Summary
The development of boronic probes enabled reliable detection and quantitative analysis of hydrogen peroxide, other nucleophilic hydroperoxides, hypochlorite and peroxynitrite. The major product, in which boronate moiety of the probe is replaced by the hydroxyl group, is however common for all those oxidants. Here, we describe how ortho-isomer of mitochondria-targeted phenylboronic acid can be used to detect and differentiate peroxynitrite-dependent and independent probe oxidation. This method highlights detection and quantification of both the major, phenolic product and the minor, peroxynitrite-specific cyclic and nitrated products of probe oxidation.
Keywords: Hydrogen peroxide, Peroxynitrite, Mitochondria-targeted probes, Boronic probes, o-MitoPhB(OH)2, HPLC-MS
1. Introduction
Boronate-based probes were developed over the last decade for detection of hydrogen peroxide (H2O2) and peroxynitrite (ONOO−) in biological systems (1–3). Mitochondria-targeted boronate probes were developed to monitor hydrogen peroxide in mitochondria (4–8). Boronates react directly and stoichiometrically with both H2O2 and ONOO− (9,10). This is in contrast to more classical fluorogenic probes (e.g. dichlorodihydrofluorescein, DCFH and dihydrorhodamine, DHR), which need the catalyst (e.g. iron, heme proteins) or react with the products of ONOO− decomposition (11–13). Additionally, in contrast to boronates, both DCFH and DHR form intermediate radical species that reduce oxygen to superoxide radical anion (11,14). However, most boronates lack specificity needed to distinguish between hydrogen peroxide, other nucleophilic hydroperoxides and peroxynitrite in a complex biological system, as discussed below (2,9,15–17). In a simple system when bolus amounts of the reactants are quickly mixed this limitation can be overcome due to significant differences of the reaction rate constants. The rate constant of the reaction of boronates with peroxynitrite is six orders of magnitude higher than of the analogous reaction with hydrogen peroxide (2,9). For example, under the conditions when the reaction with ONOO− is completed within 100 ms, the reaction with H2O2 was not completed even after 12 h (9). The situation is, however, different when the oxidants react with the probe in biological systems, with the oxidants continuously produced during the incubation with the probe. Therefore, we proposed the use of specific inhibitors of oxidants formation and/or specific scavengers of the oxidants to differentiate between different species responsible for oxidation of boronate probes in cells (2,18). We have previously reported a formation of both major, phenolic product (o-MitoPhOH) and minor, nitrated product (o-MitoPhNO2) during the reaction of peroxynitrite with ortho-isomer of mitochondria-targeted phenylboronic acid (o-MitoPhB(OH)2, Fig. 1) (19,20). More recently, we have identified an additional, more abundant minor product, cyclo-o-MitoPh (Fig. 1) (21,22). These minor products are specific for ONOO− reaction and are not produced by any other oxidant. Therefore the proposed method provides also an opportunity to distinguish between two biological nitrating pathways: peroxynitrite-dependent and myeloperoxidase/hydrogen peroxide/nitrite-dependent (21). Here, we show how this unique chemistry can be utilized to selectively detect peroxynitrite in cellular systems (23,24). The method is based on probing the oxidants (ONOO− and H2O2) in cells by incubation of cells with o-MitoPhB(OH)2 followed by extraction and HPLC-MS/MS analysis of the products formed (21,25). We describe the protocol for probe preparation, processing of biological samples and HPLC-MS-based analysis of the products formed.
