Abstract
Nowadays, Coronavirus disease (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is one of the most important health problems. The dynamics and nature of humoral responses are relevant to determine the efficacy of both, diagnostic tests and developed vaccines. Since the role of IgA in the COVID-19 disease is not fully understood, we have systematically reviewed the scientific literature on antibody IgA immunity to SARS-CoV-2 to determine if IgA could be useful as a diagnostic tool or as a biomarker of severity. We systematically reviewed 736 abstracts and identified 38 manuscripts relevant to include in the meta-analysis. The seroprevalence of IgA in SARS-CoV-2 PCR (+) confirmed patients was 86.47% (CI: 5.27–178.21). Furthermore, we found out that IgA can be produced on the first days of infection (10 days) and IgA is detected until 75 days after symptomatic onset in some studies. We also observe that IgA production is stronger in severe patients compared with mild or asymptomatic patients. Our research noticed a possible association between IgA and protection; however, the possible role of IgA as a biomarker of protection or severity remains unclear.
Keywords: COVID-19, SARS-CoV-2, IgA, Susceptibility
Graphical Abstract
1. Introduction
Near the end of 2019, cases of an unknown upper respiratory tract infection began appearing in Wuhan, Hubei Providence, China (Li et al., 2020). By early January 2020, it was determined that these infections were caused by a novel coronavirus SARS-CoV-2 (Severe Acute Respiratory Syndrome-CoronaVirus-2) inducing the disease named COVID-19 (Coronaviridae Study Group of the International Committee on Taxonomy of, 2020, Zhou et al., 2020). The average incubation period of COVID-19 has a mean of 7.8 days, with a median of 5.01 days (Zaki and Mohamed, 2021) and the response from the host vary from asymptomatic, mild symptomatic and present also severe symptoms such as acute respiratory distress syndrome (ARDS) and multi-organ dysfunction. Mucosal surfaces are key participants in the SARS-CoV-2 infection, and therefore a host mucosal immune-defense could be protective. At these mucosal surfaces, IgA, in the form of secretory IgA (S-IgA), is the predominant immunoglobulin while in the serum, monomeric IgA is the second most abundant Ig class with a concentration of about 2 mg/mL (Mestecky et al., 1986). S-IgA contributes to immune exclusion, a process by which the adsorption of pathogens to mucosal surfaces is prevented through agglutination. The multiple antigen binding sites of S-IgA permits an efficient blocking activity (de Sousa-Pereira and Woof, 2019).
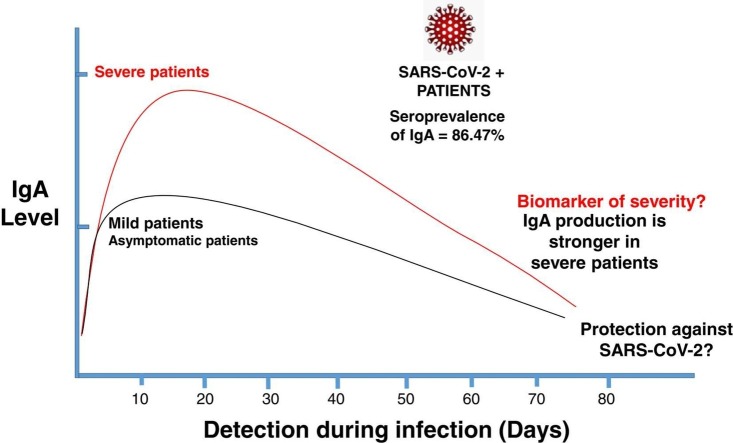
Several studies have correlated SARS-CoV-2–specific serum IgA titers with the severity of COVID-19; the patients with severe disease presented substantially high specific serum IgA antibody levels after symptom onset (Cervia et al., 2021). Conversely, the SARS-CoV-2 specific mucosal IgA response seems to correlate with protection, as in some health workers with negative serum antibody titers, SARS-CoV-2 -specific IgA with virus-neutralizing capacity was detected in mucosal fluids (tears, nasal fluid and saliva). It is important to highlight that mucosal secretory IgA is able to neutralize viruses within the intracellular epithelial cells (Bidgood et al., 2014). Remarkably, increased mucosal S protein–specific IgA titers were detected in the youngest individuals compared with older individuals, this might explain its better capacity to resolve SARS-CoV-2 infection than older people (Cervia et al., 2021). The above-mentioned studies present a background regarding the possible protective role that IgA can play against SARS-CoV2 infections, both in serum and mucosal secretions.
