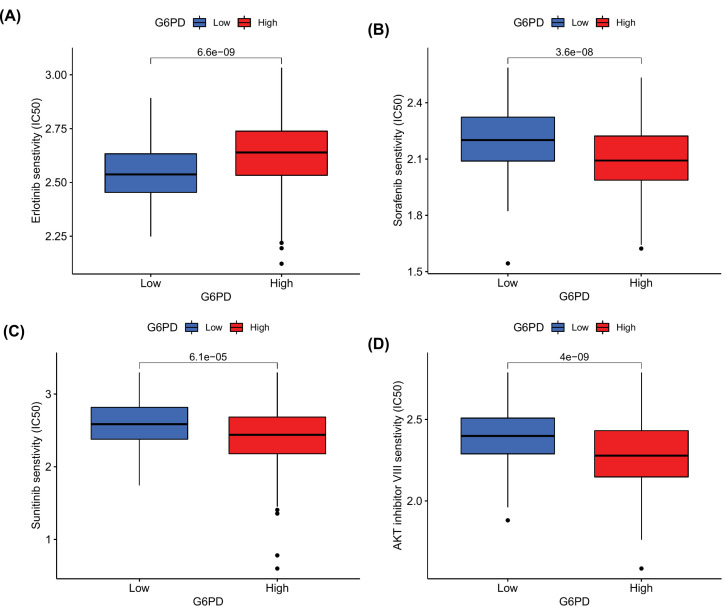
We used drug sensitivity analysis to predict the sensitivity of multiple target drugs in different subgroups of populations. Interestingly, only erlotinib was more effective in the low G6PD population (Figure 5A). Sorafenib, sunitinib, and AKT inhibitors VIII showed better therapeutic efficacy in the HCC high G6PD group (Figure 5B,D). The prediction of sensitivity of these drugs can provide useful guidance for the clinical treatment of HCC patients.
Figure 5. Sensitivity of drugs in different G6PD subgroups.
(A) Erlotinib (B) Sorafenib (C) Sunitinib (D) AKT inhibitor VIII.
G6PD expression in HCC is negatively regulated by miR-122-5p
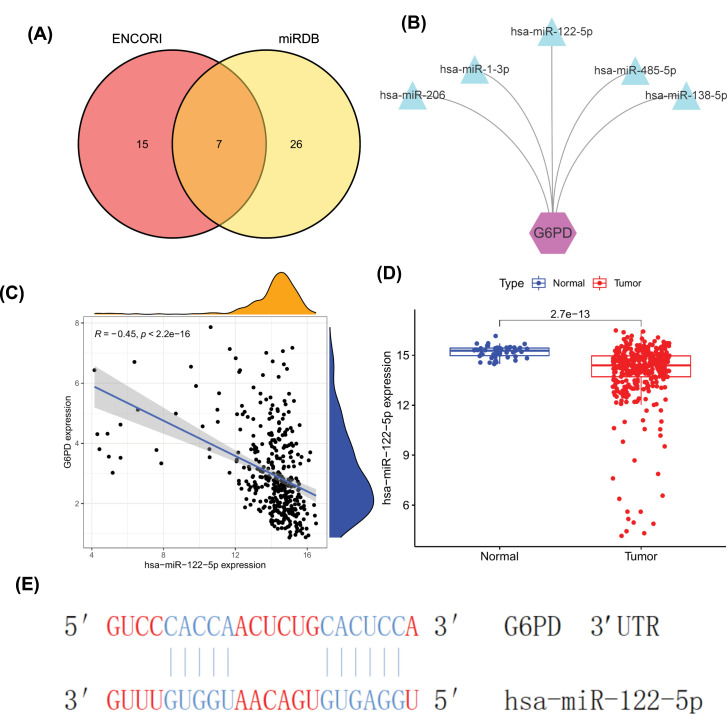
To clarify the regulatory mechanism underlying G6PD overexpression in HCC, we explored the miRNAs and lncRNAs that are implicated in the regulatory mechanism of ceRNAs. We predicted 22 miRNA-G6PD pairs using the ENCORI database and 33 pairs from the miRDB database. The intersection of the two databases was obtained using a Venn graph to obtain 7 target miRNAs (Figure 6A). Screening miRNAseq data for HCC allowed us to identify five pairs of miRNA-G6PD relationships (Figure 6B). Co-expression analysis results showed that only miR-122-5p was negatively correlated with G6PD expression (r = −0.45, P<2.2e-16; Figure 6C). Meanwhile, differential expression analysis showed that miR-122-5p was significantly down-regulated in HCC tumours (P=2.7e-13; Figure 6D). Finally, the binding site map of G6PD targeted regulation by miRNA was drawn using the Targetscan, and its binding class was 7mer-A1 (Figure 6E). In addition, high expression of G6PD was found in the Li-7 cell line of HCC and targeted binding to miR-122-5p was predicted (source = 86, Table 2). In summary, we predicted an interaction between G6PD and miR-122-5p and observed a negative correlation between their expression levels. In summary, we predicted that there was a negative regulatory relationship between G6PD and miR-122-5p.
Figure 6. Prediction of key upstream miRNAs.
