Abstract
As humans age multiple forms of biological decay ensue, and many aspects of human biology can be measured to determine how far biological machinery has drifted from homeostasis. Research has led to aging clocks being developed that claim to predict biological age as opposed to chronological age. Aging could be regarded as a measured loss of homeostatic biological equilibrium that augments biological decay in fully developed tissues. Measuring aspects of how far various elements of biology have drifted from a youthful state may allow us to make determinations on a subject's health but also make informed predictions on their biological age. As we see across human physiology, many facets that maintain human health taper off such as nicotinamide adenine dinucleotide, glutathione, catalase, super oxide dismutase, and more. Extracellular vesicle density also tapers off during age combined with epigenetic drift, telomere attrition, and stem cell exhaustion, whilst genomic instability and biological insults from environment and lifestyle factors increase. Measuring these types of biomarkers with aging clocks may allow subjects to understand their own health more accurately and enable subjects to better focus on their efforts in the pursuit of longevity and, in addition, allow healthcare practitioners to deliver better health advice.
Keywords: aging clocks, epigenetic clock, telomeres
Graph indicating where maturation starts, which (for the purpose of this article) is where aging starts. After maturation, a steady decline happens across numerous biological systems, which is shown as a percentage of how effective those systems (as an average) remain throughout age. Measuring aging with this method (known as “Ray’s Way”) offers a baseline for aging, which starts at organismal maturation with the steady descent toward senescence. This method allows comparisons with the point of maturation for better data resolution.

1. INTRODUCTION
What is biological age? How can it be measured? There are many definitions to what aging actually is; however, for the purposes of this article, aging will be considered as
a measure of how far specific biological machinery has drifted from its optimal baseline state that was once present at the moment of an organism's full growth.
Using the above metric, it becomes possible to clearly measure how far a subject's biology has drifted from homeostasis and therefore calculate a biological age and risk of death. Measuring biomarkers of aging before an organism has reached full maturation may be a flawed methodology due to various hormones, enzymes, epigenetic markers, and other biological products that may be at extremely heightened levels prior to organismal maturation, so measuring or comparing datasets to pre‐maturation levels could most likely result in misinterpretation of the data (Figure 1).
FIGURE 1.
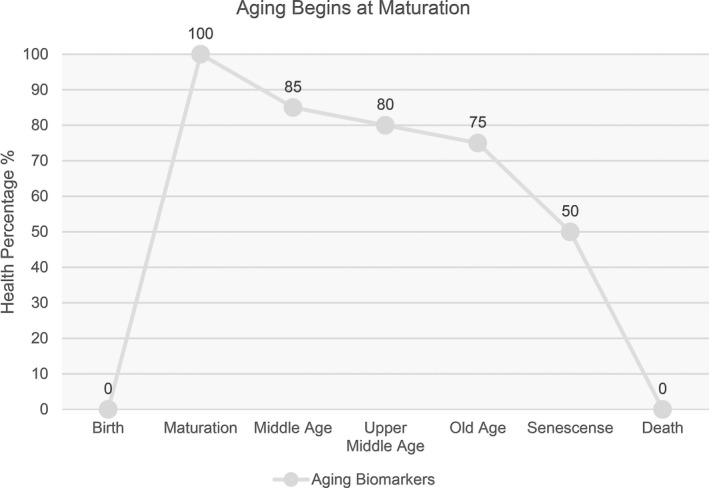
Graph indicating where maturation starts, which (for the purpose of this paper) is where aging starts. After maturation, a steady decline happens across numerous biological systems, which is shown as a percentage of how effective those systems (as an average) remain throughout age. Measuring aging with this method offers a baseline for aging, which starts at organismal maturation with the steady descent toward senescence. This method allows comparisons with the point of maturation for better data resolution
2. DISCUSSION
Aging clocks can be devised from any biological system that changes during age. Measuring the amount of variation in those biological systems may allow scientists to peer into how far an organism has drifted from youthful function or how close they are to mortality. Aging clocks specifically aim to inform subjects of their biological age, but many of these clocks deliver no information on how long a subject may have left. However, in cases where the data can deliver information on impending death (unless some type of intervention is taken), then those clocks are also mortality timers that can better serve a subject's decision making or the advice from a healthcare practitioner. Three categories exist, aging clocks that deliver biological age, aging clocks and mortality timers that deliver biological age and information to predict death, and mortality timers that only offer information that can be used to predict the onset of disease or death.
