Significance
Angiogenesis, the formation of new blood vessels, contributes fundamentally to embryonic development, tissue homeostasis, and wound healing. Basic fibroblast growth factor (FGF2) is recognized as the first proangiogenic molecule discovered and it facilitates angiogenesis by activating FGF receptor 1 (FGFR1) signaling in endothelial cells. However, the roles of FGFR and the FGF/FGFR signaling axis in angiogenesis remain unclear. Here, we report that, upon reversible, posttranslational, small ubiquitin-like modifier modification (SUMOylation), FGFR1 regulates angiogenesis by coordinating endothelial angiogenic signaling. Mechanistically, FGFR1 SUMOylation maintains the balance in the competitive recruitment of the adaptor protein FRS2α between FGFR1 and VEGFR2 receptor complexes. VEGFA/VEGFR2 signaling primarily operates under hypoxic conditions and FGF/FGFR1 signaling is more important under normoxic conditions.
Keywords: SUMOylation, FGFR1, FRS2a, angiogenesis
Abstract
Angiogenesis contributes fundamentally to embryonic development, tissue homeostasis, and wound healing. Basic fibroblast growth factor (FGF2) is recognized as the first proangiogenic molecule discovered, and it facilitates angiogenesis by activating FGF receptor 1 (FGFR1) signaling in endothelial cells. However, the precise roles of FGFR and the FGF/FGFR signaling axis in angiogenesis remain unclear, especially because of the contradictory phenotypes of in vivo FGF and FGFR gene deficiency models. Our previous study results suggested a potential role of posttranslational small ubiquitin-like modifier modification (SUMOylation), with highly dynamic regulatory features, in vascular development and disorder. Here, we identified SENP1-regulated endothelial FGFR1 SUMOylation at conserved lysines responding to proangiogenic stimuli, while SENP1 functioned as the deSUMOylase. Hypoxia-enhanced FGFR1 SUMOylation restricted the tyrosine kinase activation of FGFR1 by modulating the dimerization of FGFR1 and FGFR1 binding with its phosphatase PTPRG. Consequently, it facilitated the recruitment of FRS2α to VEGFR2 but limited additional recruitment of FRS2α to FGFR1, supporting the activation of VEGFA/VEGFR2 signaling in endothelial cells. Furthermore, SUMOylation-defective mutation of FGFR1 resulted in exaggerated FGF2/FGFR1 signaling but suppressed VEGFA/VEGFR2 signaling and the angiogenic capabilities of endothelial cells, which were rescued by FRS2α overexpression. Reduced angiogenesis and endothelial sprouting in mice bearing an endothelial-specific, FGFR1 SUMOylation-defective mutant confirmed the functional significance of endothelial FGFR1 SUMOylation in vivo. Our findings identify the reversible SUMOylation of FGFR1 as an intrinsic fine-tuned mechanism in coordinating endothelial angiogenic signaling during neovascularization; SENP1-regulated FGFR1 SUMOylation and deSUMOylation controls the competitive recruitment of FRS2α by FGFR1 and VEGFR2 to switch receptor-complex formation responding to hypoxia and normoxia angiogenic environments.
Angiogenesis, the formation of new blood vessels from preexisting vessels, contributes fundamentally to embryonic development, tissue homeostasis, and wound healing. The complex process of neovascularization is fine-tuned by a series of angiogenic factors in endothelial cells (ECs). In particular, vascular endothelial growth factors (VEGFs) and fibroblast growth factors (FGFs) serve as major regulators that stimulate ECs to migrate, proliferate, and maturate to form new vessels in both embryonic and postnatal angiogenesis. Induced by hypoxia, VEGFA has been recognized as a key driver that initiates and maintains angiogenesis by binding to its cognate receptor VEGFR2 in ECs, while VEGFR1 acts as a decoy receptor of VEGFA to negatively regulate angiogenesis. Loss of either VEGFA or VEGFR2 during development results in failure of blood vasculature formation (1, 2). VEGFA/VEGFR2 binding activates the intracellular phospholipase Cγ (PLCγ)-extracellular regulated protein kinases (ERK)1/2 signaling pathway, which exhibits a central role in vascular development and adult arteriogenesis (3, 4).