Figure 1. Scheme of oxidation of o-MitoPhB(OH)2.
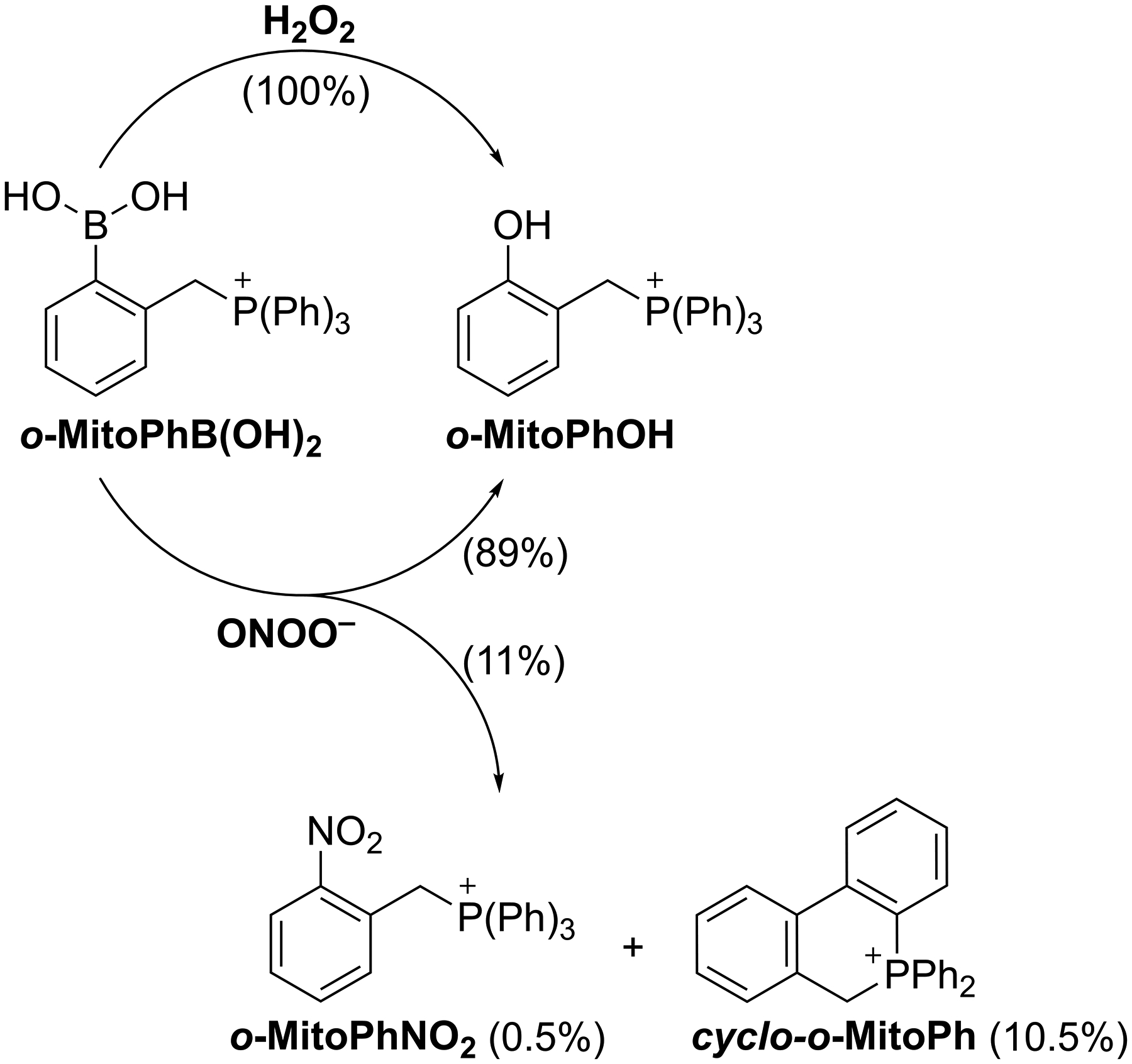
o-MitoPhB(OH)2 is oxidized to o-MitoPhOH by both ONOO− and H2O2. However, cyclo-o-MitoPh and o-MitoPhNO2 are formed only in reaction between o-MitoPhB(OH)2 and ONOO− as the minor products, with the yields of 10.5% and 0.5%, respectively (21).
2. Materials
2.1. Components for the synthesis of o-MitoPhB(OH)2
2 g of triphenylphosphine.
200 ml of anhydrous diethyl ether.
1 g of 2-(bromomethyl)phenylboronic acid.
100 ml of dichloromethane
Argon gas.
2.2. Cell incubation components
2.3. Cell extraction components
Cell lysis buffer:10 ml DPBS containing 0.1% Triton X100 spiked with 1 μM mixture of internal standards, place on ice (can be stored at 4 °C) (see Note 2).
100 ml of ice-cold acetonitrile containing 0.1% (v/v) formic acid.
10 ml of ice-cold acetonitrile containing 0.1% (v/v) formic acid and 1 μM mixture of internal standards, place on ice (can be stored for a short term at 4 °C).
Protein assay reagent (Bradford reagent).
BSA in lysis buffer: 20 mg/ml BSA. Prepare a series of BSA solutions by serial dilutions with the final concentrations of 0.5, 1.0, 1.5, 2, 3, 4, 5, 7 and 10 mg/ml. Keep the solutions on ice.
2.4. HPLC analysis components
HPLC mobile phase: 0.1 % formic acid in water (mobile phase A) and 0.1% formic acid in acetonitrile (mobile phase B) (see Note 3).
Solution of o-MitoPhNO2 (50 mM) in DMSO (see Note 4).
Solution of o-MitoPhB(OH)2 (50 mM) in DMSO.
Solution of o-MitoPhOH (50 mM) in DMSO (see Note 4).
Solution of cyclo-o-MitoPh (50 mM) in DMSO (see Note 5).
Water:acetonitrile (3:1) mixture containing 0.1% (v/v) formic acid, spiked with 1 μM mixture of the internal standards. This will be a solvent for preparation of standards for HPLC calibration.
Mixture of the standards of o-MitoPhB(OH)2, o-MitoPhOH and o-MitoPhNO2 and cyclo-o-MitoPh (0.1 mM each) in water:acetonitrile (3:1) solvent containing 0.1 % formic acid spiked with 1 μM mixture of the internal standards. Prepare serial dilutions down to 1 nM concentration. Use the solvent for standards prepared in previous step.
3. Methods
3.1. Preparation of o-MitoPhB(OH)2
The preparation of the o-MitoPhB(OH)2 probe should be carried out inside a hood with well-working ventilation!
Prepare a solution of triphenylphosphine (1.048 g, 4.0 mmol) in anhydrous diethyl ether (30 ml).
Add 2-(bromomethyl)phenylboronic acid (0.856 g, 4.0 mmol) with constant stirring under argon atmosphere.
Stir the reaction mixture for 46 h (using magnetic stirrer) at room temperature under argon atmosphere.
A crystalline precipitate of bromide salt of o-MitoPhB(OH)2 should be obtained.
Filter the suspension. Keep the solid and discard the filtrate.
Wash the solid with anhydrous diethyl ether (30 ml). Filter the suspension again and keep the solid.
Dissolve the solid in dichloromethane (in a minimum volume, < 5 ml) and precipitate the compound back by addition of diethyl ether (1:10). Filter the powder and keep the solid. Repeat this step once more.