The purpose of this study is to clarify whether IgA can serve as a diagnostic marker or it has a protective role against SARS-CoV-2 and, if enhancing this immunoglobulin could be beneficial for future treatments. Here, we conducted a systematic review and meta-analysis of the available published data to show the seroprevalence of IgA on COVID-19 patients and we discussed based on the published data the possible role as an a early diagnostic tool or as biomarker of protection or severity. Our analysis shows that IgA is produced more effectively in patients after severe disease; we also found out that IgA production is mainly 10 days after the symptomatic onset. We can hypothesized the protective role of IgA in SARS-CoV-2 infected patients and its key role in the early phases of COVID-19. As far as we know, this is the first meta-analysis and systematic review with accurate data related to the important role of the IgA in COVID-19 patients and the feasibility of the new therapies enhancing serum or mucosal IgA responses.
2. Materials and methods
2.1. Data retrieved
A systematic review of published data was conducted using PubMed to identify cases of specific IgA production against SARS-CoV-2. We used the following query terms in PubMed research papers: SARS-CoV-2 and IgA OR COVID-19 and IgA. We used an additional screening with different filters included.
Filter 1: ((Coronavirus and IgA [TIAB] OR SARS-CoV-2 and IgA [TIAB] OR COVID-19 and IgA [TIAB] NOT review [Publication type]).
Filter 2: ((SARS-CoV-2 and IgM [TIAB] OR COVID-19 and IgM [TIAB] NOT review [Publication type] NOT prepreprint)).
Filter 3: ((SARS-CoV-2 and IgG [TIAB] OR COVID-19 and IgG [TIAB] NOT review [Publication type] NOT prepreprint)).
Filter 4: ((SARS-CoV-2 and IgA [TIAB] OR COVID-19 and IgA [TIAB] NOT review [Publication type] NOT prepreprint)).
Filter 5: ((SARS-CoV-2 and antibodies [TIAB] OR COVID-19 and Antibodies [TIAB] NOT review [Publication type] NOT prepreprint)). Up to August 2021, we reviewed a total of 736 articles from PubMed and 4 papers of the additional screening after duplicate elimination; these articles were analyzed to eliminate non-SARS-CoV-2 related IgA papers. The following exclusion criteria were applied to articles: a) do not reported in the English language, b) used for validation data or kit development, c) with no patient/disease information or hospitalization data, d) reported as reviews, e) preprints, f) in which there was not mentioned antibodies or IgG or IgA or IgM in the abstract, g) meta-analysis h) patients with no PCR test, i) preclinical data, and j) letters to the editor or opinion letters. A total of 38 articles up to August 2021 were retrieved for the systematic review and the full texts were screened according to the PRISMA statements (Liberati et al., 2009). The articles were analyzed to determine the role of IgA in COVID-19 disease severity (possible IgA protective role and whether it occurs in severe, mild or asymptomatic patients.), we also analyzed IgA antibodies (which detect different proteins of the virus), temporality of the IgA induction after the PCR diagnostics (to know if the IgA is produced early or late) and the seroprevalence of IgA against SARS-CoV-2 in PCR positive patients.
2.2. Risk of bias assessment
The NIH Study Quality Assessment tool was used to assess the risk of bias and study quality for Systematic Reviews and Meta-Analyses (https://www.nhlbi.nih.gov/health-topics/study-qualityassessment-tools). Studies were initially rated as having good, fair, or poor quality, and ratings were discussed to reach consensus. The complete data and papers included in the meta-analysis were retrieved by a standardized method for two authors (AMC and VRR) independently, inconsistencies were resolved by consensus with a third reviewer (NRI).
2.3. Meta-analysis
We performed a metanalysis to determine the seroprevalence of IgA in COVID-19 patients using the Microsoft Excel spreadsheet described by Neyeloff (2012) with confidence intervals (CIs) of 95%, the forest plot was performed too. We used the random-effects model according to DerSimonian and Laird (2015) and we also used the inconsistency index (I2) reported by George et al. (2019) to evaluate the heterogeneity. Low, moderate, substantial, and considerable heterogeneity are represented by cut-offs of < 30 %, 30–59 %, 60–75 %, and > 75 % respectively.
