Abstract
Scope:
The plasma metabolomics profiles of protein intake has been rarely investigated. We aimed to identify the distinct plasma metabolomics profiles associated with overall intakes of protein as well as with intakes from animal and plant protein sources.
Methods and Results:
Cross-sectional analysis using data from 1,833 participants at high risk of cardiovascular disease. Plasma metabolomics analysis was performed using LC-MS. Associations between 385 identified metabolites and the intake of total, animal protein (AP) and plant protein (PP), and plant-to-animal ratio (PR) were assessed using elastic net continuous regression analyses. A double 10-cross-validation (CV) procedure was used and Pearson correlations coefficients between multi-metabolite weighted models and reported protein intake in each pair of training-validation datasets were calculated. A wide set of metabolites was consistently associated with each protein source evaluated. These metabolites mainly consisted of amino acids and their derivatives, acylcarnitines, different organic acids and lipid species. Few metabolites overlapped among protein sources (i.e. C14:0 SM, C20:4 carnitine, GABA and allantoin) but none of them towards the same direction. Regarding AP and PP approaches, C20:4 carnitine and dimethylglycine were positively associated with PP but negatively associated with AP. However, allantoin, C14:0 SM, C38:7 PE plasmalogen, GABA, metronidazole and trigonelline (N-methylnicotinate) behaved contrary. Ten-CV Pearson correlations coefficients between self-reported protein intake and plasma metabolomics profiles ranged from 0.21 for PR to 0.32 for total protein.
Conclusions:
Different sets of metabolites were associated with total, animal and plant protein intake. Further studies are needed to assess the contribution of these metabolites in protein biomarkers’ discovery and prediction of cardiometabolic alterations.
Keywords: LC-MS, lipidomics, metabolites, metabolomics, protein
Graphical Abstract
The plasma metabolomics profiles of protein intake has been rarely investigated. The aim of this study was to identify the circulating metabolomics profiles in relation to the consumption of total protein, animal protein, plant protein, and plant-to-animal protein ratio. We identified different plasma metabolomics profiles, which exhibited moderate correlations with the self-reported protein intake. Further studies are needed to assess the contribution of these metabolites in protein biomarkers’ discovery and prediction of metabolic alterations.
1. INTRODUCTION
Diets with a relatively high content in total dietary protein have been recommended for body weight (BW) control in the overall population [1] and glycemic control in subjects with type 2 diabetes (T2D) [2, 3]. However, the potential long-term health benefits and risks of these diets have been partially explored [4]. Current evidence supports the idea that cardiovascular disease (CVD) risk can be reduced by adhering to a dietary pattern rich in plant sources of protein compared with the typical western diet which includes a high intake of animal-based protein foods that are processed and high in saturated fat [5]. In the context of the PREDIMED study, we have previously assessed the effect of long-term high-protein consumption (including its sources and the animal-to-plant ratio) on BW changes and different causes of death [6]. We showed an U-shape relationship between total protein (TP) consumption and both total mortality and BW changes, together with specific associations depending on protein source towards beneficial effects associated with plant protein consumption. However, the overall differential impact of protein sources (i.e. animal or plant) and/or their relative proportion on health is still inconclusive and difficult to isolate [7].
Once ingested, both sources of protein share metabolic pathways. However, plant and animal sources have a distinct amino acid composition. In general, plant-based proteins are lower in essential amino acids (particularly methionine, lysine, and tryptophan) but provide higher amounts of arginine, glycine, alanine, and serine (non-essential amino acids) [8].
Nowadays, urinary excretion of urea nitrogen is widely used as an adequate biomarker of total protein (TP) intake, although it suffers from imprecision, collection error and can only provide information for TP intake, without any consideration from the food source (e.g. animal or plant protein) [9]. In fact, to obtain the most accurate measurements, individuals should maintain a constant daily intake and be in nitrogen balance. Therefore, further research is needed to identify novel reliable biomarkers of dietary intake of TP – and its different sources – that may be measurable in plasma/serum. Although it is more invasive for the patient, it is relatively easier to obtain compared to urine (less burdensome for study participants) and not prone to error due to incomplete urine collection [10].
Metabolomics is an emerging field aiming to comprehensively measure metabolites and low-molecular-weight molecules in a biological specimen [11]. To date, few studies have focused in the identification of metabolites associated with TP intake [12–15] compared to those specifically focused on meat intake (reviewed in [16]). In fact, current evidence for these associations comes indirectly from studies evaluating diet quality indexes [12] or diets varying in glycemic index (GI)/carbohydrate content [13]. Only two RCTs have explored the metabolomics differences in subjects consuming a diet with different amount of protein [14, 15]. However, no previous study has explored the systemic plasma metabolomics profiles associated with the level of protein intake as well as intakes from animal and plant-sources of protein in a large sample of subjects.
Taking advantage of a comprehensive plasma metabolomics analysis, we hypothesized that distinct plasma metabolites profiles are associated with the level of protein intake as well as the source of proteins, mainly animal and plant food sources. Therefore, the main aim of the present study was to describe the set of metabolites associated with the intake of TP, animal protein (AP), plant protein (PP), and plant-to-animal protein ratio (PR), which could help us to understand in the future the relationship between diet and cardiometabolic health. Moreover, we aimed to define a set of metabolites overlapping and unique to each protein approach.
2. MATERIAL AND METHODS
This study is a cross-sectional analysis of baseline data from two nested case-cohort studies on cardiovascular disease (CVD) and T2D (NIH-NHLBI-5R01HL118264 and NIH-NIDDK-1R01DK102896) [17, 18] within the PREDIMED study (ISRCTN35739639). The PREDIMED study is a large clinical trial carried out in Spain, aiming to assess the effects of the traditional Mediterranean diet (MedDiet) on the primary prevention of CVD in a population at high risk of CVD [19]. Participants were men (55–80 years) and women (60–80 years) without CVD at baseline and fulfilling at least one of the two following criteria: presence of T2D or three or more major cardiovascular risk factors: current smoking, hypertension, high low-density lipoprotein (LDL)-cholesterol, low high-density lipoprotein (HDL)-cholesterol, overweight or obesity, and family history of premature CVD. The trial protocol was in accordance with the Helsinki Declaration and was approved by the institutional review boards of all the centers involved. All participants provided written informed consent.
2.1. Assessment of population characteristics and dietary habits
Body mass index (BMI) was calculated as weight divided by height squared (kg/m2). Waist circumference (WC) was measured midway between the lowest rib and the iliac crest using an anthropometric tape. Dietary habits at baseline were evaluated using a validated, 137-item, semi-quantitative food frequency questionnaire (FFQ) [20]. Daily food and nutrient intakes were estimated from the FFQ by multiplying the frequency of consumption by the average portion size. Participants also filled out a general questionnaire on lifestyle habits, medication use and concurrent diseases, and a validated Spanish version of the Minnesota Leisure Time Physical Activity Questionnaire [21].
2.2. Protein intake assessment
The validity and reproducibility of the FFQ for the measurements of the different macronutrients have been previously reported [20, 22]. Pearson correlation coefficients for total protein were 0.55 (unadjusted) and 0.50 (energy-adjusted) between intakes reported in the FFQ and intakes reported in repeated food records. The intraclass correlation coefficient (ICC) between total protein intake was 0.71 (unadjusted) and 0.67 (energy-adjusted) [20]. In our study, the level of protein intake was assessed as the percentage of energy (E%) derived from protein. AP was mainly derived from meat, poultry, fish and dairy products, whereas PP was derived from legumes, cereals and nuts. Percentages of energy from AP and PP were also calculated. Finally, we also derived the plant-to-animal protein ratio. Due to the semi-quantitative basis of the FFQ, we additionally created categories of protein consumption based on extreme tertiles (T): T3 versus T1.