Leave the solid to dry. A white powder should be obtained with the melting point of 225–226 °C. Test the identity and purity of the synthesized probe (see Note 6).
3.2. Cell incubation with the probe
Prepare the cells according to the experimental conditions to be tested for peroxynitrite formation (see Note 1).
Add o-MitoPhB(OH)2 to obtain the final concentration of 50 μM (see Notes 7 and 8).
Incubate the cells for 1 h.
Collect an aliquot of the medium (0.1 ml) in 1.5 ml microcentrifuge tube and freeze in liquid nitrogen.
Remove the rest of medium and wash the cells twice with ice-cold DPBS.
Add 1 ml of ice-cold DPBS and harvest the cells, place the cell suspension in 1.5 ml microcentrifuge tube and spin down the cells by quick centrifugation. Remove the supernatant and freeze the cell pellet in liquid nitrogen.
Frozen cell pellets and media can be stored at −80 °C for at least one week before analysis.
3.3. Extraction of the products
3.3.1. Cell pellets
Preload one set of 1.5 ml microcentrifuge tubes with 0.1 ml of 0.1% formic acid in acetonitrile and place on ice.
Preload a second set of 1.5 ml microcentrifuge tubes with 0.1 ml of 0.1% formic acid in water and place on ice.
Prepare a clear-bottom 96-well plate for protein assay and place on ice.
Place the tubes with frozen cell pellets on ice.
Add 0.2 ml of the lysis buffer spiked with a mix of internal standards (1 μM each) and lyse the cells by 10 syringe strokes using a 0.5 ml insulin syringe with the needle 28 Gauge × 0.5 inch (0.36 mm × 13 mm).
Transfer 0.1 ml of the cell lysates into the tubes containing ice-cold 0.1% formic acid in acetonitrile, vortex for 10 s and place on ice. Transfer 3 × 2 μl of the cell lysate aliquots into 3 wells on 96-well plate for the protein assay.
Incubate the mixtures of cell lysates with acetonitrile for 30 min on ice.
During incubation measure the protein concentration in the cell lysates using Bradford assay and plate reader with absorption detection (see Note 9).
Vortex the tubes again for 5 s and centrifuge for 30 min at 20,000g at 4 °C.
Place the tubes back on ice and transfer 0.1 ml aliquots of the supernatants into the second set of tubes, containing 0.1% formic acid in water.
Vortex the tubes for 5 s and centrifuge for 15 min at 20,000g at 4 °C.
Transfer 0.15 ml of the supernatants into HPLC vials preloaded with conical inserts, seal the vials and place on ice. Once all solutions have been transferred, place the vials in HPLC autosampler pre-cooled to 4 °C.
3.3.2. Media
Preload one set of 1.5 ml microcentrifuge tubes with 0.1 ml of 0.1% formic acid in water and place on ice.
Place the tubes with frozen media on ice.
Add 0.1 ml of the ice-cold 0.1% formic acid in acetonitrile spiked with internal standards (1 μM each) to each tube, vortex for 10 s and place on ice.
Incubate the mixtures of media with acetonitrile for 30 min on ice.
Vortex the tubes again for 5 s and centrifuge for 30 min at 20,000g at 4 °C.
Place the tubes back on ice and transfer 0.1 ml aliquots of the supernatants into the tubes containing 0.1% formic acid in water.
Vortex the tubes for 5 s and centrifuge for 15 min at 20,000g at 4 °C.
Transfer 0.15 ml of the supernatants into HPLC vials preloaded with conical inserts, seal the vials and place on ice. Once all solutions have been transferred, place the vials in HPLC autosampler pre-cooled to 4 °C.
3.4. HPLC-MS/MS analysis of the extracts
Install the column Kinetex Phenyl-Hexyl 50 mm × 2.1 mm, 1.7 μm (Phenomenex) in the HPLC-MS/MS system. The column should be equipped with a UHPLC column filter or guard column to extend the column lifetime.
Equilibrate the column with the mobile phase (75% of mobile phase A and 25% mobile phase B).
Setup the HPLC-MS/MS method and detection parameters according to Tables 1 and 2, respectively (see Note 10).
Test the system by three injections of standards (10 μM) for the reproducibility of retention times and peak intensities for all analytes and internal standards, as shown in Fig. 2.
Run the analysis of the batch of samples.
Include the system and column wash with water:methanol (1:1) mixture at the end of batch.
Quantify each analyte based on the specific MRM transitions and calibration curves constructed in the concentration range relevant to the samples analyzed (see Note 11).
When appropriate, normalize the concentrations of analytes to the protein levels in cell lysates, as determined by Bradford method.
Increase in peak intensities of both o-MitoPhOH, cyclo-o-MitoPh, and o-MitoPhNO2 (Fig. 3) indicates formation of ONOO−, while formation of o-MitoPhOH, but not o-MitoPhNO2 (Fig. 4) indicates the presence of other oxidants, most commonly H2O2 (see Note 12).
Table 1.
HPLC method parameters
| Flow rate: | 0.5 ml/min | ||
| Gradient: | 0 min | 75% A | 25% B |
| 2 min | 68.3% A | 31.7% B | |
| 4 min | 0% A | 100% B | |
| 4.5 min | 0% A | 100% B | |
| 5 min | 75% A | 25% B | |
| Diverter valve: | 0 min | waste | |
| 1.0 min | detector | ||
| 4.0 min | waste |
Table 2.
