Abstract
Lymphomas are heterogeneous tumors with striking genetic diversity and variable outcomes even within pathologic diagnoses. Treatment response assessment relies on radiologic and nuclear scans, which cannot detect disease at the molecular level. Molecular tumor analyses require invasive tissue biopsies that cannot accurately capture spatial tumor heterogeneity within each patient. Circulating tumor DNA (ctDNA) is a minimally invasive and highly versatile biomarker that overcomes fundamental limitations of imaging scans and tissue biopsies and may aid clinical decision-making in lymphoma. In this review, we highlight the key established principles regarding ctDNA in lymphoma and emphasize the important research questions and future directions.
Significance:
ctDNA is an emerging biomarker for lymphomas that noninvasively provides genotypic information and can measure the effectiveness of treatment by detecting the presence of minimal residual disease. Key principles have emerged related to ctDNA for lymphoma, but further studies are needed to standardize its use and establish clinical utility.
Introduction
Lymphomas are a heterogeneous group of tumors with striking underlying genetic diversity and a wide range of clinical presentations. Aggressive B-cell lymphomas are mostly curable with combination chemotherapy and immunotherapy, whereas indolent lymphomas are generally incurable but often achieve durable remissions that require lifelong monitoring. Prognostic tools for lymphoma are principally based on pretreatment characteristics and do not incorporate information that emerges during treatment. Indeed, lymphomas undergo clonal evolution during and after treatment that is not easily captured with readily available clinical tools (1). The criteria that determine the quality of treatment response rely on imaging scans such as computerized tomography (CT) and FDG-positron emission tomography (PET) scans, which lack sensitivity to detect minimal residual disease (MRD) or clonogenic selection (2). Indeed, genomic profiling currently requires invasive tissue biopsies, which are limited by sampling error and do not capture spatial tumor heterogeneity or treatment-emergent clonal evolution (3–5). Further, tissue biopsies are problematic for numerous anatomic sites such as the central nervous system and deep abdominal compartments.
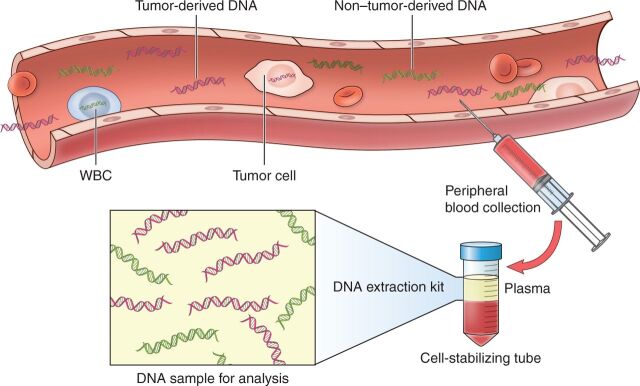
Liquid biopsies are being developed across numerous cancer types to overcome these fundamental limitations of currently available clinical tools (6). Cell-free DNA (cfDNA) refers to the circulating double-stranded DNA fragments that are freely released into the peripheral blood after normal physiologic processes of apoptosis, proliferation, and necrosis (Fig. 1; refs. 7, 8). In patients with cancer, the proportion of tumor-derived cfDNA, termed circulating tumor DNA (ctDNA), is the most well-studied form of liquid biopsy for lymphoma (9, 10). A number of studies have investigated the role of ctDNA for monitoring treatment and detecting early molecular relapse in aggressive lymphomas, including diffuse large B-cell lymphoma (DLBCL) and Hodgkin lymphoma (HL; refs. 11–21). These studies have laid the foundation for clinical applications, but critical questions remain regarding sample timing and assay methodology (22). First, the optimal methods of collection and processing need standardization, and analytical techniques should be harmonized across clinical settings (23, 24). Second, the absence of detectable MRD by ctDNA after therapy needs to be established as a surrogate for survival across lymphomas and prospectively assessed as a clinical trial endpoint. Third, genetic information from ctDNA needs to be actionable and accessible in real time for clinical applications. In this review, we focus on the key foundational principles that have been established through studies of ctDNA in DLBCL and HL and identify key research questions that need to be addressed before ctDNA can emerge as a reliable clinical decision-making tool in lymphoma (Table 1).
Figure 1.
Tumor-derived cfDNA can be noninvasively detected from peripheral blood as a liquid biopsy. As a result of the physiologic processes of apoptosis, proliferation, and secretion, degraded DNA fragments are constantly released into the peripheral blood as cfDNA; cfDNA has a short half-life of only 1 to 2 hours before being cleared from plasma by the liver, spleen, and kidney. Patients with cancer have higher total body levels of cfDNA than healthy individuals, and the tumor-derived proportion of cfDNA is termed ctDNA—the most well-studied form of liquid biopsy for lymphoma. Liquid biopsies allow for noninvasive monitoring from peripheral blood collection in specialized cell-stabilizing tubes to minimize contamination from lysis of normal white blood cells (WBC). DNA can be extracted from plasma and analyzed for the presence of tumor-specific markers including immunoglobin gene rearrangements and lymphoma-specific genetic aberrations.
Table 1.
Key established principles and remaining questions on ctDNA in lymphoma
| Biological principles |
|
| Processing and analytical principles |
|
| Clinical principles |
|
| Technical questions |
|
| Analytical questions |
|
| Clinical questions |
|
Technical and Preanalytical Considerations: Specimen Collection and Processing
The amount of cfDNA in plasma corresponds to a few thousand genomic equivalents of highly fragmented DNA per milliliter. The estimated size of cfDNA varies from ∼40 to 200 base pairs (bp), with a peak at approximately 166 bp (25). The size of cfDNA fragments, which is characteristic of caspase-dependent DNA cleavage, is due to variable length of intranucleosomal linker DNA and predominantly corresponds to mononucleosomes. Consequently, it is assumed that the bulk of cfDNA is released by cells undergoing apoptosis. Epigenomic foot printing through both nucleosome occupancy inferred from cfDNA fragmentation patterns and methylation profiling point to the hematopoietic lineages as a major source of cfDNA in healthy subjects (25, 26).
Distinguishing ctDNA from background cfDNA released by normal hematopoietic cells may be confounded by biological signals arising from clonal hematopoiesis. Clonal hematopoiesis is a common aging-related phenomenon where somatic mutations in hematopoietic stem cells are clonally propagated to their progeny. Deep next-generation sequencing (NGS) used in ctDNA mutation recovery can identify biological signals from clonal hematopoiesis in up to 90% of patients with cancer and must be distinguished from tumor-associated mutations. Consistently, a large fraction of cfDNA mutations recovered in patients with cancer have features consistent with clonal hematopoiesis (27), which confounds accurate ctDNA quantification and mutational signature recovery. Joint analysis of cfDNA and matched normal leukocytes distinguishes background mutations due to clonal hematopoiesis and allows accurate variant interpretation and recovery of lymphoma-associated mutations (27).
