ABSTRACT
Acinetobacter baumannii is an opportunistic pathogen of growing concern, as isolates are commonly multidrug resistant. While A. baumannii is most frequently associated with pulmonary infections, a significant proportion of clinical isolates come from urinary sources, highlighting its uropathogenic potential. The type II secretion system (T2SS) of commonly used model Acinetobacter strains is important for virulence in various animal models, but the potential role of the T2SS in urinary tract infection (UTI) remains unknown. Here, we used a catheter-associated UTI (CAUTI) model to demonstrate that a modern urinary isolate, UPAB1, requires the T2SS for full virulence. A proteomic screen to identify putative UPAB1 T2SS effectors revealed an uncharacterized lipoprotein with structural similarity to the intimin-invasin family, which serve as type V secretion system (T5SS) adhesins required for the pathogenesis of several bacteria. This protein, designated InvL, lacked the β-barrel domain associated with T5SSs but was confirmed to require the T2SS for both surface localization and secretion. This makes InvL the first identified T2SS effector belonging to the intimin-invasin family. InvL was confirmed to be an adhesin, as the protein bound to extracellular matrix components and mediated adhesion to urinary tract cell lines in vitro. Additionally, the invL mutant was attenuated in the CAUTI model, indicating a role in Acinetobacter uropathogenesis. Finally, bioinformatic analyses revealed that InvL is present in nearly all clinical isolates belonging to international clone 2, a lineage of significant clinical importance. In all, we conclude that the T2SS substrate InvL is an adhesin required for A. baumannii uropathogenesis.
KEYWORDS: Acinetobacter, adhesin, infection, invasin, pathogenesis, type II secretion system, urinary tract infection, virulence
INTRODUCTION
Acinetobacter spp. belonging to the Acinetobacter calcoaceticus–Acinetobacter baumannii (ACB) complex are opportunistic pathogens that cause diverse infections, with A. baumannii representing the most common species associated with human infection (1–3). While A. baumannii infections can be both community and hospital acquired, nosocomial infections in critically ill patients are common, with mortality rates reported as high as 84.3% in individuals requiring mechanical ventilation (4–7). These high mortality rates can be largely attributed to increasing multidrug resistance among A. baumannii isolates (6, 8). In fact, A. baumannii exhibits the highest prevalence of multidrug resistance among Gram-negative pathogens, leading the World Health Organization to classify the bacterium as a highest priority pathogen for research and development of new treatments (8, 9). The respiratory tract is the most common site of infection by A. baumannii, with approximately 40% of the isolates derived from pulmonary infections (2, 3, 10, 11). It should be noted though that 20% of A. baumannii isolates come from urinary tract infections (UTIs), highlighting the bacterium’s uropathogenic capacity (10, 11). However, despite the public health relevance of A. baumannii as a uropathogen, few virulence factors required specifically for UTI have been defined (10).
Bacterial virulence relies on the interaction of bacterially derived proteins with the infected host. Gram-negative bacteria have evolved to encode complex secretion systems to transport proteins across the bacterial envelope (12). The type II secretion system (T2SS) is widely distributed in Gram-negative bacteria and has been implicated in the virulence of several pathogens (13–15). T2SS effectors are translocated from the cytoplasm to the outer membrane or into the extracellular milieu in two steps. First, the effector is transported across the inner membrane into the periplasm via the general secretory (Sec) pathway or the twin arginine translocation (TAT) pathway (16, 17). Second, the effector is extruded through the outer membrane secretin (designated GspD herein) via the assembly of a pseudopilus (18–22). In Acinetobacter, the T2SS is responsible for secretion of several proteins, including the lipases LipA, LipH, and LipAN, the protease CpaA, and the γ-glutamyltransferase GGT (23–28). For A. baumannii strain ATCC 17978, a meningitis isolate from 1951, mutation of the T2SS results in attenuation in bacteremia and pneumonia models of infection (23, 29). Additionally, mutation of the T2SS in Acinetobacter nosocomialis strain M2 and A. baumannii strain AB5075, musculoskeletal isolates from 1996 and 2008, respectively, results in decreased bacterial burden in a pneumonia model of infection (24, 28). Furthermore, the Acinetobacter effectors LipAN of strain ATCC 17978 and CpaA of strain M2 are required for full virulence in a pneumonia model, and GGT of strain AB5075 is required for full virulence in a bacteremia model (23, 27, 28). While the T2SS and associated effectors clearly have roles in pathogenesis in multiple models of infection, the potential function of the T2SS in Acinetobacter uropathogenesis has not been investigated. Additionally, T2SS-dependent secretomes can differ significantly between strains, and these dissimilarities can lead to strain-dependent differences in virulence potential mediated by the T2SS (24, 28, 29). Therefore, evaluation of the T2SS in diverse clinical isolates could reveal previously unrecognized effectors that may serve as novel Acinetobacter virulence factors.
Intimate interaction of bacteria with host tissues at the site of infection is often required for pathogenesis. Indeed, adherence to and/or invasion of host epithelial cells, mediated by bacterial adhesins, is an essential process for the virulence of several pathogens (30). Many adhesins are displayed on the bacterial surface in a type V secretion system (T5SS)-dependent manner (31). These T5SSs, also termed autotransporters (ATs), are proteins consisting of two general components, a β-barrel domain, which is inserted into the outer membrane, and a passenger domain, which uses the β-barrel for transportation to the outer surface of the bacteria (32–34). The intimin-invasin family of ATs specifically are adhesins that consist of an N-terminal β-barrel domain and a C-terminal passenger domain containing multiple immunoglobulin (IG)-like domains often capped by a lectin-like domain (35, 36). Intimin is encoded by many pathogens such as Escherichia coli and functions by binding to a type III secretion system (T3SS) effector, translocated intimin receptor (TIR), which is inserted into the host membrane (37, 38). Invasin (InvA), encoded by Yersinia enterocolitica and Yersinia pseudotuberculosis, functions by binding directly to host β1-integrins, leading to a host cytoskeletal rearrangement and subsequent internalization of the bacteria (39, 40). Importantly, deletion of genes encoding intimin-invasin family proteins results in attenuation in animal models of infection (35, 36). Despite this key role that intimin-invasin family proteins play in the pathogenesis of other bacteria, potential homologs have not been identified in Acinetobacter.
In this work, we aimed to interrogate the function of the T2SS in the recent A. baumannii urinary clinical isolate, UPAB1, using a murine catheter-associated UTI (CAUTI) model and define effectors potentially required for virulence (10). In this pursuit, we identify a novel T2SS effector/intimin-invasin family protein important for Acinetobacter uropathogenesis.
RESULTS
The UPAB1 T2SS mutant is attenuated in the CAUTI model.
To test the role of the T2SS in uropathogenesis, we generated a UPAB1 mutant strain lacking gspD, the gene encoding the outer membrane secretin that is essential for T2SS function. This strain was subsequently employed in the murine CAUTI model as previously described (10). Briefly, a small piece of silicon tubing (catheter) was inserted into the urethra, and mice were transurethrally inoculated with wild-type (WT), ΔgspD, or complemented (gspD+) strains. At 24 h postinfection, mice were sacrificed, and bacteria adhered to the catheters (Fig. 1A) and bacterial burdens in the bladders (Fig. 1B) were quantified. The ΔgspD mutant exhibited decreased binding to the catheter and colonization of the bladder, with approximately 10-fold fewer CFU recovered relative to WT. The catheter binding phenotype was completely restored by genetic complementation, and the complemented strain exhibited a trend toward higher bladder burden relative to the mutant as the P value approached significance. These results indicate that the T2SS plays a role in Acinetobacter uropathogenesis.
FIG 1.
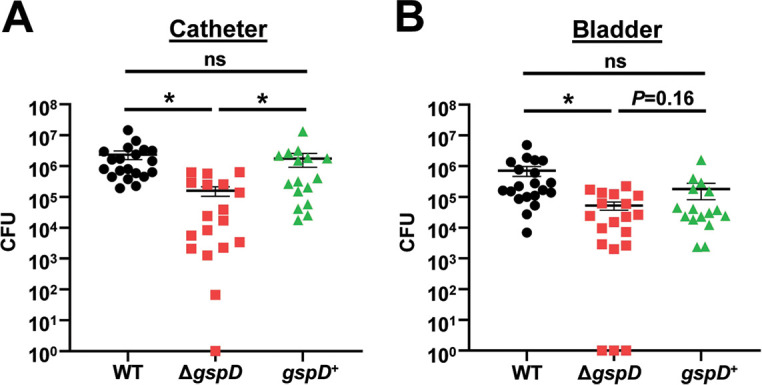
The T2SS is required for full virulence in a murine CAUTI model. Mice were implanted with a catheter followed by transurethral inoculation with UPAB1 WT, ΔgspD, or gspD+ strains. At 24 h postinfection, mice were sacrificed, and bacterial burdens on the catheter (A) and in the bladder (B) were quantified. Shown are results from at least three pooled experiments. Each data point represents an individual mouse, the horizontal line represents the mean, and the standard error of the mean (SEM) is indicated by error bars. *, P < 0.05; two-tailed Mann-Whitney U test; ns, not significant.
A proteomic analysis identifies a putative T2SS effector with structural similarity to Yersinia invasin.
