Abstract
Human infection with Crimean-Congo hemorrhagic fever virus (CCHFV), a tick-borne pathogen in the family Nairoviridae, can result in a spectrum of outcomes, ranging from asymptomatic infection through mild clinical signs to severe or fatal disease. Studies of CCHFV immunobiology have investigated the relationship between innate and adaptive immune responses with disease severity, attempting to elucidate factors associated with differential outcomes. In this article, we begin by highlighting unanswered questions, then review current efforts to answer them. We discuss in detail current clinical studies and research in laboratory animals on CCHF, including immune targets of infection and adaptive and innate immune responses. We summarize data about the role of the immune response in natural infections of animals and humans and experimental studies in vitro and in vivo and from evaluating immune-based therapies and vaccines, and present recommendations for future research.
1. Introduction
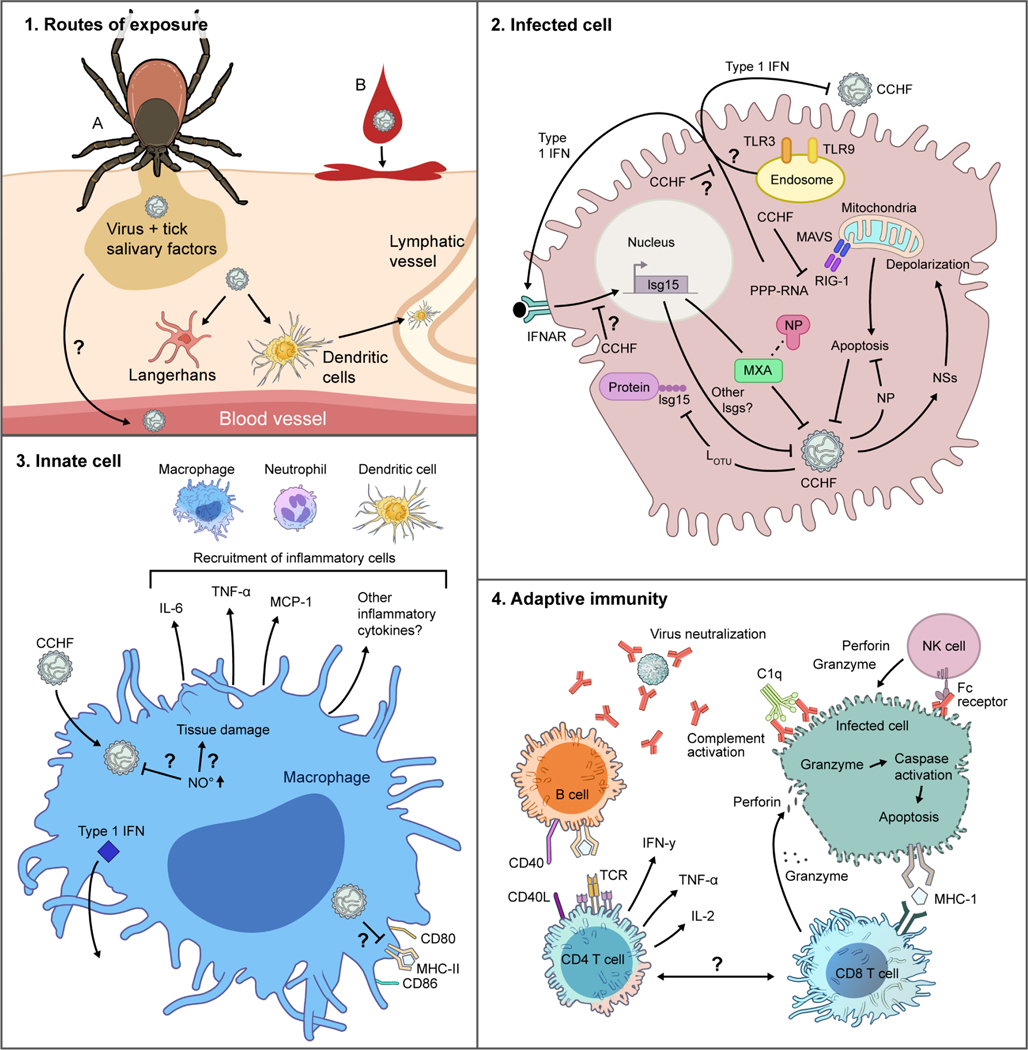
Crimean-Congo hemorrhagic fever virus (CCHFV), a tick-borne pathogen in the family Nairoviridae, can infect animals of various species, but humans appear uniquely susceptible to severe clinical disease (Hoogstraal, 1979; Spengler et al., 2016). Importantly, human infections can present as a spectrum from the absence of symptoms through mild signs to severe or fatal hemorrhagic illness (Chumakov et al., 1980; Papa et al., 2013, 2011; Sidira et al., 2012). Severity is correlated with uncontrolled viral replication in host tissues and elevated levels of cytokines; thus, disease likely results both directly from virus-mediated pathology and indirectly from immune-mediated pathologies that can lead to vascular dysfunction and, in severe cases, death. Clinical and laboratory studies have aimed to better understand the immune factors associated with and indicative of mild or severe disease; to determine correlates of protection; and to understand why humans are uniquely susceptible to severe disease from CCHFV infection.
In this report, we begin by identifying unanswered questions of CCHFV immunobiology and attempts to answer them, presenting limitations and corresponding advances in the field. We then summarize our current understanding of the role of the immune system in both protection and disease by reviewing knowledge of human CCHF and studies conducted in cell culture and animal models, evaluating innate and adaptive immune responses in the context of cell-specific, tissue- specific, and systemic infections. We then discuss what is known about responses associated with disease progression and severe outcomes; explore current development of immune-based therapies and vaccines; and conclude by providing recommendations for future research.
2. Unanswered questions
The key unanswered question for CCHF is: why are humans uniquely susceptible to clinical disease upon infection? Serological evidence indicates that CCHFV can infect numerous wild and domestic animals without overt disease. Furthermore, even in humans, CCHFV infection is often subclinical or asymptomatic. Thus, the severe, fatal illness conveyed by the name Crimean- Congo hemorrhagic fever represents a disease outcome that is likely based on both host and viral determinants that remain poorly understood. Host factors that may contribute to susceptibility and outcome include genetic predisposition, such as epigenetic factors, polymorphisms, and allelic shifts, especially in the type I interferon (IFN) signaling system. Other contributors may include age, nutrition, and microbiome of the host. What are the relative roles of these factors?
Does cellular tropism within the mammalian host contribute to disease outcome?
CCHFV exhibits broad species tropism, yet cellular binding and entry receptor(s) are unknown. Does the virus use similar receptors for entry in the tick reservoir as it does in mammalian hosts? Within varied hosts, does CCHFV exhibit similar cellular and tissue tropism?
How do hosts sense CCHFV infection?
After entering host cells, CCHFV is sensed by host proteins that trigger signaling pathways alerting the host to the pathogen and increasing expression of antiviral proteins that restrict viral replication. The ability of human clinical CCHFV isolates to cause an often rapidly fatal disease in type I IFN-deficient mice, but not IFN-expressing mice, suggests that type I IFN is a key restriction factor for CCHFV. How type I IFN controls CCHFV infection is largely unclear. Although single proteins such as RIG-I and MXA are known to sense and restrict CCHFV, many additional factors also likely contribute. Indeed, MX1, the mouse ortholog of human MXA, is non-functional (Sellers et al., 2012), demonstrating that factors beyond MX1 control the infection in laboratory strains of mice. Thus, with the exception of the mouse-adapted CCHFV model, the only mouse models of severe CCHF use mice completely deficient in type I IFN. To date, no reports have been published on CCHFV virulence in mice with more targeted deletions of innate immunity, like MAVS, ISG-15, or TLR knock-outs, or in chimeras between IFN-deficient and sufficient hosts. Furthermore, the cell types required for type I IFN response to CCHFV are unknown. Although cells like peripheral dendritic cells (DC) potently express type I IFN (Fitzgerald-Bocarsly et al., 2008), the cell types required to produce type I IFN following in vivo CCHFV infection have not been identified.
How does CCHFV antagonize host innate immune responses?
Although some antagonists of the host innate immune response have been identified, including the OTU domain- possessing deubiquitination and deISGylation functions of the L-protein (Frias-Staheli et al., 2007; Scholte et al., 2017), the virus likely encodes additional antagonists. NSs and NSm proteins of bunyaviruses pathogenic to humans have been found to exhibit conserved function antagonizing innate immunity (Leventhal et al., 2021). CCHFV possesses both NSs and NSm, yet a direct role for these proteins in antagonizing innate immunity, particularly type I IFN, has not been described. Cumulatively, our understanding of how type I IFN restricts the virus, and how the virus antagonizes host IFN responses requires improvement.
In addition to type I IFN, apoptosis can be a key innate response to viral infection (Everett and McFadden, 1999). CCHFV possesses proteins with pro-apoptotic (NSs) and anti-apoptotic (NP) functions. Furthermore, the NP contains a highly conserved DEVD motif that is cleaved by host caspase 3, indicating that interactions with host caspases may be important for the viral life cycle. In addition, suppressing caspase 3 activity improves viral yields (Karlberg et al., 2011), suggesting that host apoptosis may restrict viral replication. Several other bunyaviruses have been shown to interact with the host apoptosis machinery through their NSs and NSm proteins, and apoptosis can be a key host antiviral defense (Barber, 2001). Yet, as CCHFV encodes proteinswith both pro- and anti-apoptotic functions and retains a viral protein targeted by host caspases, it is unclear how host cell apoptosis promotes or inhibits infection. Importantly, given the common function of bunyaviral NSs and NSm in interfering with host innate immunity, is CCHFV unique among human-pathogenic members of the Bunyavirales order in possessing an NSs and NSm without antagonistic function towards the host innate immune system? Or do these proteins have yet undescribed antagonistic activity towards innate immunity?
What role does the host inflammatory response play in disease?
For many viral hemorrhagic fevers, severe disease is associated with dysregulated host inflammatory responses. Similarly, strong inflammatory responses correlate with disease outcomes in CCHFV-infected humans and animal models of CCHF, suggesting that inflammation may contribute to poor outcome. However, these studies have only been correlative for infection, and the field lacks mechanistic insights into how inflammatory conditions contribute to disease outcome. How does cellular tropism in vivo contribute to resultant inflammatory responses? Does in vivo infection of macrophages and DC lead to inflammatory cytokine production? Does the severe inflammatory response contribute to dysregulated, inappropriate, or suppressed adaptive immune responses? Are other cell types, such as neutrophils and natural killer (NK) cells, involved in control or pathogenesis of CCHFV infection? Can treatment with immunomodulatory drugs to promote protective inflammatory responses and limit pathogenic ones effectively improve outcome?
How does adaptive immunity control CCHFV infection?
Although innate immunity is key in controlling acute CCHFV infection, successful control likely also requires an effective adaptive immune response. Low or absent antibody responses are frequently associated with poor disease outcome in CCHFV-infected humans, and mice lacking adaptive immunity succumb to infection. However, our understanding of the adaptive immune responses that control acute CCHFV infection is limited. Is the low or absent antibody response in fatal cases of CCHFV merely a correlate of a failed immune response or are antibodies required to control the infection? What antibody effector functions are required? Human survivors often have poorly neutralizing antibodies, suggesting that functions beyond simple neutralization of viral particles may be needed. Although some data from infected IFNAR−/− mice suggest that T-cells and IFNγ are required to control acute infection in mice, whether these play a role in IFN-competent hosts remains to be determined. What effector functions of T-cells are necessary to control virus infection: T-helper activity towards B-cells? Cytotoxic CD8 T-cells? What cell types are important in priming antiviral T-cells? Does CCHFV possess proteins that antagonize processes necessary for effective adaptive immunity, such as antigen processing and presentation? Cytotoxic T-cells and NK cells can kill targeted cells through perforin and granzyme, Fas-FasL signaling, or secretion of cytokines like IFN-γ or tumor necrosis factor α (TNF-α) (Andersen et al., 2006). All of these pathways promote apoptosis in target cells. Does the anti-apoptotic function of CCHFV NP inhibit these effector functions?
Is immunity to CCHFV durable and broadly protective?
As a tick-borne pathogen, CCHFV is endemic in many regions of Eurasia and Africa, and people and animals may be repeatedly exposed to the virus throughout their lifetimes. To our knowledge, reinfection has not been reported. However, the long-term immune responses to CCHFV are poorly studied. Is reinfection possible? What host responses limit reinfection? Does recovery from one strain provide protection against the numerous diverse strains found throughout the world? CCHFV exhibits remarkable sequence diversity across endemic geographic regions. The contribution of this diversity to human disease and the evolutionary pressures driving this divergence are almost completely unknown.
How does the tick vector contribute to disease?
A primary route of CCHFV infection is through bites of infected ticks. Although ticks of the genus Hyalomma are considered the principal reservoir (Spengler and Estrada-Peña, 2018), others such as those in the Rhipichepahlus genus may serve as reservoirs and vectors of distinct clades of CCHFV (Papa et al., 2002). An often- overlooked factor in modeling vector-borne pathogens is the role of the vector in disease outcome. During feeding, ticks release numerous proteins with diverse impact on the host, including immunosuppressive and anti-inflammatory functions that prevent the host from reacting to the feeding tick (Šimo et al., 2017). The roles of these tick-derived factors on CCHFV infection of mammalian hosts and early immune responses are unknown.
3. Attempts to address unanswered questions
A key limitation of our understanding of CCHF has been the lack of suitable animal models. For decades after CCHFV discovery, the only animal model was newborn mice, which are extremely sensitive and useful for virus isolation, but are of limited utility for understanding immunobiology. Only in 2010 was the first report of a lethal mouse model using IFNAR−/− mice published (Bereczky et al., 2010). This model and others using type I and type II IFN deficiency or transient blockade of the type I IFN receptor have proved invaluable in studies investigating CCHFV therapeutics, such as vaccines and antiviral drugs, but are poorly suitable for studying immunobiology (Marín-Lopez et al., 2019), since they exhibit severely altered innate (and likely adaptive) immunity. Additionally, the often-rapid lethality of infection in these mice precludes study of later phases of disease, such as induction of adaptive immune and memory responses.
Several attempts have been made to advance mouse models, including humanized mice. NSG-SGM3 humanized mice infected with CCHFV develop a lethal disease characterized by liver and brain pathology (Spengler et al., 2017). The unique central nervous system (CNS) pathology observed in these mice enables study of CCHFV-associated CNS disease, an understudied area of CCHFV immunobiology. Additionally, transient depletion of the type I IFN response using antibody-mediated blockade of the type I IFN receptor has been used to improve vaccine studies by allowing immunization of immunocompetent mice (Lindquist et al., 2018; Suschak et al., 2021) while also permitting investigation of CCHFV pathogenesis in knock-out mouse strains without requiring back-crossing of IFN receptor-deficient strains to strains of interest (Lindquist et al., 2018).
The recent development of a mouse-adapted CCHFV variant has also enabled studies in immunocompetent mice (Hawman et al., 2021). This variant also allows investigations using the multitude of genetically manipulated mice available without the need for suppressing type I IFN. Significant sex-linked and strain-linked differences in disease were observed when using mouse- adapted CCHFV. Male mice develop a severe clinical disease that correlates with high viral loads, inflammatory cytokine production, and significant liver pathology. In contrast, although early viral loads are similar between male and female mice, female mice develop a milder clinical disease that correlates with earlier control of viral loads and reduced inflammatory responses. Resistance to mouse-adapted CCHFV by female mice was dependent on type I IFN, adaptive immunity, and age. Thus, multiple host factors contribute to control of mouse-adapted CCHFV in this model. Furthermore, though several laboratory mouse strains were susceptible to clinical disease after infection with mouse-adapted CCHFV, 129SV1 mice were resistant, demonstrating that genetic differences may also confer resistance to mouse-adapted CCHFV (Hawman et al., 2021). This model therefore affords the opportunity to mechanistically investigate how distinct host responses contribute to disease outcome.
Ongoing studies using the Collaborative Cross resource and studies probing the host responses to CCHFV in mice susceptible and resistant to severe CCHF are investigating how host responses and genetic diversity contribute to disease outcome. The identification of adaptive mutations in several viral proteins suggests that these proteins possess key functions during viral pathogenesis in type I IFN-competent hosts (Hawman et al., 2021) The availability of reverse genetics systems for CCHFV (Bergeron et al., 2015) will enable mechanistic investigations of how these mutations function to overcome host restriction factors. Beyond improving mammalian models of infection, recent progress has been made to model the contribution of the tick reservoir in the viral life cycle (Xia et al., 2016). In addition, the availability of a reverse genetics system will allow for mechanistic dissection of protein function in viral replication and pathogenesis (Bergeron et al., 2015).