The concentration of cfDNA in blood varies significantly. It ranges between 0 and 100 (median ∼5) ng/mL in healthy subjects and is usually elevated, though with great variability, in patients with lymphoma, ranging from 5 to 100 (median ∼25) ng/mL (28). There is no broad consensus or standardization regarding reporting the concentration of cfDNA or ctDNA in plasma. Hence, different units of measurement have been used across various studies. Considering that 3.3 pg is the mass of a single haploid copy of the human genome (hGE), the typical cfDNA concentration is ∼1,500 hGE/mL in healthy subjects and ∼7,500 hGE/mL in lymphoma patients. In patients with aggressive lymphomas, the fraction of ctDNA constitutes a median ∼5% of cfDNA in plasma at the time of diagnosis. Therefore, in most patients with lymphoma, only a small part of total cfDNA consists of ctDNA, though the ctDNA fraction is highly dependent on tumor bulk and histology. Indeed, ctDNA fraction is higher in patients with larger tumor volumes and/or aggressive lymphomas compared with patients with low tumor volume and/or indolent lymphomas. Given the typically low ctDNA fraction, high-sensitivity wet lab approaches and bioinformatics pipelines are needed for the accurate quantification and mutation profiling of ctDNA.
Preanalytical variables that reduce the ctDNA fraction affect the sensitivity of laboratory tests aiming at identification, quantification, and qualification of ctDNA. Elevated cfDNA content in plasma is not specific to lymphoma (or malignancies in general) and may occur after exercise, inflammation, and tissue damage (i.e., trauma or infarction) as well as during pregnancy. In patients with lymphoma, such concomitant conditions may decrease the ctDNA fraction. The ctDNA fraction is also affected by blood sampling techniques and collection tubes (29). Indeed, the collection of serum stimulates a release of necrotic DNA from blood cells. This mechanism could account for the lower ctDNA fractions found in serum compared with plasma samples. cfDNA is quickly cleared from plasma by organs such as the liver, spleen, and kidney. Therefore, cfDNA has a short half-life, which varies from several minutes to 1 to 2 hours. This effect has direct implications on the collection timing following lymphoma treatment when used as a surrogate biomarker for tumor reduction. Plasma collected in EDTA tubes must be isolated from the blood cellular component within 6 hours to avoid the dilution effect of ex vivo cfDNA release by white blood cell lysis, which also decreases the ctDNA fraction. Cell-stabilizing tubes avoid white blood cell lysis, thus preserving the ctDNA fraction up to 96 hours after collection. Recommendations and unresolved issues regarding proper preanalytical handling of cfDNA have been discussed in a position paper of the American Society of Clinical Oncology and College of American Pathologists (30).
Analytical Techniques and Target Considerations
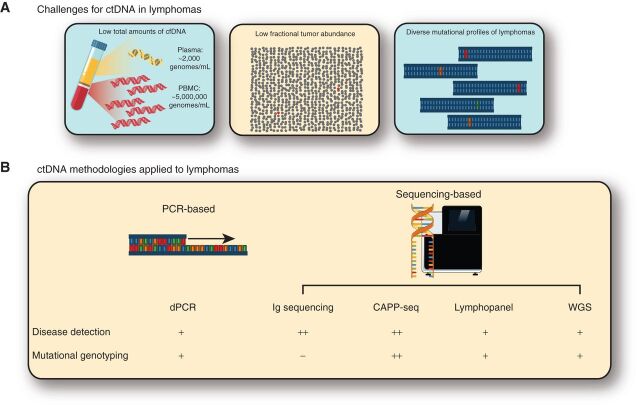
Although there are numerous potential applications of ctDNA for lymphoma, they largely fall into one of two categories: disease detection and mutational genotyping. The choice of liquid biopsy technique depends on which application is of interest. Moreover, although many techniques have been studied, an optimal ctDNA assay has multiple challenges including low total and fractional (often less than 0.5%) amounts of ctDNA, and the diversity of mutations across lymphomas (Fig. 2). Therefore, a useful ctDNA assay in lymphoma must efficiently utilize all available cfDNA, have a low background error profile to detect low-level signals, and identify the relevant mutations.
Figure 2.
Current challenges for ctDNA in lymphomas across common methodologies. A, Analysis of ctDNA in lymphoma has many challenges, including some shared in other cancer types and some more specific to lymphoma. These include: (i) a significantly lower amount of total DNA in the plasma compared with peripheral blood mononuclear cells (PBMC; left), (ii) a low proportion of tumor-derived molecules in the plasma compared with all molecules (middle), and (iii) the diverse mutational profiles of lymphomas. B, To overcome these challenges, several different approaches have been taken. These include both PCR-based and sequencing-based approaches. Each of these methods has distinct performance characteristics for both disease detection or mutational genotyping, which is qualitatively shown here. For additional details on performance between different methods for disease detection, see Table 2. CAPP-seq, cancer personalized profiling by deep sequencing; dPCR, digital PCR; WGS, whole-genome sequencing.
Methods for ctDNA Disease Detection: PCR-Based versus NGS-Based Platforms
Both PCR-based and NGS-based ctDNA methods have been applied in lymphomas, most commonly for detection of disease (Table 2). Although digital PCR (dPCR) methods assessing hotspot mutations have been used in proof-of-concept studies to track the MYD88 L265P (31, 32) and XPO1 E571K variants (33), few mutations occur with sufficient frequency to make dPCR broadly applicable. Therefore, NGS-based methods are favored for detecting and tracking ctDNA in lymphoma. These methods can assess a variety of genomic alterations, including the rearranged immunoglobulin loci (i.e., IGH, IGL, and IGK; refs. 11, 12, 34), single-nucleotide variations (SNV) and tumor-specific mutations from targeted sequencing (13, 20), and even copy-number alterations from low-pass whole-genome sequencing (WGS; ref. 16). To allow for detection of low tumor fractions, detection of Ig rearrangements through Ig high-throughput sequencing (IgHTS) was applied in initial studies for MRD detection (11, 12, 35). While showing initial promise due to its broad applicability, as most tumors contain a rearranged IGH, IGK, or IGL receptor, and low technical background error profile (36), MRD analysis is limited by the amount of total cfDNA. Indeed, the typical patient has only ∼2,000 genomes or cells worth of DNA per mL of plasma (37), so an assay that follows only one mutation—such as dPCR or IgHTS—cannot have greater sensitivity than 1:2,000 from a single millimeter of plasma, despite superior technical background error rates, when compared with higher cfDNA concentrations. Although methods to multiplex a few independent loci have emerged for both dPCR and amplicon-based Ig sequencing, the sensitivity of these methods remains fundamentally limited by the number of available DNA molecules.
Table 2.
Various methodologies applied to ctDNA in lymphomas
| Method | Technology | Sensitivity for ctDNA | Sensitivity limited by |
|---|---|---|---|
| dPCR | PCR-based | ∼1:10,000 | Amount of available DNA |
| Ig sequencing | NGS-based | ∼1:10,000 | Amount of available DNA |
| CAPP-seq | NGS-based | ∼2.5:100,000 | Error profile |
| Lymphopanel | NGS-based | ∼1:1,000 | Error profile |
| WGS | NGS-based | ∼1:100 | Error profile |
Abbreviations: CAPP-seq, cancer personalized profiling by deep sequencing; dPCR, digital PCR; WGS, whole-genome sequencing.