To identify T2SS effectors in UPAB1, we performed a proteomic analysis of the supernatant of WT and ΔgspD strains. Proteins with decreased abundance with the mutant strain relative to WT were considered putative T2SS effectors (see Table S1 in the supplemental material). As expected, we identified orthologs of proteins found in previous Acinetobacter T2SS effector screens, including the most differentially identified protein, a 5′-methylthioadenosine (MTA)/S-adenosylhomocysteine (SAH) nucleosidase, as well as lipases, a CSLREA domain-containing protein, GGT, and the glycoprotease CpaA (24, 28, 29). However, we additionally identified several proteins not found in previous analyses, several of which were annotated as hypothetical proteins of unknown function.
Putative T2SS-dependent secretome of UPAB1. Download Table S1, DOCX file, 0.04 MB (37.9KB, docx) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
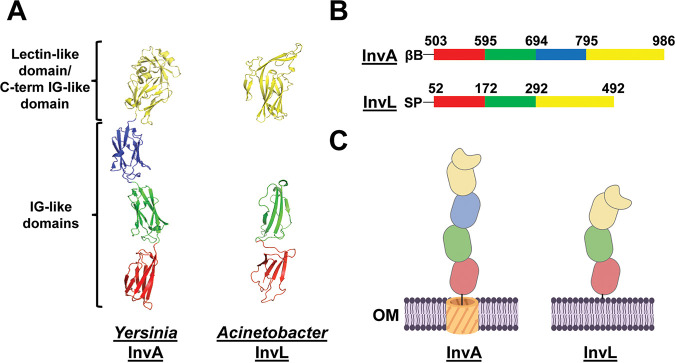
The most differentially abundant protein of unknown function in the supernatant of WT and ΔgspD strains was D1G37_RS04395. D1G37_RS04395 is a 492-amino-acid, 51.92-kDa protein predicted by SignalP 5.0 to contain an N-terminal lipoprotein secretion signal (41). Interestingly, a fold recognition analysis of D1G37_RS04395 with Phyre2 revealed the invasin InvA of Y. pseudotuberculosis as the best match (99.7% confidence; 72.0% coverage) (Fig. S1) (42, 43). Submission of D1G37_RS04395 to I-TASSER similarly predicted structural similarity to InvA (2.03 normalized Z score; 77.0% coverage) (44–46). Alphafold2 was then used to generate a model of D1G37_RS04395 to compare to the known structure of InvA (Fig. 2A) (43, 47). D1G37_RS04395 is predicted to have three IG-like domains and is capped with a C-terminal lectin-like domain that is intimately associated with the most C-terminal IG-like domain. This is comparable to the passenger domain of InvA, with the exception that InvA contains an additional IG-like domain (43). A key difference between D1G37_RS04395 and InvA, however, is that InvA is an autotransporter (T5SS) attached to the membrane by an N-terminal β-barrel domain, whereas D1G37_RS04395 is a putative T2SS effector predicted to be attached to the membrane as a lipoprotein (Fig. 2B and C) (35, 36). Of note, we performed alignments of the C-terminal 440 amino acids of InvL and 483 amino acids of InvA using LALIGN software, as these were the regions of predicted homology from our structural analysis. This revealed an overlap of 285 amino acids with 22.5% identity and 51.2% similarity. It should be noted though that the IG-like and lectin domains of the intimin-invasin family fold similarly but have little sequence identity (36, 48). Therefore, extensive similarity at the amino acid sequence level between the proteins was not expected. Overall, given specifically the predicted structural similarity of D1G37_RS04395 to the passenger domain of InvA, we will refer to this protein herein as InvL (invasin-like protein).
FIG 2.
InvL has predicted structural similarity to InvA of Yersinia. (A) The crystal structure of the passenger domain of Yersinia InvA (PDB entry 1CWV) is shown on the left, and the AlphaFold2 predicted structure of the analogous region of InvL is pictured on the right. (B) Amino acids corresponding to subdomains of the passenger domain of InvA and the predicted homologous region of InvL. (C) Graphic depiction of the structure of InvA and the predicted structure of InvL; created with BioRender.com. Red, green, and blue colors denote individual IG-like domains, and yellow denotes the C-terminal IG-like domain in intimate contact with the lectin-like domain. βB, β-barrel domain; SP, signal peptide; OM, outer membrane.
Phyre2 predicts that D1G37_RS04395 has structural similarity to Yersinia InvA. Results from the Phyre2 analysis of the amino acid sequence of full-length D1G37_RS04395 are shown. Download FIG S1, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
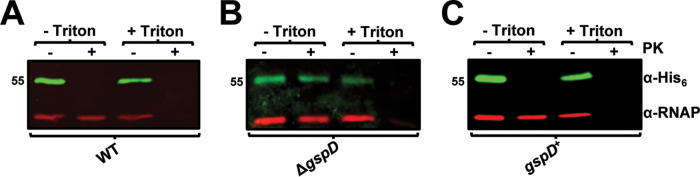
The T2SS is required for InvL secretion and surface exposure.
To confirm that secretion of InvL is dependent on a functional T2SS, WT, ΔgspD, and gspD+ strains were transformed with a plasmid expressing invL with a His6 tag (pBAV-Apr::invL-his6). Whole-cell lysate and supernatant fractions from the bacteria were then assessed by immunoblotting for the presence of InvL-His6 (Fig. 3). InvL was found in the supernatant of WT and gspD+ strains, whereas the ΔgspD mutant failed to secrete the protein. This confirmed that InvL is a T2SS substrate. Immunoblot analyses demonstrated that, in addition to being released into the supernatant, InvL was cell associated (Fig. 3). As intimin-invasin family proteins function as surface-localized adhesins, we hypothesized that cell-associated InvL is surface localized. To test this, proteinase K susceptibility assays wherein surface-exposed proteins are degraded and intracellular proteins are left intact were performed with bacteria expressing invL-his6. In WT UPAB1, InvL-His6 was completely degraded with proteinase K treatment, whereas the intracellular control, RNA polymerase (RNAP), was not (Fig. 4A). On the contrary, treatment with Triton X-100 and proteinase K resulted in degradation of both InvL-His6 and RNAP. These results indicate that InvL is surface localized, consistent with its putative role as an adhesin. Interestingly, proteinase K failed to degrade InvL-His6 expressed in the ΔgspD mutant, indicating the protein did not reach the surface of the bacteria (Fig. 4B). Alternatively, InvL-His6 was readily degraded by proteinase K treatment of gspD+ (Fig. 4C), demonstrating that the T2SS is required for InvL surface localization.
FIG 3.
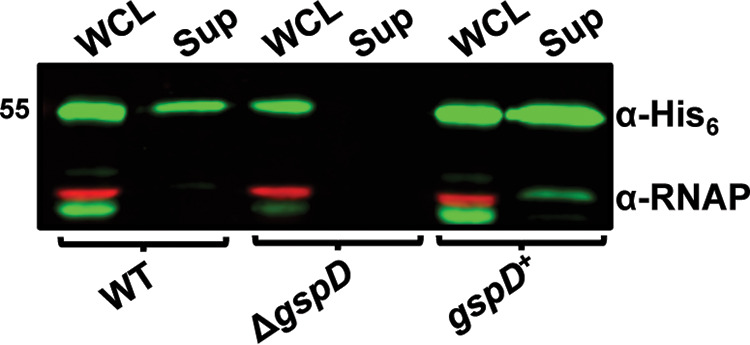
The T2SS is required for InvL secretion. Whole-cell lysate (WCL) and supernatant (Sup) fractions from WT, ΔgspD, and gspD+ UPAB1 cultures harboring pBAV-Apr::invL-his6 were probed for InvL (α-His6) and RNAP by immunoblotting. Numbers to the left indicate molecular weight in kDa. At least two biological replicates were performed, yielding similar results, and a representative blot from one replicate is shown.
FIG 4.
The T2SS is required for InvL surface localization. WT (A), ΔgspD (B), and gspD+ (C) UPAB1 cells harboring pBAV-Apr::invL-his6 were treated with proteinase K (PK; +) or left untreated (−) in the presence or absence of Triton X-100. Cells were then probed for InvL (α-His6) and RNAP by immunoblotting. Numbers to the left indicate molecular weight in kDa. At least two biological replicates were performed, yielding similar results, and a representative blot from one replicate is shown.
As InvL is predicted to be a lipoprotein, its identification in the supernatant is somewhat surprising. To further examine the form in which InvL is secreted, insoluble and soluble fractions from the supernatant of WT UPAB1 expressing invL-his6 were separated by ultracentrifugation. Immunoblot analysis of these fractions revealed that InvL was primarily found in the insoluble fraction (Fig. S2). This localization could be due to the presence of InvL in either lysis by-products or possibly outer membrane vesicles (OMVs). The physiological relevance of OMVs in Acinetobacter pathobiology remains to be investigated and will be the focus of subsequent work.