2.3. Plasma metabolomics
Fasting (for ≥8 hours) plasma EDTA samples were collected from subjects and stored at −80°C. Samples for each participant were randomly ordered and analyzed using two liquid chromatography tandem mass spectrometry (LC-MS) methods to measure polar metabolites and lipids as described previously [23–25]. Briefly, amino acids (AA) and other polar metabolites were profiled a Shimadzu Nexera X2 U-HPLC (Shimadzu Corp.) coupled to a Q-Exactive mass spectrometer (ThermoFisher Scientific). Metabolites were extracted from plasma (10 μL) using 90 μL of 74.9:24.9:0.2 (vol/vol/vol) of acetonitrile/methanol/formic acid containing stable isotope-labeled internal standards [valine-d8 (Sigma-Aldrich) and phenylalanine-d8 (Cambridge Isotope Laboratories)]. The samples were centrifuged (10 min; 9000 × g; 4°C), and the supernatants were injected directly on to a 150 × 2-mm, 3-μm Atlantis HILIC column (Waters). The column was eluted isocratically at a flow rate of 250 μL/min with 5% mobile phase A (10 mmol ammonium formate/L and 0.1% formic acid in water) for 0.5 min followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 min. MS analyses were carried out using electrospray ionization in the positive-ion and full-scan spectra were acquired over 70–800 m/z. Lipids were profiled using a Shimadzu Nexera X2 U-HPLC (Shimadzu Corp.; Marlborough, MA) coupled to an Exactive Plus orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Lipids were extracted from plasma (10 μL) using 190 μL of isopropanol containing 1,2-didodecanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids; Alabaster, AL) as an internal standard. Lipid extracts (2 μL) were injected onto a 100 × 2.1 mm, 1.7 μm ACQUITY BEH C8 column (Waters; Milford, MA). The column was eluted isocratically with 80% mobile phase A (95:5:0.1 vol/vol/vol 10mM ammonium acetate/methanol/formic acid) for 1 minute followed by a linear gradient to 80% mobile-phase B (99.9:0.1 vol/vol methanol/formic acid) over 2 minutes, a linear gradient to 100% mobile phase B over 7 minutes, then 3 minutes at 100% mobile-phase B. MS analyses were carried out using electrospray ionization in the positive ion mode using full scan analysis over 200–1100 m/z. Raw data were processed using Trace Finder version 3.1 and 3.3 (Thermo Fisher Scientific) and Progenesis QI (Nonlinear Dynamics; Newcastle upon Tyne, UK). All polar metabolite identities were determined using reference standards in keeping with the Metabolomics Standard Initiative “Level 1” designation [26]. Since reference standards are not available for all lipids, representative lipids from each lipid class were used to characterize retention time and mass to charge ratio patterns. Since the chromatographic method does not discretely resolve all isomeric lipids from one another and the mass spectrometry data do not provide specific information on acyl group composition or position in complex lipids, lipid identities are reported at the level of lipid class, total acyl carbon content, and total double bond content. To enable assessment of data quality and to facilitate data standardization across the analytical queue and sample batches, pairs of pooled plasma reference samples were analyzed at intervals of 20 study samples. One sample from each pair of pooled references served as a passive QC sample to evaluate the analytical reproducibility for measurement of each metabolite while the other pooled sample was used to standardized at using a “nearest neighbour” approach as previously described [27]. Standardized values were calculated using the ratio of the value in each sample over the nearest pooled plasma reference multiplied by the median value measured across the pooled references. Each method generated a table of results, consisting of metabolites in rows and study samples in columns. These tables were merged into a single table prior to analyses.
2.4. Statistical analysis
Baseline characteristics of study participants were described as means and standard deviations (SD) for quantitative variables, and percentages for categorical variables. Missing values of individual metabolites correspond to those determinations that were below the limit of detection. In individual metabolites with less than 20% of missing values we imputed them using the random forest imputation approach (“missForest” function from the “randomForest” R package) as it has been previously recommended in metabolomics studies [28, 29]. Importantly, different alternatives (e.g., zero value or half of the lower limit of detection) to this approach were found to generate consistent results as was previously reported by our research consortium [30]. Next, to conduct the multivariate analysis, metabolomics data was first centered and scaled using the standard deviation as the scaling factor (i.e. autoscaling) [31]. Due to the high dimensionality and collinear nature of the data, Gaussian (i.e. continuous) regression with elastic net penalty (implemented in the “glmnet” R package) was used to build a model for TP, AP, PP and PR intake. The elastic net regression combines the penalties from the Lasso - which drops some metabolite out of the model and assign a larger coefficient to one of the correlated metabolites whereas the rest are nearly zeroed - and Ridge - which keeps all the metabolites into the model and assign similar coefficients to correlated metabolites – regressions, potentially leading to a model which is both simple and predictive [32, 33].
We performed a 10 cross-validation (CV) approach, splitting the sample into training (90% of the sample) and validation set 10 independent times, and then within the training set we performed a further 10-fold CV to find the optimal value of the tuning parameter [λ (lambda)] that yielded the minimum mean-squared error (MSE). The values minMSE and minMSE + 1 standard error (SE) were calculated using argument s = “lambda.min” or s = “lambda.1se” in the cv.glmnet function (“glmnet” R package), respectively. In order to report the coefficients from each CV iteration, the lambda selection in the elastic net continuous regression was evaluated. We selected s = “lambda.min” as it gives the minimum mean CV error and s = “lambda.1se” - largest value of lambda such that error is within 1 SE of the minimum - was not deriving a model for some approaches. Apart from considering the lambda value, we evaluated the alpha parameter from 0 (i.e., Ridge regression) to 1 (i.e., Lasso regression) in 0.1 increments to test the best scenario for our data. In case of the four approaches, alpha=0.6 was the model with best predicting accuracy in the validation sets. Weighted models were constructed for each training-validation dataset pair (90% training and 10% validation) using solely the coefficients for the metabolites obtained from each elastic net regression in the training set. Ten-CV Pearson correlation coefficients (95% confidence interval [CI]) were derived considering each protein intake variable and its corresponding multi-metabolite model within each training-validation dataset. For reproducibility purposes, regression coefficients are reported using 10 iterations of the 10-CV elastic regression approach in the whole dataset. We ran a principal component analysis (PCA) using the mean elastic net continuous regression’s coefficients from the metabolites consistently selected (i.e., 9–10 times) in each of the approaches. A zero value was assigned whenever a particular metabolite was not found by a specific approach. Coefficients were centered and scaled prior to PCA analysis.
Sensitivity analysis were performed using an elastic net logistic regression employing extreme tertiles (T3 vs T1) of protein intake instead of treating the exposures using continuous data. Moreover, additional sensitivity analysis adding relevant covariates (e.g., age, sex, smoking status, case/control status) or food groups showed no alteration in the coefficients obtained in each model (i.e. not selected in each respective model). All the analyses were performed using R v.3.4.2 statistical software. These analyses were based on consistency among CV runs, and therefore any P-value is derived.
3. RESULTS
A total of 1,833 PREDIMED study participants (778 men and 1,055 women) were included in the present study. Figure 1 shows the flow chart of study participants. Characteristics of the participants are summarized in Table 1 for the whole number of subjects and divided by extreme tertiles of TP intake (T1 with n=613 and T3 with n=606). This analysis includes 42.4% of male participants with a median age of 67 years [IQR: 62, 72], a BMI of 29.69 kg/m2 [27.43, 32.24] and a prevalence of 26.8% of T2D. Values from protein intake are as follows: 16.29 E% [14.52, 18.25] for TP, 10.84 E% [9.16, 12.87] for AP, 5.29 E% [4.7, 6.05] for PP and 0.49 [0.39, 0.62] for PR.
Figure 1.
Flow-chart of study participants.
*, Unrealistic energy intake is defined as out of the range 800–4000 Kcal/day in males and 500–3500 Kcal/day in females. ┼, subjects with a set of ≥20% of metabolites with missing values. Abbreviations: CVD, cardiovascular disease; FFQ, food frequency questionnaire; T2D, type 2 diabetes.
Table 1.
