Abstract
Anthracnose, or leaf-curl disease of anemone, caused by Colletotrichum sp., has been reported to occur in Australia, western Europe, and Japan. Symptoms include tissue necrosis, corm rot, leaf crinkles, and characteristic spiral twisting of floral peduncles. Three epidemics of the disease have been recorded in Israel: in 1978, in 1990 to 1993, and in 1996 to 1998. We characterized 92 Colletotrichum isolates associated with anthracnose of anemone (Anemone coronaria L.) for vegetative compatibility (72 isolates) and for molecular genotype (92 isolates) and virulence (4 isolates). Eighty-six of the isolates represented the three epidemics in Israel, one isolate was from Australia, and five isolates originated from western Europe. We divided these isolates into three vegetative-compatibility groups (VCGs). One VCG (ANE-A) included all 10 isolates from the first and second epidemics, and 13 of 62 examined isolates from the third epidemic in Israel, along with the isolate from Australia and 4 of 5 isolates from Europe. Another VCG (ANE-F) included most of the examined isolates (49 of the 62) from the third epidemic, as well as Colletotrichum acutatum from strawberry, in Israel. Based on PCR amplification with species-specific primers, all of the anemone isolates were identified as C. acutatum. Anemone and strawberry isolates of the two VCGs were genotypically similar and indistinguishable when compared by arbitrarily primed PCR of genomic DNA. Only isolate NL-12 from The Netherlands, confirmed as C. acutatum but not compatible with either VCG, had a distinct genotype; this isolate represents a third VCG of C. acutatum. Isolates from anemone and strawberry could infect both plant species in artificial inoculations. VCG ANE-F was recovered from natural infections of both anemone and strawberry, but VCG ANE-A was recovered only from anemone. This study of C. acutatum from anemone illustrates the potential of VCG analysis to reveal distinct subspecific groups within a pathogen population which appears to be genotypically homogeneous by molecular assays.
The filamentous fungal plant pathogen Colletotrichum causes anthracnose diseases on many crops worldwide (2). Anthracnose or leaf-curl disease of anemone (Anemone coronaria L.) was first recorded in Australia in the mid-1970s (T. Woodcock and W. S. Washington, Abstr. Australas. Plant Pathol. 8:10, 1978). Since then it has occurred in other flower-producing areas, such as western Europe and Japan. Symptoms include tissue necrosis, corm rot, leaf crinkles, and characteristic spiral twisting of floral peduncles. The incitant of the disease is variably referred to as C. acutatum Simmonds, C. gloeosporioides (Penz.) Pens. & Sacc., or both (3, 5, 11, 20, 32, 34).
The first outbreak of anthracnose of anemone in Israel in 1978 was attributed to the import from Europe of corms contaminated with C. gloeosporioides (H. Vigodsky-Haas, Y. Ozeri, B. Kirshner, and M. Ruben, Abstr. Phytoparasitica 11:242–243, 1983). Following fumigation of infested fields and discontinuation of corm import, no disease was recorded in the subsequent decade. A second outbreak occurred in 1990, reached epidemic levels during 1991 and 1992, and then declined in 1993 (M. Gokkes, I. S. Ben-Ze'ev, E. Levy, O. Ben-Gal, and Y. Shabtay, Abstr. Phytoparasitica 21:148, 1993). Although isolates recovered during the second outbreak were thought to be C. gloeosporioides, a preliminary vegetative compatibility test suggested their relation to C. acutatum from Europe (T. Katan and E. Shabi, Abstr. Phytoparasitica 22:90, 1994). The most recent outbreak of anemone anthracnose began in 1996, and various levels of the disease have been observed since that time (S. Freeman, E. Shabi, Y. Nizani, and T. Katan, Abstr. Phytoparasitica 26:155–156, 1998).
Anthracnose of strawberry is another disease caused by Colletotrichum in an annual crop in Israel (13). This disease first occurred in Israel in 1995 and has spread rapidly to many nurseries and production fields. The pathogen, diagnosed as C. acutatum, causes typical anthracnose symptoms as well as root necrosis and stunting of affected plants. Vegetative compatibility and molecular analysis have shown that isolates from various sources, and associated with both symptoms, all belong to a single clonal population (13). Although anthracnose diseases of both anemone and strawberry also occur in Europe (3, 8, 11, 20, 34), it is not known whether the incitant Colletotrichum strains are genetically related or whether cross-infections occur under field conditions.
Both C. acutatum and C. gloeosporioides are collective species, encompassing various groups of strains and biotypes. Traditional methods for discriminating between species of Colletotrichum rely primarily on morphology and host preference. The use of morphological characteristics is complicated by environmental influences on these characteristics and the existence of intermediate forms. Confusion between C. acutatum and C. gloeosporioides also is compounded by the broad host range of these species and the occurrence of more than one species on a given host (1, 2, 4, 15). Species-specific PCR primers, designed for the taxonomic identification of Colletotrichum species, have been used to distinguish between C. acutatum and C. gloeosporioides (7, 12). Likewise, other methods based on randomly amplified polymorphic DNA (RAPD) (17), arbitrarily primed PCR (18), nuclear and mitochondrial DNA restriction fragment length polymorphisms (14), and sequence analyses of the internal transcribed spacer (ITS) regions of ribosomal DNA (16) have been used to estimate intraspecific genetic diversity in Colletotrichum (14, 15). Vegetative compatibility (28) is another approach used to determine the genetic relatedness among and between populations of this genus (6, 9, 25, 26, 36).