Overcoming Low Amounts of cfDNA
In theory, there are two ways to overcome the low amount of cfDNA found in plasma for MRD detection. First, an assay could simply use more plasma as input material. For example, starting with 10 mL of plasma instead of 1 mL increases the number of available genomes of DNA from 2,000 to 20,000, based on a typical cfDNA concentration. This concept, analogous to increasing “depth” of sequencing, has practical limitations, as obtaining more than 10 mL of plasma is challenging in routine clinical care. Alternatively, an assay could assess multiple mutations or loci to increase the number of available cfDNA fragments to assess for tumor content. Several assays to track multiple SNVs have been applied to ctDNA in solid tumors (38, 39), although the most frequent assay applied to lymphoma is targeted hybrid capture sequencing via cancer personalized profiling by deep sequencing (CAPP-seq; refs. 13, 20). Through CAPP-seq, many mutations—often over 100—can be leveraged for disease detection in B-cell lymphomas. Indeed, B-cell lymphomas are excellent targets for the CAPP-seq approach, as several regions of the genome are frequently and recurrently mutated by activation-induced cytidine deaminase (AICDA), making them suitable tumor reporters. In contrast to IgHTS or other single allele–based methods, where sensitivity is limited by the number of available molecules, CAPP-seq is limited by the background error rate of the assay. As such, a number of error-suppression methods, such as molecular barcoding and in silico error suppression, have been applied to lower the background rate, resulting in sensitivity of ∼2.5 parts per 100,000 (40).
Even lower background rates could further improve sensitivity for ctDNA MRD, as the number of mutations available due to AICDA and other mutational mechanisms allows for hundreds of thousands or millions of cfDNA fragments to be evaluated for tumor mutations. Such approaches to improving the sensitivity for MRD detection represent the next key advance in liquid biopsies for lymphoma, allowing disease detection at the time of lowest disease burden, such as the end of induction therapy. One approach for improving the limit of detection for ctDNA is phased variant enrichment and detection by sequencing (PhasED-seq; ref. 41). Although most ctDNA methods track SNVs (i.e., single mutations on a cfDNA molecule), PhasED-seq tracks multiple somatic alterations seen on the same cfDNA molecule (i.e., two or more mutations on a single molecule). Multiple phased variants, where two or more mutations occur in cis (i.e., on the same strand of DNA), occur in stereotyped portions of the genome in B-cell malignancies, likely due to on-target and aberrant somatic hypermutation driven by activation-induced deaminase. By detecting multiple mutations and individual cfDNA fragments, PhasED-seq lowers the background signal arising from technical or biological sources. This allows disease detection down to one part per million or lower in both technical and biological samples, including improved disease detection in plasma from patients with DLBCL both during and after completion of first-line therapy. Importantly, this improved detection sensitivity directly led to superior stratification of patient outcomes and identification of patients with relapsing disease after an initial radiographic remission (41). Although this initial report is promising, additional studies of PhasED-seq will be required to confirm its superior performance for MRD detection. Additional studies in diverse lymphoma subtypes with varying levels of activation-induced deaminase will also be useful to understand the best-use cases for this technology.
Mutational Genotyping
An additional advantage of CAPP-seq, PhasED-seq, and other targeted sequencing approaches is the ability to move beyond simple disease detection and perform mutational profiling and molecular genotyping. In the last 10 years, significant strides have been made in identifying prognostic and even predictive molecular subtypes of lymphomas. This began with the molecular cell of origin (COO) in DLBCL from gene expression (42), but more recently prognostically significant mutations in follicular lymphoma (FL; ref. 43), chronic lymphocytic leukemia (44), as well as genetic subtypes of DLBCL (45–47) have been identified. Indeed, targetable genetic alterations such as EZH2 mutations in FL (48) are emerging that can be detected by ctDNA (49). By assessing a broader panel of genes and loci, targeted sequencing with error suppression can directly assess these alterations from a liquid biopsy rather than requiring tumor tissue. This is particularly true for SNVs and small insertions and deletions, although methods for copy-number alteration detection are also in development (50, 51).
Key Studies in DLBCL
First-line combination chemotherapy cures ∼60% of DLBCL cases, but disease that relapses or is refractory to first-line therapy is difficult to cure (52). Biomarkers that reliably identify patients at high risk for treatment failure before, during, or after therapy allow for risk-adapted approaches designed to improve the cure rate. Complete eradication of disease is required to cure DLBCL, yet the current response criteria do not incorporate markers that detect the presence of MRD at the end of therapy, as has become standard in other hematologic diseases (53–56).
Multiple studies have explored the role of ctDNA in DLBCL and have established key principles regarding its potential as a tumor-specific biomarker that provides unique prognostic information before and throughout therapy. These studies have utilized different methods for ctDNA detection and have focused on different lymphoma-specific targets. Despite these technical differences, the key findings from these studies have shown striking similarities and have informed our current understanding of the potential clinical utility of ctDNA across lymphoma subtypes (57).
ctDNA as a Baseline Prognostic Marker in DLBCL
The prognosis for patients with DLBCL is related to overall tumor burden, the underlying condition of the patient, and tumor biology. Quantitative levels of baseline ctDNA have been shown to be surrogates of overall tumor burden in DLBCL. In a study of 126 patients with DLBCL, the baseline level of ctDNA for immunoglobulin receptor gene sequences was significantly associated with baseline international prognostic index (IPI) scores (P < 0.0001) and lactate dehydrogenase levels (P < 0.0001; ref. 11). Further, patients with early-stage DLBCL had significantly lower median ctDNA levels than those with advanced-stage disease (P = 0.014; ref. 11). A separate study using the same assay for ctDNA confirmed that baseline levels of ctDNA were correlated with total metabolic tumor volume (TMTV) as measured on baseline FDG-PET scans and determined that the cell-free component of the peripheral blood (i.e., plasma) was a better source of circulating DNA than the cellular component in DLBCL (12). Another key observation from these studies was that identification of a trackable clonotype required interrogation of the baseline tumor in most cases (11). This critical baseline validation step is affected by the quality of the tissue biopsy and amount of DNA input; hence, it is most successful when ≥60 ng of DNA can be extracted from baseline tissue biopsies (12). The largest study of ctDNA in DLBCL used the targeted sequencing approach CAPP-seq and analyzed the prognostic role of baseline ctDNA levels in 217 patients across 6 different treatment centers. The authors demonstrated that baseline ctDNA was associated with both event-free survival (EFS) and overall survival (OS) in patients receiving first-line or salvage therapy (14). Importantly, on multivariable analysis, the concentration of baseline ctDNA predicted EFS better than traditional prognostic variables including COO, IPI score, or TMTV on FDG-PET scans (14).