Characteristics of study subjects and according to extreme tertiles (T1 and T3) of total protein intake.
| Characteristics | All subjects (n=1,833) | T1 (n=613) | T3 (n=606) |
|---|---|---|---|
| Sociodemographic and medication variables | |||
| Age (years) | 67 [62, 72] | 68 [62, 72] | 67 [62, 71] |
| Male sex, N (%) | 778 (42.4%) | 355 (57.9%) | 155 (25.6%) |
| Body mass index (kg/m2) | 29.69 [27.43, 32.24] | 29.4 [27.26, 31.92] | 29.88 [27.5, 32.33] |
| Waist circumference (cm) | 100 [93, 107] | 101 [95, 107] | 99 [92, 106] |
| Cholesterol (mg/dL) | 209.44 [187.12, 235.32] | 210.34 [187.99, 236.44] | 210 [186.86, 236.21] |
| Triglycerides (mg/dL) | 116.76 [89.05, 157.03] | 120.97 [91.84, 163.11] | 113.12 [85.97, 151.3] |
| HDL-C (mg/dL) | 50.39 [43.97, 57.77] | 49.41 [43.48, 56.98] | 51.67 [44.67, 59.59] |
| Type 2 Diabetes, N (%) | 492 (26.8%) | 131 (21.4%) | 197 (32.5%) |
| Hypercholesterolemia, N (%) | 1408 (76.8%) | 471 (76.8%) | 469 (77.4%) |
| Hypertension, N (%) | 1599 (87.2%) | 531 (86.6%) | 527 (87%) |
| Family history of CVD, N (%) | 451 (24.6%) | 132 (21.5%) | 172 (28.4%) |
| Smoking, N (%) [yes] | 287 (15.7%) | 134 (21.9%) | 61 (10.1%) |
| Cardiac medication, N (%) | 164 (8.9%) | 59 (9.9%) | 47 (7.9%) |
| Hypotensive medication, N (%) | 1382 (75.4%) | 459 (75%) | 462 (76.5%) |
| Cholesterol lowering medication, N (%) | 852 (46.5%) | 273 (44.6%) | 291 (48.1%) |
| Nutritional variables | |||
| Total protein intake (% energy/d) | 16.29 [14.52, 18.25] | 13.84 [12.9, 14.53] | 19.19 [18.26, 20.37] |
| Animal protein intake (% energy/d) | 10.84 [9.16, 12.87] | 8.5 [7.29, 9.38] | 13.77 [12.7, 15.12] |
| Plant protein intake (% energy/d) | 5.29 [4.7, 6.05] | 5.24 [4.65, 5.89] | 5.44 [4.73, 6.18] |
| Plant-to-animal protein ratio | 0.49 [0.39, 0.62] | 0.63 [0.51, 0.79] | 0.39 [0.32, 0.48] |
| P14 questionnaire | 9 [7, 10] | 9 [7, 10] | 9 [8, 10] |
| Total protein intake (g/d) | 90.66 [77.75, 105.3] | 84.54 [72.33, 99.96] | 96.93 [82.98, 110.11] |
| Total carbohydrate intake (g/d) | 231.34 [187.85, 279.6] | 259.61 [215.91, 318.92] | 203.53 [164.83, 240.9] |
| Fat (g/d) | 97.8 [78.43, 115.32] | 106.62 [91.54, 126.01] | 81.74 [66.78, 101.56] |
| MUFA (g/d) | 49.03 [36.88, 58.56] | 55.18 [45.71, 63.91] | 38.45 [31.5, 50.46] |
| SFA (g/d) | 24.5 [19.47, 30.18] | 26.17 [21.62, 31.99] | 21.86 [17.44, 27.37] |
| PUFA (g/d) | 14.47 [11.22, 19.04] | 16.7 [12.82, 21.62] | 12.36 [9.43, 15.84] |
| Total energy intake (Kcal/d) | 2229.77 [1907.69, 2617.85] | 2477.15 [2138.42, 2874.52] | 1992.23 [1684.98, 2296.31] |
| Vegetable intake (g/d) | 311 [233, 405] | 288 [219.5, 368.67] | 332.67 [245.96, 437.21] |
| Legume intake (g/d) | 16.57 [12.57, 25.14] | 16.57 [12.57, 25.14] | 16.57 [12, 25.14] |
| Grain intake (g/d) | 216.43 [166.14, 291.21] | 236.33 [176.79, 309.79] | 192.02 [148.71, 260.36] |
| Dairy intake (g/d) | 326.31 [228.1, 550] | 275.71 [207.14, 449.52] | 367.74 [265.8, 599.49] |
| Meat intake (g/d) | 130.57 [97.71, 164.86] | 108.57 [75.1, 140.48] | 149.05 [120, 186.61] |
| Fish intake (g/d) | 97.14 [65.43, 129.24] | 81.33 [53.33, 112.86] | 110.76 [80.29, 145.38] |
| Nuts intake (g/d) | 6.29 [0, 14.86] | 6.29 [2, 17.14] | 4.29 [0, 12.86] |
| Egg intake (g/d) | 25.71 [8.57, 25.71] | 25.71 [8.57, 25.71] | 25.71 [8.57, 25.71] |
Data shows median [IQR] or number (%). CVD, cardiovascular disease; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; SFA, saturated fatty acids.
3.1. Multi-metabolite model and correlation with protein intake assessments
From the 399 metabolites originally annotated, 11 metabolites were removed due to high number of missing values (i.e. >20%) and 3 metabolites were removed as being internal standard, thus 385 metabolites were finally included in all the analysis. Figures 2 and 3 show the mean coefficient value (and SD) for the set of metabolites consistently selected (9–10 times) in the 10 CV for the four different protein intake measurements. Table 2 summarizes the number of metabolites found in each approach (positive or negative) and the Pearson correlation between multi-metabolite model and each protein intake assessment. Supplementary Table 1 shows the sensitivity analysis using the argument “lambda.1se”. Values for metabolites’ mean, SD and the times being selected in each iteration are shown in Supplementary Table 2. As may be observed, the “lambda.1se” argument generated models with a reduced number of metabolites except for a null model in case of PR.
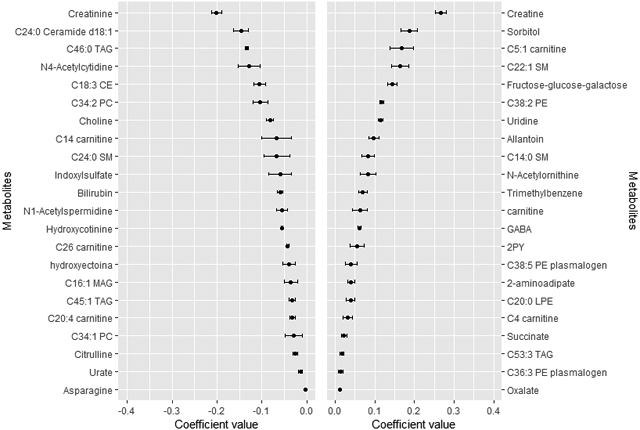
Figure 2.