Even though aging clocks have become popular, the term mortality timer should also be used (where applicable) across the industry to deliver sobering information that may assist in changing poor decision making related to a subject's lifestyle.
Many aging clocks and mortality timers exist, such as blood biomarkers (proteomics), epigenetic mechanisms, extracellular vesicles, immune system factors, telomere length, glycomic levels, grip strength, blood vessel health, and many more biological systems could be used to determine a subject's age. However, the accuracy to predict age, overall health, or impending demise may be lacking.
An examination is performed herein on the abilities of aging clocks and their limitations across human biology. A consensus shared among longevity researchers is that if you can prevent or attenuate biological facets from entering senescence, then you may be able to slow, stop, or reverse aspects of aging. For aging clocks to deliver accuracy, a metric must first be established to determine biomarkers that are regarded as either youthful, healthy, or disease resistant at specific chronological ages. These data can then be translated to a mean average that aging clocks can utilize as a guide.
One of the first aging clocks first appeared in 2011 by Hunnam et al. as cytosine phosphate guanine (CpG) islands were found to indicate various aspects of poor health. 1 This deoxyribose nucleic acid (DNA) methylation clock was further refined by Steve Horvath to include more tissues and CpG patterns. 2 Telomere measurements are also used to determine biological age, as shorter telomeres appear to indicate more genomic instability and replicative senescence. 3 Telomere attrition and the effects on health was first revealed through a series of scientific breakthroughs during the 1980s by Elizabeth Blackburn, Joseph Gall, and Carol W. Greider. 4 Though whether shorter telomeres are a product of aging, a driver of aging, or both remains unclear. The glycome has become another major contender in the aging clock space, as glycosylation signatures shift with age. 5 , 6 More recently, exosomes may also become an interesting method of measuring age and health, as it has been discovered that exosomes released from the hypothalamus wane with age. 7 Blood markers have also become a primary indicator of predicting age and mortality by measuring proteins in the blood: a proteomic clock. 8 An immuno‐clock may also be feasible by analyzing various innate and adaptive immune cells that also include T cells and B cells that are known to taper off during age. 9 Sirtuin expression may also indicate biological age, as the sirtuin genes that have been implicated in longevity declines with age; thus a sirtuin clock may deliver valuable data on how an individual is aging. 10 Many other methods and systems exist that can also be measured to determine age, disease risk, and impending or imminent death.
3. EPIGENETIC CLOCK—DNA METHYLATION
Epigenetic clocks can reveal information about lifestyle habits and environmental factors along with genetic and epigenetic expression that is able to resolve vast amounts of data about an individual's biology. 11 Epigenetic clocks are not designed to resolve data on poor genomic integrity induced by short telomeres or environmental factors or pinpoint circulating blood plasma exosomes, glycomic signatures but may identify biological insults from chemicals such as cigarette smoking. 12 , 13 , 14
Epigenetic clocks are regulated by methylation points across the genome, and as humans age, these methylation markers may change their formation or loci, which in turn affects gene expression. Many subjects may want to maintain youthful gene expression, so even epigenetic diets have been proposed. 15 However, maintaining youthful epigenetic configurations may be possible, as Fahy et al. has demonstrated that winding back these epigenetic methylation markers is also possible, meaning that earlier gene expression could be regained. 16 This presents a unique view for an aging clock, because if subjects can measure their ability to control facets of aging, and observe the resetting to earlier methylation patterns, then this may hold motivation for subjects to maintain a healthier lifestyle.
There does appear to be evidence that epigenetic clocks can predict the early onset of disease including cancer, metastasis, and mortality. 17 , 18 It was found by Zheng et al. that older epigenetic age, relative to chronological age, heightened the potential for cancer incidence within 3 to 5 years. 18
The telomere length can also be extracted from DNA methylation data. 19 According to Lu et al., DNA methylation data is a more accurate marker of age than telomeric data and was able to predict time to death, time to coronary heart disease, and time to congestive heart failure more accurately than traditional telomere measurements. Epigenetic clocks appear to remain invaluable, as they can deliver data about our genes, such as gene expression, disease penetrance, and an epigenetic (biological) age. 20 However, the same meta‐analyses by Fransquet et al. determined that a 5 year increase in DNA methylation age was implicated with an 8% to 15% elevated risk in mortality and furthermore that the small number of studies used and heterogeneity in the study's approach required further investigation to deem whether DNA methylation could be used as a clinical biomarker. 20 Moreover, it should be noted that this is only one study and conclusions on the efficacy of DNA methylation as an aging clock or mortality timer cannot be immediately drawn.