MS/MS detection parameters
| Analyte | Dominant MRM transition | Reference MRM transition | Reference MRM to dominant MRM intensities ratio | Retention time (min) |
|---|---|---|---|---|
| o-MitoPhB(OH)2 | 397.00>135.00 | 397.00>379.05 | 0.8 | 1.61 |
| o-MitoPhB(OH)2-d15 | 412.20>117.10 | 412.20>135.10 | 0.55 | 1.57 |
| o-MitoPhOH | 369.00>107.10 | 369.00>183.05 | 0.4 | 1.95 |
| o-MitoPhOH-d15 | 384.10>278.10 | 384.10>107.10 | 1.0 | 1.90 |
| o-MitoPhNO2 | 397.90>262.05 | 397.90>351.10 | 0.25 | 2.13 |
| o-MitoPhNO2-d15 | 413.10>277.15 | 413.10>113.00 | 0.85 | 2.08 |
| Cyclo-o-MitoPh | 351.10>183.05 | 351.10>165.05 | 0.7 | 2.15 |
| o-MitoPhCH3 | 367.00>105.05 | 367.00>79.05 | 0.1 | 2.78 |
Figure 2. HPLC-MS/MS detection of o-MitoPhB(OH)2, o-MitoPhOH, cyclo-o-MitoPh and o-MitoPhNO2.
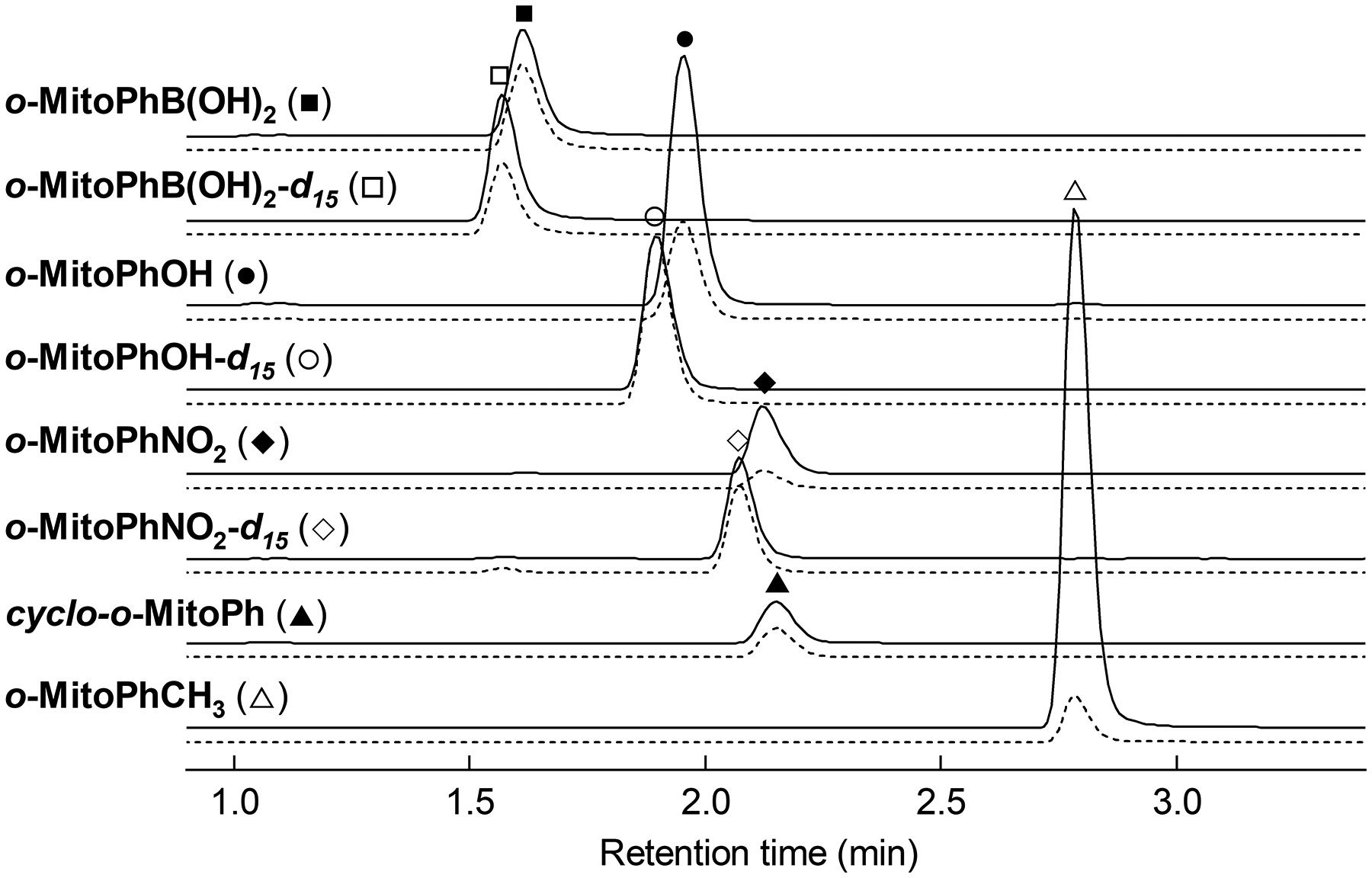
The chromatograms have been obtained with the described method by injection of 20 μl of the mixture of o-MitoPhB(OH)2, o-MitoPhOH, cyclo-o-MitoPh and o-MitoPhNO2 (solid symbols) and corresponding internal standards (open symbols) (1 μM each). For each compound two MRM transitions are shown: dominant (solid lines) and reference (dashed lines). HPLC-MS/MS traces were recorded using the parameters shown in Table 2. The traces have not been scaled and thus represent the actual intensities of each MRM transition in the equimolar mixture.