3. Results
3.1. Data recovered from the systematic review
The systematic procedure for the seroprevalence of IgA in COVID-19 patients is presented in Supplementary Figure 1. We identified 736 PubMed scientific articles filtered by their title and abstract according to the inclusion and exclusion criteria (Section 2.1). After excluding irrelevant studies we identified a total of 2 576 subjects in 38 full-text articles (with experimental data), see Supplementary Table 1
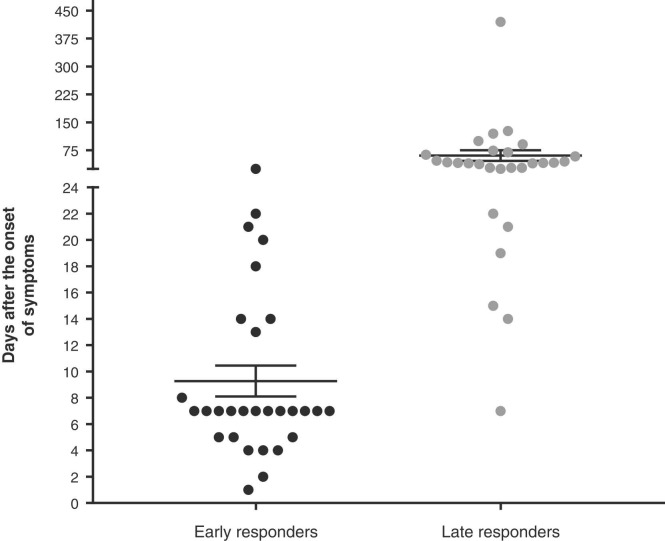
To determine the time for the IgA response to occur, scientific articles were classified according to the first and last time the immunoglobulin was measured (see Fig. 1, articles with unspecified times of IgA response were not included). The vast majority of the literature has reported an IgA response within the first 10 days of the disease (early responders appeared between 2 to 7 days after the symptoms onset) and it remained detectable up to 75 days after the beginning of the illness. Another important aspect revealed in the present meta-analysis is that the severity of the symptoms seems to be related with an increase in the IgA production and, interestingly, IgA might be appearing as early as IgM or IgG in the studies systematically analyzed (Supplementary Table 1).
Fig. 1.
IgA responses in patients with SARS-CoV-2 infection. Graph of the early and late times in which patients with SARS-CoV-2 infection developed/maintained IgA responses.
We added a commentary in the discussion section about the possible roles of the IgA in the analyzed studies including the possible role of this immunoglobulin in protection and viral clearance, as a biomarker of severity and finally its possible utility as a diagnostic tool in early times of infection. It is necessary to mention that our conclusions might be limited/biased due to the fact that the results come from different assays using different antigen preparations and tested under different assay conditions.
3.2. Seroprevalence of IgA in COVID-19 PCR (+) confirmed patients
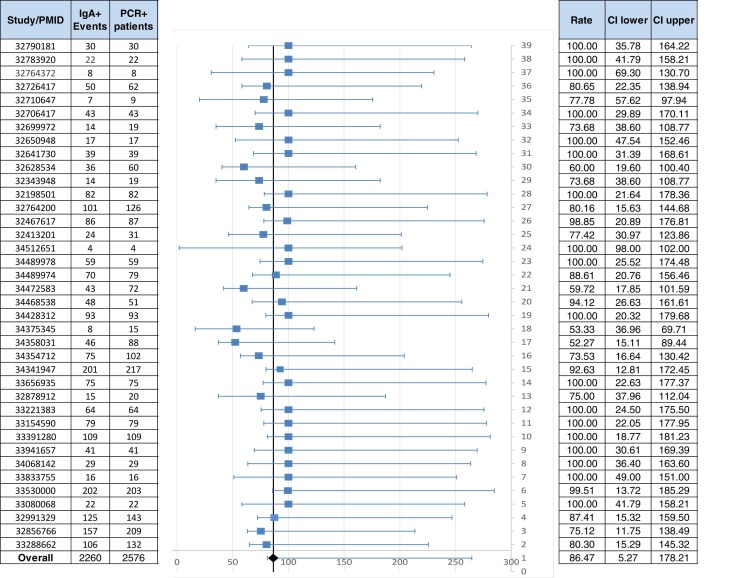
We analyzed 38 papers with a total of 2 576 COVID-19 patients confirmed by PCR test (PCR (+)); based on the meta-analysis we obtained a Q test = 31.62 giving an I2 between 0 % and 40 % which means that uncertainty might not be important, the obtained value means a moderate heterogeneity in the analyzed articles and then, the random effect model was used to combine the reported results of the studies. The seroprevalence of IgA in COVID-19-PCR (+) confirmed patients was 86.47 % (CI: 5.27–178.21) ( Fig. 2); additionally we performed the meta-analysis by the fixed effect model with consistent results (data not shown).