Another important function of baseline ctDNA is to noninvasively capture and integrate genetic information from the tumor (13). The molecular diversity of DLBCL was initially appreciated from gene-expression profiling studies that identified subtypes based on their putative COO: germinal center B-cell, activated B-cell, and primary mediastinal B-cell, with an additional 15% to 20% of cases remaining unclassified (42, 58, 59). This DLBCL classification identifies molecular phenotypes with distinct oncogenic mechanisms and differential outcomes to therapy (58, 60). Our understanding of the molecular biology of DLBCL has since evolved beyond COO subtypes, and multiplatform genomic approaches have described distinct genetic subtypes (45–47). This further classification of DLBCL into genetic subtypes relies on genetic information that comes almost exclusively from tissue biopsies. Multiple studies of ctDNA using CAPP-seq have shown that mutations and other lymphoma-relevant genetic aberrations can be identified from ctDNA even without baseline tumor tissue (13, 18, 20). Relevant genotypic information from CAPP-seq including COO classification was concordant with the tissue nearly 90% of the time, and most translocations of BCL2, BCL6, and MYC detected by fluorescence in situ hybridization (FISH) were detected from ctDNA. These studies also showed that unique genetic alterations can be identified from ctDNA even when they are not identified within the tissue biopsy, suggesting that ctDNA may be a better overall representation of genetic aberrations across multiple anatomic sites (13, 18, 20). Other studies of ctDNA in DLBCL using more restricted target panels have confirmed that genotypic information from tumors is readily available from ctDNA (61, 62). Taken together, such studies establish baseline ctDNA as an independent prognostic variable in DLBCL, which serves the dual purpose of capturing aggregate genotypic information.
ctDNA as a Marker of Treatment Response in DLBCL
End-of-Therapy Response Assessment
The current response criteria for DLBCL rely almost exclusively on imaging scans, such as CT and FDG-PET scans, to determine complete response (2). However, up to ∼15% of DLBCL will ultimately relapse despite achieving an FDG-PET–negative complete response using these criteria, and a proportion of patients with positive scans will never progress (63). Indeed, the limitations of FDG-PET scans were further demonstrated in a recent randomized study in which the visual interpretation of the end-of-therapy FDG-PET scan had no association with clinical outcomes (64). Thus, the fundamental limitation of imaging scans to detect low levels of residual disease makes it particularly important to develop biomarkers with high sensitivity for MRD. As of yet, the prognostic value of MRD is not clearly established for DLBCL as it is for other hematologic malignancies such as acute lymphocytic leukemia, chronic lymphocytic leukemia, or multiple myeloma (53, 55, 56). A clinically validated marker of MRD in DLBCL could further distinguish patients who have achieved a “complete molecular remission” from those with measurable residual disease below the detection limit of imaging.
Interim Response Assessment
Quantitative ctDNA also serves as a dynamic measure of treatment efficacy because levels often change rapidly after the initiation of therapy. In one study in DLBCL, the level of ctDNA for immunoglobulin receptor gene sequences was assessed after each cycle of first-line therapy, and patients without detectable ctDNA after two cycles had a superior five-year progression-free survival (PFS) compared with patients who remained ctDNA positive (80.2% vs. 41.7%; P < 0.0001; ref. 11). A larger and more detailed study of CAPP-seq during therapy for DLBCL showed that a 2-log reduction in ctDNA after one cycle of therapy (“early molecular response”) as well as 2.5-log reductions after two cycles (“major molecular response”) were more predictive of EFS [hazard ratio (HR) 8.58 (3.3–22.32)] and OS [HR 4.15 (1.17–15.57)] after first-line therapy compared with baseline ctDNA levels, IPI, or results on interim PET scans (14). To address the complex interplay of prognostic variables present at baseline with treatment-emergent variables such as response to therapy, a sophisticated personalized outcome prediction model called the “continuous individualized risk index” (CIRI) was developed (65). In this mathematical model designed to predict clinical outcomes, the standard baseline variables such as the IPI score, TMTV on PET scans, and COO phenotype and exploratory variables including ctDNA levels set a pretreatment probability of survival that is updated with the results of interim imaging scans, achievement of molecular response, and the response at the end of induction therapy based on conventional criteria. As compared with each individual component, CIRI demonstrated a superior ability to predict outcomes at 24 months compared with conventional prognostic markers. Taken together, these data highlight the potential of ctDNA as a dynamic biomarker that can track MRD throughout therapy to enhance existing clinical criteria.
ctDNA after Therapy for Early Disease Relapse and Clonal Evolution in DLBCL
Serial monitoring of ctDNA after therapy for DLBCL may also serve as a minimally invasive surveillance strategy to identify asymptomatic relapse and to characterize acquired resistance and clonal evolution. Surveillance monitoring with CT scans after therapy for DLBCL has proven neither effective for improving clinical outcomes nor cost-effective when applied to all patients in first remission (66, 67). However, the lack of demonstrable benefit from these studies does not establish that all surveillance strategies are futile. Indeed, there is intuitive appeal for early detection of disease relapse because both chemotherapy and immunotherapy are more effective at stages of lower tumor burden (68, 69).
In one study, 107 patients with DLBCL who achieved complete remission after first-line therapy were monitored with periodic CT scans along with prospective collected paired blood samples that were retrospectively analyzed for ctDNA (11). Overall, 17 patients relapsed after remission, and 15 (88%) of these patients had detectable ctDNA at the time of or prior to clinical relapse detected by CT scan. Further, the median (range) lead time of ctDNA detection compared with clinical relapse was 3.5 (0–200) months (11). Of the 90 patients who did not relapse after remission, ctDNA was persistently negative at almost all time points. In two (2%) cases, an isolated low-level positive ctDNA result was repeated and was negative. Another study reported that ctDNA became detectable in all 11 (100%) patients at time of relapse and in 8 (73%) patients prior to radiographic relapse with a median lead time of >2 months (13). Other studies have found similar patterns with longitudinal sampling, suggesting that either resistant tumors remain positive for ctDNA at the end of therapy or the presence of ctDNA reappears prior to disease relapse (20). These data suggest that ctDNA may be an effective surveillance strategy for patients at high risk for treatment failure. Interim results of a prospective study testing ctDNA as a surveillance strategy in 401 patients with DLBCL who achieved an FDG-PET–negative complete response have been presented (70). Patients had serial peripheral blood samples collected every 3 months for 2 years after treatment that were analyzed for MRD with an NGS-based assay for immunoglobin receptor sequence rearrangements and translocations of Bcl1/Bcl2-IgH. In the 43 patients who relapsed, a trackable sequence was identified in 39 (91%) cases, and the ctDNA was positive before or at the time of relapse in 22 (56%) of these cases, and 10 (26%) were positive more than 3 months before clinical detection. Although these data suggest that more sensitive assays are required for surveillance, it is also possible given the low detection rate compared with other published series, that specimen handling and storage diminished sensitivity (11, 12). Another function of ctDNA monitoring after therapy is to study clonal evolution and to better characterize resistant disease. One study using CAPP-seq in DLBCL showed that mutations can appear in ctDNA at disease relapse that were not present in the original tumor (20). Another study showed that mutations that predict histologic transformation to DLBCL from an underlying indolent lymphoma are present in ctDNA prior to clinical recognition (13). Taken together, these data highlight the capacity of ctDNA as a surveillance tool in DLBCL for the detection of disease prior to radiologic recurrence and to track genomic clonal evolution. It remains unproven if earlier detection of DLBCL ∼3 months prior to relapse translates into improved clinical outcomes. Enhanced detection of persistent disease at the end of therapy may lengthen the lead time from previously published studies and more precisely identify a group of patients who are not in molecular remission despite achieving radiographic remission (41).
