Significance
We present an atomic-resolution structure of the human mitochondrial kinase NADK2 bound with its natural substrate. NADK2 is an essential enzyme required for biosynthesis of a key reactive oxygen-neutralizing metabolite, NADP(H). Loss-of-function mutations in the human NADK2 gene result in microcephaly, hypotonia, and death at a young age. Here, we use structural biology and biochemistry to elucidate the molecular mechanism of this enzyme. Our work shows that NADK2 is structurally different from other NAD kinases (NADKs) due to the presence of a unique tetramer disruptor/dimerization element conserved in mitochondrial kinases of animals (EMKA). EMKA transforms molecular assembly of NADK2 and alters regulation of this NADK. Our work thereby reveals unanticipated functional properties of human NADK2 arising from its unusual structure.
Keywords: NADK2, NADK, cooperative, structure, dimer
Abstract
All kingdoms of life produce essential nicotinamide dinucleotide NADP(H) using NAD kinases (NADKs). A panel of published NADK structures from bacteria, eukaryotic cytosol, and yeast mitochondria revealed similar tetrameric enzymes. Here, we present the 2.8-Å structure of the human mitochondrial kinase NADK2 with a bound substrate, which is an exception to this uniformity, diverging both structurally and biochemically from NADKs. We show that NADK2 harbors a unique tetramer disruptor/dimerization element, which is conserved in mitochondrial kinases of animals (EMKA) and absent from other NADKs. EMKA stabilizes the NADK2 dimer but prevents further NADK2 oligomerization by blocking the tetramerization interface. This structural change bears functional consequences and alters the activation mechanism of the enzyme. Whereas tetrameric NADKs undergo cooperative activation via oligomerization, NADK2 is a constitutively active noncooperative dimer. Thus, our data point to a unique regulation of NADP(H) synthesis in animal mitochondria achieved via structural adaptation of the NADK2 kinase.
The NAD kinase (NADK) supplies all living cells with a vitally important redox metabolite, NADP(H) (NADP+ and its coupled reduced form NADPH) (1). NADK functions by adding 2′-phosphate to NAD(H) (NAD+ and its coupled reduced form NADH), a cofactor required for ATP synthesis and energy metabolism (2, 3). The addition of the single phosphate changes the physiological role of NAD(H), converting it into a functionally different cofactor NADP(H), which is essential for biosynthesis of cholesterol, production of fatty acids, and neutralization of reactive oxygen species as a mechanism that mitigates oxidative damage and stress (4).
Bacterial cells synthesize NADP(H) using a single NADK, whereas eukaryotic cells have separate pools of NADP(H) in the cytosol and mitochondria. A dedicated NADK is used to maintain each pool (5, 6). Cytosolic NADKs from bacteria and eukaryotes are well characterized, including the detailed understanding of their structural mechanisms (7, 8). Less is known about NADKs that synthesize mitochondrial NADP(H). In yeasts, the mitochondrial NADK was discovered in 2003 by tracking a gene, POS5, required for stability of mitochondrial DNA (9). Subsequently, a plant counterpart of mitochondrial NADK, the kinase NADK2 from chloroplast, was identified (10). Mitochondrial NADKs of animals remained unknown until 2012, when a mitochondrially localized protein, C5ORF33, was shown to phosphorylate NAD(H) and was renamed NADK2 (11).
The identification of vertebrate NADK2 revealed that NADPH synthesis inside mitochondria is essential in animals. In mice, homozygous knockouts of NADK2 caused embryonic and preweaning lethality with complete penetrance (12). In humans, at least four patients with genetic NADK2 defects have been described. In two cases, NADK2 defects were moderate and involved either a splice site mutation, leading to suboptimal splicing, or a loss of the start codon, leading to low levels of NADK2 translation, presumably due to the presence of an alternative start codon, CUG (13, 14). These patients were alive at 10 and 16 y of age but had uncoordinated movement, reduced vision, neural abnormalities, and microcephaly. In two remaining patients, genetic defects in NADK2 were severe and resulted in encephalopathy and lethality at 4 mo and 5 y of age (14, 15).
While knowledge of cellular and physiological functions of NADK2 is growing, mechanistic understanding of this kinase remains poor due to scarce molecular data and the absence of a protein structure. Available biochemical evidence predicts that NADK2 could be different from all (more than 70) structurally characterized NADKs from bacteria and eukaryotes. Whereas these kinases assemble into structurally similar tetramers (16), NADK2 was reported to form a dimer, rather than a tetramer, in solution (11). To decipher self-assembly of NADK2 and build a molecular understanding of this kinase, we determined its structure and carried out a structure-guided functional analysis.