Fig. 2.
Seroprevalence of IgA (+) in SARS-CoV-2 infected patients confirmed by PCR. A total of 38 scientific papers with 2 576 PCR (+) confirmed patients were analyzed. The first column shows the PMID, the second and third columns show the number of IgA patients in PCR (+) confirmed cases and the number of PCR (+) patients respectively; the incidence is labeled as overall. CI: confidence interval.
4. Discussion
COVID-19 disease is caused by the SARS-CoV-2 virus and currently represents one of the most important health threats worldwide, the control of the virus spread implies several challenges including the creation and validation of good molecular and serologic diagnostic tests, the development of possible treatments once the infection is established, a better knowledge of immune responses before, during and after the virus exposure and finally, the efficacy of the available vaccines. Almost all the challenges mentioned are related to the production of antibodies, mainly IgM and IgG, both have been widely used to measure the efficacy of vaccines and to develop diagnostic tests. However, the role of IgA (which is mainly a mucosal antibody) in the diagnosis, prognosis, treatment of COVID-19 infection and/or vaccine efficacy and illness severity remains not fully described.
Several studies have shown that IgM, IgG, and IgA antibodies against the SARS-CoV-2 Spike and RBD proteins can be detected in saliva (derived from acute and convalescent COVID-19 patients) (Sheikh-Mohamed et al., 2022), however, exacerbated systemic and mucosal IgA responses had been implicated in other pathologies too, for example in IgA nephropathy (Layward et al., 1993).
It has been recently reported that serum IgG and mucosal IgA responses against Spike and Receptor Binding Domain (RBD) are unevenly induced by SARS-CoV-2 mRNA vaccination and mucosal secretory IgA responses are associated with protection against subsequent infection. The authors found out that one immunization was able to induce systemic and mucosal antibody responses as well as anti-Spike/RBD associated secretory component in most participants. Besides, participants who experienced breakthrough infections with SARS-CoV-2 variants had lower levels of vaccine-induced serum anti-Spike/RBD IgA than mRNA vaccinated participants, whereas IgG levels were comparable between groups (Sheikh-Mohamed et al., 2022).
Here, based in a systematic review and meta-analysis, we propose that IgA in serum has an important role in the early diagnosis of SARS-CoV-2 infection as some papers suggest elsewhere. In a study published by Chen (Huang et al., 2020), the IgA specificity (96.6 %) was higher than the one for IgM or IgG; in another study, Guo et al. (2020) analyzed samples in the early stages of COVID-19 patients finding a 92.7 % of specific SARS-CoV-2 IgA while IgG and IgM were detected in only 77.9% and 85.4%, respectively. Additionally, the study performed by Huang et al. (2020), demonstrated the presence of IgA in the 100% of the analyzed patients becoming more suitable than IgM or IgG in the early diagnosis of SARS-CoV-2 infection.
Here, we found an IgA seroprevalence of 86.47 % in PCR (+) confirmed patients which supports the role of IgA as a diagnostic tool together with IgG, and its possible role in the early diagnosis of COVID-19 as useful as IgM. Some authors also proposed that IgA could be useful in the later stages of COVID-19; the hypothesis for this is that IgA may have a role in bridging the serological gap of the disease (Infantino et al., 2021). The same authors reported that anti-SARS-CoV-2 IgA concentration increases rapidly and is higher than the observed for IgM and IgG in all the analyzed time points of the study. In another report, Orth and coworkers found out a significant increase and a higher detection rate of SARS-CoV-2-specific IgA at the 2nd week after symptoms onset (Orth-Holler et al., 2020) making it detectable earlier than IgG (which appeared until the third week after onset of symptoms). Due to all the data mentioned before, the possible prognostic significance of IgA needs to be clarified (Infantino et al., 2021). We propose that IgA could be used along with IgG for the SARS-CoV-2 diagnostics in long periods after symptomatic onset since (in this review) we found out that IgA can be detected up to 75 days after symptomatic onset.