In 2018, Haddock et al. described the first immunocompetent animal model of CCHF (Haddock et al., 2018). Cynomolgus macaques infected with a human clinical CCHFV isolate faithfully recapitulate many aspects of human CCHF, including the full spectrum of disease outcomes (Cross et al., 2020; Haddock et al., 2018; Smith et al., 2019). This model has been used for evaluation of antivirals and vaccines (Hawman et al., 2021a; Hawman et al., 2020) and will likely contribute to further pre-clinical development of critically needed CCHF therapeutics. However, the ethical and practical considerations of non-human primate (NHP) research restrict the usefulness of this model for initial investigations into CCHFV immunobiology. Furthermore, although infection in cynomolgus macaques closely models human disease, both mild and severe, the spectrum of disease outcomes means that large numbers of animals are required for studies seeking to identify mechanisms of disease outcome. Thus, improved small animal models are also needed for CCHFV immunobiology research.
Importantly, as research on CCHFV immunobiology continues to progress, studies using animal models will need to be integrated with human clinical studies to correlate findings with human disease. Collaborations between bench, clinical, and field research are needed to obtain a more complete understanding of CCHFV immunobiology. Ultimately, better understanding will improve the development of safe and effective antiviral therapeutics and vaccines.
4. Current knowledge of CCHFV immunobiology
4.1. Immune cell kinetics
Leukopenia, the depletion of leukocytes such as mononuclear phagocytes, correlates with CCHF severity in humans (Joubert et al., 1985a; Swanepoel et al., 1989), and is also observed in infected mice and nonhuman primates. Monocytopenia, frequently seen in acute viral infection, was observed early in the course of infection in >90% of cynomolgus macaques challenged with either Afghanistan2009 or Hoti CCHFV strains (Cross et al., 2020). In Stat-1−/− mice that succumbed to disease 4–5 days post infection (dpi), increased macrophage levels correlated with onset of clinical signs. F4/80 and CD11b macrophage populations in the spleen remained stable before increasing 5- and 4-fold, respectively, 3 dpi (Bente et al., 2010). The kinetics of activated macrophages differed in the mice, initially rising and then decreasing 3 dpi. Interestingly, increased numbers of neutrophils and Ly6Chi inflammatory macrophages in the spleen correlated with peak clinical disease in CCHFV-infected IFNAR−/− mice, recapitulating human convalescence (Hawman et al., 2019). While overall macrophage numbers may increase over the course of disease it is unclear whether these macrophages exert antiviral activity or support continued viral replication by acting as targets for infection.
In some animal models, B- and T-cell responses to CCHFV begin soon after infection (Haddock et al., 2018; Hawman et al., 2021b; Hawman et al., 2021; Hawman et al., 2019). In IFNAR−/− mice infected with a human clinical CCHFV isolate, CD4+ and CD8+ T-cells in the liver and spleen expanded in number and expressed surface activation markers by 8 dpi (Hawman et al., 2021b), demonstrating prompt T-cell activation. Furthermore, by 8 dpi, both CD4+ and CD8+ T- cells in the liver and spleen were primed to produce antiviral cytokines like IFN-γ, TNF-α, and IL-2. Along with significantly increased expression of perforin in CD8+ T-cells, these findings suggested that T-cells rapidly engage in controlling infection (Hawman et al., 2021b).
4.2. Immune cells as targets of infection
Mononuclear cells have prominent roles in the pathogenesis of viral hemorrhagic fevers. Autopsy studies have identified CCHFV antigen-positive peripheral mononuclear phagocytes in various tissues, including spleen, liver, and intestines (Baskerville et al., 1981; Burt et al., 1997; Joubert et al., 1985; Negredo et al., 2017). Based on these reports from human cases, investigations into targets of CCHFV infection have focused on innate and adaptive mononuclear cells (Table 1). Overall, cells of the mononuclear phagocyte system (monocytes, macrophages, and DC) are more susceptible than other mononuclear cells like NK cells (Connolly-Andersen et al., 2009). Along with hepatocytes and vascular endothelial cells, they serve as primary target cells in humans (Baskerville et al., 1981; Burt et al., 1997; Joubert et al., 1985; Negredo et al., 2017).
Table 1.
CCHFV infection in immune-related cells
| Cell type | Source | Virus/VLP strains | Permissive | Reference |
|---|---|---|---|---|
| Peripheral blood mononuclear | Human | IbAr10200 | −/+ | (Connolly-Andersen et al., 2009) |
| Natural killer | Human | IbAr10200 | - | (Connolly-Andersen et al., 2009) |
| Monocyte | Human | IbAr10200 | −/+ | (Connolly-Andersen et al., 2009) |
| Macrophage | Murine | IbAr10200 | + | (Zivcec et al., 2018), (Zivcec et al., 2013), (Welch et al., 2020), (Welch et al., 2019) |
| Human | Afghanistan-2009 | + | (Zivcec et al., 2015) | |
| Human | Turkey-2004, Oman-1998, UG3010 |
++ | (Zivcec et al., 2015), (Garcia-Sastre et al., 2005) | |
| Human | IbAr10200 | −/+ | (Scholte et al., 2017), (Zivcec et al., 2015), (Garcia-Sastre et al., 2005) |
|
| M2 - Macrophage | Human | IbAr10200 | ++Δ | (Peyrefitte et al., 2010) |
| Kupffer | Human | Various South African Strains |
++ | (Burt et al., 1997) |
| Murine | IbAr10200, Afghanistan-2009, Turkey-2004, Oman-1998 |
+ | (Zivcec et al., 2018), (Zivcec et al., 2013), (Bente et al., 2010), (Welch et al., 2020), (Welch et al., 2019), (Tignor and Hanham, 1993), (Spengler et al., 2017), (Lindquist et al., 2018) | |
| Monkey | Hoti | ++ | (Haddock et al., 2018) | |
| Multinucleated giant Macrophages |
Murine** | Turkey-2004, Oman-1998 | + | (Spengler et al., 2017) |
| Histiocytes | Murine** | Turkey-2004 | + | (Spengler et al., 2017) |
| Dendritic cells | Human | IbAr10200 | ++Δ | (Connolly-Andersen et al., 2009), (Peyrefitte et al., 2010) |
| Murine | IbAr10200**, Oman-1998 | + | (Spengler et al., 2017), (Welch et al., 2019) | |
| Dermal dendritic cells | Human | IbAr10200 | ++Δ | (Rodriguez et al., 2018) |
| Human | AP92 | −/+Δ | (Rodriguez et al., 2018) | |
| Langerhans cells | Human | IbAr10200 | +Δ | (Rodriguez et al., 2018) |
| Human | AP92 | −/+Δ | (Rodriguez et al., 2018) | |
| Stellate cells | Murine | IbAr10200 | + | (Zivcec et al., 2013) |
| Glial cells | Murine** | Turkey-2004 | + | (Spengler et al., 2017) |
| Lymphocytes | NHP | Hoti | −/+ | (Haddock et al., 2018) |
| B-Cells | Human | IbAr10200 | - | (Connolly-Andersen et al., 2009) |
| T-Cells | Human | IbAr10200 | −/+ | (Connolly-Andersen et al., 2009) |
, strongly permissive; +, permissive; −/+ weakly permissive; -, not permissive;
, evidence of virus propagation from cell type;
, humanized mouse model; NHP, non-human primate (Cynomolgus macaque)
4.2.1. Macrophages
In culture, peripheral blood mononuclear cells (PBMC) and monocytes infected with high multiplicity of infection (MOI) of CCHFV contained only low levels of detectable S-segment genomes, and neither cell type produced detectable infectious virus in subsequent focus-forming assays (Connolly-Andersen et al., 2009). Naïve, non-polarized, monocyte-derived macrophages are also poorly permissive to infection with the highly passaged IbAr10200 CCHFV strain (Zivcec et al., 2015). Higher levels of infection with IbAr10200 (~10–50% cells) were observed in macrophages differentiated into an anti-inflammatory, M2-like polarized state after 24 hours post infection (hpi), and by 72 hpi, infection levels dropped below the limit of detection (Peyrefitte et al., 2010). Of note, cell culture studies of cell permissiveness may depend on the virus strain, and cells appear least permissive to IbAr10200. When a virus-like particle (VLP) system was used to assess differential virus entry, most efficient infection was observed with VLPs expressing the glycoprotein of Turkey and Oman CCHFV strains, followed by that of Afghanistan2009; lowest infection was observed with IbAr10200 glycoprotein (Zivcec et al., 2015). Similarly, an unpublished study found higher levels of viral progeny in monocyte-derived macrophages infected with UG3010 strain than IbAr10200 (personal correspondence A. Garrison, 2021) (Garcia-Sastre, 2010).
Despite these limited data from cell culture, human and animal model studies both indicate that monocyte and macrophage populations are early targets of infection, likely contributing to virus dissemination and initiation of immune activation. In IFNAR−/− mice, the earliest evidence of infection was in activated monocytes/macrophages (CD11b+Ly6C+) found in draining lymph nodes (Welch et al., 2019). Macrophage populations also represent a significant number of infected cells in primary target organs such as the liver; in IFNAR−/− mice, activated macrophages were the largest infected cell population in hepatic tissue in late-stage disease. Monocyte/macrophage activation is also seen in more severe human cases, indicated by elevated levels of neopterin, a serum protein generated from macrophage activation by inflammatory cytokines such as IFN-γ (Onguru et al., 2008).
Hepatic infection and damage (as shown, e.g., by elevated liver enzymes in serum) are characteristic of severe CCHF. Antigen-positive Kupffer or Kupffer-like cells have been identified in liver tissues in both humans and animal models (Burt et al., 1997; Lindquist et al., 2018; Zivcec et al., 2013). They are likely the first liver cells to become infected, subsequently spreading virus to surrounding hepatocytes and other non-parenchymal cells (Lindquist et al., 2018). Kupffer cells also positively stain for CCHFV antigen in infected neonate and IFNAR−/− mice (Bente et al., 2010; Tignor and Hanham, 1993; Zivcec et al., 2013). Depletion of Kupffer cells in CCHFV infection is independent of the type-I IFN response (Lindquist et al., 2018). Moreover, in mice with antibody- mediated disruption of type-I IFN signaling, Kupffer cell loss was associated with fatal outcome. Similar experiments in infected NOD-SCID-γ (NSG), Rag2-deficient, and perforin-deficient mice also demonstrated liver injury, suggesting that cytotoxic immune cells are dispensable for hepatic damage (Lindquist et al., 2018).
Other tissue-specific macrophage populations may also be involved in viral pathogenesis. Glial cells were principal targets of infection in terminally infected humanized mice. Along with CNS disease characterized by gliosis, meningitis, and meningoencephalitis, the presence of viral antigen in glial cells was the only notable difference between lethal (strain Turkey-200406546) and nonlethal (strain Oman-199809166) infection in mice (Spengler et al., 2017). However, while other nairoviruses cause prominent neurological disease, the clinical significance of this finding for human CCHF is unclear.
4.2.2. Dendritic cells
Tissue-resident DCs are presumed to be an entry point for CCHFV from tick bites and can be infected in culture; these include dermal DCs (dDC), and, to a lesser extent, Langerhans cells (LC). dDCs released virus at levels similar to infected macrophages (~8.0 × 104 pfu/mL), whereas LC only released virus at very low levels (~3.0 × 102 pfu/mL) (Rodriguez et al., 2018). Infected DCs are also detected in disease models. Stellate cells, morphologically consistent with DCs, stain for viral antigen in infected Stat-1−/− mice (Bente et al., 2010; Zivcec et al., 2013). In IFNAR−/− mice, Lin-CD45+Ly6G-CD11b+MHCII+ DCs are detected in blood, lymph nodes, and spleen, with highest levels seen in spleen in late-stage disease (Welch et al., 2019).
Susceptibility of DCs to infection has been investigated by several studies. Connolly- Andersen et al. (2009) found that in PBMC subsets, only monocyte-derived DCs (moDCs) upregulated RNA levels when infected with the IbAr10200 strain of CCHFV; monocytes, B-, T-, and NK cells did not. Furthermore, cytokines released into supernatants from infected moDCs activated human endothelial cells, demonstrating their functional role in downstream cellular activation (Connolly-Andersen et al., 2009). Peyrefitte et al. (2010) also found moDCs to be permissive to IbAr10200 infection and evaluated the resultant immune response (Peyrefitte et al., 2010). The group similarly reported DC activation and involvement in initiating downstream signaling events. Expression levels of surface CD40, CD83, and CD86 significantly increased upon infection in an MOI-dependent manner. CD40 rose in response to infection and/or replication of live virus; moreover, UV-inactivated virus had no effect on CD40 expression, but still upregulated CD83 and CD86. In addition to moDCs, increased transcription of CD surface proteins has also been detected in infected dDCs, specifically increases in CD40 (Rodriguez et al., 2018) and expression of CD80, CD86, CD197, and MHCII (unpublished; personal correspondence D. Bente, 2021).
4.2.3. B and T lymphocytes
Virus dissemination by infection of B- and T-cells appears limited at best. Studies in IFNAR−/− mice suggest that the earliest lymphocyte population to increase is CD4+ T-cells in the lymphatic system (Welch et al., 2019). However, this increase was not associated with evidence of viral infection in CD4+ T-cells. Connolly-Anderson et al. (2009) examined target cell populations by infecting human T-cells (CD3+/CD56-) and B-cells (CD19+) with IbAr10200 at a high MOI. Intracellular viral loads were monitored over 24 and 48 h; low levels of S-segment genome (<10,000-fold change from uninfected cells) were seen in T-cells but not B-cells. No infectious virus was detected in either T- or B-cells, and viral antigen was not seen by immunofluorescence in stained cells or by immunoblotting T- and B-cell lysates (Connolly- Andersen et al., 2009).
4.3. Interferon response to infection
4.3.1. IFN induction and anti-CCHFV activity
IFN responses are likely a critical determinant of disease severity. Polymorphisms in toll- like receptor (TLR) genes have been found to correlate with disease outcome in patients in Turkey (Arslan et al., 2015; Engin et al., 2016, 2010), although further studies are needed to determine if these findings apply to populations in other endemic regions. Serological evidence demonstrates that CCHFV can productively infect a multitude of wild and domestic animals, and while humans develop a spectrum of mild to severe disease upon infection, animals are almost uniformly refractory to clinical signs. Numerous wild and domestic species have been challenged with various strains of CCHFV, yet severe disease is typically only reported in laboratory rodents with deficient immunity, either due to age (i.e., newborn mice or rats) or targeted IFN-deficiencies. Ablating IFN signal transduction machinery by knocking out STAT-1, STAT-2, or IFN-α, -β, or -𝛾 receptors in mice and hamsters leads to fulminant disease (reviewed in Garrison et al., 2019; Ranadheera et al., 2020; Spengler et al., 2019, 2016). In addition, studies using wild-type and adaptive immune knockout mouse strains (e.g., NOD/SCID, Prf1−/−, and Rag2−/−) transiently suppressed with an anti-IFNAR1 monoclonal antibody (mAb) demonstrate that IFN is important for controlling infection and preventing disease even in animals lacking an intact adaptive immune system (Lindquist et al., 2018).
Developing models of severe disease in other immunocompetent laboratory animal species has been prohibitive. For example, cynomolgus macaques respond to infection similarly to humans, presenting a spectrum of disease severity from asymptomatic infection to, in severe cases, fever, rash, bleeding, and even death. However, recently, an immunocompetent mouse model for CCHF has been developed using a mouse-adapted CCHFV variant capable of causing severe disease in wild-type, immunocompetent mouse strains (Hawman et al., 2021). Nevertheless, in contrast to infection in WT mice, infection in IFNAR−/− mice with this mouse-adapted CCHFV variant is rapidly lethal, demonstrating that type I IFN exerts control over this viral strain (Hawman et al., 2021).
CCHFV evokes an IFN response in cell culture and in human infection (reviewed in Andersson et al., 2008; Weber and Mirazimi, 2008)). In cell culture, IFN and ISG responses are seen as early as 8 hpi and increase over time (Spengler et al., 2015). IFN-β, ISG56, and MxA induction is mediated by RIG-I recognition; interfering with RIG-I signaling reduces IFN production and ISG expression and increases viral replication (Spengler et al., 2015). IFN induction is reported in patients, and high IFN-γ levels are associated with poor outcome (Saksida et al., 2010). In NHPs, circulating IFN-γ increases early in disease, peaking around day 7. Its expression coincides with an induced antiviral state, sharp reduction in viral load (Cross et al., 2020; Haddock et al., 2018; Smith et al., 2019), and decreased genome equivalents 10 dpi in sera of animals that mounted IFN-γ responses (Cross et al., 2020; Smith et al., 2019). Additionally, treating CCHFV-infected IFNAR−/− mice with an antibody to block IFN-γ signaling significantly increased mortality (Hawman et al., 2021b). In immunocompetent mice infected with CCHFV, IFN-α was induced within 24 hpi (Hawman et al., 2021). Interestingly, IFN-β induction was only seen in male mice infected with the mouse-adapted CCHFV variant and correlated with more severe disease outcome in these mice (Hawman et al., 2021).