InvL localizes primarily to the insoluble fraction of bacterial supernatant. WT UPAB1 harboring pBAV::invL-his6 was grown to early stationary phase, and WCL, sup, sup insoluble, and sup soluble fractions were isolated. Fractions were then probed for InvL (α-His6) and RNAP by immunoblotting. Numbers to the left indicate molecular weight in kDa. Two biological replicates were performed, yielding similar results, and a representative blot from one replicate is shown. Download FIG S2, TIF file, 0.9 MB (965.9KB, tif) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
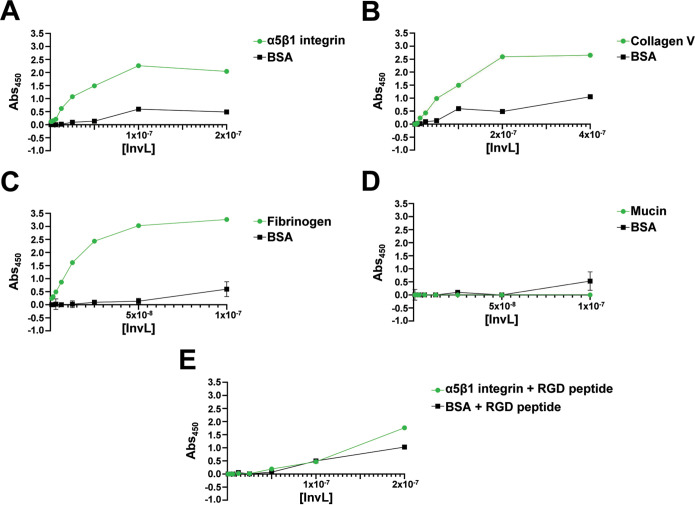
InvL binds to ECM components.
We next examined whether InvL could bind to extracellular matrix (ECM) components. First, we used enzyme-linked immunosorbent assays (ELISAs) to investigate if InvL binds to α5β1 integrin (Fig. 5A) and collagen V (Fig. 5B). These molecules are the binding partners of InvA and an E. coli homolog, FdeC, respectively (39, 49, 50). We determined that InvL bound to α5β1 integrin and collagen V with dissociation constants (Kds) of 0.38 nM and 15 μM, respectively. On the contrary, InvL did not appreciably bind the negative control, bovine serum albumin (BSA), at the tested concentrations. Since UPAB1 colocalizes with fibrinogen deposited on the catheter during CAUTI, we assessed if InvL interacts with fibrinogen (10). We found that InvL bound to fibrinogen with the highest affinity of all ECM components tested (Kd = 0.19 nM) (Fig. 5C). InvL, similar to InvA and FdeC, did not exhibit any significant binding to mucin, another common ECM component (Fig. 5D).
FIG 5.
InvL binds to multiple ECM components. ELISAs were performed to assess binding of InvL to α5β1 integrin (A), collagen V (B), fibrinogen (C), mucin (D), or α5β1 integrin in the presence of RGD-containing peptide (E). BSA served as a negative binding control. Shown are the results from two biological replicates, and SEM is indicated by error bars.
The interaction between InvA and β1 integrins is competitively inhibited by arginine-glycine-aspartate (RGD)-containing peptides, a motif required for the interaction of fibronectin with β1 integrin (51–53). RGD-containing peptides also abrogated binding of InvL to α5β1 integrin (Fig. 5E), indicating that InvL and InvA bind β1 integrins via similar mechanisms. Together, these results demonstrate that InvL binds α5β1 integrin, collagen V, and fibrinogen in vitro.
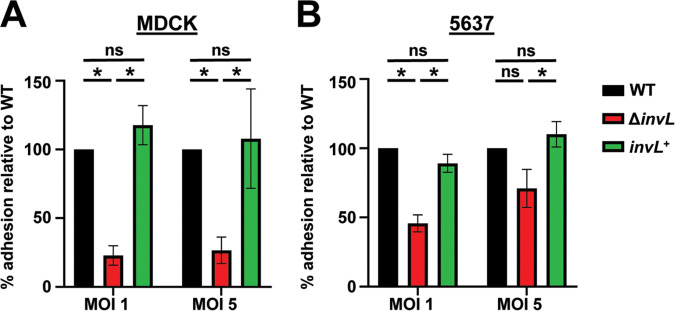
InvL facilitates UPAB1 adhesion to bladder and kidney epithelial cells.
To examine if InvL plays a role in binding to epithelial cells relevant to the CAUTI model, we determined adhesion of UPAB1 WT, ΔinvL, and invL+ strains to kidney (MDCK; Fig. 6A) and bladder (5637; Fig. 6B) cells. The ΔinvL strain exhibited significantly decreased adhesion to both cell types relative to WT at a multiplicity of infection (MOI) of 1, and a similar trend toward decreased binding by the ΔinvL strain was observed at an MOI of 5. This phenotype was reversed in the invL+ strain, confirming a role for InvL in urinary tract epithelial adhesion.
FIG 6.
InvL facilitates adhesion to epithelial cells in vitro. UPAB1 WT, ΔinvL, and invL+ strains were used in adhesion assays with MDCK (A) and 5637 (B) epithelial cells. The mean from six replicates is shown, and error bars represent the SEM. *, P < 0.05; one-way ANOVA, Tukey’s test for multiple comparisons.
Because Yersinia InvA can mediate epithelial cell invasion, we performed antibiotic protection assays to assess if InvL performs a similar function (54–58). However, internalization was not observed in any of the cell lines tested, and immunofluorescence assays following epithelial cell infection confirmed UPAB1 is not invasive (Fig. S3). These results are not surprising given that few publications exist regarding Acinetobacter internalization in epithelial cells, and internalization results are highly variable depending on the strain and cell line tested (59–64). Altogether, our results demonstrate a role for InvL in binding to diverse epithelial cell lines, but a possible role in invasion in other strains or cells types cannot be ruled out.
A. baumannii is adhered extracellularly to 5637 epithelial cells. 5637 cells were infected with WT UPAB1 at an MOI of one, incubated for one h, and immunofluorescence microscopy was used to visualize intracellular versus extracellular bacteria. Nuclei are stained blue, actin is stained red, intracellular bacteria are stained green (rarely observed), and extracellular bacteria are double stained green and far-red. Two biological replicates were examined, and a representative image from one biological replicate is shown. The scale bar equals 20 μm. Download FIG S3, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
InvL is required for full virulence in the CAUTI model.
Given the role of InvL as an adhesin, we tested if InvL contributes to Acinetobacter virulence in the murine CAUTI model. Mice were implanted with a catheter followed by transurethral inoculation with WT, ΔinvL, or invL+ strains. At 24 h postinfection, mice were sacrificed, and bacteria adhered to the catheters (Fig. 7A) and bacterial burdens in the bladders (Fig. 7B) were quantified. The ΔinvL mutant had a significant reduction in catheter binding, exhibiting approximately a 100-fold decrease relative to the WT strain, and this phenotype was partially reversed by genetic complementation. ΔinvL also had a significant defect in bladder colonization relative to the WT strain, with more than 10-fold reduced CFU recovered. This phenotype was also partially complemented with the invL+ strain. The reason for partial complementation in the CAUTI model is unknown, but this result could be due to possible differential expression of invL in the complemented strain relative to WT bacteria. Nevertheless, these results indicate an important role for InvL in A. baumannii uropathogenesis.
FIG 7.
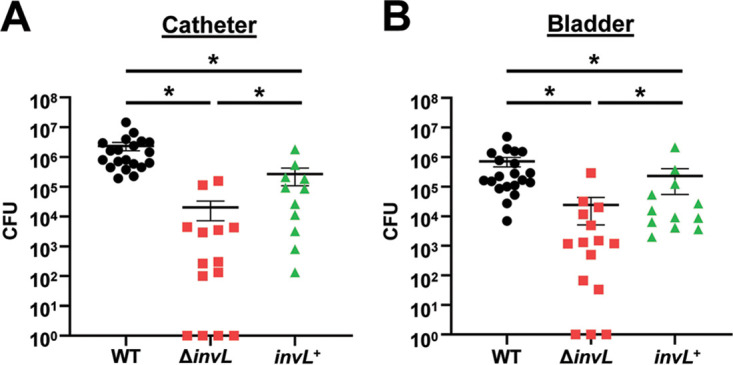
InvL is required for full virulence in a murine CAUTI model. Mice were implanted with a catheter followed by transurethral inoculation with UPAB1 WT, ΔinvL, or invL+ strains. At 24 h postinfection, mice were sacrificed and bacterial burden on the catheter (A) and in the bladder (B) were quantified. Shown are results from at least three pooled experiments. Each data point represents an individual mouse, the horizontal line represents the mean, and the SEM is indicated by error bars. *, P < 0.05; two-tailed Mann-Whitney U test.
InvL is encoded by ACB complex strains and has high prevalence among international clone 2 isolates.