Results
Structure of Human NADK2 with Bound NAD+ Reveals the Conserved Kinase Fold Bearing an Atypical Insert Domain.
NADK2 is localized to the mitochondria as a propeptide with an N-terminal mitochondrial targeting sequence (MTS) (11, 17). Using the MitoFates server, we determined that human NADK2 matures via proteolytic cleavage at amino acid H47 (SI Appendix, Fig. S1A) (18). Accordingly, we truncated the MTS and produced homogeneous mature NADK2 protein (amino acids 47 to 442; SI Appendix, Fig. S1B). High-throughput crystallization screening yielded small cocrystals of the NADK2•NAD+ complex. Following optimization, these crystals diffracted to 2.8 Å on the microfocusing X-ray beamline (SI Appendix, Table S1 and Methods). Electron density for the bound NAD+ substrate was observed at the expected active-site location (SI Appendix, Figs. S2 and S3).
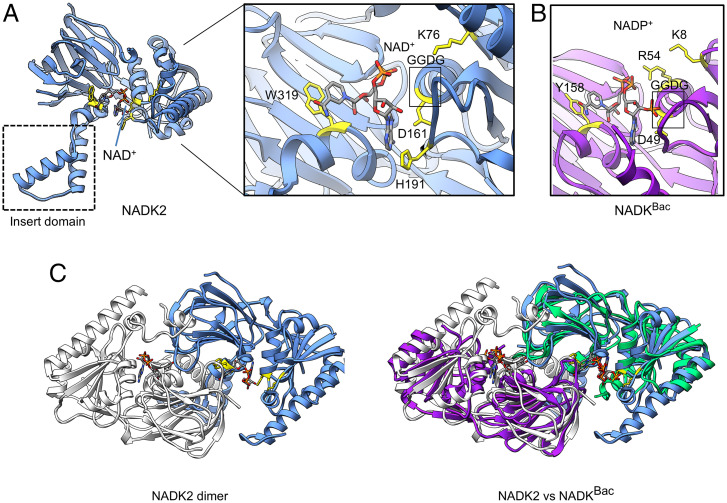
The structure of NADK2 has the same overall fold as in structurally characterized cytosolic NADKs, including the closely related ortholog from the archaea Archaeoglobus fulgidus (all-atom RMSD = 2.2 Å; Fig. 1 A and B) (8). The GGDG sequence motif bearing the catalytic side chain (D161 in human NADK2) is found in the expected position (11, 19). The NAD+ substrate adopts the same compact configuration as the nicotinamide dinucleotide bound in NADK (Fig. 1A), although specific protein-substrate contacts are somewhat different. In human NADK2, the nicotinamide and adenosine groups of NAD+ form π-π stacking interactions with the aromatic residues W319 and H191, respectively. In A. fulgidus NADK, the interaction with W319 is replaced with tyrosine, whereas the amino acid equivalent to H191 is absent due to the lack of a corresponding protein loop. The negatively charged diphosphate group of nicotinamide bound to NADK is recognized by two positively charged residues, R54 and K8, whereas in NADK2 only the cationic amino acid K76 is located near the diphosphate (Fig. 1 A and B). Further examination of the crystal structure revealed that the largest difference between all previously characterized NADKs and the mitochondrial kinase NADK2 is the presence of an inserted domain comprised of two alpha helices (Fig. 1A).
Fig. 1.
Structure of human NADK2. (A) Ribbon representation of the human NADK2 structure. (Inset) Active site with bound NAD+ substrate. Key amino acids that are involved in substrate recognition are highlighted in yellow. The dotted box shows a unique subdomain that is observed in NADK2 and absent from all previously characterized cytosolic NADKs. (B) Close-up view of the active site in a related archaeal NADK with bound product NADP+ (PDB: 1SUW). Amino acids involved in NADP+ recognition are highlighted in yellow. (C) Structure of the NADK2 homodimer observed in crystal packing (Left). Superimposition of dimers formed by human NADK2 and by bacterial NADK (PDB: 1SUW) (Right). Structures were visualized using UCSF Chimera (24).
The Insert Domain Structurally Supports the NADK2 Dimer but Blocks the Tetramer.