Overall, CCHFV is sensitive to IFN treatment and virus growth is attenuated in the presence of an intact IFN response. However, the ISGs responsible for inhibiting replication in vivo are not well understood. In IFN-producing cells, recombinant viruses with impaired IFN antagonism (due to inactivation of the protease domain within the RNA-dependent RNA polymerase) were significantly attenuated compared to wild-type virus; in cells with little or no IFN response, no such difference was observed (Scholte et al., 2017). MxA, induced exclusively by IFN-α and IFN-β, is a key mediator of the IFN-induced response against a wide range of viruses. When Vero-E6 cells constitutively expressing MxA were infected with CCHFV, progeny virus levels were reduced up to 1,000-fold compared to control cells, and accumulation of viral genomes was blocked (Andersson et al., 2004). MxA was found to colocalize with CCHFV NP in the perinuclear regions of infected cells, an interaction that may prevent viral RNA replication and inhibit production of new infectious virus particles (Andersson et al., 2004).
Pre-treating Huh-7 or HUVEC (human umbilical vein endothelial cells) for 24 h with increasing concentrations of IFN-α led to increased MxA expression and reduced accumulation ofviral NP (Andersson et al., 2006). Pre-treating these cells with type III IFN (IFN-λ1) also inhibited CCHFV. Treatment with IFN-α alone or in combination with IFN-λ1 was evaluated in A549 and HuH7 cells. Alone, both IFNs inhibited the virus; however, when given in combination, IFN-λ and IFN-α clearly antagonized each other in both cell lines, resulting in reduced activation of pivotal ISGs (i.e., MxA and 2ʹ−5ʹ-OAS) and decreased antiviral activity (Bordi et al., 2015). Notably, while treatment prior to infection can inhibit replication, IFN does not significantly affect established infection (Andersson et al., 2008): even only 1 hour after infection, IFN cannot prevent viral replication, whereas pre-treatment with the same dose significantly inhibits CCHFV replication. No attempts to use IFN to treat CCHF in humans have been reported, but it has been suggested for use in conjunction with ribavirin, based on treatment approaches reported for other viral agents, such as hepatitis C (Ergonul et al., 2007).
4.3.2. Virus-mediated IFN and ISG antagonism
Immune modulators are found in CCHFV S- and L-segments, specifically the NP and ovarian tumor (OTU)-like cysteine protease domain within the L-protein. One limited study investigated the effect of plasmid-based NP expression on IFN induction in human embryonic kidney cells (HEK293T/17) and suggested strain differences in IFN-β promoter induction as support for NP-mediated immune evasion (Fajs et al., 2014). However, in the absence of other investigations, the role of NP in IFN antagonism, including potential strain differences, remains unknown. More detailed work has been performed to investigate L-segment-mediated antagonism; several studies have indicated that the OTU-like cysteine protease domain is involved in cleaving both ubiquitin and ISG15 and in reducing the RIG-I/MAVS response (Frias-Staheli et al., 2007; Van Kasteren et al., 2012). Specifically, OTU catalytic activity is essential for CCHFV replication and deubiquitinase activity of the L-protein is critical for suppressing IFN expression in vitro (Scholte et al., 2017).
Immune antagonists have not yet been characterized in the M-segment. However, the M- segment encodes a single polyprotein, GPC, which yields multiple structural and non-structural glycoproteins that could potentially serve as immunomodulators (Altamura et al., 2007; Bergeron et al., 2015, 2007; Erickson et al., 2007; Freitas et al., 2020; Haferkamp et al., 2005; Sanchez et al., 2006, 2002; Vincent et al., 2003; Welch et al., 2020). GPC is co-translationally synthesized within the endoplasmic reticulum (ER), where proprotein convertases and N-linked glycosylases modify the immature (precursor; Pre) structural Pre-GN and Pre-GC glycoproteins. After matriculation through the Golgi compartment and further modifications by convertase and O- linked glycosylase, a mature CCHFV particle is thought to be formed with a heterodimeric form of GN and GC. The fates of the other related glycoprotein products, including mucin-like domain, GP38, GP85, GP160, NSM, and other small fragment components, are largely unknown. However, immune responses generated towards nonstructural components of the Pre-GN, such as the GP38 molecule, have been shown to be protective in vivo, supporting their role in CCHF antiviral defense (Ahmed et al., 2005; Golden et al., 2019; Mishra et al., 2020).
Among members of the Bunyavirales order, IFN antagonism is often exhibited by non- structural (NS) proteins encoded on the S and M segments (Leventhal et al., 2021). Although CCHFV encodes both NSs and NSm, their role in antagonizing host interferon responses has not been reported. However, adapting the virus to cause disease in mice with intact type I IFN through sequential passage was associated with coding changes in both NSs and NSm (Hawman et al., 2021), suggesting that these proteins may have pathogenic function in type I IFN-competent hosts.
4.4. Role of other cytokines in CCHFV infection
Cytokine studies have predominantly been conducted using serum and plasma samples (Table 2–4), with additional limited data derived from PBMC preparations or tissue samples from animal studies. Data from human patients are still relatively limited but appear to correlate well with data from animal models of disease, supporting the use of these models in pathogenesis and therapeutic studies. For example, similarly to humans, modestly elevated serum interleukin 15 (IL- 15) is reported in cynomolgus macaques (Cross et al., 2020; Haddock et al., 2018; Smith et al., 2019), and elevated TNF-α is reported in mice (Bente et al., 2010; Hawman et al., 2019). The congruity in data also includes analytes that do not change significantly during infection in both humans and animal models (e.g., IL-12 and IL-13) (Bente et al., 2010; Papa et al., 2016b).
Table 2.
Pro-inflammatory cytokine response to CCHFV: Interleukins
| Cytokine | Infected cell/species | Lethal | Sample | Level in acute infection | Reference |
|---|---|---|---|---|---|
| IL-1α | Cell (dDC, LC) | − | Cord blood | NSF | (Rodriguez et al., 2018) |
| IL-1β | Human (Turkey) | Y | Serum | + | (Papa et al., 2016b) |
| Mouse (STAT-1, IFNAR) |
Y/N | Plasma/serum, liver/spleen | + | (Bente et al., 2010), (Hawman et al., 2019), (Welch et al., 2019) | |
| Cell (DC, dDC, LC) | − | PBMC/cord blood | NSF | (Connolly-Andersen et al., 2009), (Rodriguez et al., 2018) |
|
| IL-2 | Human (Turkey) | Y | Serum | NSF | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| IL-3 | Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) |
| IL-6 | Human (Turkey, Albania) | Y/N | Serum | +* | (Ergonul et al., 2006), (Papa et al., 2006), (Ergönül et al., 2017), (Papa et al., 2016b) |
| NHP | Y/N | Serum | + | (Haddock et al., 2018), (Cross et al., 2020), (Smith et al., 2019) | |
| Mouse (STAT-1, IFNAR) |
Y | Plasma/Serum | + | (Bente et al., 2010), (Hawman et al., 2019) | |
| Cell (Mø, DC, dDC, LC) | − | PBMC/cord blood | + | (Peyrefitte et al., 2010), (Connolly-Andersen et al., 2009), (Peyrefitte et al., 2010), (Rodriguez et al., 2018) | |
| IL-7 | Cell (dDC, LC) | − | Cord Blood | NSF | (Rodriguez et al., 2018) |
| IL-8 | Human (Turkey) | Y/N | Serum | +, +/− | (Ergönül et al., 2017), (Papa et al., 2016b) |
| Cell (DC, dDC) | − | PBMC/cord blood | + | (Peyrefitte et al., 2010), (Rodriguez et al., 2018) | |
| Cell (DC, LC) | − | PBMC/cord blood | NSF | (Connolly-Andersen et al., 2009), (Rodriguez et al., 2018) |
|
| IL-9 | Human (Turkey) | Y/N | Serum | + | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| IL-12 | Human (Turkey, Kosovo) |
Y/N | Serum | NSF | (Saksida et al., 2010), (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | NSF | (Hawman et al., 2019) | |
| Cell (dDC, LC) | − | Cord blood | NSF | (Rodriguez et al., 2018) | |
| IL-13 | Human (Turkey) | Y/N | Serum | NSF | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | NSF | (Hawman et al., 2019) | |
| IL-15 | Human (Turkey) | Y/N | Serum | + | (Papa et al., 2016b) |
| NHP | Y/N | Serum | + | (Haddock et al., 2018), (Cross et al., 2020), (Smith et al., 2019) | |
| Cell (dDC, LC) | − | Cord blood | NSF | (Rodriguez et al., 2018) | |
| IL-17A | NHP | Y/N | Serum | + | (Haddock et al., 2018) |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| IL-18 | NHP | N | Serum | + | (Cross et al., 2020), (Smith et al., 2019) |
DC, dendritic cell; dDC dermal dendritic cell; IFNAR, type I interferon (IFN) receptor-deficient mice; LC, Langerhans cell; Mø, macrophage; −, not applicable; STAT-1, STAT1-deficient mice; NHP, non-human primate (all NHP data are from Cynomolgus macaques); NSF, no significant findings; +, elevated; +/−, can be elevated; Y, reported in lethal disease; Y/N, reported in lethal and non-lethal disease; N, reported in non-lethal disease;
, significantly higher in fatal CCHF than in non-fatal CCHF.
Table 4.
Anti-inflammatory cytokine response to CCHFV
| Cytokine | Infected cell/species | Lethal disease | Sample | Level in acute infection | Reference |
|---|---|---|---|---|---|
| IL-1RA | Human (Turkey) | Y | Serum | +/− | (Papa et al., 2016b) |
| NHP | Y/N | Serum | + | (Haddock et al., 2018), (Smith et al., 2019), (Cross et al., 2020) | |
| IL-4 | Human (Turkey) | Y/N | Serum | NSF | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| IL-5 | Human (Turkey) | Y/N | Serum | (−) | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| IL-10 | Human (Turkey) | Y/N | Serum | + | (Ergonul et al., 2006) |
| Human (Albania) | Y | Serum | + | (Papa et al., 2006) | |
| Human (Kosovo) | Y | Serum | + | (Saksida et al., 2010) | |
| Human (Turkey) | Y/N | Serum | + | (Papa et al., 2016b) | |
| Mouse (STAT-1, IFNAR) |
Y/N | Plasma/serum | + | (Bente et al., 2010), (Hawman et al., 2019) | |
| Cell (DC) | − | PBMC | Slightly elevated or NSF | (Connolly-Andersen et al., 2009), (Peyrefitte et al., 2010) |
|
| Cell (dDC, LC) | − | Cord Blood | NSF | (Rodriguez et al., 2018) | |
| IL-19 | Cell (DC) | − | PBMC | NSF | (Connolly-Andersen et al., 2009) |
| G-CSF | Human (Turkey) | Y/N | Serum | +/− | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | (−) | (Hawman et al., 2019) |
DC, dendritic cell; dDC dermal dendritic cell; IFNAR, type I interferon (IFN) receptor deficient mice; LC, Langerhans cell; Mø, macrophage; −, not applicable; STAT-1, STAT1-deficient mice; NHP, non-human primate (all NHP data are from Cynomolgus macaques); NSF, no significant findings; +, elevated; +/−, can be elevated; (−), decreased; Y, reported in lethal disease; Y/N, reported in lethal and non-lethal disease; N, reported in non-lethal disease;
, significantly higher in fatal CCHF than in nonfatal CCHF.
Overall, severe or fatal disease is associated with a pro-inflammatory response (Tables 2 and 3), which is thought to lead to vascular dysfunction, disseminated intravascular coagulation (DIC), organ failure, and shock (Saksida et al., 2010). In human patients in Turkey, Albania, and Kosovo, increases in pro-inflammatory cytokines (TNF-α, IL-8, IL-9, IL-15, IP-10, and monocyte chemoattractant protein 1 [MCP-1]) were associated with disease severity and poor outcomes (Ergönül et al., 2017; Ergonul et al., 2006; Papa et al., 2016b, 2006; Saksida et al., 2010). Of those, increased TNF-α has been most consistently reported in a variety of investigations, including immune cell culture, mouse models, and several human case series reports. In contrast, RANTES decreased in fatal cases and was inversely correlated with disease severity (Papa et al., 2016b). However, this finding is not recapitulated in animal models: a transient elevation in RANTES was seen during the course of infection in a non-lethal IFNAR−/− mouse model, and elevated levels of RANTES correlated with disease severity in wild-type mice (Bente et al., 2010; Hawman et al., 2021; Hawman et al., 2019).
Table 3.
Pro-inflammatory cytokine response to CCHFV: Other cytokines
| Cytokine | Infected cell/species | Lethal | Sample | Level in acute infection | Reference |
|---|---|---|---|---|---|
| IP-10 | Human (Turkey) | Y/N | Serum | + | (Papa et al., 2016b) |
| Mø | − | PBMC | + | (Peyrefitte et al., 2010) | |
| TNF-α | Human (Turkey, Albania, Kosovo) | Y/N | Serum | + | (Ergonul et al., 2006), (Papa et al., 2006), (Saksida et al., 2010), (Papa et al., 2016b) |
| Mouse (STAT-1, IFNAR) | Y/N | Plasma/serum | + | (Bente et al., 2010), (Hawman et al., 2019) | |
| Cell (Mø, DC, dDC, LC) | − | PBMC/cord blood | + | (Connolly-Andersen et al., 2009), (Peyrefitte et al., 2010), (Rodriguez et al., 2018) | |
| MCP-1 | Human (Turkey) | Y/N | Serum | + | (Papa et al., 2016b), (Ergönül et al., 2017) |
| NHP | Y/N | Serum | + | (Haddock et al., 2018), (Smith et al., 2019), (Cross et al., 2020) | |
| Mouse (STAT-1) | Y | Plasma | + | (Bente et al., 2010) | |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| Cell (Mø, dDC, LC) | − | PBMC/cord blood | + | (Peyrefitte et al., 2010), (Rodriguez et al., 2018) | |
| MIP-1a | Human (Turkey) | Y/N | Serum | +/− | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| Cell (Mø) | − | PBMC | + | (Peyrefitte et al., 2010) | |
| MIP-1b | Human (Turkey) | Y/N | Serum | +/− | (Papa et al., 2016b) |
| NHP | Y/N | Serum | + | (Haddock et al., 2018) | |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| RANTES | Human (Turkey) | Y/N | Serum | (−) | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| Cell (Mø) | − | PBMC | + | (Peyrefitte et al., 2010) | |
| CD154 | NHP | N | Serum | + | (Smith et al., 2019) |
| VEGF | Human (Turkey) | Y/N | Serum | +/− | (Papa et al., 2016b) |
| NHP | N | Serum | + | (Smith et al., 2019) | |
| GM-CSF | Human (Turkey) | Y/N | Serum | NSF | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | (−) | (Hawman et al., 2019) | |
| PDGF | Human (Turkey) | Y/N | Serum | +/− | (Papa et al., 2016b) |
| Eotaxin | Human (Turkey) | Y/N | Serum | +/− | (Papa et al., 2016b) |
| Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) | |
| KC | Mouse (IFNAR) | N | Serum | + | (Hawman et al., 2019) |
DC, dendritic cell; dDC dermal dendritic cell; IFNAR, type I interferon (IFN) receptor-deficient mice; LC, Langerhans cell; Mø, macrophage; −, not applicable; STAT-1, STAT1-deficient mice; NHP, non- human primate (all NHP data are from Cynomolgus macaques); NSF, no significant findings; +, elevated;
+/−, can be elevated; Y, reported in lethal disease; Y/N, reported in lethal and non-lethal disease; N, reported in non-lethal disease;
, significantly higher in fatal CCHF than in nonfatal CCHF.
The relationship between IL-6 expression and disease severity is not clear; in sera from patients in Turkey, increased IL-6 positively correlated with DIC, with highest levels seen in fatal cases (Ergönül et al., 2017; Ergonul et al., 2006). Similarly, increased IL-6 levels were seen in CCHFV-infected mice (Bente et al., 2010; Hawman et al., 2021; Hawman et al., 2019). However, a study in Albania found elevated IL-6 in both mild and severe cases (Papa et al., 2006), suggesting that more analysis is needed to determine the relationship between IL-6 levels and disease outcome.
Finally, increased levels of other pro-inflammatory cytokines that have not been as extensively studied are also generally associated with disease. For example, MIP-1α and MIP-1β were both elevated in one fatal case (Papa et al., 2016b), and slight increases of these factors were observed in animal models (Bente et al., 2010; Haddock et al., 2018). Similarly, elevated VEGF, G-CSF, PDGF, and eotaxin levels have been described in an individual case (Papa et al., 2016b). In wild-type mice infected with a mouse-adapted CCHFV variant, disease severity correlated with increased levels of IL-1β, IL-5, IL-6, G-CSF, KC, MCP-1, MIP-1α, MIP-1β, and RANTES (Hawman et al., 2021), suggesting that these pro-inflammatory cytokines contribute to disease severity.
Descriptions of anti-inflammatory responses during CCHFV infection are fewer. IL-10 has been evaluated in patients, animals, and cultured immune cells (Table 4). In patients, one study found that serum levels negatively correlated with DIC scores (Ergonul et al., 2006), whereas in other studies, levels differed significantly between fatal and non-fatal cases and increased expression was associated with poor outcome (Papa et al., 2016b; Saksida et al., 2010). These latter studies led to the hypothesis that severe CCHF could be the result of delayed, downregulated immune response caused by IL-10, leading to increased viral replication and dissemination (Saksida et al., 2010). Elevated IL-10 was also seen in the lethal Stat-1−/− mouse model (Bente et al., 2010). Interestingly, in a non-lethal IFNAR−/− model, serum IL-10 levels were slightly (but not significantly) elevated 14 dpi (Hawman et al., 2019). Data on other anti-inflammatory cytokines are limited: IL-5 levels decrease with disease severity (Papa et al., 2016b), and a cell culture investigation of IL-19, which can result in increased IL-10 when present during DC maturation, found no correlation between IL-19 and IL-10 levels in DCs (Connolly-Andersen et al., 2009).