Using vegetative compatibility, molecular, and pathogenicity assays, the objectives of the present study were (i) to identify the Colletotrichum species responsible for anemone anthracnose in Israel, (ii) to estimate the genetic diversity among Colletotrichum populations from anemone in Israel and their relatedness to representative C. acutatum isolates from Europe and to C. acutatum from strawberry in Israel, and (iii) to determine if isolates from anemone and strawberry can infect the alternate hosts.
MATERIALS AND METHODS
Fungal cultures.
The monoconidial Colletotrichum cultures used in this study are listed in Table 1. Colletotrichum isolates from anemone in Israel were isolated from infected plant material between 1979 and 1998. Three C. acutatum anemone isolates from Israel, ANE-4 (CABI accession no. IMI384686), ANE-27 (CABI accession no. IMI384685), and NL-12 (CABI accession no. IMI384684) representing distinct VCGs, were deposited in the culture collection of CABI Bioscience, Egham, United Kingdom. Six foreign C. acutatum isolates from anemone were included for comparison: IMI-223120 from Australia (Woodcock and Washington, Abstr. Australas. Plant Pathol.) and five isolates from Europe (United Kingdom, Italy, and The Netherlands). DNA from the previously characterized strawberry isolates of C. gloeosporioides CG-272-1 and CG-315-1 (21, 33), C. fragariae CF-63-1 (18), and C. acutatum TUT-110A and TUT-137A (13) were used as references in this study. All isolates were grown on modified Mathur's medium (MS; 0.1% yeast extract–0.1% Bacto Peptone–1% sucrose–0.25% MgSO4 · 7H2O–0.27% KH2PO4–2% agar supplemented with 25 mg of ampicillin in 1 liter of sterile distilled water) (35) in the dark at 25°C.
TABLE 1.
Isolates of C. acutatum, C. fragariae, and C. gloeosporioides from anemone and strawberry used in this study
| Species and isolatea | Origin (site) | Yr | Hostb | Organ | Tested byc:
|
||
|---|---|---|---|---|---|---|---|
| Mol | VCG | Path | |||||
| C. fragariae (CF-63-1) | Mississippi | 1981 | S | Crown | + | ||
| C. gloeosporioides | |||||||
| CG-272-1 | Florida | Unknown | S | Leaf | + | ||
| CG-315-1 | Nova Scotia, Canada | Unknown | S | Crown | + | ||
| C. acutatum | |||||||
| CA-310-1 | North Carolina | Unknown | S | Leaf | + | ||
| CA-330-1 | Tennessee | Unknown | S | Fruit | + | ||
| TUT-5954 | Israel (Plant Clinicd) | 1994 | S | Crown | + | + | |
| TUT-110A | Israel (Kadima) | 1995 | S | Fruit | + | + | + |
| TUT-137A | Israel (Even Yehuda) | 1995 | S | Root | + | + | + |
| IMI-223120e | Australia | 1978 | A | Stem | + | + | + |
| IT-23f | Italy | Unknown | A | Unknown | + | + | |
| IT-22g | Italy | 1985 | A | Unknown | + | + | |
| UK-31g | United Kingdom | 1985 | A | Unknown | + | + | |
| NL-4g | The Netherlands | 1987 | A | Unknown | + | + | |
| NL-12g | The Netherlands | 1989 | A | Unknown | + | + | |
| ANE-HV83Ch | Israel | 1979 | A | Unknown | + | + | + |
| ANE-1Wi | Israel | 1990 | A | Corm | + | + | |
| ANE-5j | Israel | 1991 | A | Unknown | + | + | |
| ANE-45K | Israel (Zelafon) | 1993 | A | Corm | + | + | |
| ANE-8Ek | Israel (Zipori) | 1993 | A | Unknown | + | + | |
| ANE-27Al | Israel (Zekaria) | 1996 | A | Leaf | + | + | + |
| ANE-41Am | Israel (Bene Zion) | 1996 | A | Sepal | + | + | |
| ANE-43Am | Israel (Bene Zion) | 1996 | A | Corm | + | + | |
| ANE-52An | Israel (Zipori) | 1996 | A | Flower | + | + | |
| ANE-61Ao | Israel (Zekaria) | 1997 | A | Corm | + | + | + |
| ANE-75AAp | Israelq (Zekaria) | 1998 | A | Leaf | + | + | |
| ANE-77Ap | Israel (Zekaria) | 1998 | A | Corm | + | + | |
| ANE-86Ap | Israel (Zekaria) | 1998 | A | Stem | + | + | |
Representative isolates originating from different sites, plots, and infected tissues.
S, strawberry; A, anemone.
Isolates were tested (+) by molecular analyses (Mol), vegetative compatibility grouping (VCG), and/or pathogenicity assays (Path).