(A) Veen diagram shows overlapping miRNAs in two databases. MiRNAs regulating G6PD were predicted by ENCORI and miRDB databases, respectively. (B) Network diagram of 5 pairs of miRNAs combined with G6PD. (C) Scatter plot of correlation between miR-122-5p and G6PD expression (R = −0.45, P<2.2e−16). (D) Difference expression of miR-122-5p between HCC and normal tissues. Low expression of miR-122-5p in HCC (P=2.7e-13). (E) Map of binding sites of miR122-5p/G6PD.
CeRNA network to control G6PD levels
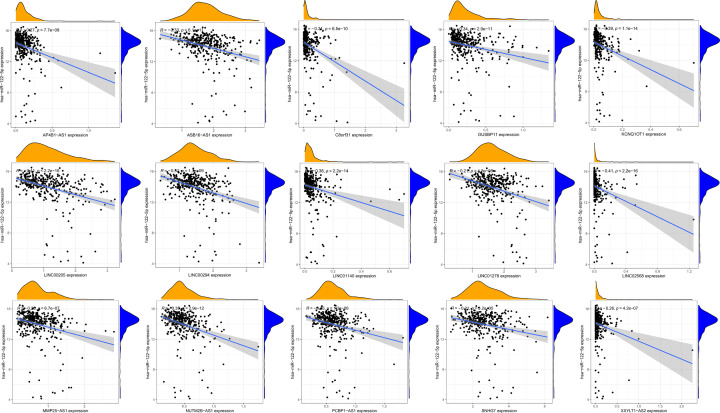
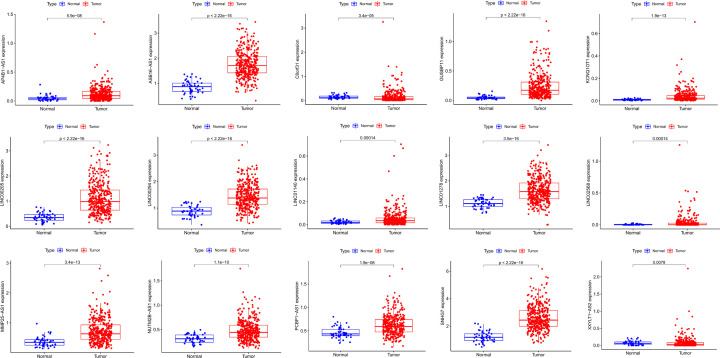
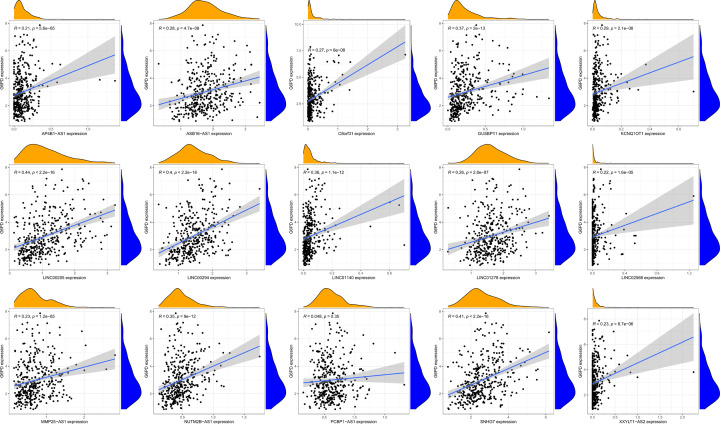
To explore the effect of miR-122-5p down-regulation in HCC, the ENCORI database was used to predict lncRNAs that target miR-122-5p. In total, 101 lncRNA-miRNA pairs were identified. Next, we analysed the differential expression of 101 lncRNAs in TCGA-LIHC samples. Subsequently, we conducted a co-expression analysis of target lncRNA and miR-122-5p. We identified 15 lncRNAs with expression patterns correlated with those of miR-122-5p (Table 4) and built a scatter diagram showing these correlations (Figure 7). Additionally, we compared the expression levels of the 15 identified lncRNAs in HCC tumour tissues with those in normal tissues (Figure 8) and found that all the identified lncRNAs were highly expressed in HCC tumours. Finally, we performed a correlation analysis of the expression levels of the 15 lncRNAs and those of G6PD in HCC (Figure 9); expression levels of 14 lncRNAs showed a positive correlation with G6PD expression (Table 5).
Table 4. Co-expression analysis of target lncRNAs and hsa-miR-122-5p.
| lncRNA | miRNA | cor | pvalue |
|---|---|---|---|
| AP4B1-AS1 | hsa-miR-122-5p | -0.274893 | P<0.001 |
| ASB16-AS1 | hsa-miR-122-5p | -0.333474 | P<0.001 |
| C8orf31 | hsa-miR-122-5p | -0.314033 | P<0.001 |
| GUSBP11 | hsa-miR-122-5p | -0.338875 | P<0.001 |
| KCNQ1OT1 | hsa-miR-122-5p | -0.387491 | P<0.001 |
| LINC00205 | hsa-miR-122-5p | -0.411210 | P<0.001 |
| LINC00294 | hsa-miR-122-5p | -0.308163 | P<0.001 |
| LINC01140 | hsa-miR-122-5p | -0.383268 | P<0.001 |
| LINC01278 | hsa-miR-122-5p | -0.210268 | 0.000048 |
| LINC02568 | hsa-miR-122-5p | -0.413327 | P<0.001 |
| MMP25-AS1 | hsa-miR-122-5p | -0.255792 | 0.000001 |
| NUTM2B-AS1 | hsa-miR-122-5p | -0.354476 | P<0.001 |
| PCBP1-AS1 | hsa-miR-122-5p | -0.249049 | 0.000001 |
| SNHG7 | hsa-miR-122-5p | -0.214748 | 0.000032 |
| XXYLT1-AS2 | hsa-miR-122-5p | -0.259494 | P<0.001 |
Figure 7. Scatter plot of correlation between 15 lncRNAs and miR-122-5p, respectively.