Given the putative role of immune cells in early infection and virus dissemination, more detailed studies were performed to investigate cytokine expression in macrophages and DCs. Infected M2-like macrophages produce elevated pro-inflammatory cytokines IFN-α, TNF-α, MCP-1, IP-10, IL-6, RANTES, and MIP-1α 24–72 hpi, suggesting that, once activated by CCHFV, these cells remain activated without hindering the antiviral state (Peyrefitte et al., 2010). These data, along with earlier susceptibility studies, indicate that M2-like macrophages may be infected by CCHFV, replicate the virus, and subsequently disseminate it while producing a host of pro- inflammatory cytokines and chemokines. In infected DCs, pro-inflammatory TNF-α and IL-6, along with anti-inflammatory IL-10, were elevated 48 hpi, whereas no differences in expression of IL-8, IL-19, and IL-1β were seen compared to uninfected cells (Connolly-Andersen et al., 2009). IL-6 and MCP-1 were elevated in both infected dDC and LC, while IL-8 and TNF-α were only elevated in dDC and LC, respectively (Rodriguez et al., 2018).
Finally, to determine the effect of cytokines secreted from DCs, supernatants from infected moDCs were applied to HUVEC. Cellular activation was measured by determining cell surface and/or transcriptional levels of intracellular adhesion molecule 1 (ICAM1), E-selectin, and vascular cell adhesion molecule 1 (VCAM1), and levels of pro-inflammatory cytokines IL-6 and IL-8 were compared to mock and lipopolysaccharide (LPS)-treated controls (Connolly-Andersen et al., 2011, 2009). DC-derived supernatants caused TNF-α mediated endothelial cell activation. Altogether, this suggests that vasculature may be activated by pro-inflammatory cytokines, specifically TNF-α, or directly by viral output from infected antigen-presenting cells (APC) (Connolly-Andersen et al., 2011).
4.5. Apoptosis and the TNF superfamily of death receptors and ligands
Across the Bunyavirales order, multiple viruses have been shown to interact with host apoptosis machinery (Leventhal et al., 2021), suggesting that modulating apoptosis may be a key step in the replication cycle. CCHFV causes ER stress, disrupting ER homeostasis and protein folding in cells (Fraisier et al., 2014), a process that induces FasL expression and has been attributed to creation of glycoproteins required for egress of maturing virions (Lasecka and Baron, 2014). Infection also induces TNF-α and FasL-mediated apoptosis in cell culture (Karlberg et al., 2015) and, as noted earlier, elevated levels of TNF-α are present in sera and correlate with fatal outcomes in human disease (Ergonul et al., 2006; Papa et al., 2006; Saksida et al., 2010). Secretion of these factors can lead to hepatic injury and has been implicated in cell loss during CCHFV infection (Karlberg et al., 2015; Rodrigues et al., 2012). Additionally, expression of the NSS protein, a cryptic ambisense product of the NP open reading frame (Barnwal et al., 2016), has been shown to induce apoptosis via the intrinsic pathways. NSs co-localizes with mitochondria within infected tissue culture cells and can disrupt mitochondrial membranes, which is also thought to lead to apoptosis (Barnwal et al., 2016). Conversely, monomeric NP has been shown to delay or inhibit apoptosis via a conserved caspase-3 cleavage motif (Carter et al., 2012; Karlberg et al., 2011; Wang et al., 2012), suggesting a dynamic role for virus-mediated apoptosis in the establishment of infection and in severe outcomes. While the virus may induce apoptosis, additional data also indicate that apoptosis is not a clear feature of severe disease. In a 2015 study, Güven et al. measured serum markers for apoptosis induction (M-30, perforin, caspase-3, and soluble FasL) in 49 patients <18 years of age. Serum values of perforin, caspase-3, and M-30 positively correlated with liver enzyme levels but negatively correlated with platelet counts. Notably, M-30 levels did not differ significantly between any of the groups. Levels of perforin, caspase-3, and soluble FasL were significantly higher in the severe and non-severe CCHF groups than in healthy controls, but no significant difference was seen between severe and non-severe groups. (Güven et al., 2015).
4.6. Complement system
Limited data indicate that the complement system is involved in protective humoral immunity to CCHFV: a complement knockout (C3−/−) mouse study evaluating a mAb demonstrated that functional complement activity is required for monoclonal antibody-mediated protection following CCHFV challenge (Golden et al., 2019). This study demonstrates that protective effects of antibodies, elicited through vaccination or infection, may require complement. C3 and C4 levels were significantly lower in fatal human cases, suggesting that the classical complement pathway may play an important role in survival (Ozturk et al., 2012). In addition, a study of pediatric cases found that elevated levels of factor Bb, the fragment of complement factor B that results from activation of the alternative pathway, were associated with increased disease severity indicated by elevated liver enzymes and thrombocytopenia, suggesting that the alternative pathway may also contribute to disease pathogenesis (Sancakdar et al., 2014).
4.7. Humoral immunity
Studies of humoral immunity are summarized in Table 5. Anti-CCHFV IgM antibodies are detected as early as 4–5 days after symptom onset and more frequently 7–8 days post-onset (Burt et al., 1994; Ergunay et al., 2014; Shepherd et al., 1989). IgG antibodies are also typically detected on days 7–9. IgG titers have been observed to increase, peaking 2–5 months after the onset of illness and remaining high for up to 3 years. In contrast, IgM titers begin to decline around day 20 and become undetectable 3–5 months after disease onset.
Table 5.
Summary of CCHFV humoral immunity data from human and animal studies.
| Source | N | Location | Date* | IgM | IgG | NAbs | Parameters associated with survival | Reference |
|---|---|---|---|---|---|---|---|---|
| Human | 7 | Bulgaria | 1989 | NR | NR | − | − | (Vassilenko et al., 1990) |
| Human | 8 | Bulgaria | 2011 | + | + | + | IgG, Non-NAbs | (Mousavi-Jazi et al., 2012) |
| Human | 11 | Bulgaria | 2013–4 | + | + | − | IgG | (Papa et al., 2016a) |
| Human | 5 | India | 2015 | + | − | − | (IgM not associated with survival) |
(Yadav et al., 2016) |
| Human | 12 | India | 2011–9 | + | − | − | IgM | (Gambhir and Rathod, 2019) |
| Human | 24 | Kosovo | 2001, 2003, 2005 |
+ | + | − | IgG, (IgM not associated with survival) |
(Duh et al., 2007) |
| Human | 3 | Pakistan | 2016 | − | + | − | IgG | (Yaqub et al., 2019) |
| Human | 7 | Russia | 1961–4 | NR | NR | − | − | (Leshchinskaya, 1965) |
| Human | 1 | Senegal | 2003 | + | + | − | IgM/IgG | (Nabeth et al., 2004) |
| Human | 7 | South Africa | 1984 | NR | NR | − | − | (Van Eeden et al., 1985) |
| Human | 35 | South Africa | 1981–7 | + | + | + | IgM/IgG, Non-NAbs | (Shepherd et al., 1989) |
| Human | 7 | South Africa | 1984 | NR | NR | − | Abs (unspecified) | (Shepherd et al., 1985) |
| Human | 101 | South Africa | 1981–92 | + | + | − | IgM/IgG | (Burt et al., 1994) |
| Human | 2 | Spain | 2016 | + | + | − | IgM/IgG | (Negredo et al., 2017) |
| Human | 20 | Turkey | 2012 | + | + | − | Anti-NP IgM/IgG, (Anti-GPC IgM/IgG not associated with survival) |
(Ergunay et al., 2014) |
| Human | 31 | Turkey | 2012–4 | − | + | − | IgG | (Kaya et al., 2014) |
| Human | 1 | Uganda | 2015 | + | + | − | IgM/IgG | (Balinandi et al., 2018) |
| Human | 7 | UAE | 1979–80 | NR | NR | − | Abs (unspecified) | (Suleiman et al., 1980) |
| Human | 1 | UK | 2014 | + | + | − | IgM/IgG | (Lumley et al., 2014) |
| Cynomolgus macaque | 12 | NA | 2018 | + | + | + | − | (Haddock et al., 2018) |
| Suckling mice | 5−8 | NA | 2005 | − | + | + | NAbs, Non-NAbs | (Bertolotti-Ciarlet et al., 2005) |
| IFNAR−/− mice | 23 | NA | 2019 | − | + | + | Non-Nabs | (Golden et al., 2019) |
| IFNAR−/− mice | 5−8 | NA | 2019 | + | + | + | Non-Nabs | (Hawman et al., 2019) |
| IFN-I deficient (Ab) FcR−/− mice |
8 | NA | 2019 | − | + | + | Non-NAb in the absence of FcR | (Golden et al., 2019) |
| IFN-I deficient (Ab) C3−/− mice |
8 | NA | 2019 | − | + | + | Non-NAb + complement | (Golden et al., 2019) |
| Rag1−/− mice | 5−8 | NA | 2021 | + | + | − | IgM/IgG | (Hawman et al., 2021) |
Date of sampling; N, sample size; NA, not applicable; NR, not reported; +, detected; −, not detected.
While neutralizing antibodies (NAbs) are often present by day 10, titers can vary greatly among patients. In some instances, titers can be high (1:256), but reports more commonly indicate only low levels of NAbs that rapidly decline 19 weeks after onset of illness (Shepherd et al., 1989). Ergunay et al. (2014) evaluated kinetics of antibodies specific for CCHFV NP and GPC, finding anti-NP IgM antibodies as the initial serological marker during human infection and becoming detectable 2–3 days after disease onset; GPC-IgM (4–6 days) and IgG antibodies (5–6 days) were detected and increased thereafter (Ergunay et al., 2014). Similarly, sera from CCHF survivors showed reactivity against NP, suggesting that NP is a target of humoral responses during infection (Karaaslan et al., 2021).
In fatal human cases, virus-specific antibodies are generally undetectable, while survivors reliably generate CCHFV-specific antibody responses (Balinandi et al., 2018; Burt et al., 1994; Gambhir and Rathod, 2019; Kaya et al., 2014; Lumley et al., 2014; Nabeth et al., 2004; Negredo et al., 2017; Papa et al., 2016a; Shepherd et al., 1989, 1985; Yaqub et al., 2019). The magnitude of antibody responses associated with positive disease outcome is not known; recent NHP and murine studies found no correlation between antibody titers or neutralization and disease severity (Fels et al., 2021; Haddock et al., 2018; Hawman et al., 2021; Hawman et al., 2019). In human patients, the appearance of IgM/IgG responses coincides with declining viremia (Burt et al., 1994; Duh et al., 2007; Ergunay et al., 2014; Kaya et al., 2014; Shepherd et al., 1989). More specifically, Ergunay et al. (2014) found that viremia clearance followed anti-NP IgM responses (Ergunay et al., 2014). A study of NHPs vaccinated with DNA encoding the NP and GPC antigens similarly showed that anti-NP IgG were dominant and likely contributed to protection from CCHFV challenge (Hawman et al., 2021a). In non-lethal mouse models, IgM and IgG responses appeared by 6 dpi (Hawman et al., 2021; Hawman et al., 2019).
Although data support that an antibody response is an indicator of improved outcomes, the role of antibodies in controlling acute infection is unclear. In IFNAR−/− mice, depleting B-cells did not affect disease progression (Hawman et al., 2021b), though the depletion regimen did not completely abrogate specific antibody responses (Hawman et al., 2021b). As noted above, antibody titers and neutralizing activity did not correlate with disease severity or outcome in CCHFV-infected NHPs (Haddock et al., 2018). Furthermore, adoptive transfer of sera from immunized mice showed that transfer of both CD3+ T-cells and sera was necessary for clinical benefit (Dowall et al., 2016a), suggesting that antibodies alone are insufficient to control acute infection. However, the role of antibody response in protection may differ between naïve and vaccinated individuals.
Whether patients who succumb to disease fail to develop IgM/IgG responses altogether or simply die before they can mount a detectable response (prior to day 7) is also unknown. Additionally, viremia in fatally infected patients may be so high that any antibodies produced become bound to virus or antigens in immune complexes and therefore are not detected by traditional serologic assays. Anecdotal support for this hypothesis comes from a fatal case in South Africa, in which the formation of immune complexes was detected in the absence of a specific antibody response, along with persistent high-titer viremia (Shepherd et al., 1985).
4.8. Cellular immunity
Early T-cell activation and induction of pro-inflammatory responses correlate with severe disease, as in other viral hemorrhagic fevers such as Ebola (Ergonul et al., 2006; Papa et al., 2006; Saksida et al., 2010). The association between cytokine expression and poor outcomes in CCHF suggests that the balance of the early inflammatory state is an important factor in progression toward severe disease. While high viral loads (>108 copies/mL) in patients are associated with fatal outcome (Cevik et al., 2007; Duh et al., 2007; Papa et al., 2007; Saksida et al., 2010), viral loads were found to decrease independently of IgG levels during the first week of infection, indicating that cellular immunity likely plays a significant role in viral control and clearance (Duh et al., 2007).
Limited work has been done examining the role of T-cell subsets in protection against CCHFV. Early studies showed that total numbers of helper (TH) T-cells are not significantly different between severe and non-severe cases (Akinci et al., 2009; Yilmaz et al., 2008), but one study observed elevated cytotoxic T-cell levels in fatal cases (Akinci et al., 2009). Furthermore, a study examining a DNA vaccine found that a predominantly TH1 response might be important for efficient protection (Hinkula et al., 2017). Regulatory T (TReg) cells, key producers of IL-10, increased in acutely infected patients compared to healthy controls, but not all cells demonstrated immunosuppressive activity (Gazi et al., 2018). Therefore, the role of TReg cells during viral pathogenesis remains unclear. In infected mice, T-cells are rapidly activated to proliferate and produce antiviral cytokines (Hawman et al., 2021b; Hawman et al., 2021; Hawman et al., 2019) and CCHFV-specific T-cell activity is sustained for weeks post-infection in mice that survive (Hawman et al., 2019).
A study of human survivors detected CD8+ T-cell responses up to 13 years after acute infection and determined that T-cell epitopes were predominantly located on the NP, with two instances of reactivity to GC-derived peptides; none of the epitopes appeared to be immunodominant (Goedhals et al., 2017). Cellular immunity directed against NP was also observed in vaccinated mice and rabbits and in human CCHF survivors (Karaaslan et al., 2021). Importantly, these findings suggest that GP-based vaccine candidates may generate suboptimal CD8+ T-cell responses. Indeed, a GP-based subunit vaccine failed to confer protection despite inducing NAbs (Kortekaas et al., 2015). However, a recombinant NP vaccine similarly failed to provide protection despite evidence of humoral and cellular responses (Dowall et al., 2016b). Given that both humoral and cellular immunity appear to be important for protection, characterizing T-cell epitopes has major implications for vaccine design. The interaction between these two responses may also be crucial, as non-neutralizing Abs have been shown to confer protection in vivo (Bertolotti-Ciarlet et al., 2005), potentially acting through mechanisms such as antibody-dependent cellular cytotoxicity.
Cytotoxic adaptive immune cells, including CD8+ T-cells, have been documented in fatal human fatal infections (Akinci et al., 2009; Yilmaz et al., 2008). One in vivo study tested wild- type mice and mice (e.g., NOD/SCID and Prf1 and Rag2 knockout mice) deficient in various adaptive responses (cytotoxic T-cell, B-cell, and/or NK cell populations). Transcripts for both NK and CD8+ T-cells were present in infected livers; however, liver injury was indistinguishable between wild-type and adaptive-deficient mice (Lindquist et al., 2018). This supports a conclusion that CCHFV replication leading to intrinsic liver cell death contributes more to hepatic injury than cytotoxic effector cells (Lindquist et al., 2018). However, in another model, depleting either CD4+ or CD8+ T-cells resulted in significantly increased mortality in infected mice, demonstrating that these cell types were required for survival (Hawman et al., 2021b). Nevertheless, the mechanism by which these T-cells contribute to survival remains to be determined. Depleting CD4 T-cells abrogated the host IFN-γ response to infection and blocking IFN-γ signaling resulted in uniform lethality in IFNAR−/− mice (Hawman et al., 2021b), suggesting that IFN-γ produced by T-cells may be one mechanism by which these cells control the infection. However, as other cell types can produce IFN-γ (Schroder et al., 2004), further studies are needed to determine the T-cell effector functions required for control of CCHFV infection.