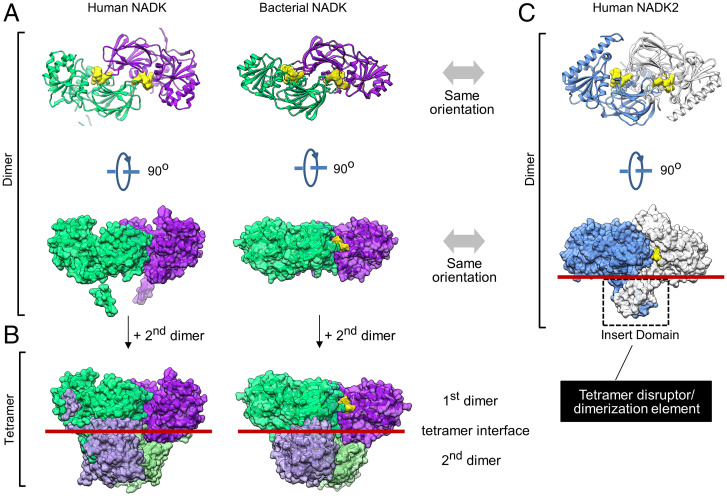
To understand the role of the inserted domain, we examined crystal packing of NADK2. In the crystal, two NADK2 monomers come in close contact, burying 4,681 Å2 of the interface surface area. The resulting symmetrical homodimer creates two composite active sites and resembles the dimers formed by bacterial (2,474-Å2 interface surface burial) and eukaryotic cytosolic (5,466-Å2 interface surface burial) NADKs (Figs. 1C and 2A and SI Appendix, Fig. S4). For all previously characterized NADKs, including the yeast mitochondrial NADK POS5 (16), the homodimers undergo further self-association to form a dimer of dimers (tetramer), which is observed in crystals and in solution (20) (Fig. 2 A and B).
Fig. 2.
Quaternary structure NADK2 compared to quaternary structures of NADKs. (A) Dimers in the crystal packing of human cytosolic NADK (PDB: 3PFN) and bacterial NADK (PDB: 1SUW). The yellow surface shows bound NADP+ (bacterial NADK) or modeled NADP+ (human NADK) positioned in the active site by aligning bacterial and human structures. (B) Tetramers observed in the crystal packing of human and bacterial NADKs. The tetramers are formed by dimerization of two homodimers in B. Red lines mark the tetramerization interface. (C) Ribbon and molecular surface representations of the NADK2 dimer observed in the crystal (present work). Dimeric insert domains from two neighboring protein copies are marked with dashed lines. The red line shows the position equivalent to the tetramerization interface in NADK.
Human NADK2 breaks this hitherto universally observed trend and does not form the tetramer (Figs. 1A and 2C). We observed that the insert domains of two neighboring NADK2 molecules cross-dimerize, nearly doubling the homodimer interface size by adding 2,109 Å2 of surface area burial. While the dimeric insert domain structurally stabilizes the dimer, it creates steric bulk at the tetramer interface and prevents higher-order oligomers of NADK2 (Fig. 2 B and C).
The NADK2 Tetramer Disruptor/Dimerization Insert Domain Is Widespread in Animal Mitochondria.
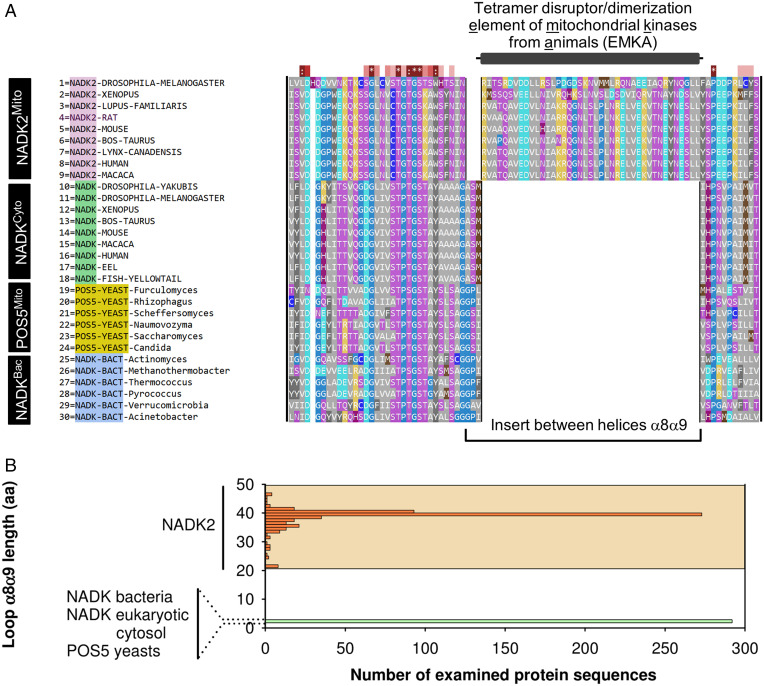
To understand which NADKs contain the insert domain, we used the MUltiple Sequence Comparison by Log-Expectation (MUSCLE) aligner and crystal structures to prepare an accurate protein sequence alignment of NADKs (21) (Fig. 3A). Our analysis showed that the insert is encoded within alpha helices α8 and α9 of human NADK2 annotation. This domain is absent from nonmitochondrial NADKs and the yeast mitochondrial kinase POS5. It is present only in mitochondrial kinases of metazoans and is conserved from humans to drosophila (Fig. 3A).