From our analysis it can be observed that the participation of IgA might be beneficial in COVID − 19 patients because IgA (+) individuals seem to have middle symptoms or they are asymptomatic, however, as we stated before, these effects should be taken with cautious because many of the results from the different analyzed articles come from different assays with different antigens preparations and tested under different conditions. Besides, the patients clinical data, sample taking and time of infection also played as important factors which can influence the outcome of the IgA response. In a study published by Fourati et al, they observed an inverse correlation between the viral load (from nasopharyngeal swabs) and the titers of IgA in serum and patients who died at day-28 displayed significantly lower titers of specific anti-S IgA compared with patients still alive at 28 days (Fourati et al., 2020). In another study, Bransetter et al showed that IgA indicates mucosal protection against the virus infection while IgG is a good indicator for immunological memory, some authors have speculated that in patients not severely affected by a SARS-CoV-2 infection, IgA plays a more prominent role but data on the topic is still inconclusive (Brandstetter et al., 2020). Furthermore, IgA production in severe critical patients has been reported by Rijkers five days after the beginning of the symptomatology in hospitalized COVID-19 patients (along with IgG), however, its role in disease progression remains unclear (Rijkers et al., 2020). We can take into consideration the IgG implication in the protective role of IgA as a synergic protective mechanism, or it could be possible that one of these antibodies has a more important protective role. Moreover, Garcia-Basteiro et al., found out that IgA responses can be detected and peak earlier than IgM and IgG, which is consistent with previous reports (Garcia-Basteiro et al., 2020; Hsueh et al., 2004; Sterlin et al., 2021), however, they did not find correlation with protection because lower IgA levels were found in participants without symptoms (Wilder-Smith et al., 2005). Additionally, Sterlin et al., 2021 reported early SARS-CoV-2–specific humoral responses dominated by IgA antibodies contributing to virus neutralization; virus neutralization is more closely related with IgA rather than IgM or IgG in the first weeks after symptom onset. However they found that early SARS-CoV-2–specific IgA response is not associated with COVID-19 severity (Sterlin et al., 2021). We proposed that IgA might be more useful in combination with IgG instead of the IgG-IgM mix as the same authors (Garcia-Basteiro et al., 2020) found that all severe and critical COVID-19 patients had higher IgA and IgG levels, while only some severe and critical patients had elevated levels of IgM.
Associations between nasal IgA responses, virus neutralization at the mucosa, and less severe disease suggest the importance of assessing mucosal immunity in larger natural infection cohorts.
Further studies must be directed to address whether differences in functions may exist between serum and mucosal IgA responses. This is important to clearly establish the precise roles of mucosal versus systemic IgA responses in protection or in immunopathology in COVID-19.
CRediT authorship contribution statement
VRR collected and analyzed the data and helped writing the manuscript, KAMP compiled and analyzed the data, RRSG actualized the data and organized the tables and figures, DRAA helped with the critical revision of the manuscript and data, LMF performed the final revision and the final approval of the manuscript, NRI designed the study, revised the data, helped write the manuscript, performed the final revision and the final approval of the manuscript.
Declaration of Competing Interest
All authors have nothing to disclose.