Received from Shelef Plant Clinic, Hod Hasharon, Israel.
Received from the International Mycological Institute, deposited by T. Woodcock in 1978.
Received from M. L. Gullino, University of Torino, Torino, Italy.
Received from A. W. Doornik, Bulb Research Center, Lisse, The Netherlands.
Isolated by the late H. Vigodsky-Haas (31) and recovered from cold storage in 1992.
Additional isolates: ANE-2C, -3, and -4.
Additional isolate: ANE-6C.
Additional isolate: ANE-9H.
Additional isolates originating from the southern growing region in Israel: ANE-11A, -12A, -13A, -14A, -15A, -16A, -17A, -18A, -19A, -20A, -21A, -22A, -23A, -24A, -25A, and -26A.
Additional isolates originating from the central growing region in Israel: ANE-28A, -29A, -30A, -31A, -32A, -33A, -34A, -35A, -36A, -37A, -38A, -39A, -40A, -42A, -44A, -46A, -47A, -48A, and -60A.
Additional isolates originating from the northern growing region in Israel: ANE-49A, -50A, -51A, -53A, -54A, -55A, -56A, -57A, -58A, and -59A.
Additional isolates originating from the southern growing region in Israel: ANE-62A, -63A, -64A, -65A, -66A, -67A, -68A, -69A, -70A, -71A, -79A, -80A, -81A, -82A, and -83A.
Additional isolates originating from the southern growing region in Israel: ANE-72A, -73A, -74A, -76A, -78A, -84A, -85A, and -87A.
Diseased plants growing from corms imported from The Netherlands.
Vegetative-compatibility grouping.
Puhalla's minimal nitrate agar (MM), a Czapele's-based sucrose-salt medium containing nitrate as the nitrogen source (29), was used to identify nit mutants and for complementation (heterokaryon) tests. Chlorate media, based on MM or potato dextrose agar amended with KClO3 (15 or 15 to 25 g/liter, respectively), were used to generate nit mutants (10, 29). Plates (9 cm in diameter) of chlorate media were centrally inoculated with 4-mm3 mycelial plugs and incubated at 25°C. Fast-growing sectors emerging from the restricted colonies were transferred to MM plates (5 cm in diameter) and examined after a 4-day incubation period. Colonies with a thin expansive mycelium were considered nit mutants. Nitrite and hypoxanthine media were used for partial phenotyping of nit mutants as nit1, nit3, or NitM (6, 10). Complementation between nit mutants was tested on MM as previously described (26). Heterokaryons were usually evident within 10 to 15 days. When mutants formed a prototrophic heterokaryon, their parent isolates were assigned to the same vegetative-compatibility group (VCG).
Isolation and purification of fungal DNA.
MS medium (100 ml) without agar was placed in 250-ml Erlenmeyer flasks and inoculated with five 5-mm2 mycelial disks derived from colony margins. The cultures were grown for 5 to 6 days on a rotary shaker (150 rpm at 25°C). At 12 h before the mycelia were harvested, the cultures were fragmented by blending them for 10 s at 24,000 rpm with a tissue homogenizer (Ultra-Turrax T25, Janke & Kunkel; IKA Labortechnik), followed by continued growth for the remainder of the time. Mycelia from 100-ml liquid cultures were collected by vacuum filtration and lyophilized. DNA was extracted and purified as previously described (17), dissolved in 0.5 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) to an approximate concentration of 200 to 500 μg/ml, and diluted to a concentration of 10 to 100 ng/ml for PCR amplification.
PCR amplification.
Primers for arbitrarily primed PCR (ap-PCR) were derived from microsatellite or repeat sequences as follows: CAGCAGCAGCAGCAG (30), TGTCTGTCTGTCTGTC (17), ACTGACTGACTGACTG (18), GACACGACACGACAC (M. Gupta and P. Filner, Abstr. Int. Soc. Plant Mol. Biol., abstr. 1705, 1991), GACAGACAGACAGACA (K. Weising, F. Weigand, A. J. Driesel, A. J. Kahl, H. Zischer, and J. T. Epplen, Abstr. Nucleic Acids Res. 17:10128, 1989). In the text, these primers are designated (CAG)5, (TGTC)4, (ACTG)4, (GACAC)3, and (GACA)4, respectively. PCR primers for species-specific amplification included ITS 4 (TCCTCCGCTTATTGATATGC) coupled with the primers of C. acutatum (CaInt2) (GGGGAAGCCTCTCGCGG) and C. gloeosporioides (CgInt) (GGCCTCCCGCCTCCGGGCGG) (7). PCR reactions were performed in a total volume of 20 μl, containing 10 to 100 ng of genomic DNA, 50 mM KCl, 10 mM Tris-HCl, 0.2 mM concentrations each of dATP, dCTP, dGTP and dTTP, 1.5 mM MgCl2, 1 U of Taq DNA polymerase (Promega, Madison, Wis.), and a 1 μM concentration of primer. The reactions were incubated in a PTC-100 thermocycler (MJ Research, Inc., Watertown, Mass.), starting with 5 min of denaturation at 95°C. For ap-PCR, this was followed by 30 cycles consisting of 30 s at 95°C, 30 s at either 60°C [for (CAG)5] or 48°C [for (GACA)4, (GACAC)3, (ACTG)4, and (TGTC)4] and 1.5 min at 72°C, with a final extension of 15 min at 72°C. Species-specific PCR reactions were performed under reaction conditions for primer (CAG)5, with 0.5 μM ITS 4 primer coupled with either a 0.5 μM concentration of primer CaInt2 for C. acutatum-specific detection or a 0.5 μM concentration of primer CgInt for C. gloeosporioides-specific detection. Species-specific primer analysis and ap-PCR were performed on DNA from the U.S. (CA-310-1 and CA-330-1) and Israeli (TUT-5954) strawberry reference isolates of C. acutatum, the U.S. isolate of C. fragariae (CF-63-1), two isolates of C. gloeosporioides (U.S. and Canadian) representing discrete genotypes (CG-315-1 of genotype Cgl-1 and CG-272-1 of genotype Cgl-2), and 89 isolates of Colletotrichum associated with anthracnose of anemone (from Israel, Europe, and Australia). Amplification products were separated in agarose gels (1.8%, 15 by 10 cm) in TAE buffer (31) electrophoresed at 80 V for 2 h. All PCR experiments were repeated at least three times with identical results being reproduced.