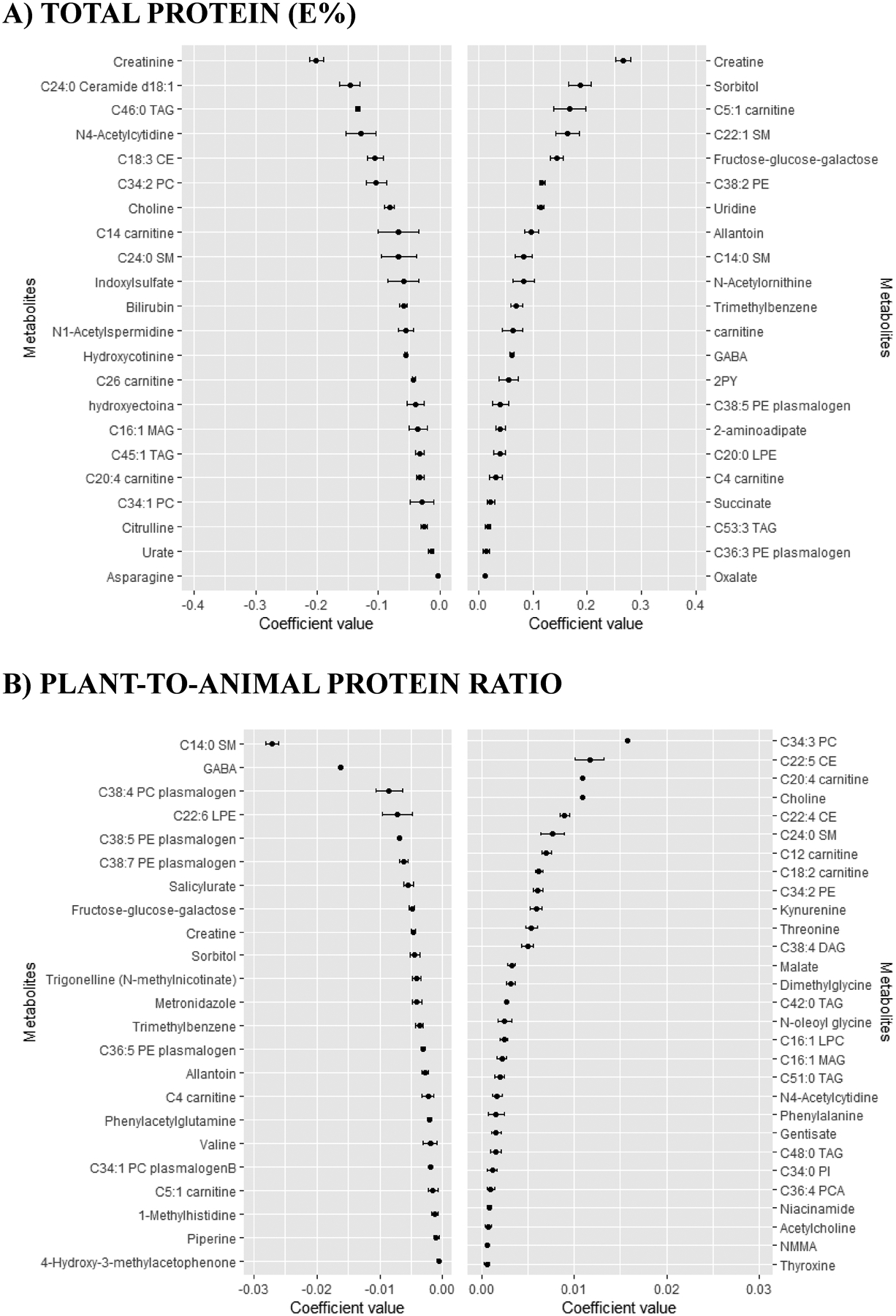
Coefficients (mean and SD) for the metabolites selected 9–10 times in the 10-cross validation of the continuous elastic regression for total protein and plant-to-animal protein ratio.
Mean and SD of the set of the metabolites selected 9–10 times in the ten times iterated 10-fold-cross validation of the elastic continuous regression procedure (using lambda.min) employing the whole dataset of subjects (n=1,833). Metabolites with negative coefficients are plotted in the left part, whereas those with positive coefficients are shown in the right part. A), Total protein (E%); B), plant-to-animal protein ratio. Abbreviations: 2PY, N-methyl-2-pyridone-5-carboxamide; CE, cholesteryl ester; CV, cross-validation; DAG, diacylglycerol; E%, energy percentage; GABA, gamma-aminobutyric acid; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; MAG, monoacylglycerol; NMMA, N-methylmalonamic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; SM, sphyngomyeline; TAG, triglyceride.
Figure 3.
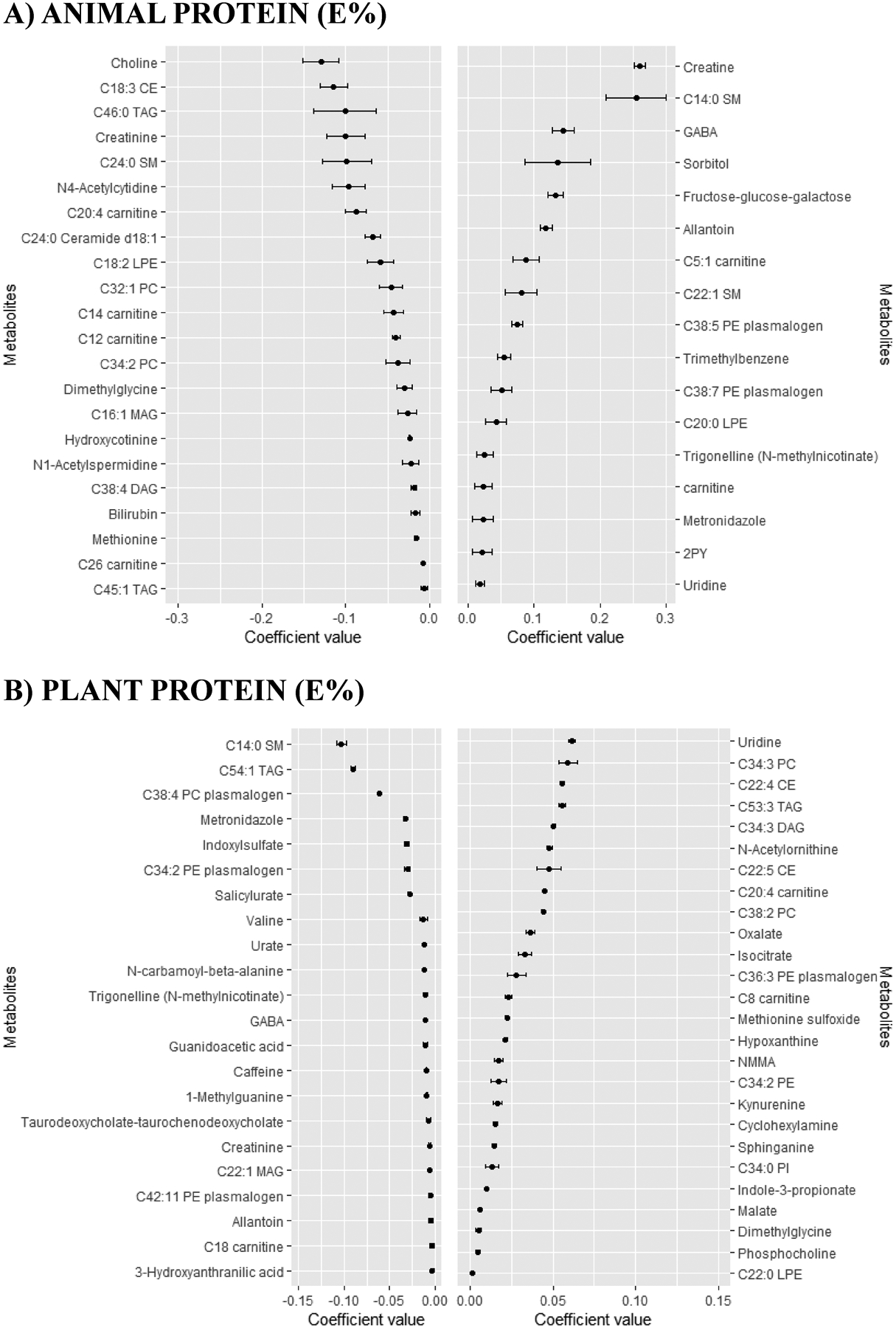
Coefficients (mean and SD) for the metabolites selected 9–10 times in the 10-CV of the continuous elastic regression for animal protein and plant protein.