Furthermore, a lower epigenetic age has also been found to have lower incidence of dementia. 21
It is also noted that different tissues age at different rates, so an epigenetic clock may not capture the condition of all organs or tissues from a blood or saliva sample, 22 though knowing disease risk, mortality, dementia risk, and more appears very useful to users of epigenetic test kits. The inclusion of composite clinical measures can greatly increase the resolution of epigenetic clocks in the tissues and cells measured. 23
Epigenetic clocks may also assist governments in expediting future anti‐aging drugs, as these clocks can deliver data within a few years as opposed to waiting decades to see the effects of new drugs on a population‐wide study.
Epigenetic clocks work by delivering an “aging score,” and this aging score, whilst incredibly accurate, may not capture the entirety of how a subject is aging, though the epigenetic clock appears to deliver accurate predictors of biological age and mortality.
4. TELOMERE ANALYSIS
Telomeres have been described as noncoding tandem repeats, using the DNA sequence “TTAGGG” that is found at the terminal ends of all mammalian chromosomes. 24 Telomere data do not currently appear to show important biological characteristics such as arterial disease, epigenetic drift, neurological decline, bone disorders, or other deep data regarding methylation or the glycocalyx; however, telomeres can clearly show a history of mitosis and a future timer on how many mitotic events remain and also indicate that disease may be imminent, as shorter telomeres are implicated in various disease. 25 Data that reveal extreme telomere attrition may also motivate subjects to begin telomerase‐boosting activities such as exercise. 26 Telomere regions are comprised of telomeric and sub‐telomeric sections that are used in estimating the overall length of telomeres. 27 Various methods exist for telomere evaluation, and this discussion will focus on quantitative fluorescence in situ hybridization (Q‐FISH) and terminal restriction fragments (TRF) analysis.
Q‐FISH is an analysis technique where a fluorescent DNA probe binds with telomeres. Q‐FISH appears to deliver an accurate analysis of telomere length. 28 TRF analysis is also proposed as an accurate technique; however, due to its limitations for scalability, TRF remains mostly for lab research projects as opposed to commercial telomere testing. 29 TRF that is analyzed via Southern blotting includes the canonical zones of telomeres; however, the noncanonical or sub‐telomeric areas may also be included, which may convolute results. 30
DNA methylation assays to determine telomere length have also been developed and are shown to outdo TRF analysis. 19 The same study shows that epigenetic clocks and telomere attrition do not share CpGs and also use independent genes that do not overlap in function, which points to epigenetic age being a separate age marker to telomeric age.
Flow‐FISH is an alteration of Q‐FISH that utilizes a flow cytometry approach that can analyze cells in suspension as opposed to using slides. Limitations of Flow‐FISH exist, however, being that cells that remain in suspension are unfixed and difficult to process, and additionally the peptide nucleic acid (PNA) probe may become bound to random cytoplasmic artifacts. 27
Many of the commercial telomere testing companies appear to calculate the average length of telomeres but do not reveal the data of the shortest telomeres, that is the percentage of the shortest telomeres, which according to Hemann et al. are the most important data to retrieve from telomere testing, 31 as critically short telomeres are implicated in genomic instability and cell survival. However, telomere testing uses blood samples, and the heterogeneity of cells in blood samples may cause fluctuations in the findings; therefore, testing more than one sample taken from different tissues may allow for a more accurate analysis to be performed. 28 , 32 Though this methodology could apply to all aging clocks and mortality timers where repeat samples could deliver variations, averaging those results may inform more accurately.