Pathogenicity assays.
Anemone corms (cv. Jerusalem) received from Yodfat Nursery (Kibbutz Yodfat, Israel) were placed in moist vermiculite and germinated in the dark at 4°C. Germinated corms were planted, one per pot (15-cm in diameter) containing peat-tuff (volcanic ash) (1:1 [vol/vol]) medium (Kekkila, Tuusula, Finland), and the emerging plants were grown in a greenhouse at 25°C under natural daylight. The foliage of 4-week-old plants were inoculated by spraying conidial suspensions (106/ml) of four anemone isolates (IMI-223120, ANE-HV83C, ANE-27A, and ANE-61A) and two strawberry isolates (TUT-110A and TUT-137A), along with a water control till runoff. To avoid splash dispersal of inoculum, plants were watered through a drip-irrigation system. Anemone plants were assessed for mortality over a 14-day period. Strawberry plants were grown and inoculated in a similar manner. Four-week-old strawberry plants (cv. Oso Grande, certified disease-free; Rahan Meristem) were used for foliar inoculation and assessed for mortality over a 9-week period. In each experiment, 20 plants (five replicates of 4 plants) of each host were inoculated with each isolate separately. Disease progress was assessed as the percentage of plant mortality. The experiments were conducted twice for each host, and the results from the two experiments were pooled for statistical analysis. The significance of pathogenicity data was determined according to standard error of means of plant mortality at each period for each isolate, and by one-way analysis of variance for final plant mortality, using the StatView (version 5.0.1, May 1999) statistical program (SAS Institute, Cary, N.C.).
RESULTS
Vegetative compatibility grouping.
Nine isolates collected in Israel during the second anthracnose epidemic in 1990 to 1993 were used for the initial study of vegetative compatibility. nit mutants from all isolates formed viable prototrophic heterokaryons with at least some of the other mutants, and the pattern of heterokaryon formation indicated that all nine isolates belonged to a single VCG. Based on their ability to form complementary heterokaryons with many mutants, four NitM mutants (ANE-1/1 and ANE-1/32 from ANE-1; ANE-4/21 and ANE-4/22 from ANE-4) were chosen as the tester strains of this VCG. nit mutants generated from isolate ANE-HV83C, which represents the late 1970s epidemic in Israel, formed complementary heterokaryons with the testers, so this isolate was assigned to the same VCG. Isolates IT-22 and IT-23 from Italy, isolate UK-31 from the United Kingdom, and isolate NL-4 from The Netherlands, all produced nit mutants that were compatible with the testers from Israel. Type culture IMI-223120 from Australia also was compatible with the testers, although its heterokaryons (inter-isolate and intra-isolate alike) developed more slowly. With the exception of isolate NL-12 from The Netherlands, all of the examined isolates obtained from anemone in 1993 or earlier and originating from various geographic locations belonged to the same VCG. By inference from their vegetative compatibility with C. acutatum isolates from Australia and Europe, the isolates from Israel also should be classified as C. acutatum.
nit mutants were generated from an additional 62 (out of 76) isolates collected from 1996 to 1998 at three sites in Israel. The mutants were paired with the tester strains of the first group, as well as in various combinations among themselves. Formation of complementary heterokaryons with the testers indicated that 13 of the isolates (ANE-72A through ANE-78A, and ANE-82A through ANE-87A) belonged to the VCG defined by the first group. The remaining 49 isolates, which were not compatible with the testers or with additional mutants of the first VCG, were compatible with one another, thus forming a second VCG of C. acutatum from anemone. Complementary nit testers (ANE-43/1 and ANE-81/12), as well as nit mutants from 12 additional isolates of the second VCG, formed complementary heterokaryons with nit mutants of three C. acutatum isolates (TUT-6C, TUT-232:1, and TUT-5954) representing the VCG population previously diagnosed as causing anthracnose and root necrosis in strawberry in Israel (13). We designated the VCG recovered exclusively from anemone VCG ANE-A (for Anemone) and the one recovered from both anemone and strawberry VCG ANE-F (for Fragaria). Complementary nit mutants of the self-compatible (28) isolate NL-12 were not compatible with either VCG ANE-A or VCG ANE-F.