Mean and SD of the set of the metabolites selected 9–10 times in the ten times iterated 10-fold-CV of the elastic continuous regression procedure (using lambda.min) employing the whole dataset of subjects (n=1,833). Metabolites with negative coefficients are plotted in the left part, whereas those with positive coefficients are shown in the right part. Abbreviations: 2PY, N-methyl-2-pyridone-5-carboxamide; CE, cholesteryl ester; CV, cross-validation; DAG, diacylglycerol; E%, energy percentage; GABA, gamma-aminobutyric acid; LPE, lysophosphatidylethanolamine; MAG, monoacylglycerol; NMMA, N-methylmalonamic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; SM, sphyngomyeline; TAG, triglyceride.
Table 2.
Pearson correlation coefficients for the different protein intake assessments.
| Assessments | Pearson correlation coefficient (95% CI) with metabolomic signature | Total metabolites consistently found to be associated* | # of metabolites with negative coefficients | # of metabolites with positive coefficients |
|---|---|---|---|---|
| Total protein (E%) | 0.32 (0.25–0.39) | 44 | 22 | 22 |
| Plant-to-animal protein ratio | 0.25 (0.20–0.30) | 52 | 23 | 29 |
| Animal protein (E%) | 0.28 (0.23–0.34) | 39 | 22 | 17 |
| Plant protein (E%) | 0.21 (0.17–0.24) | 48 | 22 | 26 |
obtained 9 or 10 times in the cross-validation procedure for the elastic net continuous approach using “lambda.min” option in the “cv.glmnet” function (“glmnet” R package).
Abbreviations: CI, confidence, interval; E%, energy percentage; NA, not available.
In the TP approach, those metabolites with the highest negative coefficient value were creatinine, C24:0 ceramide d18:1 and C46:0 triglyceride (TAG), whereas those with the highest positive coefficient value were creatine, sorbitol and C5:1 carnitine (Figure 2.A). Creatine was also the metabolite with the highest positive value in the AP approach (Figure 3.A). Uridine was the metabolite with the highest positive coefficient value in the PP approach, whereas C14:0 sphingomyelin (SM) was the metabolite with the highest negative coefficient value (Figure 3.B). In fact, C14:0 SM was also the metabolite with the highest negative coefficient value in the PR approach, whereas C34:3 phosphatidylcholine (PC) was the metabolite with the highest positive coefficient value (Figure 2.B).
Correlation between the multi-metabolomic signature and protein intake assessment differed according to the type of protein (Table 2). Of note, argument “lambda.1se” in the “cv.glmnet” function generated a reduced value of Pearson correlation and reduced number of metabolites selected that even derived a null model in case of PR approach (Supplementary Table 1). Metabolites included in the “lambda.1se” approaches were also consistently found in its respective “lambda.min” approaches (Supplementary Table 2). Pearson correlation coefficients (95% CI) sorted by increasing values were: 0.21 (0.17–0.24) for PP, 0.25 (0.20–0.30) for PR, 0.28 (0.23–0.34) for AP and 0.32 (0.25–0.39) for TP.
Sensitivity analysis using extreme tertiles of protein intake (including TP, PR, AP and PP) in the elastic net logistic regression – using “lambda-min” argument – showed comparable results in term of metabolites selected (data not shown).
Different Venn diagrams were created to display the number of unique or overlapping metabolites identified using the different protein approaches (Figure 4 and Supplementary Table 3). No overlapping metabolites were found among the four approaches when considering only positive coefficients (Figure 4.A) or negative coefficients (Figure 4.C). However, four metabolites (i.e., C14:0 SM, C20:4 carnitine, GABA and allantoin) were found in the four approaches regardless of the coefficient sign (Figure 4.B). In an attempt to differentiate the AP and PP approaches, we created individual Venn diagrams (Figure 4, D to G). Uridine was the unique metabolite with a positive value found in both AP and PP approaches (Figure 4.D). Creatinine was the unique metabolite with a negative value found in both AP and PP approaches (Figure 4.E). Only C20:4 carnitine and dimethylglycine were reported with positive coefficients in PP but negative coefficients in AP (Figure 4.F). Allantoin, C14:0 SM, C38:7 PE plasmalogen, GABA, metronidazole and trigonelline (N-methylnicotinate) were reported with negative coefficients in PP but positive coefficients in AP (Figure 4.G).
Figure 4.
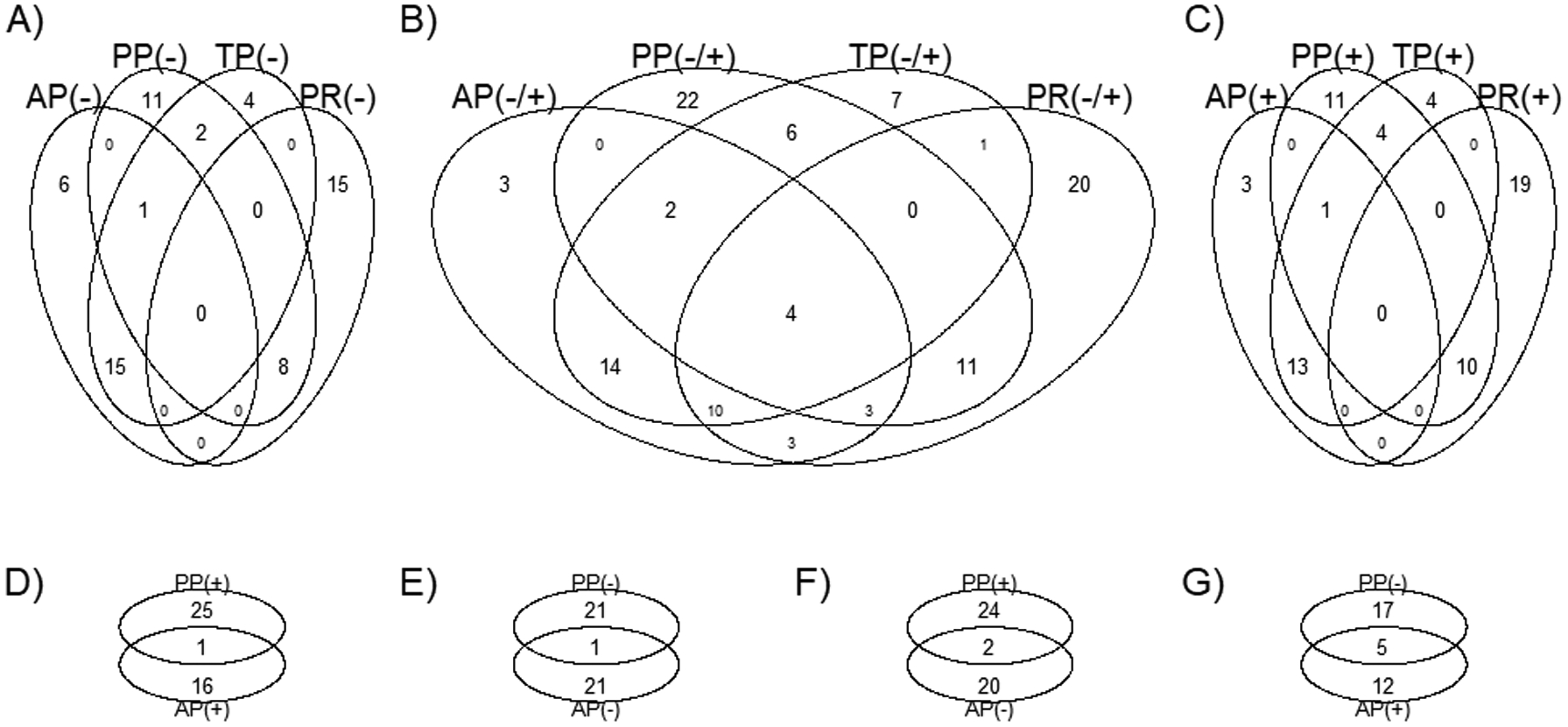
Venn diagram displaying the number of unique or overlapping metabolites identified using the different protein intake approaches by means of the elastic net continuous regression.