5. GLYCOMIC ANALYSIS
The glycome is an incredibly complex system that defines the entire collection of glycoconjugates made from carbohydrate chains that are covalently bound to lipid or protein molecules. 33 Scientists are now able to consistently measure the secreted cell‐surface glycome to peer into health and disease risk. Alterations in glycosylation have been implicated in viruses escaping detection from the immune system, pro‐cancer metastasis, standardize apoptosis, and regulate inflammatory responses according to Reily et al. The same review by Reily et al. also indicates that the structure of the glycome is also implicated in kidney capacity, health, and disease, along with autoimmune diseases, nephropathy, systemic lupus erythematosus, and inflammatory bowel disease, all incurring aberrant glycosylation. 30
Glycomic age prediction works by analyzing drift across immunoglobulin G (IgG) glycosylation, including glycans such as FA2B, FA2G2, and FA2BG2, where alterations in IgG during age can be observed. 6 Changes in glycosylation during age have been observed and replicated for over 20 years. 34 The most common post‐translational modification is the enzymatic addition of glycans to amino acid side chains, or protein glycosylation. 35
IgG glycans are efficient in mitigating inflammation, as inflammation has been implicated across biology as a possible driver of the aging process. 36 Different glycans are known to either attenuate or drive inflammation, and being able to measure these IgG signatures as either positive or negative is powerful information for those requiring a window into their biological age. 6 Both epigenetic and telomere age can be improved with exercise, and this is also seen with glycosylation. 37 Therefore, glycomic surveillance may appear to deliver a powerful window that can be used as an aging clock.
6. EXOSOMAL ANALYSIS
Exosomes are one type of extracellular vesicle that is key to carrying out multiple cellular communications between cells by suspension in cerebral spinal fluid (CSF), blood, plasma, serum, breast milk and urine (among other fluids), and various tissues. 38 , 39 Extracellular vesicles such as exosomes convey biological luggage to receiving cells such as ribonucleic acid (RNA), microRNA (miRNA), proteins, organelles, and lipids as indicated by Martins et al. 39
Even though exosomal clocks are not yet commercially available, due to the system‐wide importance that exosomes have for homeostasis, this article has sketched the concept of monitoring extracellular vesicles throughout age.
Various extracellular vesicles are implicated across multiple tissues to regenerate a myriad of disease such as liver fibrosis, cerebrovascular disease, and ischemic heart injury, among others. 40 Even though exosomes can deliver proteins and lipids, it is the ribonucleic acid (RNA) and miRNA that appears to be relevant for longevity, as the RNA or miRNA is able to produce its own proteins, which is essential for tissue repair and rejuvenation. Monitoring how ubiquitous exosomes are during age and whether there is an apparent “exosomal drift” may allow for an exosomal clock, as exosomes are also implicit in immuno‐communication. 41 , 42
Exosomes may not offer direct information on genome integrity, telomere length, stem cell function, and many other drivers of the aging process. According to Zhang et al., exosomes from the hypothalamus regulate aging via miRNAs, and these miRNAs decrease as we age, which led to the conclusion that aging is controlled by hypothalamic stem cells partly via the distribution of exosomal miRNAs. 7 This also points to the relative importance of exosomal clocks being used in conjunction with other biological measuring mechanisms such as the epigenetic or glycomic clocks to deliver a much more accurate picture of a subject's health and future prospects. 43
7. BLOOD BIOMARKERS FOR AN AGING CLOCK
Biochemical agents found in blood can also act as biomarkers that predict not only age, but impending disease, such as the non‐proteinogenic α‐amino acid homocysteine that has been implicated in the progression of heart disease. 44 Biochemical products found inside the blood are well established as markers of health and can also be used as mortality timers to predict multiple conditions and possible death. 45
Parabiosis, where scientists first stitched two rats together to share circulatory systems, has appeared to reverse hallmarks of aging. 46 , 47 The evidence points to a conclusion that blood does possibly appear to hold valuable data for an aging clock. However, human serum is complex and contains over 4000 different types of compounds among many other products, so defining exactly which biochemical markers inside human blood are associated with aging may deliver extremely accurate data for an aging clock or mortality timer. 48
Sebastiani et al. found that changes in a single biomarker may not indicate sickness or biological decline. 49 These data may imply that multiple biomarkers should first lean negatively toward a subject's health before any diagnosis is confirmed.