AP4B1-AS1, ASB16-AS1, C8orf31, GUSBP11, KCNQ1OT1, LINC00205, LINC00294, LINC01140, LINC01278, LINC02568, MMP25-AS1, NUTM2B-AS1, PCBP1-AS1, SNHG7, XXYLT1-AS2.
Figure 8. Box point composite diagram. The expression of 15 lncRNAs in HCC and adjacent normal tissues.
AP4B1-AS1, ASB16-AS1, C8orf31, GUSBP11, KCNQ1OT1, LINC00205, LINC00294, LINC01140, LINC01278, LINC02568, MMP25-AS1, NUTM2B-AS1, PCBP1-AS1, SNHG7, XXYLT1-AS2.
Figure 9. Scatter plot of correlation between 15 lncRNAs and G6PD, respectively.
AP4B1-AS1, ASB16-AS1, C8orf31, GUSBP11, KCNQ1OT1, LINC00205, LINC00294, LINC01140, LINC01278, LINC02568, MMP25-AS1, NUTM2B-AS1, PCBP1-AS1, SNHG7, XXYLT1-AS2.
Table 5. Co-expression analysis of target lncRNAs and G6PD.
| lncRNA | Gene | cor | P value |
|---|---|---|---|
| AP4B1-AS1 | G6PD | 0.207847 | <0.001 |
| ASB16-AS1 | G6PD | 0.280386 | <0.001 |
| C8orf31 | G6PD | 0.274580 | <0.001 |
| GUSBP11 | G6PD | 0.371584 | <0.001 |
| KCNQ1OT1 | G6PD | 0.287440 | <0.001 |
| LINC00205 | G6PD | 0.442060 | <0.001 |
| LINC00294 | G6PD | 0.400049 | <0.001 |
| LINC01140 | G6PD | 0.358563 | <0.001 |
| LINC01278 | G6PD | 0.264049 | <0.001 |
| LINC02568 | G6PD | 0.221990 | <0.001 |
| MMP25-AS1 | G6PD | 0.226075 | <0.001 |
| NUTM2B-AS1 | G6PD | 0.346909 | <0.001 |
| PCBP1-AS1 | G6PD | 0.048285 | 0.354189 |
| SNHG7 | G6PD | 0.408603 | <0.001 |
| XXYLT1-AS2 | G6PD | 0.231693 | <0.001 |
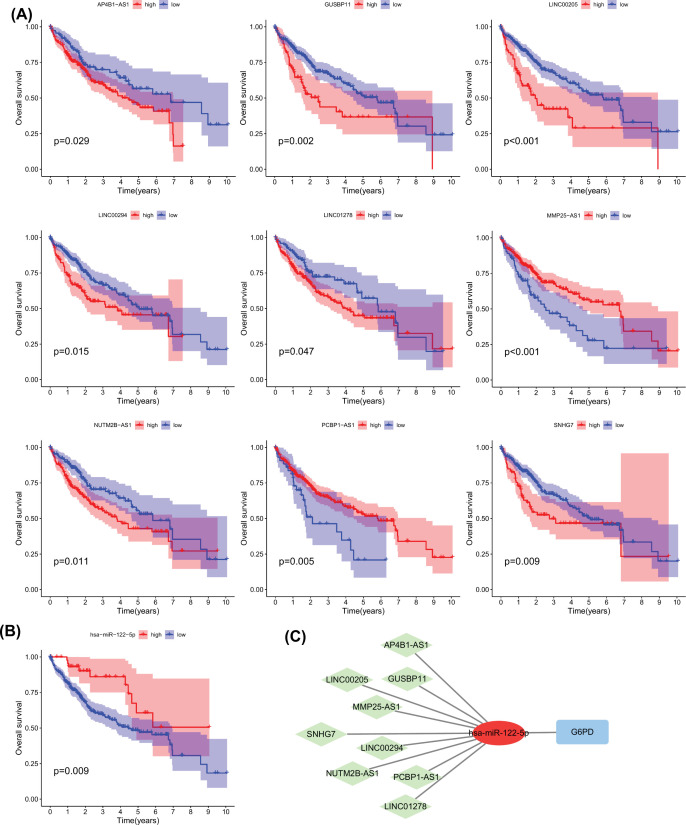
This analysis allowed us to remove the lncRNAs with large discrepancies in expression from the ceRNA network and subsequently perform a K-M survival analysis on the lncRNAs (Figure 10A). Survival analysis based on lncRNAs showed that the expression levels of nine lncRNAs were correlated with HCC survival. In addition, miR-122-5p was observed to be down-regulated in HCC, and patients with HCC with low miRNA expression had poor prognosis (Figure 10B).