A), considering only metabolites with negative coefficients; B), considering metabolites with both positive and negative coefficients; C), considering only metabolites with positive coefficients; D), considering only metabolites with positive coefficients; E), considering only metabolites with negative coefficients; F), considering only metabolites with negative coefficients in AP and positive coefficients in PP; G), considering only metabolites with positive coefficients in AP and negative coefficients in PP. Abbreviations: AP, animal protein; GABA, gamma-aminobutyric acid; PP, plant protein; PR, plant-to-animal protein ratio; SM, sphyngomyeline; TP, total protein. Supplementary Table 2 contains the metabolites belonging to each group. Four metabolites (i.e. C14:0 SM, C20:4 carnitine, GABA and allantoin) were found in the four approaches regardless of the coefficient sign (B). Any metabolite was found in the four approaches when considering only positive (A) or only negative coefficients (C). Uridine was the unique metabolite with a positive value found in both AP and PP approaches (D). Creatinine was the unique metabolite with a negative value found in both AP and PP approaches (E). C20:4 carnitine and dimethylglycine were reported with positive coefficients in PP but negative coefficients in AP (F). Allantoin, C14:0 SM, C38:7 PE plasmalogen, GABA, metronidazole and Trigonelline (N-methylnicotinate) were reported with negative coefficients in PP but positive coefficients in AP (G).
In order to identify principal components consisting of metabolites more associated with TP, AP, PP and/or PR, we additionally created a PCA based on the mean coefficients’ value from the metabolites selected by the different protein intake approaches using its respective elastic net continuous regression (Supplementary Figure 1). In this first PCA, principal component #1 accounted 53.9% of the variability, whereas the second principal component accounted 35.5% of the variability. Moreover, principal component #1 seemed useful to discriminate PP approach from TP, AP and PR approaches, whereas the second allowed the discrimination of the PR approach. In the PCA biplot we observed groups of metabolites clustered close to the four different approaches (Supplementary Figure 1). Moreover, we also reported an obvious close proximity between TP and AP approaches considering the high contribution of AP to TP intake. To solve this issue, we conducted a second PCA excluding PR approach from the PCA (Supplementary Figure 2). We showed a clear separation between TP/AP and PP approaches using the first principal component (82.1% of the variability), whereas the second component (17.9% of the variability) allowed the discrimination between the TP and AP approaches. Supplementary Table 4 shows information related to the most relevant metabolites (based on Venn diagrams and PCAs) reported in our analyses.
4. DISCUSSION
In the present analysis, we have identified a broad range of plasma metabolites associated with TP consumption and/or sources of protein using a combined CV procedure within the elastic net continuous regression. Venn diagrams and PCAs allowed the definition of clusters of metabolites associated with each protein source. The identified multi-metabolite models exhibited differing significant Pearson correlation coefficients with their intake values.
Few studies have assessed circulating plasma or serum metabolomics of diets varying in TP intake [12–15]. A total of 1,336 male Finnish smokers were used to identify biomarkers of dietary patterns (e.g. Healthy Eating Index (HEI) 2010) by using serum metabolomics [12]. Metabolites associated with TP were mainly related to free FAs (not analyzed in our study) and AA derivatives (e.g. 3-methylhistidine and creatine) [12]. Mirroring their results, we also found a positive association between TP intake and creatine. A recent 10-week RCT conducted also in elderly males consuming differing amounts of protein and using a non-targeted polar plasma metabolomics analysis showed comparable results in terms of TP intake [15]. Researchers ascribed all the modulatory effects to protein anabolism without sign of influence on other pathways related with metabolic health.
In another RCT, 21 subjects with overweight/obesity were studied during a 4-week weight stability phase according to a crossover design of 3 diets differing in protein content [13]. Among the plasma metabolites positively associated with protein, they reported alpha-hydroxybutyrate, creatine, several TAGs species and uridine, whereas those negatively associated with TP were C18:2 LPE, C40:6 PC and C56:8 TAG. We also reported a positive association between TP and uridine (also in AP and PP approaches) and creatine (also positive in AP but negative in PP). However, we only reported C53:3 TAG positively associated with TP in our study.
A recent cross-sectional study identified serum metabolites associated with dietary protein intake in 674 subjects with CKD - and differing in glomerular filtration rate - with ages ranging from 18–70 years [14]. They found 130 metabolites when comparing low-protein diet versus moderate-protein diet, and 32 metabolites when compared very-low-protein diet versus low-protein diet. Independently of the glomerular filtration rate, a total of 11 metabolites were significantly associated with TP intake including 3-methylhistidine, N-acetyl-3-methylhistidine, creatine, kynurenate and different plasmalogens. Remarkably, the half-lives of 1- and 3-methylhistidine together with other metabolites are reported to be approximately 12 hours; thus, they are solely considered short term biomarkers of red meat intake [34]. Our plasma metabolomics approach did not cover most of these metabolites. However, we found similar results in terms of positive associations of TP with creatine and with same carbon number PE and PC plasmalogens albeit with different unsaturation profile.
One limitation common to previous studies is that they have not distinguished sources of protein intake as plant/animal protein, which is important to try to understand why the effects on health are different depending on the type of protein consumed. By comparing the four different approaches we found few overlaps and many approach-specific metabolites. Most of the overlaps were found between TP and AP, probably because the high AP compared to PP intake in our population. We reported four metabolites simultaneously and positively or negatively associated with the four protein approaches. C14:0 SM, GABA and allantoin were positively associated with AP and TP, whereas negatively associated with PP and PR. The inverse scenario was exhibited by C20:4 carnitine.
C14:0 SM was previously found positively associated with TP [13] and positively associated with increasing protein consumption [14]. This SM has been recently negatively correlated with the scale of aging vigor in epidemiology (SAVE) score, thus reduced C14:0 SM values are associated with frailty [35]. Importantly, it has been negatively associated to the empirical dietary inflammatory pattern (EDIP) score, reflecting a putative anti-inflammatory role [36]. GABA was also positively associated with high TP and fat intake in a clinical trial, but the results were inconsistent with those measured in the Framingham Heart Study, where GABA was only positively correlated with carbohydrate intake [13]. It has been seen that GABA is released by β-cells in a glutamine dose-dependent manner whereas glucose induces inhibition of its release to the extracellular medium [37–39]. To increase TP intake, it is necessary to reduce the consumption of other macronutrients, such as carbohydrates, a situation that could enhance GABA production and release from beta-cells. GABA is a well-known inhibitory neurotransmitter in the brain, but it seems to be also involved in the reduction of the local immune and inflammatory responses [40]. Finally, allantoin was positively correlated with TP and AP and negatively with PR and PP. This metabolite is produced from urate in animals (excluding humans), plants and bacteria and it is considered a marker of oxidative stress. Although little is known about the association between quantity and quality of protein intake and oxidative stress, it seems that diets rich in animal-based foods lead to this condition [41], which could explain the observed associations. Interestingly, urate was found inversely associated with both TP and PP. However, sorbitol and the isomer fructose-glucose-galactose were positively associated with TP and AP, whereas negatively associated with PR. Of note, sorbitol is converted to fructose when metabolized in the liver producing biochemical effects similar to those of fructose on hepatic adenosine phosphate levels in humans, and can therefore increase uric acid production [42]. This may explain the positive association between sorbitol and fructose-glucose-galactose with TP and AP, and the negative association of urate with PP. However, high levels of serum sorbitol have been reported in individuals with T2D compared with those without the disease [43]. In our study, individuals with a higher consumption of TP were more likely to have T2D than those with a lower consumption.
Total carnitine, together with C4 and C5:1 carnitines were positively associated with TP but negatively associated with PR. Carnitine can be obtained from the diet – mainly from meat and dairy products – or endogenously synthesized from lysine and methionine. Importantly, dietary carnitine correlates with plasma concentrations and it has been reported that individuals consuming high AP diets have higher plasma carnitine levels than those consuming low amounts [44]. Carnitine participates in the transport of fatty acids (FA) for their β-oxidation in the mitochondria, a procedure where it is transformed to acylcarnitines. The accumulation of acylcarnitines could reflect alterations in the FA oxidation process, which could promote the development of metabolic diseases [45, 46]. Surprisingly, a polyunsaturated carnitine (C20:4) was found inversely associated to TP and AP, and positively to PR and PP. Further studies are needed to assess to which extend protein intake could modify carnitine-related metabolites.