Species-specific PCR analysis.
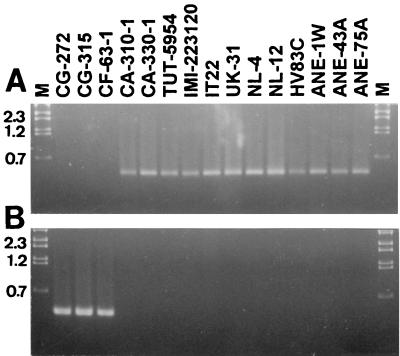
The C. acutatum-specific primers amplified DNA of C. acutatum isolates from strawberry and all anemone isolates but not that of the reference C. gloeosporioides and C. fragariae isolates (Fig. 1A). Inversely, the C. gloeosporioides-specific primers amplified DNA of C. gloeosporioides and C. fragariae isolates alone and not that of the isolates from anemone or C. acutatum from strawberry (Fig. 1B). These results place the different isolates from anemone, including isolate NL-12, in the species C. acutatum along with the strawberry reference strains.
FIG. 1.
PCR amplification products, using species-specific primers for C. acutatum (A) and C. gloeosporioides (B), of genomic DNA of reference isolates (strain origins are defined in Materials and Methods) of C. gloeosporioides, C. fragariae, and C. acutatum from strawberry in the United States, C. acutatum from strawberry in Israel, and nine Colletotrichum isolates from anemone. Lane M, DNA markers with sizes in kilobases.
ap-PCR analysis.
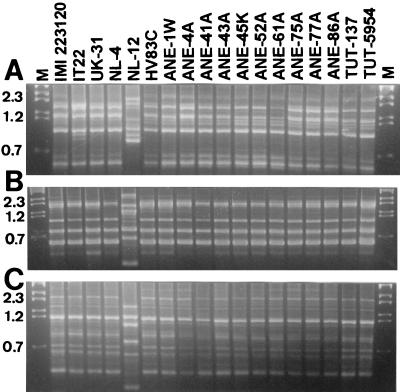
The ap-PCR banding patterns of isolate NL-12 were distinctly different from those of the other anemone and strawberry isolates. Similar ap-PCR banding patterns were observed among the anemone isolates (represented in Fig. 2 by 16 isolates) from different geographic locations and plant tissues and among the isolates from strawberry using primers (GACA)4, (CAG)5, and (GACAC)3 (Fig. 2A, B, and C, respectively) and primers (TGTC)4 and (ACTG)4 (data not shown).
FIG. 2.
Band patterns of ap-PCR-amplified genomic DNA from anemone and strawberry isolates (strain origins are defined in Materials and Methods) of C. acutatum using primers (CAG)5 (A), (GACA)4 (B), and (GACAC)3 (C). Lane M, DNA markers with sizes in kilobases.
Pathogenicity assays.
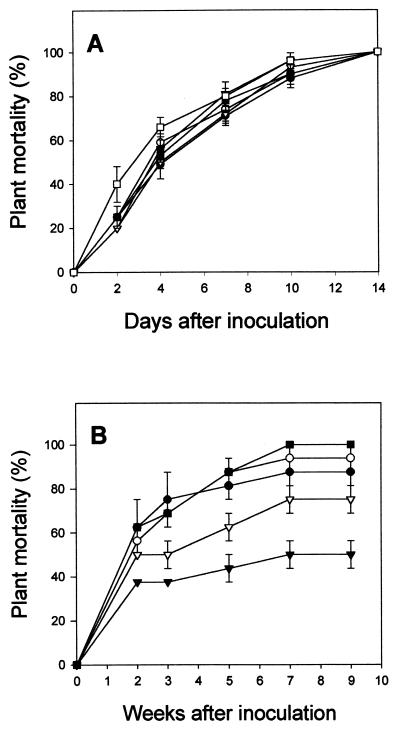
Anemone and strawberry plants were spray inoculated with conidia from both anemone and strawberry representative isolates to determine whether host specificity exists. The anemone plants developed typical leaf-curl symptoms 7 to 10 days after inoculation with anemone isolates IMI-223120, ANE-HV83C, ANE-27A, and ANE-61A. Identical symptoms also were observed on anemone plants artificially inoculated with strawberry isolates TUT-110A and TUT-137A. Disease progress in anemone was similar, without significant differences among isolates (Fig. 3A). Regardless of the isolate source, all anemone plants were killed within 14 days of inoculation. Typical anthracnose symptoms were observed on strawberry plants inoculated with anemone isolates ANE-HV83C, ANE-27A, and ANE-61A and strawberry isolates TUT-110A and TUT-137A. Mortality of strawberry plants was first observed 2 weeks after inoculation, with the experiment lasting 9 weeks. No disease was recorded in the control water-inoculated plants. The rate of disease progress varied, and plant mortality after 9 weeks was significantly different (P < 0.001) among isolates (Fig. 3B).