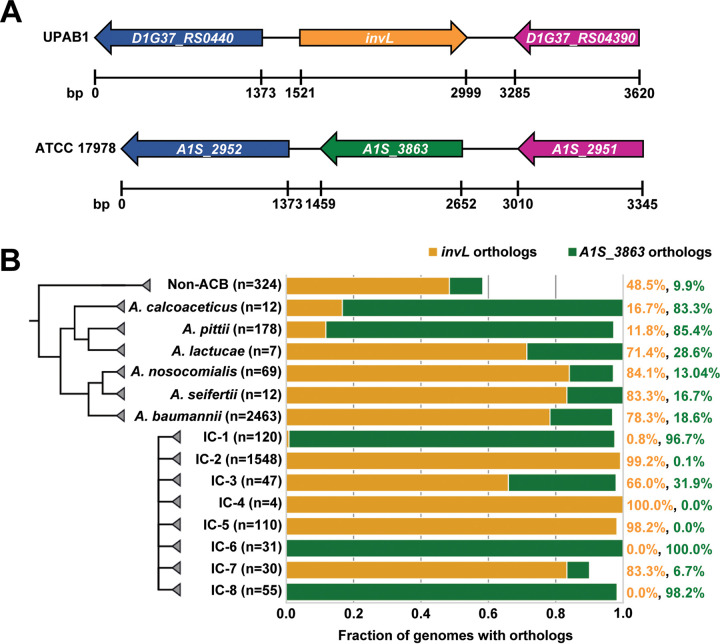
As virulence factors could differ among isolates, we assessed the distribution of invL across the Acinetobacter genus. Specifically, we performed a targeted ortholog search for invL and adjacent genes in 3,052 available genomes from the National Center for Biotechnology Information (NCBI) RefSeq Database (65). In genomes that encode InvL, the corresponding gene is flanked by genes encoding a TIGR01244 family phosphatase (D1G37_04390 in UPAB1) and a dihydrolipoyl dehydrogenase (D1G37_RS04400 in UPAB1) (Fig. 8A). While the microsynteny of the two flanking genes is conserved throughout ACB complex, only 74% (2,029/2,728) of the isolates additionally harbor the invL gene. In 23% (641/2,728), a second cluster variant exists, where the invL gene is replaced by a gene encoding a hypothetical protein (A1S_3863 in ATCC 17978). The remainder of isolates (3%; 58/2728) have no/another intervening gene. The gene represented in the second cluster variant does not appear to be homologous to InvL. First, the two proteins reside on opposite strands, which interferes with genetic exchange via bacterial recombination. Second, at the amino acid sequence level, the two proteins are only 19% identical and 31% similar, and this is substantially driven by the shared presence of an N-terminal signal peptide. Third, the length and domain architecture differ between the proteins; InvL is about 100 amino acids longer, and it harbors an invasin/intimin cell-adhesion domain (IPR008964) that is missing in the other protein. Outside the ACB complex, the gene cluster is represented in only 216/324 (67%) of the isolates. Of these, again the majority encode InvL (157/216; 72%). The second cluster variant is present in 32/216 (15%) of the isolates, and the remaining 13% have either no or another intervening gene. Of note, InvL distribution correlates with international clone (IC) number, as nearly all strains belonging to IC-2, IC-4, and IC-5 groups encode InvL (Fig. 8B). Alternatively, strains belonging to IC-1, IC-6, and IC-8 predominantly encode the second cluster variant. As IC-2 clones are among the most commonly isolated in clinical settings, the prevalence of InvL in these strains is intriguing (1–3, 66, 67). However, the fact that other international clones that are associated with human infection do not predominantly encode InvL (e.g., IC-1) implies that some other protein(s) compensates for the absence of this adhesin. In sum, we conclude that InvL is prevalent in the ACB complex and is found in nearly all IC-2 strains, which are relevant to human disease.
FIG 8.
InvL is encoded by ACB complex strains and has high prevalence in IC-2 clones. (A) InvL is encoded at a common locus in ACB complex strains. Orthologs of InvL (top) and orthologs of A1S_3863 (bottom) are encoded at a common locus between genes encoding a dihydrolipoyl dehydrogenase (D1G37_RS0440 in UPAB1 and A1S_2952 in ATCC 17978; blue) and a TIGR01244 family phosphatase (D1G37_RS04390 in UPAB1 and A1S_2951 in ATCC 17978; purple). (B) Distribution of invL and A1S_3863 in Acinetobacter. The fractions of genomes encoding InvL and A1S_3863 are reported by ACB species and international clone number. Percentage of genomes containing each ortholog by species and international clone number are indicated to the right.
DISCUSSION
Acinetobacter has emerged as a pathogen of significant clinical importance due to the increasing frequency of multidrug-resistant strains identified (6, 8, 9). Approximately 20% of isolates are derived from the urinary tract, highlighting the uropathogenic potential of A. baumannii (10, 11). Mutation of the T2SS in the uropathogenic strain UPAB1 resulted in decreased catheter-binding and lower burden in the bladder, indicative of a key role during urinary tract virulence. Through a proteomic analysis, we identified a surface-exposed T2SS effector that belongs to the intimin-invasin family, referred to here as InvL. We found that recombinant InvL binds to host ECM components and that the UPAB1 ΔinvL mutant strain has a significant defect in binding to bladder and kidney epithelial cells. Accordingly, the ΔinvL mutant was attenuated in the CAUTI model. In all, our study demonstrates the importance of the T2SS for Acinetobacter uropathogenesis and describes a novel T2SS effector, InvL, as a previously unrecognized adhesin and virulence factor that is encoded by most clinical isolates.
Interestingly, although protein modeling revealed structural similarity of InvL to Yersinia InvA, InvL is unique compared to other invasin homologs. First, while canonical invasin homologs are T5SSs, outer membrane localization of InvL is dependent on the T2SS. To our knowledge, this is the first report of an intimin-invasin family T2SS effector. Second, unlike other InvA homologs, which are anchored to the outer membrane via a large N-terminal β-barrel domain, InvL is predicted to be a lipoprotein based on the presence of a lipoprotein secretion signal (36, 41). While the T2SS is known to secrete soluble effectors into the extracellular milieu, it has also been reported to be essential for surface localization of some lipoproteins, similar to what is reported here for InvL (68). One of the best-characterized examples is the T2SS-dependent surface localization of the lipoprotein pullulanase (PulA), a starch-debranching enzyme of Klebsiella oxytoca (69–71). PulA, like many other lipoproteins, is translocated from the cytoplasm to the periplasm via the Sec pathway, where processing and acylation occur to attach the protein to the periplasmic leaflet of the inner membrane (70, 72). Whereas most outer membrane lipoproteins of Gram-negative bacteria are transported from the inner membrane to the outer membrane via the localization of lipoproteins (Lol) pathway, PulA uses the T2SS machinery (69–71). The mechanism by which the T2SS can recognize its lipoprotein effectors and transport lipidated proteins across the periplasm and outer membrane remains elusive. However, this report of InvL adds to the growing list of known T2SS effectors which are surface exposed lipoproteins (68). As several of these proteins are involved in pathogenesis and key metabolic functions (e.g., SslE of E. coli and MtrC and OmcA of Shewanella oneidensis), a better understanding of the mechanism of T2SS-dependent lipoprotein transport is needed (68, 73–78).
Interestingly, in addition to being a surface-exposed protein, we found that InvL is secreted into the supernatant. This is somewhat surprising, as this has not been reported with other members of the intimin-invasin family to our knowledge. Given that InvL localized to the insoluble fraction of the supernatant, it is possible that InvL is found in OMVs. Notably, this release of a T2SS-dependent surface-associated lipoprotein into the media is reminiscent of PulA (79, 80). However, it is unclear if PulA is associated with OMVs, and we cannot discount the possibility that the identification of PulA and InvL in the supernatant is due to their localization in bacterial lysis byproducts (68). Regardless, it is tempting to speculate that secreted InvL could serve an alternative function (e.g., modulation of host responses). However, whether InvL serves any roles other than being an adhesin is outside the scope of this manuscript and will be the subject of future work.
Similar to other members of the intimin-invasin protein family, InvL can directly bind to ECM components and facilitate cell adhesion. Specifically, InvL binds with high affinity to α5β1 integrin, collagen V, and fibrinogen. α5β1 integrin and collagen V are also bound by the intimin-invasin family proteins InvA and FdeC of Yersinia and E. coli, respectively (39, 50). Interestingly though, InvA and FdeC appear to bind to a single defined ECM component, indicating that InvL has a broader specificity. While domains/residues required for the FdeC-collagen interaction have not been defined, the interaction between InvA and β1 integrins has been extensively studied (49, 81–85). Binding of InvA to β1 integrins specifically requires the C-terminal lectin-like domain, which is present in InvL and absent from FdeC (50, 83, 85). It is tempting to speculate that the predicted InvL lectin-like domain is involved in binding to α5β1 integrin and that characteristics of Ig-like domains are responsible for recognition of collagen V. However, it should be noted that residues known to be important for the interaction between InvA and β1 integrins do not appear to be conserved in InvL (81–84). This is despite the observation that the interactions of InvL and InvA with α5β1 integrin can be similarly inhibited by RGD peptide (51–53). The low sequence identity between InvL and invasin homologs makes it difficult to accurately predict key regions of the protein required for recognition of ECM components. The structural characterization of InvL will, in the future, provide mechanistic insights into the interaction between InvL and the ECM.
Our characterization of InvL adds to the growing list of known Acinetobacter adhesins that facilitate binding to epithelial cells, which includes type IV pili, the Ata autotransporter, the FhaB/FhaC two-partner secretion system, and biofilm-associated protein (Bap) (86–96). ECM components bound by type IV pili and Bap are not known. However, Ata has been shown to bind with high affinity to collagen (types I, III, IV, and V), laminin, and glycosylated fibronectin, and FhaBC reportedly binds to fibronectin (88, 91, 92, 95). While the collagen-binding role of InvL may be redundant with that of Ata, InvL is the first published Acinetobacter adhesin to bind directly to α5β1 integrin or fibrinogen. Additionally, InvL is now the first known adhesin to be important for A. baumannii UTIs. Future work will determine the roles of these known adhesins in the multiple niches colonized by this bacterium.