Figure 10. ceRNA network.
(A) Kaplan–Meier analysis for OS of HCC patients with 9 lncRNAs expression in ceRNA network. (B) Kaplan–Meier analysis for OS of HCC patients with miR-122-5p expression in ceRNA network. (C) CeRNA networks that regulate G6PD expression.
These results showed that lncRNAs such as SNHG7, LINC00205, and GUSBP11 were significantly up-regulated in HCC and were associated with poor survival prognosis. Co-expression analysis showed that these lncRNAs were correlated with the expression of miR-122-5p and G6PD, indicating a potential regulatory relationship. Finally, a prognostic ceRNA network containing the nine lncRNAs and miR-122-5p was constructed based on G6PD expression using Cytoscape (Figure 10C). Our study suggests that G6PD expression in HCC is regulated by ceRNA, thus affecting HCC patient survival and prognosis.
Discussion
In this study, we observed that G6PD expression in HCC tissues was significantly higher than that in normal tissues. Patients with HCC with high G6PD expression showed worse prognosis and lower survival rates than those with low expression. In addition, our analyses suggest that G6PD expression is an independent prognostic risk factor for HCC and that its overexpression is correlated with cell cycle progression and the involvement of non-coding RNAs in the WNT pathway in HCC. We also found a correlation between G6PD expression and the PI3K/AKT signalling pathway in cancer, the PPARα pathway, glycolysis, and gluconeogenesis, suggesting a correlation with tumour proliferation. G6PD expression was also correlated with eight types of immune cells in the tumour immune microenvironment and positively correlated with the expression of three types of immune checkpoints. These data suggest that high G6PD expression plays an important role in the immune response and immunotherapy of HCC. Our drug sensitivity analysis showed that HCC patients with high G6PD expression were more sensitive to first-line drugs, such as sorafenib and sunitinib. Finally, we constructed a ceRNA network to study the regulation of G6PD expression in HCC tissues.
The PPP is the most active pathway in the liver. G6PD is a key enzyme that regulates the rate of the PPP and is abnormally expressed in HCC tissues, where it activates the PPP to provide NADPH and pentose for HCC cell proliferation. Pentose is a raw material used for nucleic acid synthesis in the body, and NADPH can consume excessive reactive oxygen species in abnormally proliferating tumour cells, making tumour cells resistant to oxidative stress and promoting tumour cell survival. High G6PD expression influences HCC development through a variety of pathways. It was shown that G6PD promotes cell proliferation and participates in tumourigenesis through the reprogramming signalling pathway [41]. The WNT/β-catenin pathway is abnormally activated in HCC and is involved in tumourigenesis, drug resistance, proliferation, and metastasis of tumour cells [42]. The WNT signalling pathway promotes tumour cell proliferation and differentiation, which might be caused by the polarisation of M2 macrophages [43]. Furthermore, the WNT signalling pathway could influence HCC cell proliferation by regulating the cell cycle. The abnormal regulation of cell cycle processes leads to tumourigenesis [44]. Similarly, high G6PD expression is correlated with activation of the PI3K/AKT pathway. The PI3K/AKT pathway promotes the Warburg effect by increasing enzyme activity in glycolysis as well as the membrane translocation and expression of glucose transporters [45]. Activation of the PI3K/AKT pathway ensures that the energy requirements for tumour cell proliferation and differentiation are met. The present study showed that up-regulated G6PD in HCC is correlated with activation of the aforementioned pathways to contribute to HCC occurrence and development.
Our study also revealed that the G6PD expression level is correlated with that of immune markers and immune checkpoint molecules. These results suggest that G6PD is involved in tumour immunity and contributes to the proliferation, differentiation, metastasis, and immune evasion of HCC tumour cells. G6PD not only strongly correlates with immune markers of M2 macrophages but also promotes M2 macrophage polarisation through the WNT signalling pathway [43], suggesting its involvement in the regulation of tumour-associated macrophage polarisation, favouring modification of the tumour microenvironment by tumour cells to promote their proliferation. M2 macrophages play an important role in the formation of an immunosuppressive microenvironment [46]. Notably, NK cells showed dysfunction in HCC [47], but we found that G6PD was associated with KIR2DL4, a marker of NK cells. Recent studies have found that blocking the binding of KIR2DL4 to the ligand on NK cells can restore the sensitivity of breast cancer to trastuzumab [48]. This may lead to the identification of new targets for immunotherapy in HCC. Moreover,tumour cells in HCC activate immune checkpoints to inhibit the immune respone. The use of immunotherapy to block immune checkpoint molecules was shown to be effective for treating HCC [49]. In the present study, G6PD was strongly correlated with the expression of immune checkpoint molecules, suggesting that G6PD overexpression induces immune escape of tumour cells in HCC. In conclusion, G6PD might be involved in the immune invasion of the tumour microenvironment and in the attenuated anti-tumour response in HCC treatment.