FIG. 3.
Percent mortality of anemone (A) and strawberry (B) plants spray inoculated with conidia of C. acutatum isolates from anemone (▾, ANE-HV83C; ▿, ANE-27A; ■, ANE-61A; □, IMI-223120) and strawberry (●, TUT-137A; ○, TUT-110A). In each treatment, 20 plants (five replicates of 4 plants) of each host were inoculated with each isolate separately. No disease was recorded in the control water-inoculated plants. The significance of the pathogenicity data was determined according to the standard errors of the mean of plant mortality at each day (A) or week (B) for each isolate. Where bars are missing, the error is less than 5% of the value of the point. The experiments were conducted twice for each host, and the results from the two experiments were pooled.
DISCUSSION
The main aims of this research were to identify the species of Colletotrichum responsible for anthracnose or leaf-curl disease of anemone, to determine both whether heterogeneity exists among anemone and strawberry pathogen populations and their inter-relatedness, and to examine whether these pathogen populations are host specific or whether a complex of pathogens are the causal agents of diseases occurring in both crops. We identified three VCGs in 92 Colletotrichum isolates. One VCG (designated ANE-A) included all 10 isolates from two epidemics (1978 and 1990 to 1993), and 13 of 62 isolates from a third epidemic (1996 to 1998) in Israel, along with an isolate from Australia and 4 of 5 isolates from Europe. This grouping and the chronological considerations suggest that C. acutatum isolates of VCG ANE-A in Israel and Europe are probably members of the same clone as the anemone pathogen first described in Australia (Woodcock and Washington, Abstr. Australas. Plant Pathol.). Another VCG (designated ANE-F) included most of the examined isolates (49 of 62) from the third epidemic of anemone anthracnose in Israel and was vegetatively compatible with the VCG of C. acutatum from strawberry in Israel, a pathogen considered to be of U.S. origin (13). VCG ANE-F was recovered from natural infections of both anemone and strawberry but, despite demonstrated cross-infectivity of isolates from anemone and strawberry on the two plant species in artificial inoculation, VCG ANE-A was recovered only from anemone. The involvement of VCG ANE-A in the first two outbreaks of anemone anthracnose but not in the subsequent epidemic of strawberry anthracnose (13) suggests that VCG ANE-A and VCG ANE-F differ in their host range. It may be speculated that cross-infection of anemone could have occurred during the first epidemic of strawberry anthracnose in Israel in 1995 (13), since anemone is also grown in the central Sharon region of Israel where the major cultivation of strawberry takes place. This hypothesis may explain why isolates of VCG ANE-F were only detected during the third anemone epidemic during the years 1996 to 1998 and not during the two previous outbreaks, which preceded the first outbreak of strawberry anthracnose in Israel. However, further studies are needed to estimate the relative importance of host specificity versus epidemiological factors (e.g., presence and survival of inoculum, dissemination, and cross-contamination) in anthracnose epidemics under field conditions.
Molecular tools have been successfully utilized to differentiate between species of Colletotrichum responsible for anthracnose diseases in various crops including almond, citrus, coffee, and strawberry, (14, 18, 22, 33). Based on PCR amplification with species-specific primers, all of the anemone isolates were identified as C. acutatum and were indistinguishable from reference isolates of C. acutatum from strawberry. Isolates of the two VCGs were genotypically uniform and indistinguishable when compared by ap-PCR of the genomic DNA. Only isolate NL-12 from The Netherlands, confirmed as C. acutatum with species-specific PCR (Fig. 1) but not compatible with either VCG, showed a distinct genotype (Fig. 2). This isolate represents a third VCG of C. acutatum from anemone, but its pathogenic significance is not known. C. acutatum can pathogenize various hosts, e.g., almond, apple, citrus, peach, and pecan (4, 7, 12), and we anticipate recovering additional genotypes of the anemone pathogen. Our results indicate that the former description of isolates from Israel (Gokkes et al., Abstr. Phytoparasitica; Vigodsky-Haas et al., Abstr. Phytoparasitica), and possibly also from Italy (20), as C. gloeosporioides was incorrect. Similarly, it has been reported that previously identified strains of C. gloeosporioides causing anemone anthracnose in Japan are C. acutatum (32). It is especially important to differentiate between genotypes within mixed populations for accurate diagnostics, for chemical control, and for breeding for resistance.