Key Studies in Classical HL
A salient feature of Hodgkin/Reed–Sternberg (HRS) cells is their low abundance in biopsy samples, where they usually represent only 0.1% to 2% of the tumor mass within a mixture of nonmalignant immune cells. This has been a major obstacle in the study of the biology and genetics of classical HL (cHL), prompting research to study HRS cells captured by laser microdissection or purified by flow sorting. Serendipitously, a complex plasma cfDNA profile was unexpectedly discovered by noninvasive prenatal testing technologies in a pregnant woman who was subsequently diagnosed with early-stage cHL. This proof-of-principle observation has been validated by a study that showed copy-number abnormalities in HRS cells can be identified in plasma cfDNA of ∼90% patients with early- and advanced-stage cHL with prenatal testing technologies that use NGS (16). A number of studies have identified ctDNA as a source of tumor DNA for cHL mutational profiling and potentially improve upon the limitations of tissue-based genotyping (18, 21).
ctDNA as a Baseline Prognostic Marker in cHL
Compared with healthy subjects (∼1,700 hGE/mL of plasma), patients with cHL have twofold higher levels of cfDNA (∼3,400 hGE/mL of plasma), indicating differential release of DNA by the tumor. This hypothesis was confirmed by the discovery of cHL-specific immunoglobulin gene clonotypes in the serum of 89% of patients (15). The median ctDNA level in cHL is ∼200 hGE/mL, indicating that ∼15 cfDNA fragments out of 100 are of tumor origin (71). Due to the scarcity of HRS cells, the tumor cell volume in cHL is tenfold smaller than that of other aggressive lymphomas, with cHL averaging about 15 mL (e.g., 300 mL of mass and assuming 5% HRS cells) and DLBCL averaging about 200 mL (e.g., 400 mL of mass and assuming 50% tumor cell content; ref. 19). Surprisingly, despite the lower tumor cell content in cHL compared with DLBCL, the correlation between ctDNA levels and radiologic tumor volume in cHL is strikingly similar. Specifically, cHL and DLBCL have similar ratios of ctDNA and FDG-PET–defined metabolic tumor volume (ctDNA tumor volume of ∼2 vs. ∼1.5 hGE/mL per cm3 of tumor, respectively; ref. 71). Hence, cHL appears to release a relatively greater amount of ctDNA compared with DLBCL, which may be due to the frequent polyploidy of HRS cells, suggesting that HRS cells are subject to a high cellular turnover rate. The frequent coexpression of both proliferative and apoptotic markers on HRS cells and the presence of necrosis in cHL biopsy samples seem to support the notion that active proliferation is offset by high amounts of HRS cell loss through apoptosis or necrosis (16).
Alterations of chromosome 9p24.1 lead to PD-1 ligand overexpression in cHL and are predictive of response to PD-1–blocking antibodies (72). Although the response rate to PD-1 inhibitors is high, preliminary data suggest that those with higher level 9p24.1 alterations in HRS cells have superior PFS. FISH assays specifically covering 9p24.1 on the tissue biopsy are the gold standard to determine the incidence and nature of 9p24.1 chromosomal abnormalities harbored by HRS cells (72). Compared with gold-standard FISH assays, genomic profiling of cfDNA can accurately discover and type ∼80% of copy-number abnormalities of 9p24.1 (16). Further studies could test the utility of determining 9p24.1 status from ctDNA as a predictive biomarker.
Genomic studies of somatic mutations of cHL are limited to a few studies that used laser-capture microdissection or flow sorting of HRS cells from primary tumors. Based on the exomic mutational profile of cHL from 67 such cases (73), ctDNA genotyping by targeted gene panels was able to identify 87% to 100% of biopsy-confirmed tumor mutations. Defects of the antigen-presentation machinery mark was associated with resistance to immune-checkpoint inhibitors (74), and MHC class II expression on HRS cells was identified as a potential predictor of favorable outcome after PD-1 blockade. Mutations of B2M, HLA, and CIITA are complementary genetic abnormalities associated with perturbed MHC class I and II expression in cHL (73).
ctDNA is a rich and easily accessible source of tumor DNA for cHL mutation profiling. Accordingly, targeted assays of ctDNA can be used to delineate the molecular bases of response and resistance to immunomodulatory therapy in pivotal clinical trials. On an analytical level, the recovery rate of tumor biopsy–confirmed mutations in cfDNA depends on the size of the genomic space analyzed and the genes covered by the assay. It may vary from 25% when considering one single-nucleotide variant that is highly recurrent in cHL (i.e., XPO1 E571K variant; ref. 17) to 70% when the gene panel solely covers genes that are specific for cHL, and to 90% when the gene panel broadly includes all the genes that are recurrent in mature B-cell tumors (18, 19, 21). Disease detection in 100% of cases could have a potential clinical impact, including a reduced number of “false-negative” ctDNA assays. In addition, an important factor that underlies the detection limit of all ctDNA profiling methods is the number of interrogated mutations in a tumor. Because cHL is targeted by aberrant somatic hypermutation, the recovery rate of mutations from ctDNA of patients with cHL can be maximized by integrating regions of aberrant somatic hypermutation in the genomic space covered by ctDNA (41).
ctDNA as a Marker of Treatment Response in cHL
The correlation between amounts of ctDNA in plasma and TMTV indicates that ctDNA quantification could be useful for monitoring therapy in cHL. Compared with the cellular fraction of blood, plasma has a hundredfold more tumor DNA molecules, which indicates that ctDNA assays in cHL are more informative when applied to the liquid fraction of blood than the cellular fraction.
The junction between the immunoglobulin variable, diversity, and joining genes provides a unique DNA clonotype that is shared by all malignant B cells and can be sensitively detected and quantified by NGS. Hence, the cHL-specific immunoglobulin gene clonotype can serve as a reporter for measuring ctDNA levels and residual disease under treatment. The major limitation of this approach in cHL is that, in a biopsy-free setting, tumor clonotypes cannot be identified in cfDNA, as they are admixed with and indistinguishable from the hundreds of thousands of clonotypes that are released in plasma by the normal B-cell repertoire. Before their use for monitoring in cfDNA, clonotypic sequences of cHL must first be identified in tissue biopsy. If the tissue biopsy is fresh/frozen, this step is successful in ∼70% of patients (15), with significantly lower success if formalin-fixed, paraffin-embedded (FFPE) material is used (12). As most patients have excisional lymph node biopsies stored in FFPE, this would likely be the source material in real-world applications.
In a retrospective study, the XPO1 E571K mutation was monitored in cfDNA from patients with cHL harboring this tumor reporter. Patients with detectable XPO1 E571K mutation by dPCR at the end of treatment had a shorter PFS than XPO1 E571K–negative cases (17). The limitation of ctDNA assays based on dPCR in cHL is that the tumor lacks a universal biomarker for monitoring, as no mutations occur in >90% of patients. XPO1 E571K is the sole recurrent single point mutation addressable by PCR approaches in cHL, but its frequency is limited to 10% to 20% of cases (73, 75, 76).
ctDNA assays based on disease-specific “selectors,” which are exonic and intronic targets chosen to cover regions of known recurrent mutations in a particular cancer type, allow the quantification of ctDNA based on the detection of tumor-specific mutations and can be applied to cfDNA samples without the need for precalibration from the tissue biopsy. By using ctDNA assays based on disease-specific “selectors,” three independent retrospective observational studies consistently showed a correlation between ctDNA load and disease remission (17–19).