While we have established that InvL is a virulence factor involved in Acinetobacter uropathogenesis, the mechanism(s) by which InvL participates in CAUTI is unknown. The fact that we identified fibrinogen as the highest-affinity binding partner for InvL is particularly intriguing with respect to CAUTI. Tissue damage from catheterization results in an inflammatory response that leads to increased levels of fibrinogen in the bladder that ultimately coats the catheter (97, 98). We recently showed that Acinetobacter, similar to other uropathogens such as Enterococcus faecalis and Staphylococcus aureus, colocalizes with fibrinogen on the catheter (10, 99, 100). This interaction is dependent on the virulence factors EbpA and ClfB in E. faecalis and S. aureus, respectively (98, 99). It is tempting to speculate that InvL serves a similar function with respect to Acinetobacter catheter binding. However, future work is required to examine if InvL mediates catheter binding via fibrinogen or if the ability of InvL to associate with other ECM components is responsible for its role in the CAUTI model. Additionally, future work is required to determine if InvL similarly serves as a virulence factor in other infection models (e.g., pneumonia and bacteremia). Ultimately, a better understanding of A. baumannii virulence factors such as InvL may aid in the development of novel therapeutics to combat infection by these increasingly multidrug-resistant bacteria.
MATERIALS AND METHODS
Bacterial plasmids, strains, and growth conditions.
Plasmids and strains used in this study are detailed in Table S2 in the supplemental material. Cultures were grown at 37°C using Lennox broth/agar unless otherwise noted. The following antibiotic concentrations were used when appropriate: 10 μg/mL chloramphenicol, 100 μg/mL ampicillin, 30 μg/mL apramycin, or 50 μg/mL zeocin for E. coli and 10 μg/mL chloramphenicol, 50 μg/mL apramycin, 50 μg/mL zeocin, or 300 μg/mL hygromycin B for A. baumannii.
Plasmids and strains used in this study. Download Table S2, DOCX file, 0.06 MB (57.6KB, docx) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation of constructs used in this study.
Plasmids and primers used in this study are detailed in Tables S2 and S3, respectively. Detailed information for construction of plasmids and strains used in the study is provided in the supplemental material.
Murine infection experiments.
Animal experiments were approved by the Washington University Institutional Animal Care and Use Committee. The murine CAUTI model was performed as previously described (10). Briefly, 6- to 8-week-old female C57BL/6 mice (Charles River Laboratories, Wilmington, MA) anesthetized with 4% isoflurane were implanted transurethrally with a 4- to 5-mm piece of silicon tubing (catheter). Once-passaged statically grown bacterial strains were washed twice and resuspended in phosphate-buffered saline (PBS). Mice were then transurethrally inoculated with 50 μL of the bacterial suspension containing 2 × 108 CFU. At 24 h postinfection, mice were sacrificed, and bacterial load on catheters and in bladders were quantified by serial dilution plating.
Proteomic analysis of the T2SS-dependent secretome.
Bacterial culture conditions, secreted protein enrichment, digestion of secretome samples, and liquid chromatography-mass spectrometry analysis of secretome samples were as previously described (101). Details are provided in the supplemental material.
Immunoblot analyses.
Bacterial whole-cell lysate preparation and trichloroacetic acid precipitation of supernatant fractions from mid-exponential cultures was performed as previously described (24, 104). Indicated fractions were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and proteins of interest were probed using polyclonal rabbit anti-His6 (1:2,000; Invitrogen, Waltham, MA) and monoclonal mouse anti-RNAP (1:3,500; BioLegend, San Diego, CA). IRDye-conjugated anti-mouse IgG and anti-rabbit IgG were used as secondary antibodies (1:5,000 for each; LI-COR Biosciences, Lincoln, NE), and blots were visualized with the Odyssey CLx imaging system (LI-COR Biosciences).
Proteinase K accessibility assays.
Proteinase K accessibility assays were performed as previously described, with some modification (105). Stationary-phase cultures were pelleted and resuspended in PBS supplemented with 5 mM MgCl2 to an optical density at 600 nm (OD600) of 2.5. Bacterial suspensions were treated with 200 μg/mL proteinase K (GoldBio, Olivette, MO) or sham treated with an equal volume of deionized water in the presence or absence of 2% Triton X-100 (Sigma-Aldrich, St. Louis, MO) at 37°C for 15 min. Digestion was terminated by addition of 1 mM phenylmethylsulfonyl fluoride (Amresco LLC, Salon, OH), Laemmli buffer was added to a concentration of 1×, and samples were boiled for 10 min. 10-μL samples of these preparations were separated by SDS-PAGE, and immunoblotting was performed as described above.
Expression and purification of recombinant InvL.
To generate recombinant protein for antibody generation, full-length InvL with a C-terminal His10 tag was expressed from pET-22b(+)::invL-his10 in Rosetta-gami 2(DE3) cells (Novagen, Madison, WI) using ZYM-5052 autoinducible medium for 72 h at 20°C (106). Cells were then lysed using a CF1 cell disrupter (Constant Systems Ltd., Daventry, UK). Following cell lysis, recombinant InvL was purified from the insoluble fraction by Ni-nitrilotriacetic acid affinity chromatography as previously described (107). Purified protein was then used by Antibody Research Corporation (St. Peters, MO) to generate polyclonal rabbit antibody.
To generate soluble recombinant InvL for ELISA (described below), InvL lacking the N-terminal signal sequence (as determined by SignalP 5.0) was expressed with a C-terminal His10 tag from pET-22b(+)::invL-his10(-SS) in Rosetta-gami 2(DE3) cells by addition of 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C (41). Cells were then lysed as described above, and recombinant InvL was purified from the soluble fraction using the procedure previously described with the exception that buffers were not supplemented with 0.3% N-lauryl-sarcosine (107).
ELISAs.
ELISAs were based on a previously described protocol (50). Detailed protocol information can be found in the supplemental material.
Eukaryotic cell adhesion assays.
Cell adhesion assays were performed with MDCK and 5637 cells cultured in Dulbecco's Modified Eagle Medium (DMEM) and Roswell Park Memorial Institute 1640 (RPMI-1640) (Gibco, Dublin, Ireland) supplemented with 10% heat-inactivated fetal bovine serum, respectively. 105 MDCK and 3 × 105 5637 cells (amounts determined to form confluent monolayers) were plated in 48-well Nunclon Delta cell culture–treated surface plates (ThermoFisher Scientific) overnight at 37°C with 5% CO2. Stationary-phase A. baumannii strains were pelleted at 5,432 × g for 5 min and resuspended in PBS. Following one more spin, bacteria were resuspended in the appropriate cell culture media, and 500 μL of suspension at the indicated MOI was added to the appropriate wells. Plates were then centrifuged at 200 × g and incubated at 37°C with 5% CO2 for 1 h to allow adhesion. Wells were subsequently washed three times with 500 μL PBS, cells were resuspended in 0.05% Triton X-100 in PBS, and serial dilution plating was performed to quantify bacteria. Concurrently, to quantify intracellular bacteria after the 1 h of incubation and washing, the appropriate cell culture medium supplemented with 50 μg/mL colistin was added for 1 h. After washing in PBS, cells were resuspended in 0.05% Triton X-100 in PBS, and serial dilution plating was performed to quantify bacteria.
Bioinformatic analysis of InvL distribution.
Ortholog searches were performed across a database of 3,052 Acinetobacter genomes obtained from NCBI RefSeq as previously described (65). Briefly, the collection of orthologous groups that resulted from an all versus all ortholog search using OMA was screened to associate the group that harbors a particular protein sequence of interest (108). The sequences of the associated group collectively served to train a model for the targeted ortholog search across the full database of genomes. The proteins of interest comprise the translated sequences of D1G37_RS04395 (InvL), D1G37_RS0440, D1G37_RS04390, and A1S_3863. Since the subset of genomes used by Djahanschiri et al. did not include the strain UPAB1, the orthologs identified in the strain MDR-TJ via a reciprocal best blast hit approach were used to associate orthologous groups (65). The resulting presence/absence matrix together with the genus’ phylogeny served as the input for Vicinator v0.32 (https://github.com/BIONF/Vicinator) to trace the microsynteny of the invL locus based on MDR-TJ and ATCC 17978 as the reference genomes.
Data availability.
The MS data and search results have been deposited into the PRIDE ProteomeXchange Consortium repository and can be accessed using the accession numbers PXD030460 and PXD030491 (102, 103). All other data are provided within the text or supplemental material.
Statistical methods.
All statistical analyses were performed using GraphPad Prism version 9.
Supplemental materials and methods. Download Text S1, DOCX file, 0.1 MB (124.1KB, docx) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S3, DOCX file, 0.08 MB (86.3KB, docx) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This work was supported by funding to M.F.F. (R01AI144120) and C.J.L. (T32AI007172) through the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. This study was additionally supported by a grant by the German Research Foundation (DFG) in the scope of the Research Group FOR2251 “Adaptation and persistence of A. baumannii” (grant EB-285-2/2) to I.E.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We acknowledge the Molecular Microbiology Imaging Facility at Washington University in St. Louis and Wandy Beatty for confocal microscopy assistance. We also thank Siyuan Ding and Carolina Lopez for providing eukaryotic cell lines used in the manuscript.
Contributor Information
Mario F. Feldman, Email: mariofeldman@wustl.edu.