Information concerning VCG-host relationship in C. acutatum or other Colletotrichum species is very limited (25). So far, a VCG that includes isolates from more than one host, as demonstrated for VCG ANE-F, has not been reported. In Fusarium oxysporum, the phytopathogenic species most extensively studied with respect to VCG, host specificity is evident by the fact that all members of a given VCG belong to the same forma specialis, although a forma specialis may include more than one VCG (23). In contrast, such specificity does not exist in the plant pathogen Verticillium dahliae, where isolates from various wilt-affected hosts may belong to the same VCG. Thus, V. dahliae isolates from potato, tomato, and additional hosts were found in VCG2A; isolates from cotton and four additional hosts were found in VCG2B; and isolates from potato, cotton, and other hosts were found in VCG4B (27). Although the host range of individual V. dahliae isolates may vary, the lack of host specificity at the VCG level could reflect V. dahliae's ability to attack over 400 plant species, while no more than eight VCGs and/or subgroups are presently recognized in this pathogen (24). The scarcity of comparative VCG analysis of C. acutatum populations does not enable us to decide whether C. acutatum resembles F. oxysporum, with numerous VCGs and formae speciales, or V. dahliae with its small number of VCGs, or whether it presents another model of population structure. The study of C. acutatum from anemone illustrates the potential of VCG analysis to reveal distinct subspecific groups within a pathogen population that appears to be genotypically homogeneous. The VCG approach should allow the determination of relatedness among isolates within and between populations from the same or different hosts at various geographic locations.
With respect to host range, we emphasize that conditions often employed in inoculations tests (e.g., high inoculum concentration, senescing tissues such as detached leaves or fruits, and optimized humidity and temperature) may result in artificial cross-infections that are not observed under field conditions (19), thus rendering the reliability of such assays questionable. Information based on natural infections should, therefore, be considered more reliable in determining host range and specificity and inter-relationship with the VCG.
In conclusion, we have shown that C. acutatum is the species responsible for anemone anthracnose. We identified two main primary VCGs, which share a single predominant molecular genotype, among the anemone and strawberry populations of C. acutatum from various sources. Host infectivity in artificial inoculation was not affected by isolate source. Therefore, the potential exists for cross-infection of anemone isolates on strawberry and strawberry isolates on anemone. However, whether the VCG ANE-A population can infect strawberry under field conditions remains an open question.
ACKNOWLEDGMENTS
This research was supported in part by grant no. IS-2825-97 from BARD, The U. S.-Israel Binational Agricultural Research and Development Fund; grant 132-1029-98 from the Chief Scientist, Israel Ministry of Agriculture; and the Ornamental Plants Production and Marketing Board, awarded to S. Freeman.
We thank A. W. Doornik, Lisse, The Netherlands; M. L. Gullino, University of Torino, Torino, Italy; and Ruth Cohn (formerly of the Shelef Plant Clinic) for providing isolates of Colletotrichum from anemone.
Footnotes
Contribution number 516/00 from the ARO, Institute of Plant Protection.
REFERENCES
- 1.Alahakoon P W, Brown A E, Sreenivasaprasad S. Cross-infection potential of genetic groups of Colletotrichum gloeosporioides on tropical fruits. Physiol Mol Plant Pathol. 1994;44:93–103. [Google Scholar]
- 2.Bailey J A, Jeger M J, editors. Colletotrichum: biology, pathology and control. Wallingford, United Kingdom: CAB International; 1992. [Google Scholar]
- 3.Barker I, Pitt D. Detection of the leaf curl pathogen of anemones in corms by enzyme-linked immunosorbent assay (ELISA) Plant Pathol. 1988;37:417–422. [Google Scholar]
- 4.Bernstein B, Zehr E I, Dean R A, Shabi E. Characteristics of Colletotrichum from peach, apple, pecan, and other hosts. Plant Dis. 1995;79:478–482. [Google Scholar]
- 5.Boerema G H, Hamers M E C. Check-list for scientific names of common parasitic fungi. Series C. Fungi on bulbs: ‘additional crops’ belonging to the Araceae, Begoniaceae, Compositae, Oxalidaceae and Ranunculaceae. Netherlands J Plant Pathol. 1990;96(Suppl. 1):1–23. [Google Scholar]
- 6.Brooker N L, Leslie J F, Dickman M B. Nitrate non-utilizing mutants of Colletotrichum and their use in studies of vegetative compatibility and genetic relatedness. Phytopathology. 1991;81:672–677. [Google Scholar]
- 7.Brown A E, Sreenivasaprasad S, Timmer L W. Molecular characterization of slow-growing orange and Key lime anthracnose strains of Colletotrichum from citrus as C. acutatum. Phytopathology. 1996;86:523–527. [Google Scholar]
- 8.Buddie A G, Martínez-Culebras P, Bridge P D, García M D, Querol A, Cannon P F, Monte E. Molecular characterization of Colletotrichum strains derived from strawberry. Mycol Res. 1999;103:385–394. [Google Scholar]