Neurofilament light chain (NfL) has also become a target in blood, as higher levels of NfL inside the blood may indicate damage to the nervous system. 50 NfL proteins are found to increase in aging mice, whereas fasting reduces NfL levels. NfL is well established and a profound biomarker for neurodegenerative decline, which can predict presymptomatic to symptomatic states, as well as predicting end of life, and therefore can be used as a mortality timer. 51 Proinflammatory cytokines are also indicated in multiple pathologies across aging, notably interleukin 6 (IL‐6), interleukin 1 (IL‐1), tumor necrosis factor‐α, and C‐reactive protein, which have all been correlated with an increased risk in mortality in aged subjects. 36
Blood tests also allow for screening of multiple cancers; however, these data are not included in this discussion as it poses little benefit for aging clocks but may possibly be used as a mortality timer. 52
Blood tests are able to look at hundreds of biomarkers if required, with some of the most common being liver, kidney, bilirubin, total cholesterol, c‐reactive protein, homocysteine, hemoglobin A1C, complete blood count, and lipids, among many others. 53 This discussion does not aim to list them all, though since blood testing is relatively simple and ubiquitous in the developed world, an aging clock from blood tests may prove an extremely powerful solution for an aging population.
8. CONCLUSION
Whilst this assessment into the procedures of how to effectively measure age is not exhaustive, many other iterations of biology to determine biological age may also be possible, such as a sirtuin clock, immune system clock, metabolomic, or even a microbiome clock.
From analyzing various aging clocks to mortality timers, it becomes evident that even though some aging clocks may deliver vast amounts of biological data, no single clock can deliver all biological data across all tissues from the generic samples collected with commercial kits.
All clocks appear to have limitations on the data they can deliver, albeit the data are extremely extensive from some aging clocks such as epigenetic, blood tests, and glycomic clocks. The findings of this article are (in no order of effectiveness) that epigenetic, glycomic, and blood/serum biomarkers are the three most powerful clocks that can be used, as not only can a biological age prediction be made, along with disease potential to disease penetrance, but they can also function as rough mortality timers.
An impending mortality diagnosis is a sobering prediction to any subject, so clocks that can also deliver rough time of death also function as extreme motivation for subjects to change their lifestyle habits as a matter of urgency.
Aging clocks will continue to evolve as new biomarkers are found; however, any biological machinery that wanes with age can be used to elucidate data regarding biological age.
9. LIMITATIONS OF STUDY
None.
CONFLICTS OF INTEREST
Raymond D. Palmer is Chief Science Officer of Full Spectrum Biologics, Science of Aging; host of The Longevity Experts television show; and author of The Anti‐Aging Toolkit. He holds multiple patents in biotech.
AUTHOR CONTRIBUTIONS
Raymond D Palmer is the sole author of this work.
ACKNOWLEDGEMENTS
Thank you to Steve Horvath and Gordan Lauc for providing insight into their work.
Palmer RD. Aging clocks & mortality timers, methylation, glycomic, telomeric and more. A window to measuring biological age. Aging Med. 2022;5:120–125. doi: 10.1002/agm2.12197
REFERENCES
- 1. Hannum G, Guinney J, Zhao L, et al. Genome‐wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359‐367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Victorelli S, Passos JF. Telomeres and cell senescence ‐ size matters not. EBioMedicine. 2017;21:14‐20. doi: 10.1016/J.EBIOM.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corey DR. Telomeres and telomerase: from discovery to clinical trials. Chem Biol. 2009;16(12):1219‐1223. doi: 10.1016/J.CHEMBIOL.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miura Y, Endo T. Glycomics and glycoproteomics focused on aging and age‐related diseases–Glycans as a potential biomarker for physiological alterations. Biochim Biophys Acta. 2016;1860(8):1608‐1614. doi: 10.1016/J.BBAGEN.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 6. Dall’Olio F, Vanhooren V, Chen CC, Slagboom PE, Wuhrer M, Franceschi C. N‐glycomic biomarkers of biological aging and longevity: a link with inflammaging. Ageing Res Rev. 2013;12(2):685‐698. doi: 10.1016/J.ARR.2012.02.002 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y, Kim MS, Jia B, et al. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature. 2017;548(7665):52‐57. doi: 10.1038/nature23282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lehallier B, Gate D, Schaum N, et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat Med. 2019;25(12):1843‐1850. doi: 10.1038/S41591-019-0673-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lutshumba J, Nikolajczyk BS, Bachstetter AD. Dysregulation of systemic immunity in aging and dementia. Front Cell Neurosci. 2021;15:652111. doi: 10.3389/FNCEL.2021.652111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kilic U, Gok O, Erenberk U, et al. A remarkable age‐related increase in SIRT1 protein expression against oxidative stress in elderly: SIRT1 gene variants and longevity in human. PLoS One. 2015;10(3):e0117954. doi: 10.1371/JOURNAL.PONE.0117954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71(C):3‐39. doi: 10.1016/B978-0-12-380864-6.00001-8 [DOI] [PubMed] [Google Scholar]
- 12. Joehanes R, Just AC, Marioni RE, et al. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9(5):436‐447. doi: 10.1161/CIRCGENETICS.116.001506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee KWK, Pausova Z. Cigarette smoking and DNA methylation. Front Genet. 2013;4:132. doi: 10.3389/FGENE.2013.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco‐smoking‐related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88(4):450‐457. doi: 10.1016/J.AJHG.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3(4):503‐518. doi: 10.2217/epi.11.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fahy GM, Brooke RT, Watson JP, et al. Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell. 2019;18(6):e13028. doi: 10.1111/ACEL.13028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148(1‐2):46‐57. doi: 10.1016/j.cell.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng Y, Joyce BT, Colicino E, et al. Blood epigenetic age may predict cancer incidence and mortality. EBioMedicine. 2016;5:68‐73. doi: 10.1016/J.EBIOM.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu AT, Seeboth A, Tsai P‐C, et al. DNA methylation‐based estimator of telomere length. Aging (Albany NY). 2019;11(16):5895. doi: 10.18632/AGING.102173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, Ryan J. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta‐analysis. Clin Epigenet. 2019;11(1):1‐17, doi: 10.1186/S13148-019-0656-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Degerman S, Josefsson M, Nordin Adolfsson A, et al. Maintained memory in aging is associated with young epigenetic age. Neurobiol Aging. 2017;55:167‐171. doi: 10.1016/J.NEUROBIOLAGING.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 22. Thompson MJ, Chwiałkowska K, Rubbi L, et al. A multi‐tissue full lifespan epigenetic clock for mice. Aging (Albany, NY). 2018;10(10):2832‐2854. doi: 10.18632/aging.101590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573. doi: 10.18632/AGING.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moyzis RK, Buckingham JM, Cram LS, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85(18):6622. doi: 10.1073/PNAS.85.18.6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith L, Luchini C, Demurtas J, et al. Telomere length and health outcomes: An umbrella review of systematic reviews and meta‐analyses of observational studies. Ageing Res Rev. 2019;51:1‐10. doi: 10.1016/J.ARR.2019.02.003 [DOI] [PubMed] [Google Scholar]
- 26. Nomikos NN, Nikolaidis PT, Sousa CV, Papalois AE, Rosemann T, Knechtle B. Exercise, telomeres, and cancer: ‘the exercise‐telomere hypothesis. Front Physiol. 2018;9:1798. doi: 10.3389/FPHYS.2018.01798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Montpetit AJ, Alhareeri AA, Montpetit M, et al. Telomere length: a review of methods for measurement. Nurs Res. 2014;63(4):289. doi: 10.1097/NNR.0000000000000037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hultdin M, Grönlund E, Norrback K‐F, Eriksson‐Lindström E, Roos G, Just T. Telomere analysis by fluorescence in situ hybridization and flow cytometry. Nucleic Acids Res. 1998;26(16):3651‐3656. doi: 10.1093/NAR/26.16.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mender I, Shay JW. Telomere restriction fragment (TRF) analysis. Bio‐Protocol. 2015;5(22):e1658. doi: 10.21769/bioprotoc.1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kimura M, Stone RC, Hunt SC, et al. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat Protoc. 2010;5(9):1596‐1607. doi: 10.1038/nprot.2010.124 [DOI] [PubMed] [Google Scholar]
- 31. Hemann MT, Strong MA, Hao L‐Y, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107(1):67‐77. doi: 10.1016/S0092-8674(01)00504-9 [DOI] [PubMed] [Google Scholar]