Vaughn S. Cooper, University of Pittsburgh
REFERENCES
- 1.Vijayakumar S, Biswas I, Veeraraghavan B. 2019. Accurate identification of clinically important Acinetobacter spp.: an update. Future Sci OA 5:FSO395. doi: 10.2144/fsoa-2018-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerqueira GM, Peleg AY. 2011. Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63:1055–1060. doi: 10.1002/iub.533. [DOI] [PubMed] [Google Scholar]
- 3.Sarshar M, Behzadi P, Scribano D, Palamara AT, Ambrosi C. 2021. Acinetobacter baumannii: an ancient commensal with weapons of a pathogen. Pathogens 10:387. doi: 10.3390/pathogens10040387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dexter C, Murray GL, Paulsen IT, Peleg AY. 2015. Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology and pathogenesis. Expert Rev Anti Infect Ther 13:567–573. doi: 10.1586/14787210.2015.1025055. [DOI] [PubMed] [Google Scholar]
- 5.Cisneros JM, Rodriguez-Baño J. 2002. Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin Microbiol Infect 8:687–693. doi: 10.1046/j.1469-0691.2002.00487.x. [DOI] [PubMed] [Google Scholar]
- 6.Ibrahim S, Al-Saryi N, Al-Kadmy IMS, Aziz SN. 2021. Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol Biol Rep 48:6987–6998. doi: 10.1007/s11033-021-06690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inchai J, Pothirat C, Bumroongkit C, Limsukon A, Khositsakulchai W, Liwsrisakun C. 2015. Prognostic factors associated with mortality of drug-resistant Acinetobacter baumannii ventilator-associated pneumonia. J Intensive Care 3:9. doi: 10.1186/s40560-015-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giammanco A, Calà C, Fasciana T, Dowzicky MJ. 2017. Global assessment of the activity of tigecycline against multidrug-resistant Gram-negative pathogens between 2004 and 2014 as part of the tigecycline evaluation and surveillance trial. mSphere 2:e00310-16. doi: 10.1128/mSphere.00310-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tacconelli E, WHO Pathogens Priority List Working Group, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 10.di Venanzio G, Flores-Mireles AL, Calix JJ, Haurat MF, Scott NE, Palmer LD, Potter RF, Hibbing ME, Friedman L, Wang B, Dantas G, Skaar EP, Hultgren SJ, Feldman MF. 2019. Urinary tract colonization is enhanced by a plasmid that regulates uropathogenic Acinetobacter baumannii chromosomal genes. Nat Commun 10:2763. doi: 10.1038/s41467-019-10706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagińska N, Cieślik M, Górski A, Jończyk ME. 2021. The role of antibiotic resistant A. baumannii in the pathogenesis of urinary tract infection and the potential of its treatment with the use of bacteriophage therapy. Antibiotics (Basel) 10:281. doi: 10.3390/antibiotics10030281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rapisarda C, Fronzes R. 2018. Secretion systems used by bacteria to subvert host functions. Curr Issues Mol Biol 25:1–42. doi: 10.21775/cimb.025.001. [DOI] [PubMed] [Google Scholar]
- 13.Cianciotto NP. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol 13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology (Reading) 149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- 15.Sandkvist M. 2001. Type II secretion and pathogenesis. Infect Immun 69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pugsley AP, Kornacker MG, Poquet I. 1991. The general protein-export pathway is directly required for extracellular pullulanase secretion in Escherichia coli K12. Mol Microbiol 5:343–352. doi: 10.1111/j.1365-2958.1991.tb02115.x. [DOI] [PubMed] [Google Scholar]
- 17.Voulhoux R, Ball G, Ize B, Vasil ML, Lazdunski A, Wu LF, Filloux A. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J 20:6735–6741. doi: 10.1093/emboj/20.23.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nivaskumar M, Francetic O. 2014. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta 1843:1568–1577. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Vignon G, Köhler R, Larquet E, Giroux S, Prévost MC, Roux P, Pugsley AP. 2003. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J Bacteriol 185:3416–3428. doi: 10.1128/JB.185.11.3416-3428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchik VE, Robert-Baudouy J, Condemine G. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J 16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson TL, Abendroth J, Hol WGJ, Sandkvist M. 2006. Type II secretion: from structure to function. FEMS Microbiol Lett 255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 22.Douzi B, Ball G, Cambillau C, Tegoni M, Voulhoux R. 2011. Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates. J Biol Chem 286:40792–40801. doi: 10.1074/jbc.M111.294843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson TL, Waack U, Smith S, Mobley H, Sandkvist M. 2015. Acinetobacter baumannii is dependent on the type II secretion system and its substrate LipA for lipid utilization and in vivo fitness. J Bacteriol 198:711–719. doi: 10.1128/JB.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding CM, Kinsella RL, Palmer LD, Skaar EP, Feldman MF. 2016. Medically relevant Acinetobacter species require a type II secretion system and specific membrane-associated chaperones for the export of multiple substrates and full virulence. PLoS Pathog 12:e1005391. doi: 10.1371/journal.ppat.1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Florencia Haurat M, Scott NE, di Venanzio G, Lopez J, Pluvinage B, Boraston AB, Ferracane MJ, Feldman MF. 2020. The glycoprotease CpaA secreted by medically relevant Acinetobacter species targets multiple O-linked host glycoproteins. mBio 11:e02033-20. doi: 10.1128/mBio.02033-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urusova DV, Kinsella RL, Salinas ND, Haurat MF, Feldman MF, Tolia NH. 2019. The structure of Acinetobacter-secreted protease CpaA complexed with its chaperone CpaB reveals a novel mode of a T2SS chaperone-substrate interaction. J Biol Chem 294:13344–13354. doi: 10.1074/jbc.RA119.009805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elhosseiny NM, Elhezawy NB, Sayed RM, Khattab MS, el Far MY, Attia AS. 2020. γ-Glutamyltransferase as a novel virulence factor of Acinetobacter baumannii inducing alveolar wall destruction and renal damage in systemic disease. J Infect Dis 222:871–879. doi: 10.1093/infdis/jiaa262. [DOI] [PubMed] [Google Scholar]
- 28.Elhosseiny NM, El-Tayeb OM, Yassin AS, Lory S, Attia AS. 2016. The secretome of Acinetobacter baumannii ATCC 17978 type II secretion system reveals a novel plasmid encoded phospholipase that could be implicated in lung colonization. Int J Med Microbiol 306:633–641. doi: 10.1016/j.ijmm.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Elhosseiny NM, Elhezawy NB, Attia AS. 2019. Comparative proteomics analyses of Acinetobacter baumannii strains ATCC 17978 and AB5075 reveal the differential role of type II secretion system secretomes in lung colonization and ciprofloxacin resistance. Microb Pathog 128:20–27. doi: 10.1016/j.micpath.2018.12.039. [DOI] [PubMed] [Google Scholar]
- 30.Niemann HH, Schubert WD, Heinz DW. 2004. Adhesins and invasins of pathogenic bacteria: a structural view. Microbes Infect 6:101–112. doi: 10.1016/j.micinf.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Meuskens I, Saragliadis A, Leo JC, Linke D. 2019. Type V secretion systems: an overview of passenger domain functions. Front Microbiol 10:1163. doi: 10.3389/fmicb.2019.01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leo JC, Grin I, Linke D. 2012. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci 367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan E, Chauhan N, Udatha D, Leo JC, Linke D. 2016. Type V secretion systems in bacteria. Microbiol Spectr 4:e10.1128/microbiolspec.VMBF-0009-2015. doi: 10.1128/microbiolspec.VMBF-0009-2015. [DOI] [PubMed] [Google Scholar]
- 34.Bernstein HD. 2015. Looks can be deceiving: recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol Microbiol 97:205–215. doi: 10.1111/mmi.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leibiger K, Schweers JM, Schütz M. 2019. Biogenesis and function of the autotransporter adhesins YadA, intimin and invasin. Int J Med Microbiol 309:331–337. doi: 10.1016/j.ijmm.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Leo JC, Oberhettinger P, Schütz M, Linke D. 2015. The inverse autotransporter family: intimin, invasin and related proteins. Int J Med Microbiol 305:276–282. doi: 10.1016/j.ijmm.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt MA. 2010. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol 12:1544–1552. doi: 10.1111/j.1462-5822.2010.01518.x. [DOI] [PubMed] [Google Scholar]
- 38.Frankel G, Phillips AD, Trabulsi LR, Knutton S, Dougan G, Matthews S. 2001. Intimin and the host cell–is it bound to end in Tir(s)? Trends Microbiol 9:214–218. doi: 10.1016/s0966-842x(01)02016-9. [DOI] [PubMed] [Google Scholar]
- 39.Isberg RR, Leong JM. 1990. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 60:861–871. doi: 10.1016/0092-8674(90)90099-Z. [DOI] [PubMed] [Google Scholar]
- 40.Chauhan N, Wrobel A, Skurnik M, Leo JC. 2016. Yersinia adhesins: an arsenal for infection. Proteomics Clin Appl 10:949–963. doi: 10.1002/prca.201600012. [DOI] [PubMed] [Google Scholar]