Fig. 3.
Bioinformatics analyses of insert conservation in NADKs. (A) Multiple sequence alignment of animal NADK2, cytosolic NADKs from animals and bacteria, and mitochondrial NADKs from yeasts (POS5). Sequence alignment was conducted using MUSCLE and visualized in SEQMOL-Kd. Hydrophobic residues are colored gray. (B) Bioinformatics analysis of insert size between alpha helices α8-α9 across ∼300 nonredundant NADK2 sequences compared to ∼300 nonredundant sequences of NADKs (Methods). The insert in NADKs has a uniform length of 3 amino acids, whereas the insert in NADK2 is longer and exhibits length variability, shown by orange shading.
To establish an accurate phylogenetic boundary between NADKs that form tetramers and lack the insert domain and kinases that form dimers and have the insert domain, we extended our analysis to hundreds of sequences. Toward this end, we built a regular expression that can find the α8-α9 insert in protein sequences of NADKs (Methods). Our regular expression analysis revealed that all NADKs belong to two distinct groups based on insert size. The first group contains kinases from bacteria, cytosol of eukaryotes, and mitochondria of yeasts. All members of this group have an insert of 3 amino acids. The second group contains kinases from animal mitochondria. These kinases stand out due to a much larger insert of 22 to 47 amino acids. Our analysis shows that the tetramer disruptor/dimerization insert domain is a conserved element of mitochondrial NADKs from animals (EMKA) (Fig. 3B).
EMKA Promotes NADK2 Homodimerization and Activation.
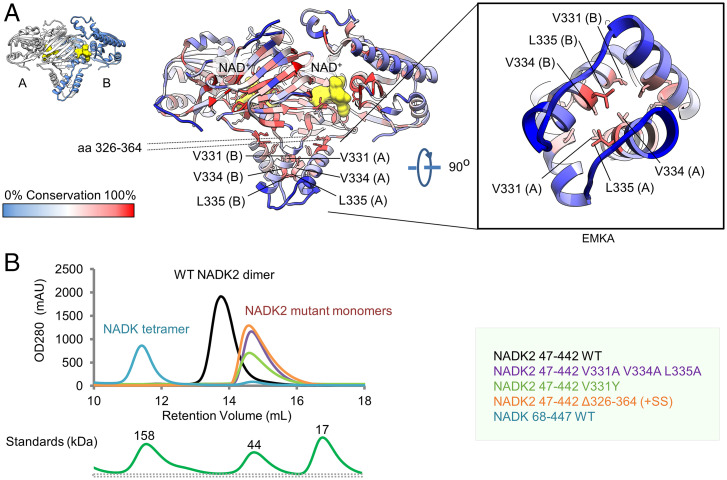
To determine whether the presence of EMKA has functional consequences, we tested the effect of this element on self-association and enzymatic activity of NADK2 by mutagenesis. To identify functionally important amino acids, we colored the crystal structure of NADK2 by amino acid conservation (Fig. 4A). High conservations were observed for active-site residues interacting with NAD+ and for amino acids located inside the hydrophobic core. A cluster of conservation was also observed at the EMKA/EMKA dimer interface, indicating that EMKA homodimerization is conserved in NADKs from animal mitochondria. The identified amino acids were selected for site-directed mutagenesis and functional analyses.
Fig. 4.
Probing the role of EMKA in stabilizing the quaternary structure of NADK2 in solution. (A) Structure of the NADK2 homodimer colored by amino acid conservation and prepared using SEQMOL-Kd and PyMOL. Key conserved residues predicted to stabilize the EMKA/EMKA dimerization interface are marked. The NAD+ substrate is colored yellow. (B) Quaternary states of human NADK and NADK2 in solution profiled by size exclusion chromatography. NADK migrates as a tetramer with small but detectable quantities of monomer. Wild-type (WT) NADK2 migrates as a homogeneous dimer. NADK2 EMKA mutants migrate as homogeneous monomers. Migration of protein standards is provided for size reference. OD280, optical density at wavelength 280 nm; mAU, milli absorbance units.