An unmet medical need in cHL is the early and accurate identification of patients who are refractory to chemotherapy, as they are candidates for treatment modification to maximize the chances of cure, as well as good-risk patients, as they are candidates for treatment de-escalation to reduce complications of therapy. The ideal test for response-adapted treatment approaches needs to be reliable and have a strong correlation with long-term treatment outcome. Interim PET/CT performed after two chemotherapy cycles is a clinical-grade tool that is used to detect residual disease in cHL and modify treatment accordingly. However, interim FDG-PET results are inconsistent with clinical outcomes, mainly due to false-positive findings in ∼20% to 30% of patients and may result in overtreatment.
ctDNA assays based on disease-specific “selectors” combined with FDG-PET imaging may significantly improve the accurate early identification of chemorefractory disease and provide proof of concept of ctDNA as a novel precision-medicine biomarker in cHL. More precisely, a drop of >100-fold or >2-log drop in ctDNA after two chemotherapy courses, a threshold proposed for DLBCL, is associated with complete response and cure in cHL (18). Conversely, a drop of <2-log in ctDNA after two chemotherapy courses is associated with progression and inferior survival. Indeed, cured patients with an interim false-positive FDG-PET have a >2-log drop in ctDNA, whereas relapsing patients with an interim false-negative FDG-PET have <2-log drop in ctDNA.
ctDNA after Therapy for Clonal Evolution in cHL
Genotyping of longitudinal ctDNA samples collected before first-line treatment at the time of relapse and during salvage therapy with transplantation, brentuximab vedotin, and/or nivolumab has identified clonal evolution patterns in cHL. In cHL treated with ABVD (doxorubicin hydrochloride, bleomycin sulfate, vinblastine sulfate, and dacarbazine) or brentuximab vedotin, an ancestral clone that is resistant to chemotherapy propagates over successive disease progression. No single gene mutation or pathway alteration identified at relapse has systematically recapitulated the chemorefractory phenotype of cHL (18). In patients treated with checkpoint inhibitors, the longitudinal tracking of circulating DNA mutations identifies two different patterns of clonal evolution associated with sensitivity (clonal reshaping) or resistance (clonal persistence) to checkpoint blockade. Among patients maintaining a partial response with nivolumab, ancestral clones were cyclically suppressed and replaced by novel clones harboring new mutations (18). Such a pattern might be interpreted as drug-promoted selection against cancer neoantigens through the development of mutations that evade treatment sensitivity, whereas patients with primary resistance to checkpoint blockade have persistence of the baseline mutation profile. At variance with solid tumor patients, resistance to checkpoint blockade in cHL is not associated with acquisition of B2M mutations in the immune-evasion pathway (77).
Future Directions
It is critical to note that the methodologies and technologies that have been studied in lymphomas remain in their infancy, and this is a rapidly changing field; emerging technologies or alternative methods may provide additional or orthogonal information. As an example, methylation-based ctDNA analyses may emerge as important noninvasive biomarkers across cancer types, including lymphoma (78). Indeed, we are likely to see additional and novel questions that can be addressed by ctDNA across a variety of clinical situations.
In addition to technological improvements in assays that detect and quantify ctDNA, it is anticipated that the value of ctDNA will also be explored in other lymphoma subtypes, including indolent lymphomas, peripheral T-cell lymphomas, and lymphomas that are difficult to biopsy including those involving the central nervous system and intravascular large B-cell lymphoma (79–84). FL is an indolent B-cell lymphoma with marked variation in clinical behavior, including high-risk subsets that progress shortly after diagnosis and/or relapse within a few years of initial therapy (85). The potential utility of ctDNA for baseline prognostication in FL or as a method to detect MRD at the end of induction therapy remains understudied. Further, the minimally invasive nature of ctDNA allows for serial monitoring that may capture important clonal evolution events that precede histologic transformation of FL to a more aggressive lymphoma (13).
Conclusions
ctDNA is a highly promising and versatile biomarker in lymphoma that can overcome fundamental limitations of imaging scans and tissue biopsies. Multiple landmark studies in aggressive B-cell lymphomas have established key principles, but prospective validation in clinical trials for DLBCL and HL is needed to demonstrate clinical utility. It will be important to establish a road map for international standardization and harmonization within lymphoma subtypes based on an accepted set of criteria. Indeed, the field lacks a consensus on the minimum number of targets required to effectively genotype tumors from ctDNA, which may be different across lymphoma subtypes. Perhaps most importantly, MRD needs to be established as a surrogate marker for OS in DLBCL and cHL and accepted as a clinical trial endpoint. As the technology continues to evolve, additional methods for assessing DNA methylation status and the use of genome-wide NGS may be important in certain clinical situations (50, 86). In a similar way to how therapies increasingly incorporate immunotherapy such as CAR-T treatments, the biological information obtained from ctDNA to accurately identify patients who do not derive benefit may be therapy specific (87). If these goals can be achieved in aggressive B-cell lymphomas, then similar principles are likely to pertain to indolent lymphomas.
Authors' Disclosures
D. Rossi reports grants and other support from Janssen, AbbVie, and AstraZeneca outside the submitted work. D.M. Kurtz reports other support from Foresight Diagnostics and personal fees from Roche and Genentech outside the submitted work, as well as patents related to methods for analysis of cell-free nucleic acids pending and licensed to Foresight Diagnostics. A.A. Alizadeh reports personal fees and other support from Roche, Gilead, and Chugai, other support from Genentech, Celgene, Janssen, FortySeven, Foresight Diagnostics, Cibermed Inc., and Pharmacyclics, and grants from Bristol Myers Squibb during the conduct of the study, as well as a patent for 20210214437 issued, licensed, and with royalties paid from FortySeven, a patent for 20210172022 pending and licensed to Foresight, a patent for 20210033608 pending, a patent for 10167514 issued and licensed to CiberMed, a patent for 10633450 issued, a patent for 20190338364 pending and licensed to CiberMed, a patent for 9605320 issued, and a patent for 20140296081 issued, licensed, and with royalties paid from Roche. No disclosures were reported by the other authors.