Figure 3. HPLC-MS/MS analysis of the extracts of RAW 264.7 macrophages activated to produce peroxynitrite.
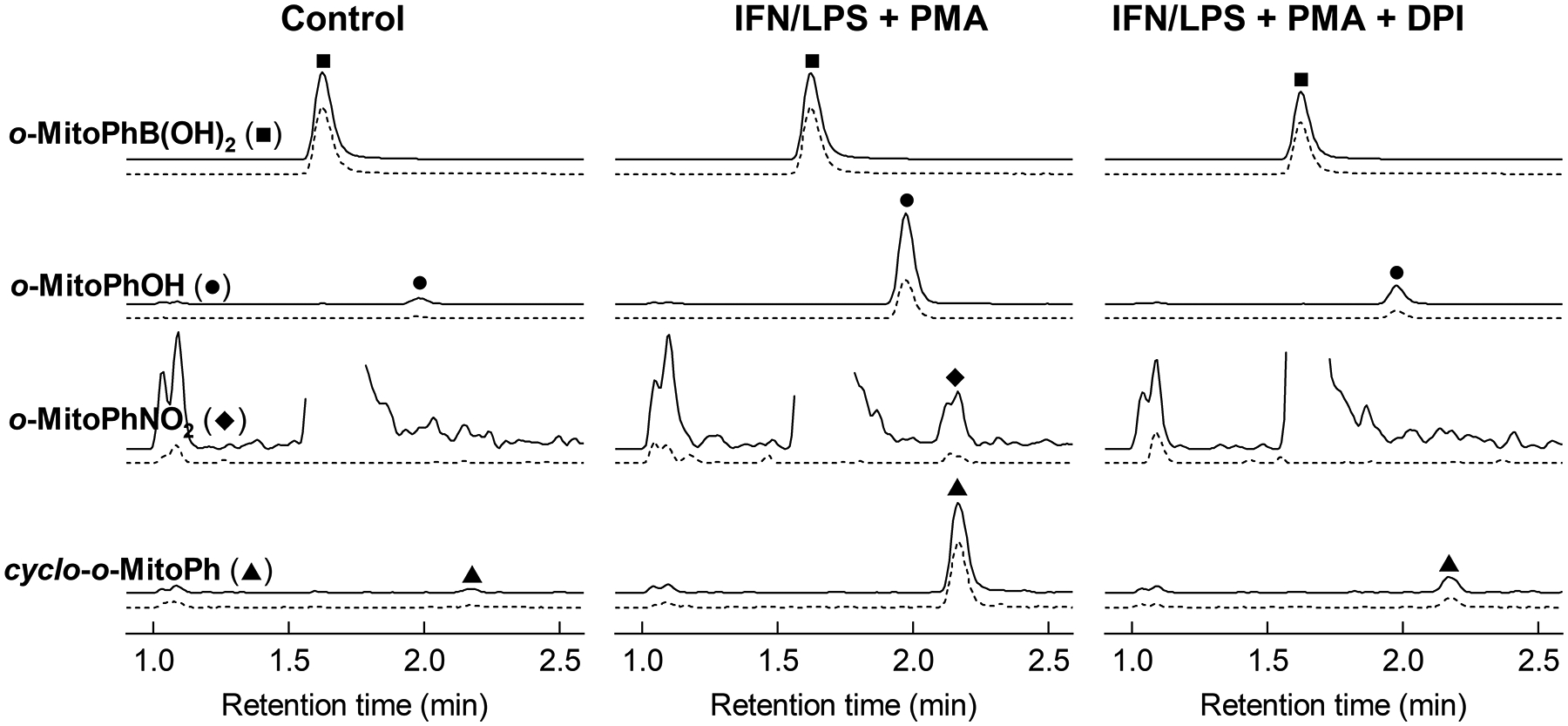
To produce peroxynitrite RAW 264.7 cells were pretreated overnight with interferon γ (IFN, 50 U/ml) and lipopolysaccharide (LPS, 1 μg/ml) followed by addition of PMA (1 μM) in the presence or absence of diphenyleneiodonium (DPI, 1 μM). During stimulation with PMA, o-MitoPhB(OH)2 (50 μM) was present and incubated for 1 h. Cell pellets were collected and processed as described in the protocol. The intensities of o-MitoPhOH were multiplied by a factor of 10 and intensities of cyclo-o-MitoPh and o-MitoPhNO2 were multiplied by 200 and 2000, respectively, to fit the same scale as of o-MitoPhB(OH)2. Solid lines represent the dominant transitions used for quantification and the dashed lines represent the reference transitions used for confirmation of peak identity.
Figure 4. HPLC-MS/MS analysis of the media of neutrophil-like cells activated to produce hydrogen peroxide.