To examine the quaternary state of NADK2 in solution, we used size exclusion chromatography. We also purified and analyzed human cytosolic NADK to provide a direct comparison with a kinase that lacks EMKA. NADK migrated as a homogeneous tetramer in equilibrium with trace amounts of monomer. In contrast, NADK2 migrated as a stable homogeneous dimer (Fig. 4B and SI Appendix, Fig. S5). EMKA deletion (a swap of 39 amino acids of EMKA for two S residues introduced to prevent structural strain) disrupted the dimer and produced a monomeric NADK2. Thus, the EMKA/EMKA interface is essential for stable self-association of NADK2. A strong disruptive effect also occurred with EMKA point mutations. Mutations V331A+V334A+L335A, removing three hydrophobic side chains at the EMKA/EMKA interface, and a point mutation, V331Y, introducing steric bulk, caused NADK2 homodimer dissociation into monomers (Fig. 4B).
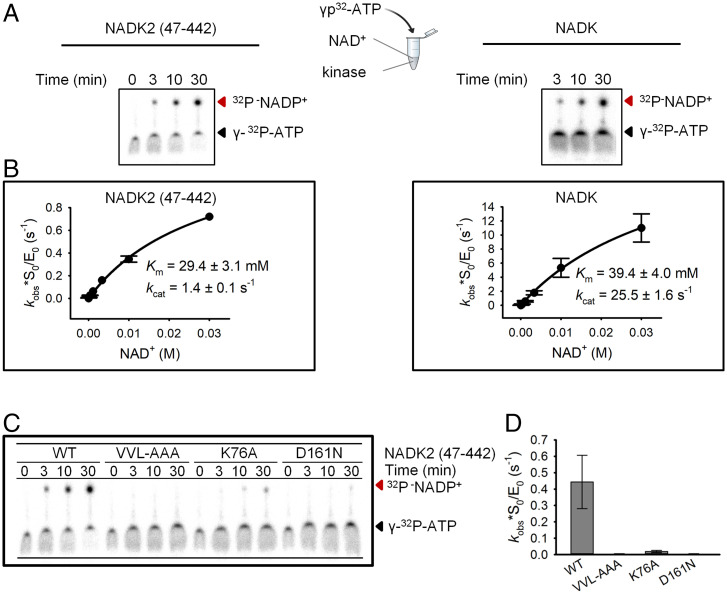
To probe the role of EMKA in NADK2 kinase activity, we used enzyme kinetics. Previously, kinase kinetics of NADK2 has been examined by a coupled colorimetric reaction with glucose-6-phosphate dehydrogenase, i.e., by an indirect readout (11, 17). To avoid quantitative uncertainties inherent to indirect assays, here we used a polyacrylamide gel electrophoresis assay, which directly monitors the formation of the radiolabeled product NADP+ (Methods and Fig. 5A). The direct assay visualized robust NADP+ synthesis by the NADK2 protein used in our work (Fig. 5A). We confirmed the previous report that NADK prefers NAD+ compared to NADH, whereas NADK2 is more promiscuous and can use NAD+ and NADH as equivalent substrates (SI Appendix, Fig. S6) (11). We determined that ATP binds to NADK and to NADK2 with similar affinity (Km = 1.7 and 2.2 mM, respectively; SI Appendix, Fig. S7A). These values also match the published values for both kinases (11).
Fig. 5.
Kinetic analysis of NADK2 and NADK by direct assay. (A) Radiolabeling-based detection of NAD+ phosphorylation by polyacrylamide gel electrophoresis (20%, 29:1). Assays were performed with 12.5 μM NADK2 mature protein (amino acids 47 to 442) or 1 μM full-length NADK. Reactions contained 8.75 mM NAD+ and trace γ-32P-ATP and were conducted at 37 °C. Substrates (ATP) and products (NADP+) are indicated by arrows. (B) Multiple-turnover analysis of substrate binding to NADK and NADK2. (C) Kinase activity profiling of WT and mutant NADK2. The dimerization interface mutant (V331A V334A L335A [VVL-AAA]), substrate-contacting mutant (K76A), and catalytic center mutant (D161N) were analyzed. (D) Quantification of the data in C and SI Appendix, Fig. S8. Error bars show SE from three experimental replicates.
Previously, the catalytic constant (kcat) for NADK2 has been determined in arbitrary units of units per milligram, and it has been suggested that NADK2 is a relatively slow enzyme exhibiting an apparent Vmax that is ∼200-fold smaller than that for NADK (11). Using the direct assay, we determined the absolute value of kcat. NADK2 is a relatively rapid enzyme, exhibiting kcat = 1.4 s−1 in the presence of trace γ-32P-ATP (Fig. 5B) and 2.2 s−1 with a physiologically more relevant background of 2 mM cold ATP•Mg2+ (SI Appendix, Fig. S7B). These values place the turnover number for NADK2 ∼20-fold closer to that of NADK (we determined kcat = 25.5 s−1; Fig. 5B) than reported previously (11).