References
- 1. Rushton CK, Arthur SE, Alcaide M, Cheung M, Jiang A, Coyle KM, et al. Genetic and evolutionary patterns of treatment resistance in relapsed B-cell lymphoma. Blood Adv 2020;4:2886–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol 2014;32:3059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Araf S, Wang J, Korfi K, Pangault C, Kotsiou E, Rio-Machin A, et al. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia 2018;32:1261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hav M, Gerdtsson E, Singh M, Colombo A, Hicks J, Kuhn P, et al. Abstract 2789: Highly multiplexed imaging mass cytometry reveals immune cell composition and spatial heterogeneity in diffuse large B cell lymphoma associated with treatment outcome. Cancer Res 2019;79:2789. [Google Scholar]
- 5. Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med 2015;21:846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kilgour E, Rothwell DG, Brady G, Dive C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell 2020;37:485–95. [DOI] [PubMed] [Google Scholar]
- 7. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659–65. [PubMed] [Google Scholar]
- 8. Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989;46:318–22. [DOI] [PubMed] [Google Scholar]
- 9. Roschewski M, Staudt LM, Wilson WH. Dynamic monitoring of circulating tumor DNA in non-Hodgkin lymphoma. Blood 2016;127:3127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rossi D, Spina V, Bruscaggin A, Gaidano G. Liquid biopsy in lymphoma. Haematologica 2019;104:648–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roschewski M, Dunleavy K, Pittaluga S, Moorhead M, Pepin F, Kong K, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol 2015;16:541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurtz DM, Green MR, Bratman SV, Scherer F, Liu CL, Kunder CA, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 2015;125:3679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scherer F, Kurtz DM, Newman AM, Stehr H, Craig AF, Esfahani MS, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 2016;8:364ra155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurtz DM, Scherer F, Jin MC, Soo J, Craig AFM, Esfahani MS, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol 2018;36:2845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oki Y, Neelapu SS, Fanale M, Kwak LW, Fayad L, Rodriguez MA, et al. Detection of classical Hodgkin lymphoma specific sequence in peripheral blood using a next-generation sequencing approach. Br J Haematol 2015;169:689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vandenberghe P, Wlodarska I, Tousseyn T, Dehaspe L, Dierickx D, Verheecke M, et al. Non-invasive detection of genomic imbalances in Hodgkin/Reed–Sternberg cells in early and advanced stage Hodgkin's lymphoma by sequencing of circulating cell-free DNA: a technical proof-of-principle study. Lancet Haematol 2015;2:e55–65. [DOI] [PubMed] [Google Scholar]
- 17. Camus V, Stamatoullas A, Mareschal S, Viailly PJ, Sarafan-Vasseur N, Bohers E, et al. Detection and prognostic value of recurrent exportin 1 mutations in tumor and cell-free circulating DNA of patients with classical Hodgkin lymphoma. Haematologica 2016;101:1094–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spina V, Bruscaggin A, Cuccaro A, Martini M, Di Trani M, Forestieri G, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood 2018;131:2413–25. [DOI] [PubMed] [Google Scholar]
- 19. Desch AK, Hartung K, Botzen A, Brobeil A, Rummel M, Kurch L, et al. Genotyping circulating tumor DNA of pediatric Hodgkin lymphoma. Leukemia 2020;34:151–66. [DOI] [PubMed] [Google Scholar]
- 20. Rossi D, Diop F, Spaccarotella E, Monti S, Zanni M, Rasi S, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood 2017;129:1947–57. [DOI] [PubMed] [Google Scholar]
- 21. Camus V, Viennot M, Lequesne J, Viailly PJ, Bohers E, Bessi L, et al. Targeted genotyping of circulating tumor DNA for classical Hodgkin lymphoma monitoring: a prospective study. Haematologica 2021;106:154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huet S, Salles G. Potential of circulating tumor DNA for the management of patients with lymphoma. JCO Oncol Pract 2020;16:561–8. [DOI] [PubMed] [Google Scholar]
- 23. Greytak SR, Engel KB, Parpart-Li S, Murtaza M, Bronkhorst AJ, Pertile MD, et al. Harmonizing cell-free DNA collection and processing practices through evidence-based guidance. Clin Cancer Res 2020;26:3104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rossi D, Kurtz DM, Roschewski M, Cavalli F, Zucca E, Wilson WH. The development of liquid biopsy for research and clinical practice in lymphomas: report of the 15-ICML workshop on ctDNA. Hematol Oncol 2020;38:34–7. [DOI] [PubMed] [Google Scholar]
- 25. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016;164:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hohaus S, Giachelia M, Massini G, Mansueto G, Vannata B, Bozzoli V, et al. Cell-free circulating DNA in Hodgkin's and non-Hodgkin's lymphomas. Ann Oncol 2009;20:1408–13. [DOI] [PubMed] [Google Scholar]
- 29. Kang Q, Henry NL, Paoletti C, Jiang H, Vats P, Chinnaiyan AM, et al. Comparative analysis of circulating tumor DNA stability in K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem 2016;49:1354–60. [DOI] [PubMed] [Google Scholar]
- 30. Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631–41. [DOI] [PubMed] [Google Scholar]
- 31. Drandi D, Genuardi E, Dogliotti I, Ferrante M, Jimenez C, Guerrini F, et al. Highly sensitive MYD88(L265P) mutation detection by droplet digital polymerase chain reaction in Waldenstrom macroglobulinemia. Haematologica 2018;103:1029–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hattori K, Sakata-Yanagimoto M, Suehara Y, Yokoyama Y, Kato T, Kurita N, et al. Clinical significance of disease-specific MYD88 mutations in circulating DNA in primary central nervous system lymphoma. Cancer Sci 2018;109:225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jardin F, Pujals A, Pelletier L, Bohers E, Camus V, Mareschal S, et al. Recurrent mutations of the exportin 1 gene (XPO1) and their impact on selective inhibitor of nuclear export compounds sensitivity in primary mediastinal B-cell lymphoma. Am J Hematol 2016;91:923–30. [DOI] [PubMed] [Google Scholar]
- 34. Scheijen B, Meijers RWJ, Rijntjes J, van der Klift MY, Mobs M, Steinhilber J, et al. Next-generation sequencing of immunoglobulin gene rearrangements for clonality assessment: a technical feasibility study by EuroClonality-NGS. Leukemia 2019;33:2227–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ching T, Duncan ME, Newman-Eerkes T, McWhorter MME, Tracy JM, Steen MS, et al. Analytical evaluation of the clonoSEQ assay for establishing measurable (minimal) residual disease in acute lymphoblastic leukemia, chronic lymphocytic leukemia, and multiple myeloma. BMC Cancer 2020;20:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Faham M, Zheng J, Moorhead M, Carlton VE, Stow P, Coustan-Smith E, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood 2012;120:5173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kurtz DM, Soo J, Co Ting Keh L, Alig S, Chabon JJ, Sworder BJ, et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat Biotechnol 2021. Jul 21 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–11. [DOI] [PubMed] [Google Scholar]
- 43. Pastore A, Jurinovic V, Kridel R, Hoster E, Staiger AM, Szczepanowski M, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol 2015;16:1111–22. [DOI] [PubMed] [Google Scholar]
- 44. International CLLIPIwg. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol 2016;17:779–90. [DOI] [PubMed] [Google Scholar]
- 45. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 2018;378:1396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 2020;37:551–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morschhauser F, Tilly H, Chaidos A, McKay P, Phillips T, Assouline S, et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol 2020;21:1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nagy A, Batai B, Balogh A, Illes S, Mikala G, Nagy N, et al. Quantitative analysis and monitoring of EZH2 mutations using liquid biopsy in follicular lymphoma. Genes (Basel) 2020;11:785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stewart P, Gazdova J, Darzentas N, Wren D, Proszek P, Fazio G, et al. Euroclonality-NGS DNA capture panel for integrated analysis of IG/TR rearrangements, translocations, copy number and sequence variation in lymphoproliferative disorders. Blood 2019;134:888. [Google Scholar]