- 41.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 42.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamburger ZA, Brown MS, Isberg RR, Bjorkman PJ. 1999. Crystal structure of invasin: a bacterial integrin-binding protein. Science 286:291–295. doi: 10.1126/science.286.5438.291. [DOI] [PubMed] [Google Scholar]
- 44.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai JC, Yen MR, Castillo R, Leyton DL, Henderson IR, Saier MH. 2010. The bacterial intimins and invasins: a large and novel family of secreted proteins. PLoS One 5:e14403. doi: 10.1371/journal.pone.0014403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leong JM, Fournier RS, Isberg RR. 1990. Identification of the integrin binding domain of the Yersinia pseudotuberculosis invasin protein. EMBO J 9:1979–1989. doi: 10.1002/j.1460-2075.1990.tb08326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nesta B, Spraggon G, Alteri C, Moriel DG, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, Fontana MR, Frankel G, Mobley HLT, Rappuoli R, Pizza M, Serino L, Soriani M. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3:e00010-12. doi: 10.1128/mBio.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pierschbacher MD, Ruoslahti E. 1984. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 52.van Nhieu GT, Isberg RR. 1991. The Yersinia pseudotuberculosis invasin protein and human fibronectin bind to mutually exclusive sites on the alpha 5 beta 1 integrin receptor. J Biol Chem 266:24367–24375. doi: 10.1016/S0021-9258(18)54238-1. [DOI] [PubMed] [Google Scholar]
- 53.Ruoslahti E. 1996. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol 12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 54.Alrutz MA, Isberg RR. 1998. Involvement of focal adhesion kinase in invasin-mediated uptake. Proc Natl Acad Sci USA 95:13658–13663. doi: 10.1073/pnas.95.23.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruce-Staskal PJ, Weidow CL, Gibson JJ, Bouton AH. 2002. Cas, Fak and Pyk2 function in diverse signaling cascades to promote Yersinia uptake. J Cell Sci 115:2689–2700. doi: 10.1242/jcs.115.13.2689. [DOI] [PubMed] [Google Scholar]
- 56.Wong KW, Isberg RR. 2003. Arf6 and phosphoinositol-4-phosphate-5-kinase activities permit bypass of the Rac1 requirement for beta1 integrin-mediated bacterial uptake. J Exp Med 198:603–614. doi: 10.1084/jem.20021363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pizarro-Cerdá J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Pizarro-Cerdá J, Cossart P. 2004. Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat Cell Biol 6:1026–1033. doi: 10.1038/ncb1104-1026. [DOI] [PubMed] [Google Scholar]
- 59.An Z, Huang X, Zheng C, Ding W. 2019. Acinetobacter baumannii outer membrane protein A induces HeLa cell autophagy via MAPK/JNK signaling pathway. Int J Med Microbiol 309:97–107. doi: 10.1016/j.ijmm.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Zhang K, Shi X, Wang C, Wang F, Fan J, Shen F, Xu J, Bao W, Liu M, Yu L. 2016. Critical role of bacterial isochorismatase in the autophagic process induced by Acinetobacter baumannii in mammalian cells. FASEB J 30:3563–3577. doi: 10.1096/fj.201500019R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bist P, Dikshit N, Koh TH, Mortellaro A, Tan TT, Sukumaran B. 2014. The Nod1, Nod2, and Rip2 axis contributes to host immune defense against intracellular Acinetobacter baumannii infection. Infect Immun 82:1112–1122. doi: 10.1128/IAI.01459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parra-Millán R, Guerrero-Gómez D, Ayerbe-Algaba R, Pachón-Ibáñez ME, Miranda-Vizuete A, Pachón J, Smani Y. 2018. Intracellular trafficking and persistence of Acinetobacter baumannii requires transcription factor EB. mSphere 3:e00106-18. doi: 10.1128/mSphere.00106-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krzymińska S, Frąckowiak H, Kaznowski A. 2012. Acinetobacter calcoaceticus-baumannii complex strains induce caspase-dependent and caspase-independent death of human epithelial cells. Curr Microbiol 65:319–329. doi: 10.1007/s00284-012-0159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asensio NC, Rendón JM, Burgas MT. 2021. Time-resolved transcriptional profiling of epithelial cells infected by intracellular Acinetobacter baumannii. Microorganisms 9:354. doi: 10.3390/microorganisms9020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Djahanschiri B, di Venanzio G, Distel JS, Breisch J, Dieckmann MA, Goesmann A, Averhoff B, Göttig S, Wilharm G, Feldman MF, Ebersberger I. 2022. Evolutionarily stable gene clusters shed light on the common grounds of pathogenicity in the Acinetobacter calcoaceticus-baumannii complex. bioRxiv doi: 10.1101/2022.01.10.475602. [DOI] [PMC free article] [PubMed]
- 66.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 67.Karah N, Sundsfjord A, Towner K, Samuelsen Ø. 2012. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updat 15:237–247. doi: 10.1016/j.drup.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 68.Rondelet A, Condemine G. 2013. Type II secretion: the substrates that won’t go away. Res Microbiol 164:556–561. doi: 10.1016/j.resmic.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 69.d'Enfert C, Ryter A, Pugsley AP. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J 6:3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pugsley AP, Poquet I, Kornacker MG. 1991. Two distinct steps in pullulanase secretion by Escherichia coli K12. Mol Microbiol 5:865–873. doi: 10.1111/j.1365-2958.1991.tb00760.x. [DOI] [PubMed] [Google Scholar]
- 71.Poquet I, Faucher D, Pugsley AP. 1993. Stable periplasmic secretion intermediate in the general secretory pathway of Escherichia coli. EMBO J 12:271–278. doi: 10.1002/j.1460-2075.1993.tb05653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Konovalova A, Silhavy TJ. 2015. Outer membrane lipoprotein biogenesis: Lol is not the end. Philos Trans R Soc Lond B Biol Sci 370:20150030. doi: 10.1098/rstb.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richardson DJ, Butt JN, Fredrickson JK, Zachara JM, Shi L, Edwards MJ, White G, Baiden N, Gates AJ, Marritt SJ, Clarke TA. 2012. The “porin-cytochrome” model for microbe-to-mineral electron transfer. Mol Microbiol 85:201–212. doi: 10.1111/j.1365-2958.2012.08088.x. [DOI] [PubMed] [Google Scholar]
- 74.Lower BH, Yongsunthon R, Shi L, Wildling L, Gruber HJ, Wigginton NS, Reardon CL, Pinchuk GE, Ubay TCD, Boily JF, Lower SK. 2009. Antibody recognition force microscopy shows that outer membrane cytochromes OmcA and MtrC are expressed on the exterior surface of Shewanella oneidensis MR-1. Appl Environ Microbiol 75:2931–2935. doi: 10.1128/AEM.02108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DiChristina TJ, Moore CM, Haller CA. 2002. Dissimilatory Fe(III) and Mn(IV) reduction by Shewanella putrefaciens requires ferE, a homolog of the pulE (gspE) type II protein secretion gene. J Bacteriol 184:142–151. doi: 10.1128/JB.184.1.142-151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi L, Deng S, Marshall MJ, Wang Z, Kennedy DW, Dohnalkova AC, Mottaz HM, Hill EA, Gorby YA, Beliaev AS, Richardson DJ, Zachara JM, Fredrickson JK. 2008. Direct involvement of type II secretion system in extracellular translocation of Shewanella oneidensis outer membrane cytochromes MtrC and OmcA. J Bacteriol 190:5512–5516. doi: 10.1128/JB.00514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baldi DL, Higginson EE, Hocking DM, Praszkier J, Cavaliere R, James CE, Bennett-Wood V, Azzopardi KI, Turnbull L, Lithgow T, Robins-Browne RM, Whitchurch CB, Tauschek M. 2012. The type II secretion system and its ubiquitous lipoprotein substrate, SslE, are required for biofilm formation and virulence of enteropathogenic Escherichia coli. Infect Immun 80:2042–2052. doi: 10.1128/IAI.06160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moriel DG, Bertoldi I, Spagnuolo A, Marchi S, Rosini R, Nesta B, Pastorello I, Mariani Corea VA, Torricelli G, Cartocci E, Savino S, Scarselli M, Dobrindt U, Hacker J, Tettelin H, Tallon LJ, Sullivan S, Wieler LH, Ewers C, Pickard D, Dougan G, Fontana MR, Rappuoli R, Pizza M, Serino L. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc Natl Acad Sci USA 107:9072–9077. doi: 10.1073/pnas.0915077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pugsley AP, Chapon C, Schwartz M. 1986. Extracellular pullulanase of Klebsiella pneumoniae is a lipoprotein. J Bacteriol 166:1083–1088. doi: 10.1128/jb.166.3.1083-1088.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kornacker MG, Boyd A, Pugsley AP, Plastow GS. 1989. Klebsiella pneumoniae strain K21: evidence for the rapid secretion of an unacylated form of pullulanase. Mol Microbiol 3:497–503. doi: 10.1111/j.1365-2958.1989.tb00196.x. [DOI] [PubMed] [Google Scholar]
- 81.Leong JM, Morrissey PE, Marra A, Isberg RR. 1995. An aspartate residue of the Yersinia pseudotuberculosis invasin protein that is critical for integrin binding. EMBO J 14:422–431. doi: 10.1002/j.1460-2075.1995.tb07018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frankel G, Lider O, Hershkoviz R, Mould AP, Kachalsky SG, Candy DC, Cahalon L, Humphries MJ, Dougan G. 1996. The cell-binding domain of intimin from enteropathogenic Escherichia coli binds to beta1 integrins. J Biol Chem 271:20359–20364. doi: 10.1074/jbc.271.34.20359. [DOI] [PubMed] [Google Scholar]