Acknowledgments
The authors wish to acknowledge the financial support of Tecnológico de Monterrey, CONACYT (Estancias Posdoctorales Vinculadas al Fortalecimiento de la Calidad del Posgrado Nacional 2019–1, Grant: 740975 and 769256 to NRI).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.micres.2022.127105.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
References
- Bidgood S.R., Tam J.C., McEwan W.A., Mallery D.L., James L.C. Translocalized IgA mediates neutralization and stimulates innate immunity inside infected cells. Proc. Natl. Acad. Sci. USA. 2014;111(37):13463–13468. doi: 10.1073/pnas.1410980111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstetter S., Roth S., Harner S., Buntrock-Dopke H., Toncheva A.A., Borchers N., Kabesch M. Symptoms and immunoglobulin development in hospital staff exposed to a SARS-CoV-2 outbreak. Pediatr. Allergy Immunol. 2020;31(7):841–847. doi: 10.1111/pai.13278. [DOI] [PubMed] [Google Scholar]
- Cervia C., Nilsson J., Zurbuchen Y., Valaperti A., Schreiner J., Wolfensberger A., Boyman O. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J. Allergy Clin. Immunol. 2021;147(2):545–557. doi: 10.1016/j.jaci.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of V. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R., Laird N. Meta-analysis in clinical trials revisited. Contemp. Clin. Trials. 2015;45(Pt A):139–145. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourati S., Hue S., Pawlotsky J.M., Mekontso-Dessap A., de Prost N. SARS-CoV-2 viral loads and serum IgA/IgG immune responses in critically ill COVID-19 patients. Intensive Care Med. 2020;46(9):1781–1783. doi: 10.1007/s00134-020-06157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Basteiro A.L., Moncunill G., Tortajada M., Vidal M., Guinovart C., Jimenez A., Dobano C. Seroprevalence of antibodies against SARS-CoV-2 among health care workers in a large Spanish reference hospital. Nat. Commun. 2020;11(1):3500. doi: 10.1038/s41467-020-17318-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J., Bajaj D., Sankaramangalam K., Yoo J.W., Joshi N.S., Gettinger S., Farrell J.J. Incidence of pancreatitis with the use of immune checkpoint inhibitors (ICI) in advanced cancers: a systematic review and meta-analysis. Pancreatology. 2019;19(4):587–594. doi: 10.1016/j.pan.2019.04.015. [DOI] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., Xiao M., Chang, Yang F., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10(12):1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Chen H., Xue M., Huang H., Zheng P., Luo W., Zhong N. Characteristics and roles of severe acute respiratory syndrome coronavirus 2-specific antibodies in patients with different severities of coronavirus 19. Clin. Exp. Immunol. 2020;202(2):210–219. doi: 10.1111/cei.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infantino M., Manfredi M., Grossi V., Lari B., Fabbri S., Benucci M., Pesce G. Closing the serological gap in the diagnostic testing for COVID-19: the value of anti-SARS-CoV-2 IgA antibodies. J. Med. Virol. 2021;93(3):1436–1442. doi: 10.1002/jmv.26422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layward L., Finnemore A.M., Allen A.C., Harper S.J., Feehally J. Systemic and mucosal IgA responses to systemic antigen challenge in IgA nephropathy. Clin. Immunol. Immunopathol. 1993;69(3):306–313. doi: 10.1006/clin.1993.1185. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gotzsche P.C., Ioannidis J.P., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339 doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestecky J., Russell M.W., Jackson S., Brown T.A. The human IgA system: a reassessment. Clin. Immunol. Immunopathol. 1986;40(1):105–114. doi: 10.1016/0090-1229(86)90073-5. [DOI] [PubMed] [Google Scholar]
- Neyeloff J.L. Meta-analyses and Forest plots using a microsoft excel spreadsheet: step-by-step guide focusing on descriptive data analysis. BMC Res. Notes. 2012;5:52. doi: 10.1186/1756-0500-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth-Holler D., Eigentler A., Weseslindtner L., Most J. Antibody kinetics in primary- and secondary-care physicians with mild to moderate SARS-CoV-2 infection. Emerg. Microbes Infect. 2020;9(1):1692–1694. doi: 10.1080/22221751.2020.1793690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers G., Murk J.L., Wintermans B., van Looy B., van den Berge M., Veenemans J., Reimerink J. Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J. Infect. Dis. 2020;222(8):1265–1269. doi: 10.1093/infdis/jiaa463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh-Mohamed S., Isho B., Chao G.Y.C., Zuo M., Cohen C., Lustig Y., Gommerman J.L. Systemic and mucosal IgA responses are variably induced in response to SARS-CoV-2 mRNA vaccination and are associated with protection against subsequent infection. Mucosal Immunol. 2022 doi: 10.1038/s41385-022-00511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa-Pereira P., Woof J.M. IgA: structure, function, and developability. Antibodies. 2019;8(4) doi: 10.3390/antib8040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claer L., Gorochov G. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13(577) doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Smith A., Teleman M.D., Heng B.H., Earnest A., Ling A.E., Leo Y.S. Asymptomatic SARS coronavirus infection among healthcare workers, Singapore. Emerg. Infect. Dis. 2005;11(7):1142–1145. doi: 10.3201/eid1107.041165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki N., Mohamed E.A. The estimations of the COVID-19 incubation period: a scoping reviews of the literature. J. Infect. Public Health. 2021;14(5):638–646. doi: 10.1016/j.jiph.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.