Creatine was the metabolite most positively associated with TP and AP, whereas negatively associated with PR. Previous clinical trials also reported creatine as a marker of TP [14]. Animal protein foods are considered the main sources of creatine [47]. Therefore, it is not surprising that low levels of creatine were observed in vegetarians [48] in a cross-sectional study, results that are supported by a clinical trial where women switching from omnivore to vegetarian diet experimented a reduction in creatine levels after 3 months of intervention [49]. Creatinine – a breakdown product of creatine phosphate in muscle – was found negatively associated with TP, AP and PP. These results are in line with previous findings which reported a negative correlation between TP intake and serum creatinine [50]. Since a positive association exists between TP intake and urinary excretion of creatinine, the reported negative association could be due to the enhanced creatinine clearance. In fact, urinary, but not serum/plasma creatinine, has been suggested as a biomarker of meat consumption [16, 51].
Some metabolites were solely identified in the PR approach or in combination with PP approach (e.g., NMMA and malate). In fact, a wide set of metabolites were positively associated with PR and not found in any other approach. It comprised: i) essential AAs such as phenylalanine and threonine; ii) AAs’ derivatives such as N-oleoyl glycine; iii) other molecules such as gentisate, acetylcholine, niacinamide and different lipid species such as C16:1 LPC, C36:4 PC-A, and saturated TAGs (C42, C48 and C51). The health implications of these findings should be further investigated.
This approach has some drawbacks that deserve comment. First, as it has been performed in older adults at high CVD risk from a Mediterranean area, the generalizability of the findings to other populations may be limited. Moreover, due to the cross-sectional design, causation cannot be inferred. Even though we included in the analysis a relatively large sample size that was analyzed using a validated FFQ, we cannot exclude misclassification bias. Moreover, we did not distinguish the different sources of animal protein, which may have a distinct impact on health. Additionally, a measure of total urinary nitrogen excretion was not available for our subjects of study, which did not allow us to assess the correlation with our metabolites. Even though elastic net regression derived a relatively simple and predictive model, we cannot completely disregard a lack of specific metabolites into the models due to putative multicollinearity. Moreover, as we only included annotated metabolites, we cannot assure that a multi-metabolite model based on untargeted metabolites will not outperform ours. Strengths of the present study include the use of a multi-metabolomics approach to analyze a wide range of metabolite compounds; we have cross-internally validated our results; and we have performed different sensitivity analysis to assess the role of other putative confounders, such as sex and dietary factors, into the selected metabolites.
In conclusion, our findings show that TP, AP, PP and PR consumption are associated with distinct sets of plasma metabolites mainly related to AAs and their derivatives, together with acylcarnitines, different organic compounds, and lipid species, which are the reflection of changes in metabolic pathways potentially implicated in disease prevention or development. Some of these metabolites have been discovered as markers of protein consumption in other epidemiologic studies. In the current study, we provided a deeper understanding of the metabolic response to protein intake providing new functional insight to its potential role in health. The extent to which the sets of metabolites associated with protein intake we identified in the study are associated with health outcomes remains to be evaluated.
Supplementary Material
Acknowledgments:
We thank the participants for their enthusiastic collaboration, the PREDIMED personnel for excellent assistance, and the personnel of all affiliated primary care centers.
Conflict of interest statement:
JS-S is a non-paid member of the Scientific Committee of the International Nut and Dried Fruit Foundation. He has received grants/research support from the American Pistachio Growers and International Nut and Dried Fruit Foundation through his Institution. He has received honoraria from Nuts for Life, Danone and Eroski. He reports personal fees from Danone. He is a member of the executive committee of the Instituto Danone Spain. Any other co-authors have a conflict of interest that is relevant to the subject matter or materials included in this Work.
Funding sources:
This study was funded by the National Institutes of Health (R01DK102896, F31DK114938), the Spanish Ministry of Health (Instituto de Salud Carlos III) and the Ministerio de Economía y Competitividad-Fondo Europeo de Desarrollo Regional (Projects CNIC-06/2007, RTIC G03/140, CIBER 06/03, PI06-1326, PI07-0954, PI11/02505, SAF2009-12304 and AGL2010-22319-C03-03) and by the Generalitat Valenciana (ACOMP2010-181, AP111/10, AP-042/11, ACOM2011/145, ACOMP/2012/190, ACOMP/2013/159 and ACOMP/213/165). This work is partially supported by ICREA under the ICREA Academia programme. Dr. PH-A is supported by a postdoctoral fellowship (Juan de la Cierva-Formación, FJCI-2017-32205). Dr. CP is recipient of the Instituto de Salud Carlos III Miguel Servet fellowship (grant CP 19/00189). Dr. MG-F was supported by American Diabetes Association grant #1-18-PMF-029.
Abbreviations:
- AA
amino acid
- CE
cholesteryl ester
- CV
cross-validation
- CVD
cardiovascular disease
- FA
fatty acid
- FFQ
food frequency questionnaire
- ICC
intraclass correlation coefficient
- LPC
lysophosphatidylcholine
- MSE
mean-squared error
- PC
phosphatidylcholine
- RCT
randomized clinical trial
- SM
sphingomyelin
- T2D
type 2 diabetes
- TAG
triglyceride
- WC
waist circumference
REFERENCES
- [1].Larsen TM, Dalskov S-M, van Baak M, Jebb SA, et al. , Diets with High or Low Protein Content and Glycemic Index for Weight-Loss Maintenance. N. Engl. J. Med 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gannon MC, Nuttall FQ, Saeed A, Jordan K, et al. , An increase in dietary protein improves the blood glucose response in persons with type 2 diabetes. Am. J. Clin. Nutr 2003, 78, 734–741. [DOI] [PubMed] [Google Scholar]
- [3].Dong J-Y, Zhang Z-L, Wang P-Y, Qin L-Q, Effects of high-protein diets on body weight, glycaemic control, blood lipids and blood pressure in type 2 diabetes: meta-analysis of randomised controlled trials. Br. J. Nutr 2013, 110, 781–9. [DOI] [PubMed] [Google Scholar]
- [4].Hu FB, Protein, body weight, and cardiovascular health., 2005. [DOI] [PubMed]
- [5].Millen BE, Abrams S, Adams-Campbell L, Anderson CA, et al. , The 2015 Dietary Guidelines Advisory Committee Scientific Report: Development and Major Conclusions. Adv. Nutr 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hernández-Alonso P, Salas-Salvadó J, Ruiz-Canela M, Corella D, et al. , High dietary protein intake is associated with an increased body weight and total death risk. Clin. Nutr 2015. [DOI] [PubMed] [Google Scholar]
- [7].Richter CK, Skulas-Ray AC, Champagne CM, Kris-Etherton PM, Plant Protein and Animal Proteins: Do They Differentially Affect Cardiovascular Disease Risk? Adv. Nutr 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Krajcovicova-Kudlackova M, Babinska K, Valachovicova M, Health benefits and risks of plant proteins. Bratisl. Lek. Listy 2005, 106, 231–4. [PubMed] [Google Scholar]
- [9].Bingham SA, Urine Nitrogen as a Biomarker for the Validation of Dietary Protein Intake. J. Nutr 2003. [DOI] [PubMed] [Google Scholar]
- [10].Jenab M, Slimani N, Bictash M, Ferrari P, et al. , Biomarkers in nutritional epidemiology: applications, needs and new horizons. Hum. Genet 2009, 125, 507–525. [DOI] [PubMed] [Google Scholar]