In contrast to VEGFs, FGFs are not classical vascular growth factors. FGFs are expressed in nearly all tissues and act as pleiotropic molecules in embryonic development, organogenesis, tissue maintenance, and metabolism by activating FGF/FGFR signaling (5). Upon the binding of the FGF ligand, FGFR is autophosphorylated and activated, and the adaptor protein FRS2 docks to the juxta membrane region of FGFR via its phosphotyrosine-binding (PTB) domain, followed by downstream signaling transduction. The dysregulation of FGF signaling has an important role in cancer development, in which it mediates tumor cell proliferation, differentiation, and survival (6). In the vasculature, FGF2 (bFGF) was recognized as the first proangiogenic molecule (7), and it is involved in vascular homeostasis and angiogenesis through prototypic FGF2/FGFR1 signaling in ECs. Beside contributing to the sprouting of new vessels under physiological or pathological states (8–11), FGFR1 signaling is also associated with the maintenance of tumor angiogenesis and EC survival during angiogenesis, as well as vascular homeostasis and EC barrier function (12–15). Nevertheless, other findings make the appreciation fuzzy and debatable. Mice with either FGF1 or FGF2 deficiency had few angiogenesis abnormalities; FGF2-knockout mice exhibited no alterations in vessel repair following mechanical injury, although there was a mild delay in wound repair. Even mice harboring double FGF1 and FGF2 knockout did not have obvious abnormalities in angiogenesis (16–19). Endothelial FGFR1 and FGFR2 deficiencies are dispensable for physiological vascular development and vascular homeostasis but are essential for pathological neovascularization after injury (20). Our recent study demonstrated that endothelial FGFR1 and global FGFR3 deficiency impaired blood and lymphatic vascular development in mice (21). Therefore, the precise role of the FGFR and FGF/FGFR signaling axis in angiogenesis needs to be better understood. On the other hand, attention is needed for the potential coordinating function of FGFR in vascular growth when considering the complex crosstalk between growth factor systems and the versatile biology of FGF in cell fate determination. Indeed, numerous studies have indicated that FGF signaling regulates other vascular growth factor pathways at multiple levels, with outcomes especially intertwined with those of VEGF signaling in the vasculature (22, 23). The failure of antiangiogenic therapeutics aiming at any single growth factor (e.g., VEGFA, FGF2) in the clinic mirrors the complexity of these interactions (24, 25). Intriguingly, in ECs, FRS2α functions as the major adaptor protein for FGFR1 and also plays pivotal roles in the maintenance of VEGFR phosphorylation, as we demonstrated previously (26). However, the potential role of FRS2α in signaling crosstalk between FGFR and VEGFR pathways is obscure. Also, it is still unknown how to regulate the binding affinity of FRS2α with different tyrosine kinase receptors.
Compared with classical posttranslational modifications (PTMs), such as phosphorylation, ubiquitination, and glycosylation, small ubiquitin-like modification (SUMOylation) is a relatively new and unique PTM. SUMOylation is characterized by highly dynamic and reversible processes, with the latter executed by the deSUMOylation protease (deSUMOylase) family of sentrin-specific proteases (SENPs) (27). Although SUMOylation substrates are mostly categorized as nuclear proteins, other studies, including our own, have demonstrated that SUMOylation targets both cytoplasmic and membrane proteins. To date, SUMOylation has been identified as being extensively involved in controlling cellular processes, including genome maintenance, gene expression, protein stability, protein subcellular localization, and signal transduction (28–31). Notably, SUMOylation displays systematic and spatiotemporal features during these regulations by coordinating other PTMs based on their complex interplay (28, 32, 33). The importance of PTMs on FGFR function has been highlighted, particularly the effects of phosphorylation on signal transduction and ubiquitination on protein stability. On the other hand, results of our previous studies implied engagement of SUMOylation in vasculature formation by targeting endothelial receptors, the critical effectors of vascular patterning and extension (34, 35). It will be interesting to determine whether SUMOylation is another PTM of FGFR with a specific role in FGFR functioning during angiogenesis. In the present study, we demonstrated that SENP1-regulated FGFR1 SUMOylation acts as an intrinsic regulatory mechanism for coordinating FGF2/FGFR1 and VEGFA/VEGFR2 pathways, the most predominant angiogenic signaling pathways in ECs, and consequent angiogenic responsiveness during neovascularization. Our results shed light on the precise role of FGFR in the vasculature.
Results
SUMO Modification of FGFR1 in ECs.