- 83.Saltman LH, Lu Y, Zaharias EM, Isberg RR. 1996. A region of the Yersinia pseudotuberculosis invasin protein that contributes to high affinity binding to integrin receptors. J Biol Chem 271:23438–23444. doi: 10.1074/jbc.271.38.23438. [DOI] [PubMed] [Google Scholar]
- 84.Khan TA, Wang X, Maynard JA. 2016. Inclusion of an RGD motif alters invasin integrin-binding affinity and specificity. Biochemistry 55:2078–2090. doi: 10.1021/acs.biochem.5b01243. [DOI] [PubMed] [Google Scholar]
- 85.Leong JM, Morrissey PE, Isberg RR. 1993. A 76-amino acid disulfide loop in the Yersinia pseudotuberculosis invasin protein is required for integrin receptor recognition. J Biol Chem 268:20524–20532. doi: 10.1016/S0021-9258(20)80757-1. [DOI] [PubMed] [Google Scholar]
- 86.Piepenbrink KH, Lillehoj E, Harding CM, Labonte JW, Zuo X, Rapp CA, Munson RS, Goldblum SE, Feldman MF, Gray JJ, Sundberg EJ. 2016. Structural diversity in the type IV pili of multidrug-resistant Acinetobacter. J Biol Chem 291:22924–22935. doi: 10.1074/jbc.M116.751099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ronish LA, Lillehoj E, Fields JK, Sundberg EJ, Piepenbrink KH. 2019. The structure of PilA from Acinetobacter baumannii AB5075 suggests a mechanism for functional specialization in Acinetobacter type IV pili. J Biol Chem 294:218–230. doi: 10.1074/jbc.RA118.005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tram G, Poole J, Adams FG, Jennings MP, Eijkelkamp BA, Atack JM. 2021. The Acinetobacter baumannii autotransporter adhesin Ata recognizes host glycans as high-affinity receptors. ACS Infect Dis 7:2352–2361. doi: 10.1021/acsinfecdis.1c00021. [DOI] [PubMed] [Google Scholar]
- 89.Hatefi Oskuei R, Darvish Alipour Astaneh S, Rasooli I. 2021. A conserved region of Acinetobacter trimeric autotransporter adhesion, Ata, provokes suppression of Acinetobacter baumannii virulence. Arch Microbiol 203:3483–3493. doi: 10.1007/s00203-021-02343-1. [DOI] [PubMed] [Google Scholar]
- 90.Weidensdorfer M, Ishikawa M, Hori K, Linke D, Djahanschiri B, Iruegas R, Ebersberger I, Riedel-Christ S, Enders G, Leukert L, Kraiczy P, Rothweiler F, Cinatl J, Berger J, Hipp K, Kempf VAJ, Göttig S. 2019. The Acinetobacter trimeric autotransporter adhesin Ata controls key virulence traits of Acinetobacter baumannii. Virulence 10:68–81. doi: 10.1080/21505594.2018.1558693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litrán T. 2012. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 194:3950–3960. doi: 10.1128/JB.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bentancor LV, Routray A, Bozkurt-Guzel C, Camacho-Peiro A, Pier GB, Maira-Litrán T. 2012. Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect Immun 80:3381–3388. doi: 10.1128/IAI.06096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Darvish Alipour Astaneh S, Rasooli I, Mousavi Gargari SL. 2014. The role of filamentous hemagglutinin adhesin in adherence and biofilm formation in Acinetobacter baumannii ATCC19606(T). Microb Pathog 74:42–49. doi: 10.1016/j.micpath.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 94.Astaneh SDA, Rasooli I, Gargari SLM. 2017. Filamentous hemagglutinin adhesin FhaB limits A. baumannii biofilm formation. Front Biosci 9:266–275. doi: 10.2741/e801. [DOI] [PubMed] [Google Scholar]
- 95.Pérez A, Merino M, Rumbo-Feal S, Álvarez-Fraga L, Vallejo JA, Beceiro A, Ohneck EJ, Mateos J, Fernández-Puente P, Actis LA, Poza M, Bou G. 2017. The FhaB/FhaC two-partner secretion system is involved in adhesion of Acinetobacter baumannii AbH12O-A2 strain. Virulence 8:959–974. doi: 10.1080/21505594.2016.1262313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brossard KA, Campagnari AA. 2012. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun 80:228–233. doi: 10.1128/IAI.05913-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Flores-Mireles AL, Walker JN, Bauman TM, Potretzke AM, Schreiber HL, Park AM, Pinkner JS, Caparon MG, Hultgren SJ, Desai A. 2016. Fibrinogen release and deposition on urinary catheters placed during urological procedures. J Urol 196:416–421. doi: 10.1016/j.juro.2016.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Flores-Mireles AL, Pinkner JS, Caparon MG, Hultgren SJ. 2014. EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice. Sci Transl Med 6:254ra127. doi: 10.1126/scitranslmed.3009384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walker JN, Flores-Mireles AL, Pinkner CL, Schreiber HL, Joens MS, Park AM, Potretzke AM, Bauman TM, Pinkner JS, Fitzpatrick JAJ, Desai A, Caparon MG, Hultgren SJ. 2017. Catheterization alters bladder ecology to potentiate Staphylococcus aureus infection of the urinary tract. Proc Natl Acad Sci USA 114:E8721–E8730. doi: 10.1073/pnas.1707572114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu W, Flores-Mireles AL, Cusumano ZT, Takagi E, Hultgren SJ, Caparon MG. 2017. Host and bacterial proteases influence biofilm formation and virulence in a murine model of enterococcal catheter-associated urinary tract infection. NPJ Biofilms Microbiomes 3:28. doi: 10.1038/s41522-017-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sycz G, di Venanzio G, Distel JS, Sartorio MG, Le NH, Scott NE, Beatty WL, Feldman MF. 2021. Modern Acinetobacter baumannii clinical isolates replicate inside spacious vacuoles and egress from macrophages. PLoS Pathog 17:e1009802. doi: 10.1371/journal.ppat.1009802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz Ş, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vizcaíno JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kinsella RL, Lopez J, Palmer LD, Salinas ND, Skaar EP, Tolia NH, Feldman MF. 2017. Defining the interaction of the protease CpaA with its type II secretion chaperone CpaB and its contribution to virulence in Acinetobacter species. J Biol Chem 292:19628–19638. doi: 10.1074/jbc.M117.808394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Graham DE, Groshong AM, Jackson-Litteken CD, Moore BP, Caimano MJ, Blevins JS. 2020. The BB0345 hypothetical protein of Borrelia burgdorferi is essential for mammalian infection. Infect Immun 88:e00472-20. doi: 10.1128/IAI.00472-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 107.Jackson-Litteken CD, Tyler Ratliff C, Kneubehl AR, Siletti C, Pack L, Lan R, Huynh TAN, Lopez JE, Blevins JS. 2021. The diadenylate cyclase CdaA is critical for Borrelia turicatae virulence and physiology. Infect Immun 89:e00787-20. doi: 10.1128/IAI.00787-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Altenhoff AM, Levy J, Zarowiecki M, Tomiczek B, Vesztrocy AW, Dalquen DA, Müller S, Telford MJ, Glover NM, Dylus D, Dessimoz C. 2019. OMA standalone: orthology inference among public and custom genomes and transcriptomes. Genome Res 29:1152–1163. doi: 10.1101/gr.243212.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Putative T2SS-dependent secretome of UPAB1. Download Table S1, DOCX file, 0.04 MB (37.9KB, docx) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phyre2 predicts that D1G37_RS04395 has structural similarity to Yersinia InvA. Results from the Phyre2 analysis of the amino acid sequence of full-length D1G37_RS04395 are shown. Download FIG S1, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
InvL localizes primarily to the insoluble fraction of bacterial supernatant. WT UPAB1 harboring pBAV::invL-his6 was grown to early stationary phase, and WCL, sup, sup insoluble, and sup soluble fractions were isolated. Fractions were then probed for InvL (α-His6) and RNAP by immunoblotting. Numbers to the left indicate molecular weight in kDa. Two biological replicates were performed, yielding similar results, and a representative blot from one replicate is shown. Download FIG S2, TIF file, 0.9 MB (965.9KB, tif) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A. baumannii is adhered extracellularly to 5637 epithelial cells. 5637 cells were infected with WT UPAB1 at an MOI of one, incubated for one h, and immunofluorescence microscopy was used to visualize intracellular versus extracellular bacteria. Nuclei are stained blue, actin is stained red, intracellular bacteria are stained green (rarely observed), and extracellular bacteria are double stained green and far-red. Two biological replicates were examined, and a representative image from one biological replicate is shown. The scale bar equals 20 μm. Download FIG S3, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids and strains used in this study. Download Table S2, DOCX file, 0.06 MB (57.6KB, docx) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download Text S1, DOCX file, 0.1 MB (124.1KB, docx) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S3, DOCX file, 0.08 MB (86.3KB, docx) .
Copyright © 2022 Jackson-Litteken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The MS data and search results have been deposited into the PRIDE ProteomeXchange Consortium repository and can be accessed using the accession numbers PXD030460 and PXD030491 (102, 103). All other data are provided within the text or supplemental material.