- [11].Clish CB, Metabolomics: an emerging but powerful tool for precision medicine. Mol. Case Stud 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Playdon MC, Moore SC, Derkach A, Reedy J, et al. , Identifying biomarkers of dietary patterns by using metabolomics. Am. J. Clin. Nutr 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Esko T, Hirschhorn JN, Feldman HA, Hsu YHH, et al. , Metabolomic profiles as reliable biomarkers of dietary composition. Am. J. Clin. Nutr 2017, 105, 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rebholz CM, Zheng Z, Grams ME, Appel LJ, et al. , Serum metabolites associated with dietary protein intake: Results from the Modification of Diet in Renal Disease (MDRD) randomized clinical trial. Am. J. Clin. Nutr 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Durainayagam B, Mitchell CJ, Milan AM, Zeng N, et al. , Impact of a High Protein Intake on the Plasma Metabolome in Elderly Males: 10 Week Randomized Dietary Intervention. Front. Nutr 2019, 6, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cuparencu C, Praticó G, Hemeryck LY, Sri Harsha PSC, et al. , Biomarkers of meat and seafood intake: An extensive literature review. Genes Nutr. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang DD, Toledo E, Hruby A, Rosner BA, et al. , Plasma ceramides, mediterranean diet, and incident cardiovascular disease in the PREDIMED trial (prevención con dieta mediterránea). Circulation 2017, 135, 2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ruiz-Canela M, Guasch-Ferré M, Toledo E, Clish CB, et al. , Plasma branched chain/aromatic amino acids, enriched Mediterranean diet and risk of type 2 diabetes: case-cohort study within the PREDIMED Trial. Diabetologia 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Estruch R, Ros E, Salas-Salvadó J, Covas M-I, et al. , Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N. Engl. J. Med 2018, 378, e34. [DOI] [PubMed] [Google Scholar]
- [20].Fernández-Ballart JD, Piñol JL, Zazpe I, Corella D, et al. , Relative validity of a semi-quantitative food-frequency questionnaire in an elderly Mediterranean population of Spain. Br. J. Nutr 2010, 103, 1808–16. [DOI] [PubMed] [Google Scholar]
- [21].Elosua R, Marrugat J, Molina L, Pons S, et al. , Validation of the Minnesota Leisure Time Physical Activity Questionnaire in Spanish men. The MARATHOM Investigators. Am. J. Epidemiol 1994, 139, 1197–1209. [DOI] [PubMed] [Google Scholar]
- [22].De La Fuente-Arrillaga C, Vzquez Ruiz Z, Bes-Rastrollo M, Sampson L, et al. , Reproducibility of an FFQ validated in Spain, in: Public Health Nutrition, 2010. [DOI] [PubMed] [Google Scholar]
- [23].Mascanfroni ID, Takenaka MC, Yeste A, Patel B, et al. , Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat. Med 2015, 21, 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].O’Sullivan JF, Morningstar JE, Yang Q, Zheng B, et al. , Dimethylguanidino valeric acid is a marker of liver fat and predicts diabetes. J. Clin. Invest 2017, 127, 4394–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rowan S, Jiang S, Korem T, Szymanski J, et al. , Involvement of a gut–retina axis in protection against dietary glycemia-induced age-related macular degeneration. Proc. Natl. Acad. Sci 2017, 114, E4472–E4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sumner LW, Amberg A, Barrett D, Beale MH, et al. , Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mayers JR, Wu C, Clish CB, Kraft P, et al. , Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gromski P, Xu Y, Kotze H, Correa E, et al. , Influence of Missing Values Substitutes on Multivariate Analysis of Metabolomics Data. Metabolites 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wei R, Wang J, Su M, Jia E, et al. , Missing Value Imputation Approach for Mass Spectrometry-based Metabolomics Data. Sci. Rep 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hernández‐Alonso P, Papandreou C, Bulló M, Ruiz‐Canela M, et al. , Plasma Metabolites Associated with Frequent Red Wine Consumption: A Metabolomics Approach within the PREDIMED Study. Mol. Nutr. Food Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, et al. , Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics 2006, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zou H, Hastie T, Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol 2005. [Google Scholar]
- [33].De Mol C, De Vito E, Rosasco L, Elastic-net regularization in learning theory. J. Complex 2009. [Google Scholar]
- [34].Cross AJ, Major JM, Sinha R, Urinary biomarkers of meat consumption. Cancer Epidemiol. Biomarkers Prev 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Marron MM, Harris TB, Boudreau RM, Clish CB, et al. , Metabolites associated with vigor to frailty among community-dwelling older black men. Metabolites 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tabung FK, Liang L, Huang T, Balasubramanian R, et al. , Identifying metabolomic profiles of inflammatory diets in postmenopausal women. Clin. Nutr 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Smismans A, Schuit F, Pipeleers D, Nutrient regulation of gamma-aminobutyric acid release from islet beta cells. Diabetologia 1997. [DOI] [PubMed] [Google Scholar]
- [38].Newsholme P, Brennan L, Rubi B, Maechler P, New insights into amino acid metabolism, β-cell function and diabetes. Clin. Sci 2005. [DOI] [PubMed] [Google Scholar]
- [39].Wang C, Kerckhofs K, Van De Casteele M, Smolders I, et al. , Glucose inhibits GABA release by pancreatic β-cells through an increase in GABA shunt activity. Am. J. Physiol. - Endocrinol. Metab 2006. [DOI] [PubMed] [Google Scholar]
- [40].Bhat R, Axtell R, Mitra A, Miranda M, et al. , Inhibitory role for GABA in autoimmune inflammation. Proc. Natl. Acad. Sci. U. S. A 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kitabchi AE, McDaniel KA, Wan JY, Tylavsky FA, et al. , Effects of high-protein versus high-carbohydrate diets on markers of β- Cell function, oxidative stress, lipid peroxidation, proinflammatory cytokines, and adipokines in obese, premenopausal women without diabetes. Diabetes Care 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lotito S, Frei B, Consumption of flavonoid-rich foods and increased plasma antioxidant capacity in humans: Cause, consequence, or epiphenomenon? Free Radic. Biol. Med 2006, 41, 1727–1746. [DOI] [PubMed] [Google Scholar]
- [43].Preston GM, Calle RA, Elevated serum sorbitol and not fructose in type 2 diabetic patients. Biomark. Insights 2010, 2010, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Steiber A, Kerner J, Hoppel CL, Carnitine: A nutritional, biosynthetic, and functional perspective. Mol. Aspects Med 2004. [DOI] [PubMed] [Google Scholar]
- [45].Guasch-Ferré M, Ruiz-Canela M, Li J, Zheng Y, et al. , Plasma acylcarnitines and risk of type 2 diabetes in a mediterranean population at high cardiovascular risk. J. Clin. Endocrinol. Metab 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mihalik SJ, Goodpaster BH, Kelley DE, Chace DH, et al. , Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Balsom PD, Söderlund K, Ekblom B, Creatine in Humans with Special Reference to Creatine Supplementation. Sport. Med 1994. [DOI] [PubMed] [Google Scholar]
- [48].Delanghe J, De Slypere JP, De Buyzere M, Robbrecht J, et al. , Normal reference values for creatine, creatinine, and carnitine are lower in vegetarians. Clin. Chem 1989. [PubMed] [Google Scholar]
- [49].Blancquaert L, Baguet A, Bex T, Volkaert A, et al. , Changing to a vegetarian diet reduces the body creatine pool in omnivorous women, but appears not to affect carnitine and carnosine homeostasis: A randomised trial. Br. J. Nutr 2018. [DOI] [PubMed] [Google Scholar]
- [50].Kesteloot HE, Joossens JV, Relationship between dietary protein intake and serum urea, uric acid and creatinine, and 24-hour urinary creatinine excretion: The birnh study. J. Am. Coll. Nutr 1993. [DOI] [PubMed] [Google Scholar]
- [51].Dragsted LO, Biomarkers of meat intake and the application of nutrigenomics. Meat Sci. 2010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