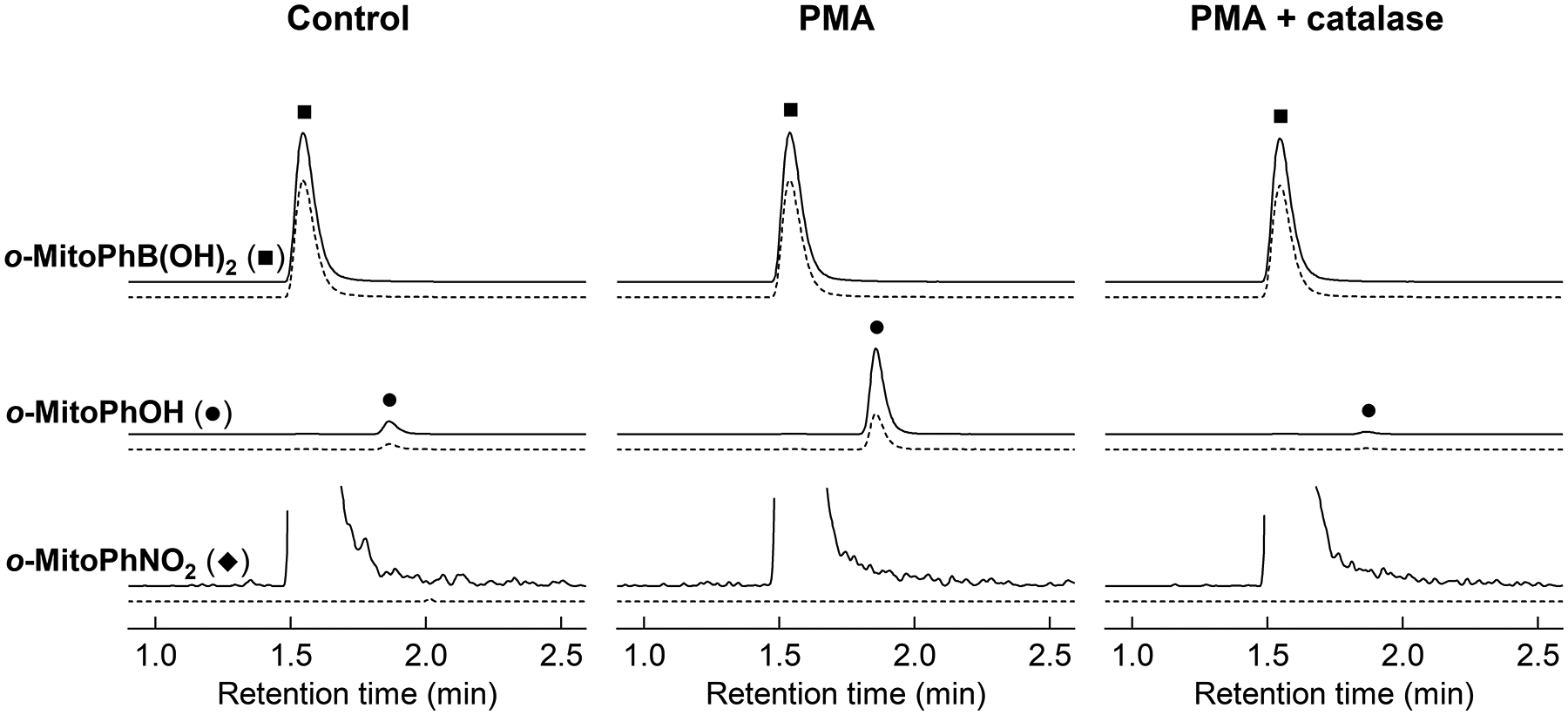
To produce hydrogen peroxide, HL60 cells differentiated for 4 days with all-trans retinoic acid were stimulated with PMA (1 μM) in the presence and absence of catalase (1 kU/ml) and co-treated with o-MitoPhB(OH)2. Aliquots of media were collected after 30 min and processed and analyzed as described in the protocol. The intensities of o-MitoPhOH were multiplied by a factor of 5 and intensities of o-MitoPhNO2 were multiplied by 1000, to fit the same scale as of o-MitoPhB(OH)2.
4. Notes
- The medium used may be selected according to experimental design, and cell culture needs. However, it is preferred that when monitoring extracellular ONOO− and/or H2O2, the components of the medium capable of scavenging those oxidants should be avoided, if possible. For example, pyruvate in the medium may efficiently compete with boronate probes for H2O2.
- Internal standards for o-MitoPhB(OH)2, o-MitoPhOH, and o-MitoPhNO2 are their isotopologs containing deuterated triphenylphosphonium moiety (Fig. 2). These are not commercially available but can be synthesized in analogous ways as o-MitoPhB(OH)2, o-MitoPhOH, and o-MitoPhNO2, (see Note 4, for description of synthesis of o-MitoPhB(OH)2 see 3.1.), but starting with commercially-available triphenylphosphine-d15. As cyclo-o-MitoPh is not synthesized from triphenylphosphine, we do not have deuterated isotopolog for use as an internal standard. Instead, we use (2-methylbenzyl)triphenylphosphonium (o-MitoPhCH3) as an internal standard for cyclo-o-MitoPh (Fig. 2).
LC-MS (preferably UHPLC-MS) grade solvents and formic acid should be used. After preparation, mobile phase should be passed through 0.2 μm filter. Prepare only the amount of mobile phase, which is necessary for the experiment. Do not store the mobile phase for longer than 2–3 days to avoid any growth of the biological matter.
The standards of the products, o-MitoPhOH and o-MitoPhNO2 are commercially available, but can be also synthesized. The phenolic product can be prepared by reaction o-MitoPhB(OH)2 with hydrogen peroxide, followed by addition of catalase to remove excess H2O2. The nitrated product can be synthesized in an analogous protocol as described for o-MitoPhB(OH)2, but starting with 2-nitrobenzyl bromide instead of 2-(bromomethyl)phenylboronic acid.