4.9. Protective B- and T-cell epitopes
Viral epitopes that elicit durable protective immune responses have been investigated in survivor and vaccinee PBMC and sera, or have been predicted by modeling or immunoinformatic approaches using in silico methods (various discriminant analyses, predictive databases, and machine learning) comparing factors such as solubility, probabilities of allergenicity, and probabilities of antigenicity within adaptive immune cell populations (Nosrati et al., 2020, 2019; Oany et al., 2015; Shrivastava et al., 2020). However, for the latter approach, predictions assessed to date appear largely unrewarding; the only study aiming to validate a CCHFV immunoinformatic approach demonstrated that epitopes selected in silico based on highest prediction scores were often some of the least reactive sites recognized by human survivor sera (Burt et al., 2013).
Serological B-cell epitope studies have largely focused on antibody responses against portions of the GPC. These studies report that sera from human survivors in areas including Turkey, Uganda, and South Africa bind to all portions of the GPC (Fritzen et al., 2018; Mishra et al., 2020; Shalitanati et al., 2018). Interestingly, despite high genetic variability between strains, serum samples from diverse geographic regions contain antibodies with common B-cell epitopes within the mucin-like domain (Fritzen et al., 2018). Other studies in animals have relied on synthetic peptides in combination with rabbit and sheep antisera to CCHFV, identifying several linear B-cell epitopes: 8 within GN (Shalitanati et al., 2018), 6 within GC (Zhang et al., 2019), 9 broad epitopes within NP (Moming et al., 2018), and 5 fine epitopes within NP (Liu et al., 2014). A mouse mAb study identified antibodies that could neutralize diverse strains (Zivcec et al., 2017), demonstrating broadly protective epitopes within the GPC. Additionally, non-neutralizing antibodies that target GP38 can be protective against lethal challenge (Bertolotti-Ciarlet et al., 2005; Golden et al., 2019), indicating that antibody responses directed against these regions of GPC can be protective. Nevertheless, the genetic diversity of the M segment and encoded GPC may be important for vaccine design, since incomplete protection is observed when vaccine- encoded GPC and challenge virus differ, at least with some vaccines (Suschak et al., 2021).
GPC-based vaccines can elicit GPC-specific T-cells (Buttigieg et al., 2014; Hawman et al., 2021a; Suschak et al., 2021), indicating that the GPC can be targeted by T-cells. However, T-cell epitope studies from human cases and animal models have demonstrated that T-cells react primarily to NP, with most of the molecule eliciting memory T-cell responses (Goedhals et al., 2017; Hawman et al., 2021a; Karaaslan et al., 2021; Mousavi-Jazi et al., 2012). As in B-cell studies, PBMCs from geographically diverse samples elicited T-cell memory responses to the NP. However, unlike B-cell epitopes, limited reactivity to GPC was seen. Only two domains within GC induce T-cell responses (Goedhals et al., 2017), while no T-cell epitopes have been observed within the GN (Goedhals et al., 2017). One of the limitations for both sets of B- and T-cell studies is the reliance on synthetic peptides used to stimulate PBMC or to bind antibodies in sera, and the potential effects of O- or N-linked glycosylation present on each mature protein of the GPC. Only one study attempted to synthetically glycosylate the peptides used to determine epitope regions within the GPC (Fritzen et al., 2018); no additional reactivity of the sera was observed after on- chip glycosylation of the peptides. While the authors caution that these experiments may not mimic the natural glycosylation pattern on the envelop proteins, the limited value from this approach to date suggests that post-translational modifications should be carefully considered in future epitope studies that examine the GPC.
5. Knowledge gained from efforts to develop immune-based therapies and vaccines
5.1. Therapeutic antibodies
Several mAb targeting the GC, such as 8A1, 11E7, 30F7, and 12A9 (curated within the Joel M. Dalrymple-Clarence J. Peters USAMRIID Antibody Collection through BEI Resources, NIAID, NIH) can neutralize wild-type virus or VLP (Ahmed et al., 2005; Bertolotti-Ciarlet et al., 2005; Zivcec et al., 2017). However, in mice, neutralization is not directly associated with protection (Fels et al., 2021; Kortekaas et al., 2015), and several non-neutralizing antibodies from the collection, such as 13G8, 10G4, and 6B12, also confer protection (Ahmed et al., 2005; Bertolotti-Ciarlet et al., 2005; Zhang et al., 2019; Zivcec et al., 2017).
Antibody treatment has not consistently improved outcomes in patients (Leshchinskaya, 1965; Suleiman et al., 1980; Van Eeden et al., 1985; Vassilenko et al., 1990). Data from two studies are consistent with antibody therapy-mediated protection. During a nosocomial outbreak in South Africa (1985), human convalescent serum was administered to 5 patients, all of whom survived, while 2 untreated patients succumbed to disease (Van Eeden et al., 1985). Similarly, in Bulgaria, 7 CCHF patients survived severe disease after receiving immune plasma, though no control group was evaluated (Vassilenko et al., 1990). However, several other attempts to utilize immune plasma to treat CCHF were less successful (Leshchinskaya, 1965; Suleiman et al., 1980). These mixed results are likely due to small sample sizes and the lack of standardization for convalescent plasma dosing. While the potential efficacy of administered plasma based on NAb titers was determined in only one report (Vassilenko et al., 1990) from Bulgaria, other studies indicate that the neutralizing activity of convalescent plasma from patients is often low (Shepherd et al., 1989).
Although not all passive transfer studies to date have shown a benefit, antibody treatment is still a promising approach as the optimal quality and characteristics of antibody-based therapeutics are determined. Bertolotti-Ciarlet et al. (2005) generated 14 mAbs targeting the PreGN [later identified as targeting GP38 domain (Golden et al., 2019)] and 14 targeting GC, and evaluated in vitro plaque reduction and protective efficacy in suckling mice when mAbs are administered 24 h before or 24 h after challenge (Bertolotti-Ciarlet et al., 2005; Golden et al., 2019). Overall, neutralization did not correlate with in vivo protection, supporting evidence from vaccination studies (section 5.2). Treatment before challenge conferred better levels of protection, and efficacy was independent of mAb neutralization titers. Treatment post infection was largely ineffective in groups receiving Gc-targeted neutralizing antibodies, whereas some protection was seen in almost all groups receiving non-neutralizing, GN-targeted mAbs. Only 6 of 12 neutralizing GC mAbs conferred ≥20% protection vs. 9 of 14 PreGn/GP38 non-neutralizing mAbs (Bertolotti-Ciarlet et al., 2005, Golden et al., 2019). A more recent study of the protective efficacy of these antibodies in neonatal and IFNAR−/− mice confirmed these results (Golden et al., 2019). Notably, the researchers identified a non-neutralizing antibody (mAb-13G8) targeting the PreGN GP38 domain that conferred 100% protection from lethal challenge, indicating that a mechanism other than virus neutralization may be an important correlate of survival and protection. This mechanism could be Fc-mediated effector functions such as ADCC, ADCP, or complement-dependent cytotoxicity (Golden et al., 2019). Recently, neutralizing antibodies identified in human survivors were found to protect mice from lethal infection when administered 24 hours prior to infection or 30 min post- exposure. When the same candidates were given therapeutically at 24 hpi, protective efficacy was largely lost (Fels et al., 2021). These outcomes are consistent with the lack of robust NAb titers seen in human survivors as well as recipients of the Bulgarian vaccine (Mousavi-Jazi et al., 2012; Shepherd et al., 1989), and suggest that antibodies may have protective functions beyond neutralizing infectious particles.
5.2. Vaccines
To date, over 30 CCHFV vaccine candidates have been evaluated, as reviewed in (Tipih and Burt, 2020). In general, all studies that assessed Ig responses found detectable levels of antibodies; however, NAbs were only generated when some form of GC (i.e., whole virion, VLP, GPC, or GC) was used as antigenic stimulation. Non-neutralizing antibodies that target portions of the PreGN molecule, specifically GP38, confer passive protection, and vaccines targeting only GP38 protect against several CCHFV strains (Golden et al., 2019; Mishra et al., 2020; Suschak et al., 2021). While this molecule was originally considered a non-structural protein (Bergeron et al., 2007; Bertolotti-Ciarlet et al., 2005; Sanchez et al., 2006), recent studies show that GP38 can be antigenically found on the surface of nascent CCHF VLP (Golden et al., 2019) and is required for production of infectious VLP (Freitas et al., 2020). As of the writing of this review, these data have yet to be validated with wild-type CCHFV.
Low levels of NAbs after vaccination with the Bulgarian inactivated CCHFV vaccine are consistent with the neutralizing responses in human cases (section 7) (Mousavi-Jazi et al., 2012). While this vaccine may have contributed to reducing the number of reported CCHF cases in Bulgaria over a 21-year period, NAb titers were low in most vaccinees, even in individuals who had received four doses (Mousavi-Jazi et al., 2012; Papa et al., 2016a).
A recent study of a DNA-based CCHFV vaccine in cynomolgus macaques yielded similar results: compared to sham-vaccinated controls, vaccinated animals effectively controlled infection in the absence of robust NAb titers (Hawman et al., 2021a). Correspondingly, several other experimental vaccines evaluated in murine models produced high NAb titers in the absence of protection (Buttigieg et al., 2014; Kortekaas et al., 2015). In addition, in humans vaccinated in Bulgaria, T-cell responses to NP correlate with the number of doses received (Mousavi-Jazi et al., 2012). T-cell responses have also been implicated in vaccines encoding NP; numerous vectored and DNA/RNA vaccines encoding NP confer up to 100% protection from lethal CCHFV challenge in immunocompromised mice (Aligholipour Farzani et al., 2019b, 2019c, 2019a; Scholte et al., 2019; Zivcec et al., 2018) and NHPs (Hawman et al., 2021a). While complete protection can be achieved with NP alone, the delivery or vector of expressing this antigen is important. For example, the modified vaccinia Ankara virus (MVA) platform provides protection with GPC alone, but not with NP alone (Buttigieg et al., 2014; Dowall et al., 2016a; Dowall et al., 2016b). Despite classic dogma that vaccine protection is achieved through NAb production, this may not be the case for CCHF. In addition, the high sequence diversity of GPC, with up to 25% differences in amino acid sequences among strains (Bente et al., 2013), suggests incomplete protection may be a concern for GPC-only vaccines when the infecting strain differs from the vaccine-encoded antigen. Thus, presence of the highly conserved NP (<5% amino acid differences among strains) in vaccines may be warranted.
6. Conclusion: goals for future research
An overview of innate and adaptive responses discussed above is provided in Figure 1 and summarized in Table 6, highlighting “knowns” and “unknowns” regarding key processes in the immunobiology of CCHF: (1) virus exposure (mucosal/tick bite); (2) cellular infection; (3) liver injury; and (4) adaptive immunity. As discussed above (Section 2), many studies have attempted to address important questions; continuing efforts to answer these questions will have importance for human health beyond CCHF. In conclusion, addressing these questions will help us understand how the virus causes disease in humans; why only some humans develop severe disease; and how viruses spill over into human populations.
Figure 1.
An overview of current knowledge of innate and adaptive responses in immunobiology of CCHFV infection (see accompanying Table 6). (1) Whether infection is via tick bite (A) or mucosal exposure (B), the initial events, including virus trafficking from the site of infection and dissemination to other organs, are poorly understood. Tissue damage is likely a combination of direct infection (2), inflammatory innate responses (3), and killing of infected cells by activated adaptive immune cells (4). Upon infection (2), many host and viral proteins are engaged to promote and limit viral replication. Host sensors that detect infection and initiate innate responses are not well understood and host proteins that act as restriction factors largely remain to be identified. Although type I IFN response is likely a key determinant of disease outcome and infection rapidly becomes resistant to type I IFN-mediated restriction, how virus blocks or resists type I IFN is largely unknown. Additionally, CCHFV possesses proteins with both pro- and anti-apoptotic function, suggesting apoptosis modulation may be a key factor in pathogenesis. Innate cell responses likely play a key early role in disease outcome (3). Infected innate cells can produce pro-inflammatory cytokines that recruit inflammatory cells, further contributing to tissue injury. Innate immune cells such as macrophages and dendritic cells can be infected, although whether they are important for controlling the virus through IFN production, antigen presentation, or reactive oxygen species production is unclear. Cells appear to only become partially activated in response to infection, and whether virus directly inhibits full activation resulting in improper antigen presentation requires further study. (4) Lastly, the role of adaptive immunity in controlling infection warrants further study. Neutralizing antibodies are not necessary for protection, yet other critical effector functions of antibody responses remain to be determined. The role of T-cells is even less clear. It is unknown if CD4 T-helper activity is required for support of B- and CD8-T-cell responses; cytotoxic activity of NK and/or CD8 T-cells is required for controlling virus; T-cells control CCHFV through production of antiviral cytokines; or virus-infected cells resist cytotoxic effector cells through actively blocking host cell apoptosis.
Table 6.
Summary of CCHFV immunobiology: knowns and unknowns
| Process/response | Known | Unknown |
|---|---|---|
| Infection | • Occurs via tick bite or mucosal exposure. | • How does route of infection affect host response? |
| Tick-borne exposure | • Tick-derived salivary factors are delivered along with CCHFV particles. • Resident tissue cells and surveilling immune cells are the likely initial targets of CCHFV replication. • Surveilling immune cells detect virus and traffic back to lymphoid tissue to initiate innate and adaptive immune responses. |
• Is initial amplification at the site of the tick bite required or can virus directly enter blood circulation to seed distant tissues? • Role of tick salivary factors in modulating the innate response to infection. • Role of resident tissue cells in initial viral amplification. • Role of leukocyte trafficking in spread of CCHFV to lymphoid tissues. |
| Cellular infection | • CCHFV inhibits RIG-I mediated sensing. • CCHFV L-protein OTU-like domain blocks ISG15 labeling of host and viral proteins. • The host MXA protein interacts with CCHFV NP to inhibit viral replication. • Established CCHFV infections are resistant to type I IFN-mediated restriction. • Virus infection elicits a pro-inflammatory cytokine response. • NSs protein can depolarize mitochondria and activate intrinsic apoptosis pathways. • NP has anti-apoptotic activity, blocking both intrinsic and extrinsic apoptosis pathways, and a highly conserved DEVD-cleavage motif that can be cleaved by host caspases. |
• Other CCHFV antagonists of innate immunity. • Other host proteins are likely involved in the restriction of CCHFV. • Relative role of cytoplasmic and endosomal sensors. • Why is CCHFV resistant to type-I IFN once infection is established? • Can established CCHFV infections block signaling from the type I IFN receptor? • Why does CCHFV possess proteins with pro- and anti-apoptotic function and retain a sequence that targets the CCHFV NP for cleavage by host caspases? • Does blockade of apoptosis by CCHFV result in inflammatory cell death such as necrosis or necroptosis? |
| Liver pathology | • Virus infection causes liver pathology characterized by hepatocellular necrosis. • Infected innate immunity cells produce inflammatory cytokines. • Pro-inflammatory cytokines drive recruitment of further inflammatory cells, leading to further tissue damage. • Macrophages and dendritic cells likely serve as key antigen-presenting cells for initiation of adaptive immunity. • CCHFV infection of innate cells causes partial activation, potentially impairing resulting stimulation of adaptive immune cells. |
• What causes tissue pathology? Direct viral killing of infected cells, killing of infected cells by adaptive immune responses (CTLs or antibody-dependent cellular cytotoxicity), and/or recruitment of inflammatory cells to infected tissues? • Does virus cause inflammatory immune signaling in infected innate cells? • How does CCHFV infection leads to incomplete activation of APCs? • What role do reactive oxygen species play in tissue damage? • What cell types are required for type I IFN response in vivo? Where does the initial type I IFN, crucial for surviving CCHFV, come from? |
| Adaptive immunity | • Deficiencies in B- and T-cell responses can lead to rapidly fatal infections in mouse models. • Both B- and T-cells likely contribute to survival in acute CCHF. • Neutralization of infectious virus may be dispensable for virus control. |
• Effector functions required of B- and T-cells for protection? • Is virus neutralization truly dispensable for protection? • Role of Fc-dependent effector functions, such as activation of complement or ADCC. • Are CD4 T-helper cells required for effective antibody and CD8 T-cell functions? • Are CD4 T-cells required for antiviral cytokine production? • Role of CD8 T-cells in infection control? Crucial for cell lysis or cytokine production? • Can CCHFV block granzyme-mediated cell killing by CTLs and NK cells? |
Why are humans uniquely susceptible to clinical disease? Humans are largely a dead-end host for CCHF; severe disease is therefore an incidental outcome of selective pressures exerted by the tick and mammalian species that serve as the reservoir and amplifying hosts. Understanding how CCHFV causes disease in humans, but not in most other vertebrates, will provide critical insight into the ability of viruses to spill over into human populations and the factors present in various hosts that prevent or promote disease. Comparative studies in susceptible and resistant models, such as the use of lethal versus non-lethal CCHFV strains in immunodeficient models or male and female mice infected with mouse-adapted CCHFV, provide an opportunity to identify how distinct host responses may contribute to disease outcome. Similarly, comprehensive investigation of CCHFV infection in cattle, sheep, goats or other resistant species will provide insight into unique host responses of reservoir species that likely exert significant selective pressures on the virus. Lastly, mouse adaptation of CCHFV provides a roadmap to the key viral proteins necessary for in vivo pathogenesis, and understanding how the virus has adapted to cause disease in mice will provide insight into how it causes disease in humans.