- 9.Correll J C, Morelock T E, Guerber J C. Vegetative compatibility and virulence of the spinach anthracnose pathogen, Colletotrichum dematium. Plant Dis. 1993;77:688–691. [Google Scholar]
- 10.Correll J C, Klittich C J R, Leslie J F. Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology. 1987;77:1640–1646. [Google Scholar]
- 11.Doornik A W. Heat treatment to control Colletotrichum acutatum on corms of Anemone coronaria. Netherlands J Plant Pathol. 1992;98:377–386. [Google Scholar]
- 12.Förster H, Adaskaveg J E. Identification of subpopulations of Colletotrichum acutatum and epidemiology of almond anthracnose in California. Phytopathology. 1999;89:1056–1065. doi: 10.1094/PHYTO.1999.89.11.1056. [DOI] [PubMed] [Google Scholar]
- 13.Freeman S, Katan T. Identification of Colletotrichum species responsible for anthracnose and root necrosis of strawberry in Israel. Phytopathology. 1997;87:516–521. doi: 10.1094/PHYTO.1997.87.5.516. [DOI] [PubMed] [Google Scholar]
- 14.Freeman S, Katan T, Shabi E. Characterization of Colletotrichum gloeosporioides isolates from avocado and almond fruits with molecular and pathogenicity test. Appl Environ Microbiol. 1996;62:1014–10205. doi: 10.1128/aem.62.3.1014-1020.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman S, Katan T, Shabi E. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Dis. 1998;82:596–605. doi: 10.1094/PDIS.1998.82.6.596. [DOI] [PubMed] [Google Scholar]
- 16.Freeman S, Minz D, Jurkevitch E, Maymon M, Shabi E. Molecular analyses of Colletotrichum species from almond and other fruits. Phytopathology. 2000;90:608–614. doi: 10.1094/PHYTO.2000.90.6.608. [DOI] [PubMed] [Google Scholar]
- 17.Freeman S, Pham M, Rodriguez R J. Molecular genotyping of Colletotrichum species based on arbitrarily primed PCR, A+T-rich DNA, and nuclear DNA analyses. Exp Mycol. 1993;17:309–322. [Google Scholar]
- 18.Freeman S, Rodriguez R J. Differentiation of Colletotrichum species responsible for anthracnose of strawberry by arbitrarily primed PCR. Mycol Res. 1995;99:501–504. [Google Scholar]
- 19.Freeman S, Shabi, E. E. Cross-infection of subtropical and temperate fruits by Colletotrichum species from various hosts. Physiol Mol Plant Pathol. 1996;49:395–404. [Google Scholar]
- 20.Gullino M L, Garibaldi A. Results of experimental trials for controlling leaf curling of anemone caused by Colletotrichum gloeosporioides. Med Fac Landouww Rijksuniv Gent. 1981;46:873–879. [Google Scholar]
- 21.Gunnell P S, Gubler W D. Taxonomy and morphology of Colletotrichum species pathogenic to strawberry. Mycologia. 1992;84:157–165. [Google Scholar]
- 22.Johnston P R, Jones D. Relationships among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia. 1997;89:420–430. [Google Scholar]
- 23.Katan T. Current status of vegetative compatibility groups in Fusarium oxysporum. Phytoparasitica. 1999;27:51–64. [Google Scholar]
- 24.Katan T. Vegetative compatibility in populations of Verticillium—an overview. In: Tjamos E C, Rowe R C, Heale J B, Fravel D R, editors. Advances in verticillium research and disease management. St. Paul, Minn: APS Press; 2000. pp. 69–86. [Google Scholar]
- 25.Katan T. Vegetative compatibility in Colletotrichum. In: Prusky D, Freeman S, Dickman M B, editors. Colletotrichum: host specificity, pathology and host-pathogen interactions. St. Paul, Minn: APS Press; 2000. pp. 45–56. [Google Scholar]
- 26.Katan T, Shabi E. Vegetative compatibility among isolates of Colletotrichum gloeosporioides from almond in Israel. Eur J Plant Pathol. 1996;102:597–600. [Google Scholar]
- 27.Korolev N, Katan J, Katan T. Vegetative compatibility groups of Verticillium dahliae in Israel: their distribution and association with pathogenicity. Phytopathology. 2000;90:529–536. doi: 10.1094/PHYTO.2000.90.5.529. [DOI] [PubMed] [Google Scholar]
- 28.Leslie J F. Fungal vegetative compatibility. Annu Rev Phytopathol. 1993;31:127–150. doi: 10.1146/annurev.py.31.090193.001015. [DOI] [PubMed] [Google Scholar]
- 29.Puhalla J E. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Can J Bot. 1985;63:179–183. [Google Scholar]
- 30.Rodriguez R J, Yoder O C. A family of conserved repetitive DNA elements from the fungal plant pathogen Glomerella cingulata (Colletotrichum lindemuthianum) Exp Mycol. 1991;15:232–242. [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 32.Sato T, Ueda S, Iijima A, Tezuka N. Re-identification of pathogens of anemone and prune anthracnose. Ann Phytopathol Soc Jpn. 1996;62:170–174. [Google Scholar]
- 33.Sreenivasaprasad S, Brown A E, Mills P R. DNA sequence variation and interrelationships among Colletotrichum species causing strawberry anthracnose. Physiol Mol Plant Pathol. 1992;41:265–281. [Google Scholar]
- 34.Tramier R, Bettachini A. Colletotrichum acutatum: maladie nouvelle de l'anemone en France. Phytiatr Phytopharm. 1980;29:121–124. [Google Scholar]
- 35.Tu J C. An improved Mathur's medium for growth, sporulation and germination of spores of Colletotrichum lindemuthianum. Microbiosis. 1985;44:87–93. [Google Scholar]
- 36.Wasilwa L A, Correll J C, Morelock T E, McNew R E. Reexamination of races of the cucurbit anthracnose pathogen Colletotrichum orbiculare. Phytopathology. 1993;83:1190–1198. [Google Scholar]