Cyclo-o-MitoPh is not commercially available should be synthesized according to the published method (21). If only small amounts of cyclo-o-MitoPh are needed, it may be also produced by reacting o-MitoPhB(OH)2 with ONOO−, followed by HPLC-based purification. The identity of the isolated fraction and the extent of contamination by o-MitoPhNO2 need to be determined. In our experience, sufficient chromatographic resolution between the peaks of cyclo-o-MitoPh and o-MitoPhNO2 can be obtained using Raptor Biphenyl column (Restek, Bellefonte, PA, USA; 100 mm × 2.1 mm, 2.7 μm).
Purity of synthesized o-MitoPhB(OH)2 should be tested by HPLC and the compound repurified, if needed. The identity should be confirmed by NMR (20,26) and HRMS (C25H23BO2P+, m/z = 397.1535) (19,20) analyses.
The concentration of o-MitoPhB(OH)2 used for probing of ONOO− should be chosen so as it does not interfere with mitochondrial function. We did not observe significant effects of 50 μM o-MitoPhB(OH)2 on the rate of oxygen consumption by RAW 264.7 cells or MiaPaCa-2 pancreatic cancer cells (25).
When exposing cells to o-MitoPhB(OH)2 it is preferred to add the medium containing the probe, rather than directly adding a solution of concentrated o-MitoPhB(OH)2 in DMSO, to avoid local exposure of cells to high concentrations of DMSO.
If plate reader with absorption detection is not available, protein measurements can be carried out using regular spectrophotometer. The volume of cell lysate needed for the assay may be higher, depending on the volume of the spectrophotometer cell.
To protect the detector, only portion of eluate is flowed into mass detector. This is achieved by using a diverter valve, which directs the flow into waste before 1 min and after 4 min after injection. Between 1 min and 4 min the flow is directed into the detector and the signals recorded.
For each compound two MRM transitions (pairs of parent ion/daughter ion) are recorded. The primary (dominant) transition is used for quantification, while the secondary (reference) transition is used for confirmation of the identity of the analyte. If the ratio of reference to dominant MRM transitions (Table 2) is outside the range allowed (typically ± 30%), the peak is rejected and not used for quantification. Due to similar m/z values of o-MitoPhB(OH)2 and o-MitoPhNO2, there is a small peak of the boronate appearing in the channel of the nitro derivative (Fig. 2, a peak at 1.55 min), which may show up as the dominant peak, when o-MitoPhB(OH)2 is in high excess comparing to o-MitoPhNO2 (Fig. 3). This peak is not however observed in the reference channel, exemplifying the usefulness of the reference transitions for peak identification.
The identity of the oxidizing species can be tested by application of specific inhibitors and/or scavengers and/or identification of ONOO-specific products (13;15). Confirmation of peroxynitrite involvement may be obtained by testing the inhibitory effects of inhibitors of nitric oxide synthase, for example L-NAME (18,20). Detection of ONOO− marker products, cyclo-o-MitoPh and o-MitoPhNO2, is sufficient to confirm the involvement of ONOO− in probe oxidation (21), as shown in Fig. 3. The inhibitory activity of catalase on the yield of o-MitoPhOH and the lack of formation of o-MitoPhNO2 (Fig. 4) indicates the involvement of H2O2 and not ONOO− in probe oxidation.
Acknowledgement
This work was supported by NIH grant R01CA208648 (B.K.). JZ was supported in part by Institutional Research Grant IRG #16-183-31 from the American Cancer Society and the MCW Cancer Center. MH was supported in part by the French National Research Agency ANR-16-CE07-0023-01. A.S. was supported by Polish National Science Center within the SONATA BIS program (Grant Number 2015/18/E/ST4/00235). RP was supported by a grant from Polish National Science Centre (NCN) within the SONATA BIS 6 program (Grant no. 2016/22/E/ST4/00549).