Does cellular tropism within the mammalian host contribute to disease outcome? We currently have limited understanding of the tropism of CCHFV in vivo. Many hemorrhagic fever viruses cause disease by infecting immune cells, leading to dysregulated host responses. Determining the cellular tropism of CCHFV in susceptible and resistant hosts may identify key cell populations that contribute to disease susceptibility and increase our understanding of how responses in these infected cells can contribute to disease outcome.
How does the host sense CCHFV infection? Despite the severe morbidity and mortality of human infection, how the host senses CCHFV and the restriction factors that limit viral replication are still poorly understood. Identifying host sensing and restricting factors will provide necessary data for understanding how CCHFV causes such a broad spectrum of disease outcomes in humans.
How does CCHFV antagonize host innate immune responses? Across human pathogenic viruses in the Bunyavirales order, the role of CCHFV proteins in mediating pathogenesis are among the least understood. Improved understanding of how the virus antagonizes specific host innate restriction factors will enable development of targeted therapeutics that block this antagonism or target the host to promote antiviral responses. Understanding how the virus interacts with the human host to cause disease will also enable study of whether the genetic diversity of CCHFV contributes to disease outcome. Are some strains more virulent than others? If so, can they be linked by similar genetic traits? This information, combined with surveilling circulating strains in endemic areas, would enable identification of particular strains that may pose an enhanced risk and direct limited public health resources to those most at risk for severe disease.
What role does the host inflammatory response play in disease? For many viral hemorrhagic fevers, dysregulated inflammatory responses contribute to severe morbidity and mortality. A detailed understanding of whether this happens in CCHF is needed. Additionally, animal studies that use therapeutic agents to modulate the host inflammatory response will enable design of therapeutic strategies that promote protection and limit damaging responses. Such strategies may improve clinical outcome in severely ill patients.
Is immunity durable and broadly protective? CCHFV is among the most widely distributed and genetically diverse hemorrhagic fever viruses. As ongoing pre-clinical development continues and eventual clinical development of vaccines begins, creating a vaccine for each individual strain or even closely related strains will be impractical. Thus, a successful vaccine must protect against all strains, but whether immunity will be broadly protective, and which correlates of vaccine-induced immunity contribute to broad protection, are poorly studied. Research is needed to address the breadth of immunity to CCHFV to direct critical vaccine development. Knowledge of the duration of immunity is also critical for determining public health strategies for deploying vaccines to at-risk populations.
Highlights.
Humans are uniquely susceptible to severe disease upon infection with Crimean-Congo hemorrhagic fever virus (CCHFV)
CCHFV pathogenesis appears to be a contribution of virus-induced pathology and dysregulated inflammatory immune responses
Both innate and adaptive immunity likely contribute to control of CCHFV infection
Recent advances in animal modeling of infection will further studies of host and viral determinants of immunity and disease
A better understanding of CCHFV immunobiology is needed to direct vaccine and antiviral advancement
Acknowledgements
We thank Drs. Dennis Bente, Aura Garrison and Anita McElroy for sharing unpublished works, Perry Rose, Rocky Mountain Laboratories Visual Medical Arts Department for figure preparations and Dr. Tatyana Klimova for assistance with editing the manuscript.
Financial support. This work was supported in part by CDC Emerging Infectious Disease Research Core Funds, the NIH/NIAID Division of Intramural Research, and an NIAID grant (1R01AI151006). Work was also supported in part by a cooperative agreement from the U.S. Department of Agriculture—Agricultural Research Service (USDA-ARS; 58-8064-9-015; T.E.S. and B.H.B.) and by appointments to the Centers for Disease Control and Prevention, administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the USDA-ARS (S.E.R. and T.J.O). ORISE is managed by Oak Ridge Associated Universities (ORAU) under contract with DOE.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Potential conflicts of interest. All authors: no reported conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed AA, McFalls JM, Hoffmann C, Filone CM, Stewart SM, Paragas J, Khodjaev S, Shermukhamedova D, Schmaljohn CS, Doms RW, Bertolotti- Ciarlet A, 2005. Presence of broadly reactive and group-specific neutralizing epitopes on newly described isolates of Crimean-Congo hemorrhagic fever virus. The Journal of General Virology 86, 3327–3336. 10.1099/vir.0.81175-0 [DOI] [PubMed] [Google Scholar]
- Akinci E, Yilmaz M, Bodur H, Ongürü P, Bayazit FN, Erbay A, Ozet G, 2009. Analysis of lymphocyte subgroups in Crimean-Congo hemorrhagic fever. International Journal of Infectious Diseases 13, 560–3. 10.1016/j.ijid.2008.08.027 [DOI] [PubMed] [Google Scholar]
- Aligholipour Farzani T, Földes K, Ergünay K, Gurdal H, Bastug A, Ozkul A, 2019a. Immunological analysis of a CCHFV mRNA vaccine candidate in mouse models. Vaccines 7, 115. 10.3390/vaccines7030115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aligholipour Farzani T, Földes K, Hanifehnezhad A, Yener Ilce B, Bilge Dagalp S, Amirzadeh Khiabani N, Ergünay K, Alkan F, Karaoglu T, Bodur H, Ozkul A, 2019b. Bovine herpesvirus type 4 (BoHV-4) vector delivering nucleocapsid protein of Crimean-Congo hemorrhagic fever virus induces comparable protective immunity against lethal challenge in IFNα/β/γR−/− mice models. Viruses 11. 10.3390/v11030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aligholipour Farzani T., Hanifehnezhad A, Földes K, Ergünay K, Yilmaz E, Hashim Mohamed Ali H, Ozkul A, 2019c. Co-delivery effect of CD24 on the immunogenicity and lethal challenge protection of a DNA vector expressing nucleocapsid protein of Crimean-Congo hemorrhagic fever virus. Viruses 11. 10.3390/v11010075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altamura LA, Bertolotti-Ciarlet A, Teigler J, Paragas J, Schmaljohn CS, Doms RW, 2007. Identification of a novel C-terminal cleavage of Crimean-Congo hemorrhagic fever virus PreGN that leads to generation of an NSM protein. Journal of Virology 81, 6632–42. 10.1128/JVI.02730-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MH, Schrama D, Thor Straten P, Becker JC, 2006. Cytotoxic T cells. The Journal of Investigative Dermatology 126, 32–41. 10.1038/sj.jid.5700001 [DOI] [PubMed] [Google Scholar]
- Andersson I, Bladh L, Mousavi-Jazi M, Magnusson K-E, Lundkvist A, Haller O, Mirazimi A, 2004. Human MxA protein inhibits the replication of Crimean-Congo hemorrhagic fever virus. Journal of Virology 78, 4323–9. 10.1128/jvi.78.8.4323-4329.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson I, Karlberg H, Mousavi-Jazi M, Martínez-Sobrido L, Weber F, Mirazimi A, 2008. Crimean-Congo hemorrhagic fever virus delays activation of the innate immune response. Journal of Medical Virology 80, 1397–404. 10.1002/jmv.21222 [DOI] [PubMed] [Google Scholar]
- Andersson I, Lundkvist A, Haller O, Mirazimi A, 2006. Type I interferon inhibits Crimean-Congo hemorrhagic fever virus in human target cells. Journal of Medical Virology 78, 216–22. 10.1002/jmv.20530 [DOI] [PubMed] [Google Scholar]
- Arslan S, Engin A, Özbilüm N, Bakır M, 2015. Toll-like receptor 7 Gln11Leu, c.4–151A/G, and +1817G/T polymorphisms in Crimean-Congo hemorrhagic fever. Journal of Medical Virology 87, 1090–5. 10.1002/jmv.24174 [DOI] [PubMed] [Google Scholar]
- Balinandi S, Patel K, Ojwang J, Kyondo J, Mulei S, Tumusiime A, Lubwama B, Nyakarahuka L, Klena JD, Lutwama J, Strӧher U, Nichol ST, Shoemaker TR, 2018. Investigation of an isolated case of human Crimean-Congo hemorrhagic fever in Central Uganda, 2015. International Journal of Infectious Diseases 68, 88–93. 10.1016/j.ijid.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN, 2001. Host defense, viruses and apoptosis. Cell Death & Differentiation 8, 113–126. 10.1038/sj.cdd.4400823 [DOI] [PubMed] [Google Scholar]
- Barnwal B, Karlberg H, Mirazimi A, Tan Y-J, 2016. The non-structural protein of Crimean-Congo hemorrhagic fever virus disrupts the mitochondrial membrane potential and induces apoptosis. Journal of Biological Chemistry 291, 582–592. 10.1074/jbc.M115.667436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville A, Satti A, Murphy FA, Simpson DI, 1981. Congo-Crimean haemorrhagic fever in Dubai: histopathological studies. Journal of Clinical Pathology 34, 871–874. 10.1136/jcp.34.8.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bente DA, Alimonti JB, Shieh W-J, Camus G, Ströher U, Zaki S, Jones SM, 2010. Pathogenesis and immune response of Crimean-Congo hemorrhagic fever virus in a STAT-1 knockout mouse model. Journal of Virology 84, 11089–11100. 10.1128/JVI.01383-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bente DA., Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M, 2013. Crimean-Congo hemorrhagic fever: History, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Research 100, 159–189. 10.1016/j.antiviral.2013.07.006 [DOI] [PubMed] [Google Scholar]
- Bereczky S, Lindegren G, Karlberg H, Akerstrom S, Klingstrom J, Mirazimi A, 2010. Crimean-Congo hemorrhagic fever virus infection is lethal for adult type I interferon receptor-knockout mice. Journal of General Virology 91, 1473–1477. 10.1099/vir.0.019034-0 [DOI] [PubMed] [Google Scholar]
- Bergeron E, Vincent MJ, Nichol ST, 2007. Crimean-Congo hemorrhagic fever virus glycoprotein processing by the endoprotease SKI-1/S1P is critical for virus infectivity. Journal of Virology 81, 13271–13276. 10.1128/JVI.01647-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron É, Zivcec M, Chakrabarti AK, Nichol ST, Albariño CG, Spiropoulou CF, 2015. Recovery of recombinant Crimean-Congo hemorrhagic fever virus reveals a function for non-structural glycoprotein cleavage by furin. PLoS Pathogens. 10.1371/journal.ppat.1004879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A, Smith J, Strecker K, Paragas J, Altamura LA, McFalls JM, Frias-Stäheli N, García-Sastre A, Schmaljohn CS, Doms RW, 2005. Cellular localization and antigenic characterization of Crimean-Congo hemorrhagic fever virus glycoproteins. Journal of Virology 79, 6152–6161. 10.1128/JVI.79.10.6152-6161.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi L, Lalle E, Caglioti C, Travaglini D, Lapa D, Marsella P, Quartu S, Kis Z, Arien KK, Huemer HP, Meschi S, Ippolito G, di Caro A, Capobianchi MR, Castilletti C, 2015. Antagonistic antiviral activity between IFN-lambda and IFN- alpha against lethal Crimean-Congo hemorrhagic fever virus in vitro. PLOS ONE 10, e0116816. 10.1371/journal.pone.0116816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt FJ, Leman PA, Abbott JC, Swanepoel R, 1994. Serodiagnosis of Crimean- Congo haemorrhagic fever. Epidemiology and Infection 113, 551–562. 10.1017/S0950268800068576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt FJ, Samudzi RR, Randall C, Pieters D, Vermeulen J, Knox CM, 2013. Human defined antigenic region on the nucleoprotein of Crimean-Congo hemorrhagic fever virus identified using truncated proteins and a bioinformatics approach. Journal of Virological Methods 193, 706–712. 10.1016/j.jviromet.2013.07.055 [DOI] [PubMed] [Google Scholar]
- Burt FJ, Swanepoel R, Shieh WJ, Smith JF, Leman PA, Greer PW, Coffield LM, Rollin PE, Ksiazek TG, Peters CJ, Zaki SR, 1997. Immunohistochemical and in situ localization of Crimean-Congo hemorrhagic fever (CCHF) virus in human tissues and implications for CCHF pathogenesis. Archives of Pathology & Laboratory Medicine 121, 839–46. [PubMed] [Google Scholar]
- Buttigieg KR, Dowall SD, Findlay-Wilson S, Miloszewska A, Rayner E, Hewson R, Carroll MW, 2014. A novel vaccine against Crimean-Congo haemorrhagic fever protects 100% of animals against lethal challenge in a mouse model. PLoS ONE 9, e91516. 10.1371/journal.pone.0091516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SD, Surtees R, Walter CT, Ariza A, Bergeron É, Nichol ST, Hiscox JA, Edwards TA, Barr JN, 2012. Structure, function, and evolution of the Crimean- Congo hemorrhagic fever virus nucleocapsid protein. Journal of Virology 86, 10914–10923. 10.1128/JVI.01555-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik MA, Erbay A, Bodur H, Eren SS, Akinci E, ener K, Onguru P, Kubar A, 2007. Viral load as a predictor of outcome in Crimean-Congo hemorrhagic fever. Clinical Infectious Diseases 45, e96–e100. 10.1086/521244 [DOI] [PubMed] [Google Scholar]
- Chumakov MP, Goldfarb LG, Reznikova O.Yu., Kondratenko VF, Myskin AA, 1980. An epidemiological model of Crimean hemorrhagic fever *. The American Journal of Tropical Medicine and Hygiene 29, 260–264. 10.4269/ajtmh.1980.29.260 [DOI] [PubMed] [Google Scholar]
- Connolly-Andersen A-M, Douagi I, Kraus AA, Mirazimi A, 2009. Crimean Congo hemorrhagic fever virus infects human monocyte-derived dendritic cells. Virology 390, 157–62. 10.1016/j.virol.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Connolly-Andersen A-M, Moll G, Andersson C, Akerstrom S, Karlberg H, Douagi I, Mirazimi A, 2011. Crimean-Congo hemorrhagic fever virus activates endothelial cells. Journal of Virology 85, 7766–7774. 10.1128/JVI.02469-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross RW, Prasad AN, Borisevich V, Geisbert JB, Agans KN, Deer DJ, Fenton KA, Geisbert TW, 2020. Crimean-Congo hemorrhagic fever virus strains Hoti and Afghanistan cause viremia and mild clinical disease in cynomolgus monkeys. PLOS Neglected Tropical Diseases 14, e0008637. 10.1371/journal.pntd.0008637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall SD, Buttigieg KR, Findlay-Wilson SJD, Rayner E, Pearson G, Miloszewska A, Graham VA, Carroll MW, Hewson R, 2016. A Crimean-Congo hemorrhagic fever (CCHF) viral vaccine expressing nucleoprotein is immunogenic but fails to confer protection against lethal disease. Human Vaccines and Immunotherapeutics 12, 519–527. 10.1080/21645515.2015.1078045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall SD, Graham VA, Rayner E, Hunter L, Watson R, Taylor I, Rule A, Carroll MW, Hewson R, 2016. Protective effects of a Modified Vaccinia Ankara- based vaccine candidate against Crimean-Congo haemorrhagic fever virus require both cellular and humoral responses. PloS one 11, e0156637. 10.1371/journal.pone.0156637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duh D, Saksida A, Petrovec M, Ahmeti S, Dedushaj I, Panning M, Drosten C, Avšič-Županc T, 2007. Viral load as predictor of Crimean-Congo hemorrhagic fever outcome. Emerging Infectious Diseases 13, 1769–1772. 10.3201/eid1311.070222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin A, Arslan S, Kizildag S, Oztürk H, Elaldi N, Dökmetas I, Bakir M, 2010. Toll- like receptor 8 and 9 polymorphisms in Crimean-Congo hemorrhagic fever. Microbes and Infection 12, 1071–8. 10.1016/j.micinf.2010.07.012 [DOI] [PubMed] [Google Scholar]
- Engin A, Arslan S, Özbilüm N, Bakir M, 2016. Is there any relationship between Toll-like receptor 3 c.1377C/T and −7C/A polymorphisms and susceptibility to Crimean Congo hemorrhagic fever? Journal of Medical Virology 88, 1690–6. 10.1002/jmv.24519 [DOI] [PubMed] [Google Scholar]