A major difference between the direct assay and the published indirect parameters emerged during measurements of Km for NAD+. Previous analysis suggested that NADK2 binds NAD+ with Km = 22 μM, which is ∼50-fold stronger compared to the value for NADK (11). Here we determined that NADK2 binds NAD+ with Km ∼30 to 60 mM (Fig. 5B and SI Appendix, Fig. S7B), which is ∼1,000-fold larger compared to the literature value (11). Follow up side-by-side measurements of Km showed that NADK2 binds NAD+ comparably to NADK.
The direct kinetics assay provided us with the tool needed to probe the role of EMKA in NADK2 kinase activity. Point mutations of the three side chains that stabilize the EMKA/EMKA interface (V331A+V334A+L335A) blocked NADK2 activity (Fig. 5 C and D). Strong inhibition of kinase activity occurred also with a single point mutant, V331Y, which sterically hinders the EMKA/EMKA interface and destabilizes the NADK2 homodimer in solution (Figs. 4B and 6A). The effects of the EMKA/EMKA interface mutations were at least as strong as the effects of active-site mutations tested as a reference. The NADK2 point mutant K76A, which removes the diphosphate-interacting positive charge, inhibited NADK2 activity by ∼10-fold (Figs. 1A and 5 C and D), whereas removal of a protonatable catalytic amino acid, D161, by an isosteric mutation, D161N, brought the kinase activity down to trace levels (Fig. 5 C and D). These results establish that EMKA is critically important for stabilizing the catalytically active homodimer of NADK2 kinase.
Fig. 6.
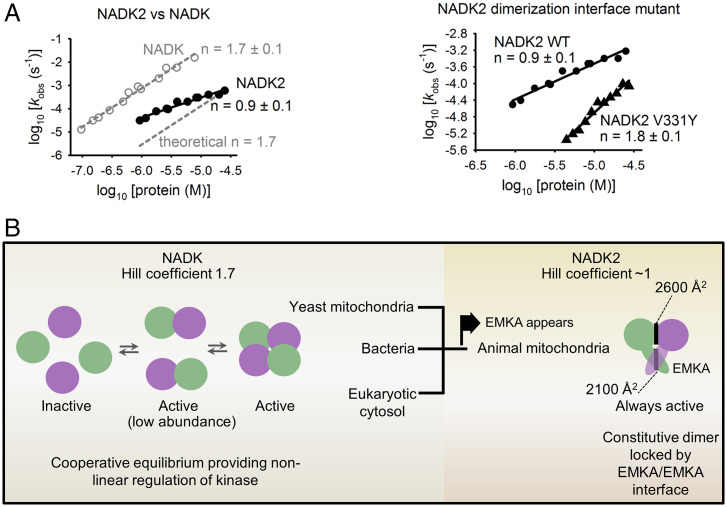
Cooperative activation of NADK versus NADK2. (A) Cooperative activation profiles for WT NADK, WT NADK2, and the NADK2 V331Y interface mutant, plotted in double-logarithm coordinates. The slope of each line gives the Hill coefficient (n). Dashed line is drawn to show a reference slope n = 1.7 (WT NADK). Reaction conditions were as in Fig. 5. Each data point was obtained from exponential fitting to an independent first-order kinetics time course. (B) Model for cooperative regulation of human NADKs. Self-association renders NADK cooperative. NADK2 from mitochondria of animals evolved a unique homodimerization element, EMKA, which stabilizes the homodimer, renders NADK2 noncooperative, and thus buffers the kinase activity against changes in NADK2 protein levels.
EMKA Determines Cooperative Regulation of NADK2.
Unlike conventional monomeric enzymes that show either linear or less-than-linear rate dependence on enzyme concentration, enzymes that form dimers and larger assemblies can undergo cooperative activation and exhibit stronger-than-linear activation at increasing enzyme concentrations (22). To determine whether NADKs exhibit cooperativity and whether different quaternary structures result in different cooperativity for NADK and NADK2, we measured the Hill coefficient (n) for self-association and activation of both human enzymes. Of note, previously described cooperativity analyses of NADKs have been limited to assessing enzyme-substrate binding equilibrium; these experiments did not provide information about kinase self-association to form a quaternary assembly (7).