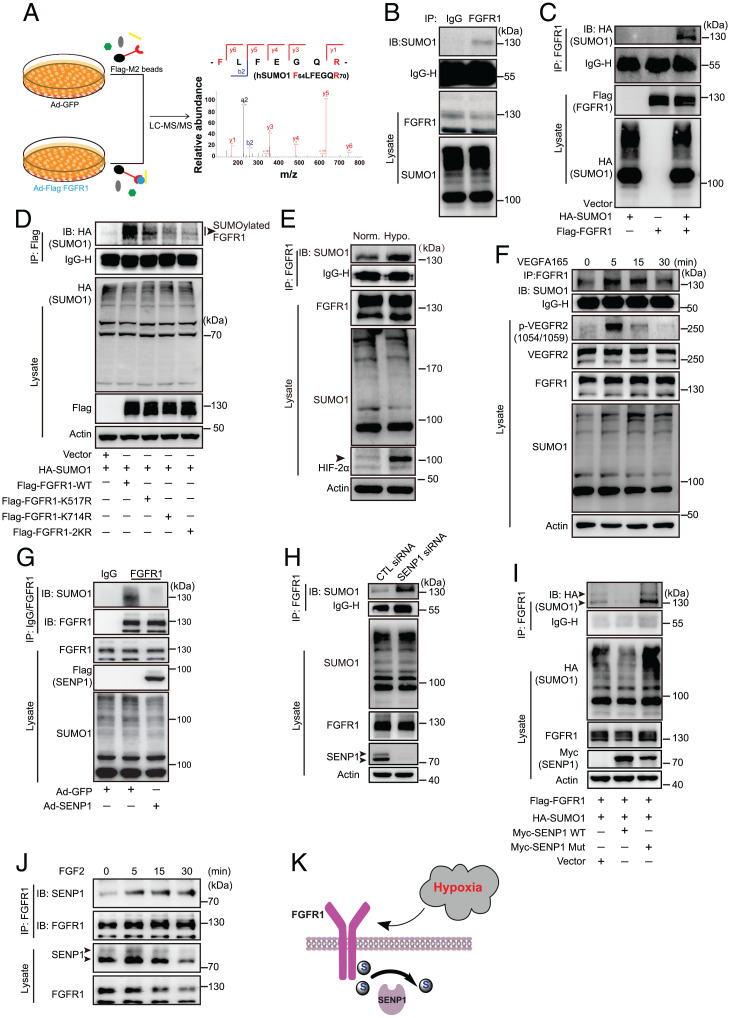
PTMs, such as phosphorylation and ubiquitination, are closely associated with the function of endothelial FGFR1 in angiogenesis and vascular homeostasis maintenance; however, the role of SUMOylation has not been determined. Therefore, whether SUMO modification on endothelial FGFR1 is involved in angiogenesis was examined. We first tested this possibility by analyzing the amino acid sequence of FGFR1 protein, using computational system-based software (SUMOsp 2.0 and SUMOplot). Two classic SUMO-binding motifs were predicted in the tyrosine kinase domain of FGFR1 (SI Appendix, Fig. S1 A and B). Meanwhile, to test the conjugation of SUMO molecule on FGFR1, FGFR1 proteins were immunoprecipitated from the denatured lysates of human umbilical vein endothelial cells (HUVECs) transduced with adenoviral Flag-FGFR1 (Ad-Flag-FGFR1) or adenoviral GFP (Ad-GFP) and analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS). According to the LC-MS/MS analysis, SUMO1 was among the FGFR1 (Flag)-immunoprecipitated proteins (Fig. 1A). More importantly, endogenous FGFR1 SUMOylation was detected in human microvascular endothelial cells (HMVECs) by immunoprecipitation (IP) (Fig. 1B). The occurrence of endogenous SUMO modification on FGFR1 was further confirmed by overexpressing SUMO1 (hemagglutinin [HA]) and FGFR1 (Flag) constructs in 293T cells (Fig. 1C). To better understand the biochemical characteristics of FGFR1 SUMOylation, SUMO-binding sites on FGFR1 were determined via bioinformatics analysis. K517 and K714, which are evolutionarily conserved among vertebrates (SI Appendix, Fig. S1C), were identified as critical SUMOylation sites on FGFR1. The IP assay demonstrated that the FGFR1 K517R or K714R mutation led to diminished levels of SUMOylated FGFR1, and the K517/714R (FGFR1-2KR) mutation showed almost no SUMOylated FGFR1 when coexpressed with SUMO1 (Fig. 1D). To preliminarily explore the physiological significance of FGFR1 SUMOylation in the proangiogenic process, HMVECs were treated with hypoxia or VEGFA, which are critical inductive factors for angiogenesis, followed by detecting FGFR1 SUMOylation by IP assays. The level of FGFR1 SUMOylation was induced by hypoxia or VEGFA (Fig. 1 E and F). More interestingly, the trends of FGFR1 SUMOylation were similar to VEGFR2 phosphorylation upon VEGFA stimulation (SI Appendix, Fig. S1 D and E), suggesting a putative role for FGFR1 SUMOylation in the angiogenic signaling and proangiogenic process.