- Ergonul O, Mirazimi A, Dimitrov DS, 2007. Treatment of Crimean-Congo hemorrhagic fever, in: Crimean-Congo Hemorrhagic Fever. Springer Netherlands, Dordrecht, pp. 245–269. 10.1007/978-1-4020-6106-6_19 [DOI] [Google Scholar]
- Ergönül Ö, Şeref C, Eren Ş, Çelikbaş A, Baykam N, Dokuzoğuz B, Gönen M, Can F, 2017. Cytokine response in crimean-congo hemorrhagic fever virus infection. Journal of Medical Virology 89, 1707–1713. 10.1002/jmv.24864 [DOI] [PubMed] [Google Scholar]
- Ergonul O, Tuncbilek S, Baykam N, Celikbas A, Dokuzoguz B, 2006. Evaluation of serum levels of interleukin (IL)–6, IL‐ 10, and tumor necrosis factor–α in patients with Crimean‐ Congo hemorrhagic fever. The Journal of Infectious Diseases 193, 941–944. 10.1086/500836 [DOI] [PubMed] [Google Scholar]
- Ergunay K, Kocak Tufan Z, Bulut C, Kinikli S, Demiroz AP, Ozkul A, 2014. Antibody responses and viral load in patients with Crimean-Congo hemorrhagic fever: a comprehensive analysis during the early stages of the infection. Diagnostic Microbiology and Infectious Disease 79, 31–36. 10.1016/j.diagmicrobio.2013.12.015 [DOI] [PubMed] [Google Scholar]
- Erickson BR, Deyde V, Sanchez AJ, Vincent MJ, Nichol ST, 2007. N-linked glycosylation of Gn (but not Gc) is important for Crimean Congo hemorrhagic fever virus glycoprotein localization and transport. Virology 361, 348–55. 10.1016/j.virol.2006.11.023 [DOI] [PubMed] [Google Scholar]
- Everett H, McFadden G, 1999. Apoptosis: an innate immune response to virus infection. Trends in Microbiology 7, 160–165. 10.1016/S0966-842X(99)01487-0 [DOI] [PubMed] [Google Scholar]
- Fajs L, Resman K, Avšič-Županc T, 2014. Crimean-Congo hemorrhagic fever virus nucleoprotein suppresses IFN-beta-promoter-mediated gene expression. Archives of Virology 159, 345–348. 10.1007/s00705-013-1816-2 [DOI] [PubMed] [Google Scholar]
- Fels JM, Maurer DP, Herbert AS, Wirchnianski AS, Vergnolle O, Cross RW, Abelson DM, Moyer CL, Mishra AK, Aguilan JT, Kuehne AI, Pauli NT, Bakken RR, Nyakatura EK, Hellert J, Quevedo G, Lobel L, Balinandi S, Lutwama JJ, Zeitlin L, Geisbert TW, Rey FA, Sidoli S, McLellan JS, Lai JR, Bornholdt ZA, Dye JM, Walker LM, Chandran K, 2021. Protective neutralizing antibodies from human survivors of Crimean-Congo hemorrhagic fever. Cell 184, 3486–3501.e21. 10.1016/j.cell.2021.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Bocarsly P, Dai J, Singh S, 2008. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine & Growth Factor Reviews 19, 3–19. 10.1016/j.cytogfr.2007.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraisier C, Rodrigues R, Vu Hai V, Belghazi M, Bourdon S, Paranhos-Baccala G, Camoin L, Almeras L, Peyrefitte CN, 2014. Hepatocyte pathway alterations in response to in vitro Crimean Congo hemorrhagic fever virus infection. Virus Research 179, 187–203. 10.1016/j.virusres.2013.10.013 [DOI] [PubMed] [Google Scholar]
- Freitas N, Enguehard M, Denolly S, Levy C, Neveu G, Lerolle S, Devignot S, Weber F, Bergeron E, Legros V, Cosset F-L, 2020. The interplays between Crimean-Congo hemorrhagic fever virus (CCHFV) M segment-encoded accessory proteins and structural proteins promote virus assembly and infectivity. PLOS Pathogens 16, e1008850. 10.1371/journal.ppat.1008850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N, Giannakopoulos N. v., Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, García- Sastre A, Virgin HW, 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host & Microbe 2, 404–416. 10.1016/j.chom.2007.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzen A, Risinger C, Korukluoglu G, Christova I, Corli Hitzeroth A, Viljoen N, Burt FJ, Mirazimi A, Blixt O, 2018. Epitope-mapping of the glycoprotein from Crimean-Congo hemorrhagic fever virus using a microarray approach. PLOS Neglected Tropical Diseases 12, e0006598. 10.1371/journal.pntd.0006598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhir RM., Rathod MM, 2019. Prognostic factors in CCHF - An Indian origin study. The Journal of the Association of Physicians of India 67, 35–37. [PubMed] [Google Scholar]
- Garcia-Sastre A, 2010. Diversity, replication, pathogenicity and cell biology of Crimean- Congo hemorrhagic fever virus. New York. [Google Scholar]
- Garrison AR, Smith DR, Golden JW, 2019. Animal models for Crimean-Congo hemorrhagic fever human disease. Viruses. 10.3390/v11070590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazi U, Yapar D, Karasartova D, Gureser AS, Akdogan O, Unal O, Baykam N, Taylan Ozkan A, 2018. The role of T reg population in pathogenesis of Crimean Congo hemorrhagic fever. Virus Research 250, 1–6. 10.1016/j.virusres.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Goedhals D, Paweska JT, Burt FJ, 2017. Long-lived CD8+ T cell responses following Crimean-Congo haemorrhagic fever virus infection. PLoS Neglected Tropical Diseases 11, e0006149. 10.1371/journal.pntd.0006149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JW, Shoemaker CJ, Lindquist ME, Zeng X, Daye SP, Williams JA, Liu J, Coffin KM, Olschner S, Flusin O, Altamura LA, Kuehl KA, Fitzpatrick CJ, Schmaljohn CS, Garrison AR, 2019. GP38-targeting monoclonal antibodies protect adult mice against lethal Crimean-Congo hemorrhagic fever virus infection. Science Advances 5, eaaw9535. 10.1126/sciadv.aaw9535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güven AS, Sancakdar E, Uysal EB, Kaya A, Oflaz MB, Karapinar H, Bolat F, Tuzcu N, Deveci K, Cevit Ö, İcagasioglu FD, 2015. Evaluation of serum perforin, caspase-3, sFasL and M-30 levels as apoptotic markers in children with Crimean-Congo hemorrhagic fever. Pediatric Infectious Disease Journal 34, 208–213. 10.1097/INF.0000000000000530 [DOI] [PubMed] [Google Scholar]
- Haddock E, Feldmann F, Hawman DW, Zivcec M, Hanley PW, Saturday G, Scott DP, Thomas T, Korva M, Avšič -Županc T, Safronetz D, Feldmann H, 2018. A cynomolgus macaque model for Crimean–Congo haemorrhagic fever. Nature Microbiology 3, 556–562. 10.1038/s41564-018-0141-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp S, Fernando L, Schwarz TF, Feldmann H, Flick R, 2005. Intracellular localization of Crimean-Congo Hemorrhagic Fever (CCHF) virus glycoproteins. Virology Journal 2, 42. 10.1186/1743-422X-2-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawman David W, Ahlén G, Appelberg KS, Meade-White K, Hanley PW, Scott D, Monteil V, Devignot S, Okumura A, Weber F, Feldmann H, Sällberg M, Mirazimi A, 2021a. A DNA-based vaccine protects against Crimean-Congo haemorrhagic fever virus disease in a Cynomolgus macaque model. Nature Microbiology 6, 187–195. 10.1038/s41564-020-00815-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawman DW, Haddock E, Meade-White K, Nardone G, Feldmann F, Hanley PW, Lovaglio J, Scott D, Komeno T, Nakajima N, Furuta Y, Gowen BB, Feldmann H, 2020. Efficacy of favipiravir (T-705) against Crimean-Congo hemorrhagic fever virus infection in cynomolgus macaques. Antiviral Research 181, 104858. 10.1016/j.antiviral.2020.104858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawman DW, Meade-White K, Haddock E, Habib R, Scott D, Thomas T, Rosenke R, Feldmann H, 2019. Crimean-Congo hemorrhagic fever mouse model recapitulating human convalescence. Journal of Virology 93. 10.1128/JVI.00554-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawman David W., Meade-White K, Leventhal S, Carmody A, Haddock E, Hasenkrug K, Feldmann H, 2021. T-cells and interferon gamma are necessary for survival following Crimean-Congo hemorrhagic fever virus infection in mice. Microorganisms 9, 279. 10.3390/microorganisms9020279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawman David W, Meade-White K, Leventhal S, Feldmann F, Okumura A, Smith B, Scott D, Feldmann H, 2021b. Immunocompetent mouse model for Crimean- Congo hemorrhagic fever virus. eLife 10. 10.7554/eLife.63906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkula J, Devignot S, Åkerström S, Karlberg H, Wattrang E, Bereczky S, Mousavi- Jazi M, Risinger C, Lindegren G, Vernersson C, Paweska J, van Vuren PJ, Blixt O, Brun A, Weber F, Mirazimi A, 2017. Immunization with DNA plasmids coding for Crimean-Congo hemorrhagic fever virus capsid and envelope proteins and/or virus-like particles induces protection and survival in challenged mice. Journal of Virology 91. 10.1128/JVI.02076-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraal H, 1979. The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. Journal of Medical Entomology 5, 307–417. [DOI] [PubMed] [Google Scholar]
- Joubert JR, King JB, Rossouw DJ, Cooper R, 1985a. A nosocomial outbreak of Crimean-Congo haemorrhagic fever at Tygerberg Hospital. Part III. Clinical pathology and pathogenesis. South African Medical Journal = Suid-Afrikaanse tydskrif vir geneeskunde 68, 722–8. [PubMed] [Google Scholar]
- Joubert JR, King JB, Rossouw DJ, Cooper R, 1985b. A nosocomial outbreak of Crimean- Congo haemorrhagic fever at Tygerberg Hospital Part Ill. Clinical pathology and pathogenesis. South African Medical Journal. [PubMed] [Google Scholar]
- Karaaslan E, Çetin NS, Kalkan-Yazıcı M, Hasanoğlu S, Karakeçili F, Özdarendeli A, Kalkan A, Kılıç AO, Doymaz MZ, 2021. Immune responses in multiple hosts to nucleocapsid protein (NP) of Crimean-Congo hemorrhagic fever virus (CCHFV). PLOS Neglected Tropical Diseases 15, e0009973. 10.1371/journal.pntd.0009973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg H, Tan Y-J, Mirazimi A, 2015. Crimean-Congo haemorrhagic fever replication interplays with regulation mechanisms of apoptosis. The Journal of General Virology 96, 538–546. 10.1099/jgv.0.000011 [DOI] [PubMed] [Google Scholar]
- Karlberg H, Tan Y-J, Mirazimi A, 2011. Induction of caspase activation and cleavage of the viral nucleocapsid protein in different cell types during Crimean-Congo hemorrhagic fever virus infection. Journal of Biological Chemistry 286, 3227–3234. 10.1074/jbc.M110.149369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya S, Elaldi N, Kubar A, Gursoy N, Yilmaz M, Karakus G, Gunes T, Polat Z, Gozel MG, Engin A, Dokmetas I, Bakir M, Yilmaz N, Sencan M, 2014. Sequential determination of serum viral titers, virus-specific IgG antibodies, and TNF- α, IL-6, IL-10, and IFN-γ levels in patients with Crimean-Congo hemorrhagic fever. BMC Infectious Diseases 14, 416. 10.1186/1471-2334-14-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortekaas J, Vloet RPM, McAuley AJ, Shen X, Bosch BJ, de Vries L, Moormann RJM, Bente DA, 2015. Crimean-Congo hemorrhagic fever virus subunit vaccines induce high levels of neutralizing antibodies but no protection in STAT1 knockout mice. Vector-Borne and Zoonotic Diseases 15, 759–764. 10.1089/vbz.2015.1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasecka L, Baron MD, 2014. The molecular biology of nairoviruses, an emerging group of tick-borne arboviruses. Archives of Virology 159, 1249–1265. 10.1007/s00705-013-1940-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchinskaya EV, 1965. Clinical picture of Crimean hemorrhagic fever [in Russian]. Inst Polio Virus Entsef Akad Med Nauk SSSR 7, 226–36. [Google Scholar]
- Leventhal SS., Wilson D, Feldmann H, Hawman DW, 2021. A Look into Bunyavirales genomes: functions of non-structural (NS) proteins. Viruses 13. 10.3390/v13020314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist ME, Zeng X, Altamura LA, Daye SP, Delp KL, Blancett C, Coffin KM, Koehler JW, Coyne S, Shoemaker CJ, Garrison AR, Golden JW, 2018. Exploring Crimean-Congo hemorrhagic fever virus-induced hepatic injury using antibody-mediated type I interferon blockade in mice. Journal of Virology 92. 10.1128/JVI.01083-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Li Yang, Zhao J, Deng F, Duan X, Kou C, Wu T, Li Yijie, Wang Y, Ma J, Yang J, Hu Z, Zhang F, Zhang Y, Sun S, 2014. Fine epitope mapping of the central immunodominant region of nucleoprotein from Crimean-Congo hemorrhagic fever virus (CCHFV). PLoS ONE 9, e108419. 10.1371/journal.pone.0108419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley S, Atkinson B, Dowall SD, Pitman JK, Staplehurst S, Busuttil J, Simpson AJ, Aarons EJ, Petridou C, Nijjar M, Glover S, Brooks TJ, Hewson R, 2014. Non-fatal case of Crimean-Congo haemorrhagic fever imported into the United Kingdom (ex Bulgaria), June 2014. Eurosurveillance 19. 10.2807/1560-7917.ES2014.19.30.20864 [DOI] [PubMed] [Google Scholar]
- Marín-Lopez A, Calvo-Pinilla E, Moreno S, Utrilla-Trigo S, Nogales A, Brun A, Fikrig E, Ortego J, 2019. Modeling arboviral infection in mice lacking the interferon alpha/beta receptor. Viruses 11, 35. 10.3390/v11010035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Moyer CL, Abelson DM, Deer DJ, el Omari K, Duman R, Lobel L, Lutwama JJ, Dye JM, Wagner A, Chandran K, Cross RW, Geisbert TW, Zeitlin L, Bornholdt ZA, McLellan JS, 2020. Structure and characterization of Crimean-Congo hemorrhagic fever virus GP38. Journal of Virology 94. 10.1128/jvi.02005-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moming A, Tuoken D, Yue X, Xu W, Guo R, Liu D, Li Y, Hu Z, Deng F, Zhang Y, Sun S, 2018. Mapping of B-cell epitopes on the N- terminal and C- terminal segment of nucleocapsid protein from Crimean-Congo hemorrhagic fever virus. PloS ONE 13, e0204264. 10.1371/journal.pone.0204264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi-Jazi M, Karlberg H, Papa A, Christova I, Mirazimi A, 2012. Healthy individuals’ immune response to the Bulgarian Crimean-Congo hemorrhagic fever virus vaccine. Vaccine 30, 6225–9. 10.1016/j.vaccine.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Nabeth P, Thior M, Faye O, Simon F, 2004. Human Crimean-Congo hemorrhagic fever, Sénégal. Emerging Infectious Diseases 10, 1881–1882. 10.3201/eid1010.040586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negredo A., de la Calle-Prieto F, Palencia-Herrejón E, Mora-Rillo M, Astray- Mochales J, Sánchez-Seco MP, Bermejo Lopez E, Menárguez J, Fernández-Cruz A, Sánchez-Artola B, Keough-Delgado E, Ramírez de Arellano E, Lasala F, Milla J, Fraile JL, Ordobás Gavín M, Martinez de la Gándara A, López Perez L, Diaz-Diaz D, López-García MA, Delgado-Jimenez P, Martín-Quirós A, Trigo E, Figueira JC, Manzanares J, Rodriguez-Baena E, Garcia-Comas L, Rodríguez- Fraga O, García-Arenzana N, Fernández-Díaz M. v., Cornejo VM, Emmerich P, Schmidt-Chanasit J, Arribas JR, 2017. Autochthonous Crimean–Congo Hemorrhagic Fever in Spain. New England Journal of Medicine 377, 154–161. 10.1056/NEJMoa1615162 [DOI] [PubMed] [Google Scholar]
- Nosrati M, Behbahani M, Mohabatkar H, 2019. Towards the first multi-epitope recombinant vaccine against Crimean-Congo hemorrhagic fever virus: A computer- aided vaccine design approach. Journal of Biomedical Informatics 93, 103160. 10.1016/j.jbi.2019.103160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosrati M, Mohabatkar H, Behbahani M, 2020. Introducing of an integrated artificial neural network and Chou’s pseudo amino acid composition approach for computational epitope-mapping of Crimean-Congo haemorrhagic fever virus antigens. International Immunopharmacology 78, 106020. 10.1016/j.intimp.2019.106020 [DOI] [PubMed] [Google Scholar]
- Oany AR, Ahmad SAI, Hossain U, Pervin T, 2015. Identification of highly conserved regions in L-segment of Crimean-Congo hemorrhagic fever virus and immunoinformatic prediction about potential novel vaccine. Advances and Applications in Bioinformatics and Chemistry 8, 1. 10.2147/AABC.S75250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onguru P, Akgul EO, Akıncı E, Yaman H, Kurt YG, Erbay A, Bayazıt FN, Bodur H, Erbil K, Acıkel CH, Cevik MA, 2008. High serum levels of neopterin in patients with Crimean–Congo hemorrhagic fever and its relation with mortality. Journal of Infection 56, 366–370. 10.1016/j.jinf.2008.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk B, Tutuncu E, Kuscu F, Gurbuz Y, Sencan I, Tuzun H, 2012. Evaluation of factors predictive of the prognosis in Crimean-Congo hemorrhagic fever: new suggestions. International Journal of Infectious Diseases 16, e89–e93. 10.1016/j.ijid.2011.06.005 [DOI] [PubMed] [Google Scholar]