- 52. Crump M, Neelapu SS, Farooq U, Van Den Neste E, Kuruvilla J, Westin J, et al. Outcomes in refractory diffuse large B-cell lymphoma: results from the international SCHOLAR-1 study. Blood 2017;130:1800–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bassan R, Bruggemann M, Radcliffe HS, Hartfield E, Kreuzbauer G, Wetten S. A systematic literature review and meta-analysis of minimal residual disease as a prognostic indicator in adult B-cell acute lymphoblastic leukemia. Haematologica 2019;104:2028–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berry DA, Zhou S, Higley H, Mukundan L, Fu S, Reaman GH, et al. Association of minimal residual disease with clinical outcome in pediatric and adult acute lymphoblastic leukemia: a meta-analysis. JAMA Oncol 2017;3:e170580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018;131:2745–60. [DOI] [PubMed] [Google Scholar]
- 56. Landgren O, Rustad EH. Meeting report: advances in minimal residual disease testing in multiple myeloma 2018. Adv Cell Gen Ther 2019;2:e26. [Google Scholar]
- 57. Bruggemann M, Kotrova M, Knecht H, Bartram J, Boudjogrha M, Bystry V, et al. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia 2019;33:2241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002;346:1937–47. [DOI] [PubMed] [Google Scholar]
- 59. Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. PNAS 2008;105:13520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008;359:2313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bohers E, Viailly PJ, Dubois S, Bertrand P, Maingonnat C, Mareschal S, et al. Somatic mutations of cell-free circulating DNA detected by next-generation sequencing reflect the genetic changes in both germinal center B-cell-like and activated B-cell-like diffuse large B-cell lymphomas at the time of diagnosis. Haematologica 2015;100:e280–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Camus V, Sarafan-Vasseur N, Bohers E, Dubois S, Mareschal S, Bertrand P, et al. Digital PCR for quantification of recurrent and potentially actionable somatic mutations in circulating free DNA from patients with diffuse large B-cell lymphoma. Leuk Lymphoma 2016;57:2171–9. [DOI] [PubMed] [Google Scholar]
- 63. Adams HJ, Nievelstein RA, Kwee TC. Prognostic value of complete remission status at end-of-treatment FDG-PET in R-CHOP-treated diffuse large B-cell lymphoma: systematic review and meta-analysis. Br J Haematol 2015;170:185–91. [DOI] [PubMed] [Google Scholar]
- 64. Schoder H, Polley MY, Knopp MV, Hall NC, Kostakoglu L, Zhang J, et al. Prognostic value of interim FDG-PET in diffuse large cell lymphoma: results from the CALGB 50303 clinical trial. Blood 2020;135:2224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kurtz DM, Esfahani MS, Scherer F, Soo J, Jin MC, Liu CL, et al. Dynamic risk profiling using serial tumor biomarkers for personalized outcome prediction. Cell 2019;178:699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thompson CA, Ghesquieres H, Maurer MJ, Cerhan JR, Biron P, Ansell SM, et al. Utility of routine post-therapy surveillance imaging in diffuse large B-cell lymphoma. J Clin Oncol 2014;32:3506–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Huntington SF, Svoboda J, Doshi JA. Cost-effectiveness analysis of routine surveillance imaging of patients with diffuse large B-cell lymphoma in first remission. J Clin Oncol 2015;33:1467–74. [DOI] [PubMed] [Google Scholar]
- 68. Hamlin PA, Zelenetz AD, Kewalramani T, Qin J, Satagopan JM, Verbel D, et al. Age-adjusted international prognostic index predicts autologous stem cell transplantation outcome for patients with relapsed or primary refractory diffuse large B-cell lymphoma. Blood 2003;102:1989–96. [DOI] [PubMed] [Google Scholar]
- 69. Dean EA, Mhaskar RS, Lu H, Mousa MS, Krivenko GS, Lazaryan A, et al. High metabolic tumor volume is associated with decreased efficacy of axicabtagene ciloleucel in large B-cell lymphoma. Blood Adv 2020;4:3268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kumar A, Westin J, Schuster SJ, Nowakowski GS, Lossos IS, Batlevi CL, et al. Interim analysis from a prospective multicenter study of next-generation sequencing minimal residual disease assessment and CT monitoring for surveillance after frontline treatment in diffuse large B-cell lymphoma. Blood 2020;136:46–7. [Google Scholar]
- 71. Jin M, Kurtz DM, Esfahani MS, Sworder BJ, Schroer-Martin S, Glover J, et al. Circulating tumor DNA as a biomarker for the noninvasive genotyping and monitoring of classical Hodgkin lymphoma. HemaSphere 2018;2:4–5. [Google Scholar]
- 72. Roemer MGM, Redd RA, Cader FZ, Pak CJ, Abdelrahman S, Ouyang J, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J Clin Oncol 2018;36:942–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wienand K, Chapuy B, Stewart C, Dunford AJ, Wu D, Kim J, et al. Genomic analyses of flow-sorted Hodgkin Reed–Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv 2019;3:4065–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schoenfeld AJ, Hellmann MD. Acquired resistance to immune checkpoint inhibitors. Cancer Cell 2020;37:443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Reichel J, Chadburn A, Rubinstein PG, Giulino-Roth L, Tam W, Liu Y, et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed–Sternberg cells. Blood 2015;125:1061–72. [DOI] [PubMed] [Google Scholar]
- 76. Tiacci E, Ladewig E, Schiavoni G, Penson A, Fortini E, Pettirossi V, et al. Pervasive mutations of JAK–STAT pathway genes in classical Hodgkin lymphoma. Blood 2018;131:2454–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Di Trani M, Rizzo E, Locatelli S, Marino F, Cristaldi V, Spina V, et al. Longitudinal assessment of circulating tumor mutational burden using a next-generation sequencing cancer gene panel: a potential biomarker of response to programmed cell death 1 (PD-1) blockade in patients with relapsed/refractory classical Hodgkin lymphoma. Blood 2019;134:131. [Google Scholar]
- 78. Beer TM, McDonnell CH, Nadauld L, Liu MC, Klein EA, Reid RL, et al. Interim results of PATHFINDER, a clinical use study using a methylation-based multi-cancer early detection test. J Clin Oncol 2021;39:3010. [Google Scholar]
- 79. Sarkozy C, Huet S, Carlton VE, Fabiani B, Delmer A, Jardin F, et al. The prognostic value of clonal heterogeneity and quantitative assessment of plasma circulating clonal IG-VDJ sequences at diagnosis in patients with follicular lymphoma. Oncotarget 2017;8:8765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lakhotia R, Melani C, Pittaluga S, Dunleavy K, Saba NS, Lucas AN, et al. Circulating tumor DNA dynamics during therapy predict outcomes in mantle cell lymphoma. Blood 2018;132:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mutter JA, Alig S, Lauer EM, Esfahani MS, Mitschke J, Kurtz DM, et al. Noninvasive detection, classification, and risk stratification of primary CNS lymphomas by CTDNA profiling. Hematol Oncol 2021;39:85–7. [Google Scholar]
- 82. Suehara Y, Sakata-Yanagimoto M, Hattori K, Nanmoku T, Itoh T, Kaji D, et al. Liquid biopsy for the identification of intravascular large B-cell lymphoma. Haematologica 2018;103:e241–e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Melani C, Pittaluga S, Yee L, Lucas A, Shovlin M, Jacob A, et al. Next-generation sequencing based monitoring of circulating-tumor DNA in untreated peripheral T-cell lymphoma. Blood 2017;130:2728.28935695 [Google Scholar]
- 84. Qi F, Cao Z, Chen B, Chai Y, Lin J, Ye J, et al. Liquid biopsy in extranodal NK/T-cell lymphoma: a prospective analysis of cell-free DNA genotyping and monitoring. Blood Adv 2021;5:2505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Casulo C, Byrtek M, Dawson KL, Zhou X, Farber CM, Flowers CR, et al. Early relapse of follicular lymphoma after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone defines patients at high risk for death: an analysis from the National LymphoCare Study. J Clin Oncol 2015;33:2516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nassiri F, Chakravarthy A, Feng S, Shen SY, Nejad R, Zuccato JA, et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat Med 2020;26:1044–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sworder B, Kurtz DM, Macaulay C, Frank MJ, Alig S, Garofalo A, et al. Circulating DNA for molecular response prediction, characterization of resistance mechanisms and quantification of CAR T cells during axicabtagene ciloleucel therapy. Blood 2019;134:550. [Google Scholar]