Many patients gained no benefit from sorafenib treatment and suffered extreme adverse events and heavy financial burdens [50]. Thus, patients who are most likely to benefit from this therapy need to be identified. The results of this study may provide guidance for the clinical use of sorafenib. Considering that HCC patients with high G6PD levels have better sensitivity to sorafenib and the role of G6PD in the immune microenvironment, the combination of immune checkpoint inhibitors (CPIs) can be considered, which may yield better efficacy. Chen et al. proposed a strategy of sorafenib combined with CPIs in the treatment of HCC in a mouse model; results revealed significantly delayed tumour growth and metastasis [51]. Also, good results have also been achieved in the study of combined CPIs. The combination of atezolizumab and bevacizumab showed a better clinical significance than sorafenib in unresectable HCC patients in China [52]. Our results also indicate that HCC populations with high G6PD levels may be resistant to erlotinib; however, the specific mechanism remains to be studied.
As G6PD is a potential oncogene in HCC, it is necessary to study the effects of abnormal G6PD activation. Recent studies have found that noncoding RNAs can not only regulate G6PD expression via different processes but also participate in HCC occurrence and development [41]. To clarify the regulatory mechanism underlying G6PD overexpression in HCC, we constructed a lncRNA-miRNA-G6PD ceRNA network, which allowed for the identification of genes affected by abnormal expression of miRNAs [53] and lncRNAs [54]. Our observations suggest the up-regulation of G6PD expression mediated by the nine lncRNAs via binding to miR-122-5p. The miR-122-5p has been shown to be a tumour suppressor gene in many tumours. MiR-122-5p inhibits the proliferation and metastasis of tumour cells by inhibiting GIT1 in gastric cancer [55]. Previous studies have shown that expression of miR-122-5p is significantly reduced in HCC and can be restored after taurine treatment to inhibit glycolytic activity and ultimately metabolic activity of HCC cells to affect cancer cell proliferation, when overexpression of miR-122-5p significantly inhibited the proliferation, migration and invasion of HCC cells [56,57]. Our study revealed nine lncRNAs up-regulated in HCC and correlated with G6PD expression. The expression levels of these nine lncRNAs were also associated with HCC survival.
Our study showed that SNHG7 is abnormally highly expressed in HCC patients, and HCC patients with high-SNHG7 have a poor prognosis. LncRNA SNHG7 can also promote the growth and proliferation of multiple tumour cells [58]. Highly expressed SNHG7 is associated with a poor prognosis in colorectal cancer [59]. A study revealed that SNHG7 promotes the development of HCC by sponging miR-122-5p to regulate FOXK2 [24]. In our ceRNA, SNHG7 could regulate G6PD through sponge miR-122-5p in HCC; this provides novel insights regarding the SNHG7/miR-122-5p regulatory model. It has also been found that LINC00205, an emerging lncRNA, is highly up-regulated in lung cancer, gastric cancer, and retinoblastoma and is associated with a poor prognosis [60–62]. Studies have proved that LINC00205 promotes the proliferation and migration of HCC cells by targeting the expression of miR-122-5p [57]. In our study, LINC00205 is the first model to regulate G6PD expression as a ceRNA molecular sponge miR-122-5p. These novel abnormal ceRNA regulatory networks proposed in this study can provide new insights into the regulatory mechanism of G6PD and serve as a potential target for HCC immunotherapy.
In summary, we systematically studied the expression, prognostic value, and biological role of G6PD in HCC and confirmed that G6PD is an oncogene in HCC. We constructed a novel lncRNA/miR-122-5p/G6PD ceRNA network, revealing the potential regulatory mechanism underlying G6PD dysregulation. Each molecule in this novel ceRNA network has a high prognostic value for HCC. However, in vitro and in vivo experiments are needed to explore the abundance of molecules in the ceRNA network and further confirm the role of G6PD and its regulatory network in HCC. Our results suggest that G6PD can be used as an immunotherapy target to improve the survival and prognosis of patients and that the ceRNA regulatory network of G6PD has potential diagnostic and therapeutic value in HCC.
Abbreviations
- ceRNA
competing endogenous RNA
- DEG
differentially expressed gene
- FC
log2 fold-change
- GSEA
gene set enrichment analysis
- G6PD
glucose-6-phosphate dehydrogenase
- HCC
hepatocellular carcinoma
- HNSC
head and neck squamous cell carcinoma
- HPA
Human Protein Atlas
- IHC
immunohistochemical
- K-M
Kaplan–Meier
- LIHC
liver hepatocellular carcinoma
- lncRNA
long non-coding RNA
- MCR
multivariate Cox proportional hazards regression
- miRNA
microRNA
- mRNA
messenger RNA
- NADPH
nicotinamide adenine dinucleotide phosphate
- ncRNA
non-coding RNA
- NES
normalize enrichment score
- NK
natural killer
- OS
overall survival
- PPP
pentose phosphate pathway
- seq
sequencing
- TCGA
The Cancer Genome Atlas
- UCEC
Uterine Corpus Endometrial Carcinoma
- UCR
univariate Cox proportional hazards regression
- UCSC
University of California Santa Cruz
Contributor Information
Yuanyuan Zhang, Email: zhyy@dali.edu.cn.
Guangming Wang, Email: wgm1991@dali.edu.cn.
Data Availability
Publicly available datasets were analyzed in this study. These data can be found here: TCGA database (https://portal.gdc.cancer.gov/) and UCSC database (http://xena.ucsc.edu/).