By varying protein concentration in the kinase assay, we found that NADK exhibits a Hill coefficient of n = 1.7 (Fig. 6A). This value exceeds n = 1 of monomeric enzymes and thereby establishes that NADK is a cooperative enzyme activated via self-association. Analogous measurements revealed that NADK2 is not cooperative and exhibits a Hill coefficient of ∼1 (Fig. 6A). To probe the role of the EMKA/EMKA interface in defining cooperativity of NADK2, we obtained a similar enzyme-activity profile for the point mutant V331Y that weakens the NADK2 dimer (Figs. 4B and 6A). The point mutation attenuated the kinase activity, as expected, but converted NADK2 into a cooperative enzyme (n = 1.8; Fig. 6A). These data identify EMKA as a suppressor of cooperativity in NADKs.
Discussion
We established that human NADK2 bears a unique regulatory element, EMKA, which blocks tetramerization and converts this kinase into a stable homodimer (Fig. 4). The difference between quaternary structures of NADK2 and NADK has functional consequences, leading to different cooperativity properties of these NADKs. Cytosolic NADK forms a tetramer and undergoes cooperative activation via self-association with a Hill coefficient of n = 1.7. This value is smaller than n = 4, which is expected for a perfectly cooperative enzyme with four subunits, indicating that NADK self-association must occur via a catalytically active dimeric NADK intermediate. In contrast, NADK2 is not cooperative and exhibits a Hill coefficient of n ∼1, as expected for a monomeric enzyme (Fig. 6A). Considering that homodimerization is required for the kinase activity of NADK2 (Figs. 4 and 5), the absence of cooperativity indicates that NADK2 forms a constitutively active homodimer that does not dissociate. This interpretation is confirmed by the mutant V331Y, which weakened the homodimer sufficiently to unlock dissociation equilibrium and converted NADK2 into a robustly cooperative enzyme (Figs. 4B and 6 A and B).
Structurally, tetrameric NADKs form two types of dimer interfaces (8). The first interface (IF1) contacts the ligand (Fig. 2A), whereas the second interface (IF2) involves only protein-protein interactions (at the tetramer interface in Fig. 2B). Both interfaces bury comparable dimer surface areas of ∼2,200 to 2,500 Å2, raising the question of their contribution to cooperative regulation. NADK2 forms only IF1, which is normally so stable that its role in cooperativity is not detected. However, if IF1 is allowed to reversibly dissociate, it can provide cooperativity, as evidenced by cooperative activation of NADK2 variants with EMKA mutations (Fig. 6A). For NADK, both interfaces must contribute to cooperativity, as the enzyme can dissociate fully to form monomers (Fig. 4B). Assembly of the NADK tetramer from these monomers requires the formation of both IF1 and IF2, which thereby participate in cooperative activation of the tetrameric kinase.
Why does EMKA deletion produce a monomeric protein, not a tetramer, as seen for cytosolic NADK? One might expect a tetramer if the only difference between NADK2 and NADK was steric bulk provided by EMKA. However, amino acids at the tetramerization interface are also vastly different between NADK and NADK2. These differences arise from the loss of evolutionary pressure for tetramerization in the constitutively dimeric kinase NADK2, leading to a loss of a functional tetramerization interface revealed by our experiments with NADK2-ΔEMKA.
Our results show that EMKA is a key element of NADK2 that defines not only the structure but also the activation dynamics of this kinase. The observation that EMKA is a relatively large domain that nearly doubles the homodimer interface size in NADK2 points to metabolic constraints creating substantial functional pressure, forcing NADKs inside animal mitochondria to evolve this element. Indeed, mitochondrial NADK2 of all animals that we profiled encodes EMKA (Fig. 3). A key functional change that occurs in NADK2 due to EMKA, the loss of cooperative activation, is predicted to have a buffering effect on the kinase output under conditions of fluctuating protein levels. For each 10-fold decrease in the kinase concentration, the activity of tetrameric NADK is weakened by 50-fold (101.7), whereas the activity of NADK2 is weakened by at most 10-fold (Fig. 6A). These estimates suggest that one hypothetical role for EMKA is to provide a mechanism ensuring continued NADK2 activity if animal mitochondria develop an NADK2 deficiency.
In summary, the activation of NADK by self-association renders this enzyme cooperative and poised for regulation. The importance of NADK regulation is underscored by the recently described phosphorylation of vertebrate NADK by the protein kinase Akt, which modulates the activity of NADK (23). During this reaction, Akt phosphorylates the N terminus of NADK, poised for modulating NADK self-association due to the major structural role of the N terminus in stabilizing the NADK quaternary state (SI Appendix, Fig. S9). In contrast to NADK, NADK2 uncoupled itself from cooperative regulation via acquisition of EMKA. Thus, NADK2 inside animal mitochondria appears to be optimized to provide sustained kinase output.