Reference List
- 1.Lippert AR, Van de Bittner GC, and Chang CJ (2011) Boronate oxidation as a bioorthogonal reaction approach for studying the chemistry of hydrogen peroxide in living systems. Acc. Chem. Res 44, 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zielonka J, Sikora A, Hardy M, Joseph J, Dranka BP, and Kalyanaraman B (2012) Boronate probes as diagnostic tools for real time monitoring of peroxynitrite and hydroperoxides. Chem. Res. Toxicol 25, 1793–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sikora A, Zielonka J, Debowska K, Michalski R, Smulik-Izydorczyk R, Pieta J, Podsiadly R, Artelska A, Pierzchala K, and Kalyanaraman B (2020) Boronate-based probes for biological oxidants: A novel class of molecular tools for redox biology. Frontiers in Chemistry 8, 580899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickinson BC, and Chang CJ (2008) A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J. Am. Chem. Soc 130, 9638–9639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocheme HM, Quin C, McQuaker SJ, Cabreiro F, Logan A, Prime TA, Abakumova I, Patel JV, Fearnley IM, James AM, Porteous CM, Smith RA, Saeed S, Carre JE, Singer M, Gems D, Hartley RC, Partridge L, and Murphy MP (2011) Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab 13, 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocheme HM, Logan A, Prime TA, Abakumova I, Quin C, McQuaker SJ, Patel JV, Fearnley IM, James AM, Porteous CM, Smith RA, Hartley RC, Partridge L, and Murphy MP (2012) Using the mitochondria-targeted ratiometric mass spectrometry probe MitoB to measure H2O2 in living Drosophila. Nat. Protoc 7, 946–958 [DOI] [PubMed] [Google Scholar]
- 7.Dickinson BC, Lin VS, and Chang CJ (2013) Preparation and use of MitoPY1 for imaging hydrogen peroxide in mitochondria of live cells. Nat. Protoc 8, 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zielonka J, Joseph J, Sikora A, Hardy M, Ouari O, Vasquez-Vivar J, Cheng G, Lopez M, and Kalyanaraman B (2017) Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem Rev 117, 10043–10120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sikora A, Zielonka J, Lopez M, Joseph J, and Kalyanaraman B (2009) Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic. Biol. Med 47, 1401–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikora A, Zielonka J, Lopez M, Dybala-Defratyka A, Joseph J, Marcinek A, and Kalyanaraman B (2011) Reaction between peroxynitrite and boronates: EPR spin-trapping, HPLC Analyses, and quantum mechanical study of the free radical pathway. Chem. Res. Toxicol 24, 687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wardman P (2007) Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic. Biol. Med 43, 995–1022 [DOI] [PubMed] [Google Scholar]
- 12.Kalyanaraman B (2011) Oxidative chemistry of fluorescent dyes: implications in the detection of reactive oxygen and nitrogen species. Biochem. Soc. Trans 39, 1221–1225 [DOI] [PubMed] [Google Scholar]
- 13.Zielonka J, and Kalyanaraman B (2018) Small-molecule luminescent probes for the detection of cellular oxidizing and nitrating species. Free Radic Biol Med 128, 3–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zielonka J, and Kalyanaraman B (2012) Methods of investigation of selected radical oxygen/nitrogen species in cell-free and cellular systems. in Principles of Free Radical Biomedicine. Volume I (Pantopoulos K, and Schipper HM eds.), Nova Science Publishers, New York. pp 201–264 [Google Scholar]
- 15.Zielonka J, Sikora A, Joseph J, and Kalyanaraman B (2010) Peroxynitrite is the major species formed from different flux ratios of co-generated nitric oxide and superoxide: direct reaction with boronate-based fluorescent probe. J. Biol. Chem 285, 14210–14216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalski R, Zielonka J, Gapys E, Marcinek A, Joseph J, and Kalyanaraman B (2014) Real-time measurements of amino acid and protein hydroperoxides using coumarin boronic acid. Journal of Biological Chemistry 289, 22536–22553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truzzi DR, and Augusto O (2017) Influence of CO2 on Hydroperoxide Metabolism. in Hydrogen peroxide metabolism in health and disease (Vissers MC, Hampton M, and Kettle AJ eds.), CRC Press, Boca Raton, FL. pp 81–99 [Google Scholar]
- 18.Zielonka J, Zielonka M, Sikora A, Adamus J, Joseph J, Hardy M, Ouari O, Dranka BP, and Kalyanaraman B (2012) Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J. Biol. Chem 287, 2984–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sikora A, Zielonka J, Adamus J, Debski D, Dybala-Defratyka A, Michalowski B, Joseph J, Hartley RC, Murphy MP, and Kalyanaraman B (2013) Reaction between peroxynitrite and triphenylphosphonium-substituted arylboronic acid isomers: identification of diagnostic marker products and biological implications. Chem. Res. Toxicol 26, 856–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zielonka J, Sikora A, Adamus J, and Kalyanaraman B (2015) Detection and differentiation between peroxynitrite and hydroperoxides using mitochondria-targeted arylboronic acid. in Methods in Molecular Biology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zielonka J, Zielonka M, Verplank L, Cheng G, Hardy M, Ouari O, Ayhan MM, Podsiadły R, Sikora A, Lambeth JD, and Kalyanaraman B (2016) Mitigation of NADPH oxidase 2 activity as a strategy to inhibit peroxynitrite formation. Journal of Biological Chemistry 291, 7029–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rios N, Radi R, Kalyanaraman B, and Zielonka J (2020) Tracking isotopically labeled oxidants using boronate-based redox probes. J Biol Chem 295, 6665–6676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zielonka J, Joseph J, Sikora A, and Kalyanaraman B (2013) Real-time monitoring of reactive oxygen and nitrogen species in a multiwell plate using the diagnostic marker products of specific probes. Methods Enzymol 526, 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy M, Zielonka J, Karoui H, Sikora A, Michalski R, Podsiadly R, Lopez M, Vasquez-Vivar J, Kalyanaraman B, and Ouari O (2018) Detection and Characterization of Reactive Oxygen and Nitrogen Species in Biological Systems by Monitoring Species-Specific Products. Antioxid Redox Signal 28, 1416–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng G, Zielonka M, Dranka B, Kumar SN, Myers CR, Bennett B, Garces AM, Dias Duarte Machado LG, Thiebaut D, Ouari O, Hardy M, Zielonka J, and Kalyanaraman B (2018) Detection of mitochondria-generated reactive oxygen species in cells using multiple probes and methods: Potentials, pitfalls, and the future. J Biol Chem 293, 10363–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison DE, Issa F, Bhadbhade M, Groebler L, Witting PK, Kassiou M, Rutledge PJ, and Rendina LM (2010) Boronated phosphonium salts containing arylboronic acid, closo-carborane, or nido-carborane: synthesis, X-ray diffraction, in vitro cytotoxicity, and cellular uptake. JBIC Journal of Biological Inorganic Chemistry 15, 1305–1318 [DOI] [PubMed] [Google Scholar]