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Natural Science Foundation of Yunnan Province under [grant number 202001BA070001-133]; Cultivating Plan Program for the Leader in Science and Technology of Yunnan Province under [grant number D-2017057]; Yunnan Natural Science Foundation of Department of Education under [grant number 2020J0590]; and Science and Technology project of Dali City under [grant number 2021KBG083]. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
CRediT Author Contribution
Pengyu Wang: Data curation, Formal analysis, Writing—original draft. Xitong Yang: Data curation, Visualisation. Dan Liu: Data curation, Visualisation. Yunhui Yang: Software, Visualisation. Yuanyuan Zhang: Resources, Writing—review & editing. Guangming Wang: Writing—review & editing.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A.et al. (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Zeng H., Chen W., Zheng R., Zhang S., Ji J.S., Zou X.et al. (2018) Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health 6, e555–e567 10.1016/S2214-109X(18)30127-X [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D. and Jemal A. (2016) Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 4.El-Serag H.B. (2011) Hepatocellular carcinoma. N. Engl J. Med. 365, 1118–27 [DOI] [PubMed] [Google Scholar]
- 5.Luo P., Wu S., Yu Y., Ming X., Li S., Zuo X.et al. (2020) Current status and perspective biomarkers in AFP negative HCC: towards screening for and diagnosing hepatocellular carcinoma at an earlier stage. Pathol. Oncol. Res. 26, 599–603 10.1007/s12253-019-00585-5 [DOI] [PubMed] [Google Scholar]
- 6.Benson A.B., D'Angelica M.I., Abbott D.E., Anaya D.A., Anders R., Are C.et al. (2021) Hepatobiliary cancers, Version 2.2021, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 19, 541–565 10.6004/jnccn.2021.0022 [DOI] [PubMed] [Google Scholar]
- 7.Payen V.L., Porporato P.E., Baselet B. and Sonveaux P. (2016) Metabolic changes associated with tumor metastasis, part 1: tumor pH, glycolysis and the pentose phosphate pathway. Cell. Mol. Life Sci. 73, 1333–1348 10.1007/s00018-015-2098-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perl A., Hanczko R., Telarico T., Oaks Z. and Landas S. (2011) Oxidative stress, inflammation and carcinogenesis are controlled through the pentose phosphate pathway by transaldolase. Trends Mol. Med. 17, 395–403 10.1016/j.molmed.2011.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R., Wang W., Yang Y. and Gu C. (2020) Exploring the role of glucose6phosphate dehydrogenase in cancer (review). Oncol. Rep. 44, 2325–2336 10.3892/or.2020.7803 [DOI] [PubMed] [Google Scholar]
- 10.Zhang C., Zhang Z., Zhu Y. and Qin S. (2014) Glucose-6-phosphate dehydrogenase: a biomarker and potential therapeutic target for cancer. Anticancer Agents Med. Chem. 14, 280–289 10.2174/18715206113136660337 [DOI] [PubMed] [Google Scholar]
- 11.Liu C.L., Hsu Y.C., Lee J.J., Chen M.J., Lin C.H., Huang S.Y.et al. (2020) Targeting the pentose phosphate pathway increases reactive oxygen species and induces apoptosis in thyroid cancer cells. Mol. Cell. Endocrinol. 499, 110595 10.1016/j.mce.2019.110595 [DOI] [PubMed] [Google Scholar]
- 12.Wang X., Liu H., Zhang X., Li X., Gu H., Zhang H.et al. (2016) G6PD downregulation triggered growth inhibition and induced apoptosis by regulating STAT3 signaling pathway in esophageal squamous cell carcinoma. Tumour Biol. 37, 781–789 10.1007/s13277-015-3861-9 [DOI] [PubMed] [Google Scholar]
- 13.Leopold J.A., Walker J., Scribner A.W., Voetsch B., Zhang Y.Y., Loscalzo A.J.et al. (2003) Glucose-6-phosphate dehydrogenase modulates vascular endothelial growth factor-mediated angiogenesis. J. Biol. Chem. 278, 32100–32106 10.1074/jbc.M301293200 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q., Han Q., Yang Z., Ni Y., Agbana Y.L., Bai H.et al. (2020) G6PD facilitates clear cell renal cell carcinoma invasion by enhancing MMP2 expression through ROSMAPK axis pathway. Int. J. Oncol. 57, 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagashio R., Oikawa S., Yanagita K., Hagiuda D., Kuchitsu Y., Igawa S.et al. (2019) Prognostic significance of G6PD expression and localization in lung adenocarcinoma. Biochim. Biophys. Acta Proteins Proteom 1867, 38–46 10.1016/j.bbapap.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 16.Zampella E.J., Bradley E.L. Jr. and Pretlow T.G. II (1982) Glucose-6-phosphate dehydrogenase: a possible clinical indicator for prostatic carcinoma. Cancer 49, 384–387 [DOI] [PubMed] [Google Scholar]
- 17.Salmena L., Poliseno L., Tay Y., Kats L. and Pandolfi P.P. (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X., Ding J., Wang X., Cheng Z. and Zhu Q. (2020) NUDT21 regulates circRNA cyclization and ceRNA crosstalk in hepatocellular carcinoma. Oncogene 39, 891–904 10.1038/s41388-019-1030-0 [DOI] [PubMed] [Google Scholar]
- 19.Samek W., Binder A., Montavon G., Lapuschkin S. and Muller K.R. (2017) Evaluating the visualization of what a deep neural network has learned. IEEE Trans. Neural Netw. Learn Syst. 28, 2660–2673 10.1109/TNNLS.2016.2599820 [DOI] [PubMed] [Google Scholar]