- 32. Svenson U, Nordfjäll K, Baird D, et al. Blood cell telomere length is a dynamic feature. PLoS One. 2011;6(6):21485. doi: 10.1371/JOURNAL.PONE.0021485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol. 2019;15(6):346‐366. doi: 10.1038/s41581-019-0129-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parekh R, Roitt I, Isenberg D, Dwek R, Rademacher T. Age‐related galactosylation of the N‐linked oligosaccharides of human serum IgG. J Exp Med. 1988;167(5):1731. doi: 10.1084/JEM.167.5.1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448. doi: 10.1038/NRM3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Michaud M, Balardy L, Moulis G, et al. Proinflammatory cytokines, aging, and age‐related diseases. J Am Med Dir Assoc. 2013;14(12):877‐882. doi: 10.1016/J.JAMDA.2013.05.009 [DOI] [PubMed] [Google Scholar]
- 37. Tijardović M, Marijančević D, Bok D, et al. Intense physical exercise induces an anti‐inflammatory change in IgG N‐glycosylation profile. Front Physiol. 2019;10:1522. doi: 10.3389/FPHYS.2019.01522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoshida M, et al. Extracellular vesicle‐contained eNAMPT delays aging and extends lifespan in mice. Cell Metab. 2019;30(2):329‐342.e5. doi: 10.1016/j.cmet.2019.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martins TS, Catita J, Rosa IM, A. B. da Cruz e Silva O, Henriques AG. Exosome isolation from distinct biofluids using precipitation and column‐based approaches. PLoS One. 2018;13(6):e0198820. doi: 10.1371/JOURNAL.PONE.0198820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferguson SW, Wang J, Lee CJ, et al. The microRNA regulatory landscape of MSC‐derived exosomes: A systems view. Sci Rep. 2018;8(1):1419. doi: 10.1038/S41598-018-19581-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prattichizzo F, Micolucci L, Cricca M, et al. Exosome‐based immunomodulation during aging: A nano‐perspective on inflamm‐aging. Mech Ageing Dev. 2017;168:44‐53. doi: 10.1016/J.MAD.2017.02.008 [DOI] [PubMed] [Google Scholar]
- 42. D’Anca M, Fenoglio C, Serpente M, et al. Exosome determinants of physiological aging and age‐related neurodegenerative diseases. Front Aging Neurosci. 2019;11:232. doi: 10.3389/FNAGI.2019.00232/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ahmadi M, Rezaie J. Ageing and mesenchymal stem cells derived exosomes: Molecular insight and challenges. Cell Biochem Funct. 2021;39(1):60‐66. doi: 10.1002/CBF.3602/FORMAT/PDF [DOI] [PubMed] [Google Scholar]
- 44. Refsum H, Ueland PM, Nygård O, Vollset SE. Homocysteine and cardiovascular disease. Annu Rev Med. 1998;49:31‐62. doi: 10.1146/ANNUREV.MED.49.1.31 [DOI] [PubMed] [Google Scholar]
- 45. Quinn JG, Tansey EA, Johnson CD, Roe SM, Montgomery LEA. Blood: tests used to assess the physiological and immunological properties of blood. Adv Physiol Educ. 2016;40(2):165‐175. doi: 10.1152/ADVAN.00079.2015 [DOI] [PubMed] [Google Scholar]
- 46. Conboy IM, Rando TA. Heterochronic parabiosis for the study of the effects of aging on stem cells and their niches. Cell Cycle. 2012;11(12):2260. doi: 10.4161/CC.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kennedy BK, Berger SL, Brunet A, et al. Aging: a common driver of chronic diseases and a target for novel interventions. Cell. 2014;159(4):709. doi: 10.1016/J.CELL.2014.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Psychogios N, Hau DD, Peng J, et al. The human serum metabolome. PLoS One. 2011;6(2):e16957. doi: 10.1371/JOURNAL.PONE.0016957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sebastiani P, Thyagarajan B, Sun F, et al. Biomarker signatures of aging. Aging Cell. 2017;16(2):329‐338. doi: 10.1111/ACEL.12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaeser SA, Lehallier B, Thinggaard M, et al. A neuronal blood marker is associated with mortality in old age. Nat Aging. 2021;1(2):218‐225. doi: 10.1038/s43587-021-00028-4 [DOI] [PubMed] [Google Scholar]
- 51. Loeffler T, Schilcher I, Flunkert S, Hutter‐Paier B. Neurofilament‐light chain as biomarker of neurodegenerative and rare diseases with high translational value. Front Neurosci. 2020;14:579. doi: 10.3389/FNINS.2020.00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cree IA. Improved blood tests for cancer screening: general or specific? BMC Cancer. 2011;11:499. doi: 10.1186/1471-2407-11-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Allan GM, Young J. Complete blood count for screening? Can Fam Phys. 2017:63(10):772. /pmc/articles/PMC5638475/ [PMC free article] [PubMed] [Google Scholar]