Methods
Human NADK2 and NADK proteins were expressed in Escherichia coli and purified to homogeneity by size exclusion fast protein liquid chromatography (FPLC). NAD(H) kinase kinetics assays were conducted at 37 °C using γ-32P-ATP radioactive substrate and gel electrophoresis, followed by gel quantification on a Typhoon phosphorimager. Initial crystallization of NADK2 was achieved using a high-throughput Mosquito robot, in 100-nL hanging drops. Crystals were optimized and grown in 0.5-μL hanging drops. Diffraction data were collected on the National Synchrotron Light Source II (NSLS-II) Frontier Microfocusing Macromolecular Crystallography (FMX) beamline. The NADK2-substrate cocrystal structure was determined using experimental phasing with selenomethionine.
Supplementary Material
Acknowledgments
This research used the FMX beamline of the NSLS-II, a US Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract DE-SC0012704. The Center for BioMolecular Structure is supported by the NIH, National Institute of General Medical Sciences, through a Center Core P30 Grant (P30GM133893) and by the DOE Office of Biological and Environmental Research (Grant KP1605010). This study was funded by NIH Grant 1R01GM110161-01 (to A.K.) and by the Vallee Foundation (to A.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200923119/-/DCSupplemental.
Data Availability
X-ray and diffraction data and coordinates have been deposited in the Protein Data Bank (PDB; accession no. 7N29). All other study data are included in the article and/or SI Appendix.
References
- 1.Li B. B., et al. , NAD kinases: Metabolic targets controlling redox co-enzymes and reducing power partitioning in plant stress and development. Front. Plant Sci. 9, 379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantó C., Menzies K. J., Auwerx J., NAD(+) metabolism and the control of energy homeostasis: A balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie N., et al. , NAD+ metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Target. Ther. 5, 227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju H. Q., Lin J. F., Tian T., Xie D., Xu R. H., NADPH homeostasis in cancer: Functions, mechanisms and therapeutic implications. Signal Transduct. Target. Ther. 5, 231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasad U. V., et al. , Cloning, expression and characterization of NAD kinase from Staphylococcus aureus involved in the formation of NADP (H): A key molecule in the maintaining of redox status and biofilm formation. Adv. Biomed. Res. 6, 97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis C. A., et al. , Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poncet-Montange G., Assairi L., Arold S., Pochet S., Labesse G., NAD kinases use substrate-assisted catalysis for specific recognition of NAD. J. Biol. Chem. 282, 33925–33934 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Liu J., et al. , Crystal structures of an NAD kinase from Archaeoglobus fulgidus in complex with ATP, NAD, or NADP. J. Mol. Biol. 354, 289–303 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Strand M. K., et al. , POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryot. Cell 2, 809–820 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai M. F., et al. , NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol. Biol. 59, 553–564 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Ohashi K., Kawai S., Murata K., Identification and characterization of a human mitochondrial NAD kinase. Nat. Commun. 3, 1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickinson M. E., et al. ; International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center, High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tort F., et al. , Lysine restriction and pyridoxal phosphate administration in a NADK2 patient. Pediatrics 138, e20154534 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Pomerantz D. J., et al. ; Collaborators of UDN, Clinical heterogeneity of mitochondrial NAD kinase deficiency caused by a NADK2 start loss variant. Am. J. Med. Genet. A. 176, 692–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houten S. M., et al. , Mitochondrial NADP(H) deficiency due to a mutation in NADK2 causes dienoyl-CoA reductase deficiency with hyperlysinemia. Hum. Mol. Genet. 23, 5009–5016 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Ando T., et al. , Structural determinants of discrimination of NAD+ from NADH in yeast mitochondrial NADH kinase Pos5. J. Biol. Chem. 286, 29984–29992 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R., MNADK, a novel liver-enriched mitochondrion-localized NAD kinase. Biol. Open 2, 432–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukasawa Y., et al. , MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics 14, 1113–1126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai S., Mori S., Mukai T., Hashimoto W., Murata K., Molecular characterization of Escherichia coli NAD kinase. Eur. J. Biochem. 268, 4359–4365 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Grose J. H., Joss L., Velick S. F., Roth J. R., Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 103, 7601–7606 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korennykh A. V., et al. , The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoxhaj G., et al. , Direct stimulation of NADP+ synthesis through Akt-mediated phosphorylation of NAD kinase. Science 363, 1088–1092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
X-ray and diffraction data and coordinates have been deposited in the Protein Data Bank (PDB; accession no. 7N29). All other study data are included in the article and/or SI Appendix.