- 20.Han T.S., Hur K., Cho H.S. and Ban H.S. (2020) Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular carcinoma. Cancers (Basel) 12, 2622 10.3390/cancers12092622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oura K., Morishita A. and Masaki T. (2020) Molecular and functional roles of MicroRNAs in the progression of hepatocellular carcinoma-a review. Int. J. Mol. Sci. 21, 8362 10.3390/ijms21218362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z., Wu W., Ou P., Wu M., Zeng F., Zhou B.et al. (2021) MiR-122-5p knockdown protects against APAP-mediated liver injury through up-regulating NDRG3. Mol. Cell. Biochem. 476, 1257–1267 10.1007/s11010-020-03988-0 [DOI] [PubMed] [Google Scholar]
- 23.Maruyama S., Furuya S., Shiraishi K., Shimizu H., Akaike H., Hosomura N.et al. (2018) miR-122-5p as a novel biomarker for alpha-fetoprotein-producing gastric cancer. World J. Gastrointest Oncol. 10, 344–350 10.4251/wjgo.v10.i10.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Z., Gao J. and Huang S. (2022) LncRNA SNHG7 promotes the HCC progression through miR-122-5p/FOXK2 axis. Dig. Dis. Sci. 67, 925–935 10.1007/s10620-021-06918-2 [DOI] [PubMed] [Google Scholar]
- 25.Yang X., Sun L., Wang L., Yao B., Mo H. and Yang W. (2019) LncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4. Biomed. Pharmacother. 118, 109386 10.1016/j.biopha.2019.109386 [DOI] [PubMed] [Google Scholar]
- 26.Grossman R.L., Heath A.P., Ferretti V., Varmus H.E., Lowy D.R., Kibbe W.A.et al. (2016) Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375, 1109–1112 10.1056/NEJMp1607591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M.I., Huber W. and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickham H. (2016) ggplot2:Elegant Graphics for Data Analysis, Springer [Google Scholar]
- 29.Goldman M.J., Craft B., Hastie M., Repecka K., McDade F., Kamath A.et al. (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sjostedt E., Zhong W., Fagerberg L., Karlsson M., Mitsios N., Adori C.et al. (2020) An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367, eaay5947 10.1126/science.aay5947 [DOI] [PubMed] [Google Scholar]
- 31.Schroder M.S., Culhane A.C., Quackenbush J. and Haibe-Kains B. (2011) survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics 27, 3206–3208 10.1093/bioinformatics/btr511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A.et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Z., Xiao M., Du D., Luo N., Liu D., Liu T.et al. (2021) DNASE1L3 as a prognostic biomarker associated with immune cell infiltration in cancer. Onco. Targets Ther. 14, 2003–2017 10.2147/OTT.S294332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siemers N.O., Holloway J.L., Chang H., Chasalow S.D., Ross-MacDonald P.B., Voliva C.F.et al. (2017) Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS ONE 12, e0179726 10.1371/journal.pone.0179726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danaher P., Warren S., Dennis L., D'Amico L., White A., Disis M.L.et al. (2017) Gene expression markers of tumor infiltrating leukocytes. J. Immunother Cancer 5, 18 10.1186/s40425-017-0215-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J.H., Liu S., Zhou H., Qu L.H. and Yang J.H. (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y. and Wang X. (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 48, D127–D131 10.1093/nar/gkz757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agarwal V., Bell G.W., Nam J.W. and Bartel D.P. (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legeay M., Doncheva N.T., Morris J.H. and Jensen L.J. (2020) Visualize omics data on networks with Omics Visualizer, a Cytoscape App. F1000 Res 9, 157 10.12688/f1000research.22280.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geeleher P., Cox N. and Huang R.S. (2014) pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 9, e107468 10.1371/journal.pone.0107468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Zhou X.Y., Lu X.Y., Chen K.D. and Yao H.P. (2021) Involvement of the circular RNA/microRNA/glucose-6-phosphate dehydrogenase axis in the pathological mechanism of hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. 20, 530–534 10.1016/j.hbpd.2021.08.013 [DOI] [PubMed] [Google Scholar]
- 42.Vilchez V., Turcios L., Marti F. and Gedaly R. (2016) Targeting Wnt/beta-catenin pathway in hepatocellular carcinoma treatment. World J. Gastroenterol. 22, 823–832 10.3748/wjg.v22.i2.823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao J., Zhang C., Jiang G.Q., Jin S.J., Wang Q., Wang A.Q.et al. (2021) Identification of hepatocellular carcinoma-related genes associated with macrophage differentiation based on bioinformatics analyses. Bioengineered 12, 296–309 10.1080/21655979.2020.1868119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang B., Xunsun Liu J.Y., Yang D., Yang L.L., Kong D.X.et al. (2012) The effect of cell cycle and expression of cyclin B1 and cyclin C protein in hepatocellular carcinoma cell line HepG2 and SMMC-7721 after of silencing beta-catenin gene. Hepatogastroenterology 59, 515–518 [DOI] [PubMed] [Google Scholar]