- Papa A, Bino S, Llagami A, Brahimaj B, Papadimitriou E, Pavlidou V, Velo E, Cahani G, Hajdini M, Pilaca A, Harxhi A, Antoniadis A, 2002. Crimean-Congo hemorrhagic fever in Albania, 2001. European Journal of Clinical Microbiology and Infectious Diseases 21, 603–606. 10.1007/s10096-002-0770-9 [DOI] [PubMed] [Google Scholar]
- Papa A, Bino S, Velo E, Harxhi A, Kota M, Antoniadis A, 2006. Cytokine levels in Crimean-Congo hemorrhagic fever. Journal of Clinical Virology 36, 272–276. 10.1016/j.jcv.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Papa A, Drosten C, Bino S, Papadimitriou Ε, Panning M, Velo E, Kota M, Harxhi A, Antoniadis A, 2007. Viral load and Crimean-Congo hemorrhagic fever. Emerging Infectious Diseases 13, 805–806. 10.3201/eid1305.061588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa A, Pappa S, Panayotova E, Papadopoulou E, Christova I, 2016a. Molecular epidemiology of Crimean-Congo hemorrhagic fever in Bulgaria-an update. Journal of Medical Virology 88, 769–773. 10.1002/jmv.24400 [DOI] [PubMed] [Google Scholar]
- Papa A, Sidira P, Kallia S, Ntouska M, Zotos N, Doumbali E, Maltezou HC, Demiris N, Tsatsaris A, 2013. Factors associated with IgG positivity to Crimean- Congo hemorrhagic fever virus in the area with the highest seroprevalence in Greece. Ticks and Tick-borne Diseases 4, 417–420. 10.1016/j.ttbdis.2013.04.003 [DOI] [PubMed] [Google Scholar]
- Papa A, Tsergouli K, Çağlayık DY, Bino S, Como N, Uyar Y, Korukluoglu G, 2016b. Cytokines as biomarkers of Crimean-Congo hemorrhagic fever. Journal of Medical Virology 88, 21–27. 10.1002/jmv.24312 [DOI] [PubMed] [Google Scholar]
- Papa A., Tzala E, Maltezou HC, 2011. Crimean-Congo hemorrhagic fever virus, Northeastern Greece. Emerging Infectious Diseases 17, 141–143. 10.3201/eid1701.100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrefitte CN, Perret M, Garcia S, Rodrigues R, Bagnaud A, Lacote S, Crance JM, Vernet G, Garin D, Bouloy M, Paranhos-Baccala G, 2010. Differential activation profiles of Crimean-Congo hemorrhagic fever virus- and Dugbe virus- infected antigen-presenting cells. Journal of General Virology 91, 189–198. 10.1099/vir.0.015701-0 [DOI] [PubMed] [Google Scholar]
- Ranadheera C, Valcourt EJ, Warner BM, Poliquin G, Rosenke K, Frost K, Tierney K, Saturday G, Miao J, Westover JB, Gowen BB, Booth S, Feldmann H, Wang Z, Safronetz D, 2020. Characterization of a novel STAT 2 knock-out hamster model of Crimean-Congo hemorrhagic fever virus pathogenesis. Scientific Reports 10, 12378. 10.1038/s41598-020-69054-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues R, Paranhos-Baccalà G, Vernet G, Peyrefitte CN, 2012. Crimean-Congo hemorrhagic fever virus-infected hepatocytes induce ER-stress and apoptosis crosstalk. PLoS ONE 7, e29712. 10.1371/journal.pone.0029712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez SE, McAuley AJ, Gargili A, Bente DA, 2018. Interactions of human dermal dendritic cells and Langerhans cells treated with Hyalomma tick saliva with Crimean-Congo hemorrhagic fever virus. Viruses 10. 10.3390/v10070381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksida A, Duh D, Wraber B, Dedushaj I, Ahmeti S, Avšič-Županc T, 2010. Interacting roles of immune mechanisms and viral load in the pathogenesis of Crimean-Congo hemorrhagic fever. Clinical and Vaccine Immunology 17, 1086–1093. 10.1128/CVI.00530-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancakdar E, Uysal EB, Güven AS, Kaya A, Filiz AK, Acibucu DO, Deveci K, Seker MM, 2014. Evaluation of complement system in children with Crimean-Congo hemorrhagic fever. European Review for Medical and Pharmacological Sciences 18, 675–9. [PubMed] [Google Scholar]
- Sanchez AJ, Vincent MJ, Erickson BR, Nichol ST, 2006. Crimean-Congo hemorrhagic fever virus glycoprotein precursor is cleaved by Furin-like and SKI-1 proteases to generate a novel 38-kilodalton glycoprotein. Journal of Virology 80, 514–525. 10.1128/JVI.80.1.514-525.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AJ, Vincent MJ, Nichol ST, 2002. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. Journal of Virology 76, 7263–7275. 10.1128/JVI.76.14.7263-7275.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholte FEM, Spengler JR, Welch SR, Harmon JR, Coleman-McCray JD, Freitas BT, Kainulainen MH, Pegan SD, Nichol ST, Bergeron É, Spiropoulou CF, 2019. Single-dose replicon particle vaccine provides complete protection against Crimean-Congo hemorrhagic fever virus in mice. Emerging Microbes & Infections 8, 575–578. 10.1080/22221751.2019.1601030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholte FEM, Zivcec M, Dzimianski J. v., Deaton MK, Spengler JR, Welch SR, Nichol ST, Pegan SD, Spiropoulou CF, Bergeron É, 2017. Crimean-Congo hemorrhagic fever virus suppresses innate immune responses via a ubiquitin and ISG15 specific protease. Cell Reports 20, 2396–2407. 10.1016/j.celrep.2017.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Hertzog PJ., Ravasi T., Hume DA., 2004. Interferon-γ: an overview of signals, mechanisms and functions. Journal of Leukocyte Biology 75, 163–189. 10.1189/jlb.0603252 [DOI] [PubMed] [Google Scholar]
- Sellers RS, Clifford CB, Treuting PM, Brayton C, 2012. Immunological variation between inbred laboratory mouse strains. Veterinary Pathology 49, 32–43. 10.1177/0300985811429314 [DOI] [PubMed] [Google Scholar]
- Shalitanati A, Yu H, Liu D, Xu W-X, Yue X, Guo R, Li Y, Deng F, Yang J, Zhang Y, Sun S, 2018. Fine mapping epitope on glycoprotein-Gn from Crimean- Congo hemorrhagic fever virus. Comparative Immunology, Microbiology and Infectious Diseases 59, 24–31. 10.1016/j.cimid.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Shepherd AJ, Swanepoel R, Leman PA, 1989. Antibody response in Crimean-Congo hemorrhagic fever. Clinical Infectious Diseases 11, S801–S806. 10.1093/clinids/11.Supplement_4.S801 [DOI] [PubMed] [Google Scholar]
- Shepherd AJ, Swanepoel R, Shepherd SP, Leman PA, Blackburn NK, Hallett AF, 1985. A nosocomial outbreak of Crimean-Congo haemorrhagic fever at Tygerberg Hospital. Part V. Virological and serological observations. South African Medical Journal = Suid-Afrikaanse tydskrif vir geneeskunde 68, 733–6. [PubMed] [Google Scholar]
- Shrivastava N, Verma A, Dash PK, 2020. Identification of functional epitopes of structural proteins and in-silico designing of dual acting multiepitope anti-tick vaccine against emerging Crimean-Congo hemorrhagic fever virus. European Journal of Pharmaceutical Sciences 151, 105396. 10.1016/j.ejps.2020.105396 [DOI] [PubMed] [Google Scholar]
- Sidira P, Maltezou HC, Haidich A-B, Papa A, 2012. Seroepidemiological study of Crimean-Congo haemorrhagic fever in Greece, 2009–2010. Clinical Microbiology and Infection 18, E16–E19. 10.1111/j.1469-0691.2011.03718.x [DOI] [PubMed] [Google Scholar]
- Šimo L, Kazimirova M, Richardson J, Bonnet SI, 2017. The essential role of tick salivary glands and saliva in tick feeding and pathogen transmission. Frontiers in Cellular and Infection Microbiology 7, 281. 10.3389/fcimb.2017.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Shoemaker CJ, Zeng X, Garrison AR, Golden JW, Schellhase CW, Pratt W, Rossi F, Fitzpatrick CJ, Shamblin J, Kimmel A, Zelko J, Flusin O,Koehler JW, Liu J, Coffin KM, Ricks KM, Voorhees MA, Schoepp RJ, Schmaljohn CS, 2019. Persistent Crimean-Congo hemorrhagic fever virus infection in the testes and within granulomas of non-human primates with latent tuberculosis. PLOS Pathogens 15, e1008050. 10.1371/journal.ppat.1008050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler JR, Estrada-Peña A, 2018. Host preferences support the prominent role of Hyalomma ticks in the ecology of Crimean-Congo hemorrhagic fever. PLoS Neglected Tropical Diseases 12, e0006248. 10.1371/journal.pntd.0006248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler JR, Estrada-Peña A, Garrison AR, Schmaljohn C, Spiropoulou CF, Bergeron É, Bente DA, 2016. A chronological review of experimental infection studies of the role of wild animals and livestock in the maintenance and transmission of Crimean-Congo hemorrhagic fever virus. Antiviral Research 135, 31–47. 10.1016/j.antiviral.2016.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler JR, Kelly Keating M, McElroy AK, Zivcec M, Coleman-McCray JD, Harmon JR, Bollweg BC, Goldsmith CS, Bergeron É, Keck JG, Zaki SR, Nichol ST, Spiropoulou CF, 2017. Crimean-Congo hemorrhagic fever in humanized mice reveals glial cells as primary targets of neurological infection. The Journal of Infectious Diseases 216, 1386–1397. 10.1093/infdis/jix215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler JR., Patel JR, Chakrabarti AK, Zivcec M, García-Sastre A, Spiropoulou CF, Bergeron É, 2015. RIG-I mediates an antiviral response to Crimean-Congo hemorrhagic fever virus. Journal of Virology 89, 10219–10229. 10.1128/JVI.01643-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler JR, Welch SR, Scholte FEM, Coleman-McCray JD, Harmon JR, Nichol ST, Bergeron É, Spiropoulou CF, 2019. Heterologous protection against Crimean- Congo hemorrhagic fever in mice after a single dose of replicon particle vaccine. Antiviral Research 170, 104573. 10.1016/j.antiviral.2019.104573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suleiman MN, Muscat-Baron JM, Harries JR, Satti AG, Platt GS, Bowen ET, Simpson DI, 1980. Congo/Crimean haemorrhagic fever in Dubai. An outbreak at the Rashid Hospital. Lancet 2, 939–41. [PubMed] [Google Scholar]
- Suschak JJ, Golden JW, Fitzpatrick CJ, Shoemaker CJ, Badger C. v., Schmaljohn CS, Garrison AR, 2021. A CCHFV DNA vaccine protects against heterologous challenge and establishes GP38 as immunorelevant in mice. npj Vaccines 6, 31. 10.1038/s41541-021-00293-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R, Gill DE, Shepherd AJ, Leman PA, Mynhardt JH, Harvey S, 1989. The clinical pathology of Crimean-Congo hemorrhagic fever. Clinical Infectious Diseases 11, S794–S800. 10.1093/clinids/11.Supplement_4.S794 [DOI] [PubMed] [Google Scholar]
- Tignor GH, Hanham CA, 1993. Ribavirin efficacy in an in vivo model of Crimean- Congo hemorrhagic fever virus (CCHF) infection. Antiviral Research 22, 309–325. 10.1016/0166-3542(93)90040-P [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipih T, Burt FJ, 2020. Crimean–Congo hemorrhagic fever virus: Advances in vaccine development. BioResearch Open Access 9, 137–150. 10.1089/biores.2019.0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden PJ, van Eeden SF, Joubert JR, King JB, van de Wal BW, Michell WL, 1985. A nosocomial outbreak of Crimean-Congo haemorrhagic fever at Tygerberg Hospital. Part II. Management of patients. South African Medical Journal = Suid-Afrikaanse tydskrif vir geneeskunde 68, 718–21. [PubMed] [Google Scholar]
- van Kasteren PB, Beugeling C, Ninaber DK, Frias-Staheli N, van Boheemen S, Garcia-Sastre A, Snijder EJ, Kikkert M, 2012. Arterivirus and Nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. Journal of Virology 86, 773–785. 10.1128/JVI.06277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilenko SM, Vassilev TL, Bozadjiev LG, Bineva IL, Kazarov GZ, 1990. Specific intravenous immunoglobulin for Crimean-Congo haemorrhagic fever. Lancet 335, 791–2. 10.1016/0140-6736(90)90906-l [DOI] [PubMed] [Google Scholar]
- Vincent MJ, Sanchez AJ, Erickson BR, Basak A, Chretien M, Seidah NG, Nichol ST, 2003. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. Journal of Virology 77, 8640–8649. 10.1128/JVI.77.16.8640-8649.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dutta S, Karlberg H, Devignot S, Weber F, Hao Q, Tan YJ, Mirazimi A, Kotaka M, 2012. Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage. Journal of Virology 86, 12294–12303. 10.1128/JVI.01627-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Mirazimi A, 2008. Interferon and cytokine responses to Crimean Congo hemorrhagic fever virus; an emerging and neglected viral zonoosis. Cytokine & Growth Factor Reviews 19, 395–404. 10.1016/j.cytogfr.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch SR., Ritter JM, McElroy AK, Harmon JR, Coleman-McCray JD, Scholte FEM, Kobinger GP, Bergeron É, Zaki SR, Nichol ST, Spengler JR, Spiropoulou CF, 2019. Fluorescent Crimean-Congo hemorrhagic fever virus illuminates 4phagocytic cell targets in Ifnar−/− mice. PLOS Pathogens 15, e1008183. 10.1371/journal.ppat.1008183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch Stephen R, Scholte FEM, Spengler JR, Ritter JM, Coleman-McCray JD, Harmon JR, Nichol ST, Zaki SR, Spiropoulou CF, Bergeron E, 2020. The Crimean-Congo hemorrhagic fever virus NSm protein is dispensable for growth in vitro and disease in ifnar−/− mice. Microorganisms 8. 10.3390/microorganisms8050775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Beck AS, Gargili A, Forrester N, Barrett ADT, Bente DA, 2016. Transstadial transmission and long-term association of Crimean-Congo hemorrhagic fever virus in ticks Shapes Genome Plasticity. Scientific Reports 6, 35819. 10.1038/srep35819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PD, Patil DY, Shete AM, Kokate P, Goyal P, Jadhav S, Sinha S, Zawar D, Sharma SK, Kapil A, Sharma DK, Upadhyay KJ, Mourya DT, 2016. Nosocomial infection of CCHF among health care workers in Rajasthan, India. BMC Infectious Diseases 16, 624. 10.1186/s12879-016-1971-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaqub T, Oneeb M, Mukhtar N, Tahir Z, Shahid F, Subhan S, Salman M, 2019. Crimean–Congo Haemorrhagic Fever: Case study analysis of a sporadic outbreak from Chakwal, Pakistan. Zoonoses and Public Health 66, 871–873. 10.1111/zph.12623 [DOI] [PubMed] [Google Scholar]
- Yilmaz M, Aydin K, Akdogan E, Sucu N, Sonmez M, Omay SB, Koksal I, 2008. Peripheral blood natural killer cells in Crimean-Congo hemorrhagic fever. Journal of Clinical Virology 42, 415–417. 10.1016/j.jcv.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Zhang J, Simayi A, Wang M, Moming A, Xu W, Wang C, Li Y, Ding J, Deng F, Zhang Y, Sun S, 2019. Fine mapping epitope on glycoprotein Gc from Crimean- Congo hemorrhagic fever virus. Comparative Immunology, Microbiology and Infectious Diseases 67, 101371. 10.1016/j.cimid.2019.101371 [DOI] [PubMed] [Google Scholar]
- Zivcec M, Guerrero LIW, Albariño CG, Bergeron É, Nichol ST, Spiropoulou CF, 2017. Identification of broadly neutralizing monoclonal antibodies against Crimean- Congo hemorrhagic fever virus. Antiviral Research 146, 112–120. 10.1016/j.antiviral.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M, Metcalfe MG, Albariño CG, Guerrero LW, Pegan SD, Spiropoulou CF, Bergeron É, 2015. Assessment of inhibitors of pathogenic Crimean-Congo hemorrhagic fever virus strains using virus-like particles. PLOS Neglected Tropical Diseases 9, e0004259. 10.1371/journal.pntd.0004259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M, Safronetz D, Scott D, Robertson S, Ebihara H, Feldmann H, 2013. Lethal Crimean-Congo hemorrhagic fever virus infection in interferon α/β receptor knockout mice is associated with high viral loads, proinflammatory responses, and coagulopathy. The Journal of Infectious Diseases 207, 1909–1921. 10.1093/infdis/jit061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M., Safronetz D., Scott DP., Robertson S., Feldmann H., 2018. Nucleocapsid protein-based vaccine provides protection in mice against lethal Crimean-Congo hemorrhagic fever virus challenge. PLOS Neglected Tropical Diseases 12, e0006628. 10.1371/journal.pntd.0006628 [DOI] [PMC free article] [PubMed] [Google Scholar]