- 45.Cheng J., Huang Y., Zhang X., Yu Y., Wu S., Jiao J.et al. (2020) TRIM21 and PHLDA3 negatively regulate the crosstalk between the PI3K/AKT pathway and PPP metabolism. Nat. Commun. 11, 1880 10.1038/s41467-020-15819-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswas S.K. and Mantovani A. (2010) Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11, 889–896 10.1038/ni.1937 [DOI] [PubMed] [Google Scholar]
- 47.Fisicaro P. and Boni C. (2022) T and NK cell-based immunotherapy in chronic viral hepatitis and hepatocellular carcinoma. Cells 11, 180 10.3390/cells11020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng G., Guo Z., Li W., Xi W., Zuo B., Zhang R.et al. (2021) Interaction between HLA-G and NK cell receptor KIR2DL4 orchestrates HER2-positive breast cancer resistance to trastuzumab. Signal Transduct. Target Ther. 6, 236 10.1038/s41392-021-00629-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greten T.F., Lai C.W., Li G. and Staveley-O'Carroll K.F. (2019) Targeted and immune-based therapies for hepatocellular carcinoma. Gastroenterology 156, 510–524 10.1053/j.gastro.2018.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song S., Bai M., Li X., Gong S., Yang W., Lei C.et al. (2022) Early predictive value of circulating biomarkers for sorafenib in advanced hepatocellular carcinoma. Expert Rev. Mol. Diagn. 22, 361–378 10.1080/14737159.2022.2049248 [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., Huang Y., Reiberger T., Duyverman A.M., Huang P., Samuel R.et al. (2014) Differential effects of sorafenib on liver versus tumor fibrosis mediated by stromal-derived factor 1 alpha/C-X-C receptor type 4 axis and myeloid differentiation antigen-positive myeloid cell infiltration in mice. Hepatology 59, 1435–1447 10.1002/hep.26790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin S., Ren Z., Feng Y.H., Yau T., Wang B., Zhao H.et al. (2021) Atezolizumab plus bevacizumab versus sorafenib in the chinese subpopulation with unresectable hepatocellular carcinoma: phase 3 randomized, open-label IMbrave150 study. Liver Cancer 10, 296–308 10.1159/000513486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y., Yang L., Chen T., Liu X., Guo Y., Zhu Q.et al. (2019) A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol. Cancer 18, 28 10.1186/s12943-019-0957-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J., Yin X., Yu X., Dai C. and Zhou F. (2019) Long noncoding RNA LINC00488 functions as a ceRNA to regulate hepatocellular carcinoma cell growth and angiogenesis through miR-330-5. Dig. Liver Dis. 51, 1050–1059 10.1016/j.dld.2019.03.012 [DOI] [PubMed] [Google Scholar]
- 55.Jiao Y., Zhang L., Li J., He Y., Zhang X. and Li J. (2021) Exosomal miR-122-5p inhibits tumorigenicity of gastric cancer by downregulating GIT1. Int. J. Biol. Markers 36, 36–46 10.1177/1724600821990677 [DOI] [PubMed] [Google Scholar]
- 56.Nabi A.A., Atta S.A., El-Ahwany E., Elzayat E. and Saleh H. (2021) Taurine upregulates miRNA-122-5p expression and suppresses the metabolizing enzymes of glycolytic pathway in hepatocellular carcinoma. Mol. Biol. Rep. 48, 5549–5559 10.1007/s11033-021-06571-y [DOI] [PubMed] [Google Scholar]
- 57.Zhang L., Wang Y., Sun J., Ma H. and Guo C. (2019) LINC00205 promotes proliferation, migration and invasion of HCC cells by targeting miR-122-5p. Pathol. Res. Pract. 215, 152515 10.1016/j.prp.2019.152515 [DOI] [PubMed] [Google Scholar]
- 58.Najafi S., Ghafouri-Fard S., Hussen B.M., Jamal H.H., Taheri M. and Hallajnejad M. (2021) Oncogenic roles of small nucleolar RNA Host Gene 7 (SNHG7) long noncoding RNA in human cancers and potentials. Front Cell Dev. Biol. 9, 809345 10.3389/fcell.2021.809345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y., Zeng C., Hu J., Pan Y., Shan Y., Liu B.et al. (2018) Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J. Hematol. Oncol. 11, 89 10.1186/s13045-018-0632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y., Hu Y., Wu Y., Zhang D. and Huang D. (2022) LINC00205 promotes tumor malignancy of lung adenocarcinoma through sponging miR-185-5p. Lab. Med. 53, 39–46 10.1093/labmed/lmab041 [DOI] [PubMed] [Google Scholar]
- 61.Huangfu L., Fan B., Wang G., Gan X., Tian S., He Q.et al. (2022) Novel prognostic marker LINC00205 promotes tumorigenesis and metastasis by competitively suppressing miRNA-26a in gastric cancer. Cell Death Discov. 8, 5 10.1038/s41420-021-00802-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang S., Long J. and Hu Y. (2020) Long noncoding RNA LINC00205 enhances the malignant characteristics of retinoblastoma by acting as a molecular sponge of microRNA-665 and consequently increasing HMGB1 expression. Biochem. Biophys. Res. Commun. 526, 396–403 10.1016/j.bbrc.2020.03.083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: TCGA database (https://portal.gdc.cancer.gov/) and UCSC database (http://xena.ucsc.edu/).