Fig. 1.
Characterization of a posttranslational modification of FGFR1, which is directly regulated by SENP1. (A) Identification of SUMO1 peptides in the Flag-FGFR1 binding complex using LC-MS/MS. HUVECs were infected with Ad-GFP or Ad-Flag-FGFR1 separately, and protein complexes associated with FGFR1 proteins were immunoprecipitated with anti-Flag magnetic beads under denaturing conditions followed by MS identification.  : magnetic beads and the conjugated anti-Flag on the beads;
: magnetic beads and the conjugated anti-Flag on the beads;  : FGFR1 proteins fused with 3*Flag tag;
: FGFR1 proteins fused with 3*Flag tag;  : potential proteins conjugated with FGFR1;
: potential proteins conjugated with FGFR1;  and
and  : the soluble proteins in cell lysates. (B) Representative blot of endogenous SUMOylated FGFR1 in HMVECs. Immunoprecipitated FGFR1 was immunoblotted to detect SUMO1. (C) Representative blot of exogenous SUMOylated FGFR1 in 293T cells transfected with HA-SUMO1, Flag-FGFR1, or HA-SUMO1 plus Flag-FGFR1. SUMO1 (HA) was immunoprecipitated followed by immunoblotting to detect SUMO1 (HA) and FGFR1 (Flag). (D) Identification of FGFR1 SUMOylation sites in 293T cells transfected with mutants bearing single lysine (K) to arginine (R) substitutions at two putative SUMOylation sites (K517 or K714) or double mutations (the FGFR1-2KR mutant). SUMO1 (HA) was immunoprecipitated followed by immunoblotting for FGFR1 (Flag). Arrowhead indicates band of interest. (E) Representative blot of SUMOylated FGFR1 under hypoxic conditions. HMVECs were treated under normoxia (norm.) or hypoxia (hypo.; 1% oxygen) for 12 h and maintained at 37 °C. SUMOylation of FGFR1 was determined by IP with FGFR1 followed by immunoblotting for detection of SUMO1. Arrowhead indicates band of interest. (F) Representative blot of SUMOylated FGFR1 after VEGFA treatment for the indicated time points. Four dishes of HMVECs were starved in basic EBM-2 medium for 4 h. VEGFA165 was then added at indicated time points (30, 15, 5, and 0 min) into corresponding dishes before cell harvesting at the same time. SUMOylated FGFR1 was determined by IP with FGFR1 followed by immunoblotting for detection of SUMO1. (G) Representative blot of endogenous SUMOylated FGFR1 in HMVECs overexpressing GFP or SENP1. (H) Representative blot of endogenous SUMOylated FGFR1 in HMVECs transfected with control (CTL) siRNA or SENP1 siRNA. Arrowhead indicates band of interest. (I) Representative blot of SUMOylated FGFR1 in 293T cells transfected with Flag-FGFR1 plus SUMO1, SUMO1, and SENP1-WT or SUMO1 and the SENP1 mutant. Arrowhead indicates band of interest. (J) Representative blot showing the association between SENP1 and FGFR1 after FGF2 stimulation. HMVECs were starved in basic EBM-2 medium for 4 h followed by FGF2 stimulation at the different time points indicated. The association between SENP1 and FGFR1 was determined by IP with FGFR1 followed by immunoblotting for detection of SENP1. Arrowhead indicates band of interest. All the representative blots were from three independent experiments. (K) Model for SENP1-regulated endothelial FGFR1 SUMOylation in response to hypoxia.
: the soluble proteins in cell lysates. (B) Representative blot of endogenous SUMOylated FGFR1 in HMVECs. Immunoprecipitated FGFR1 was immunoblotted to detect SUMO1. (C) Representative blot of exogenous SUMOylated FGFR1 in 293T cells transfected with HA-SUMO1, Flag-FGFR1, or HA-SUMO1 plus Flag-FGFR1. SUMO1 (HA) was immunoprecipitated followed by immunoblotting to detect SUMO1 (HA) and FGFR1 (Flag). (D) Identification of FGFR1 SUMOylation sites in 293T cells transfected with mutants bearing single lysine (K) to arginine (R) substitutions at two putative SUMOylation sites (K517 or K714) or double mutations (the FGFR1-2KR mutant). SUMO1 (HA) was immunoprecipitated followed by immunoblotting for FGFR1 (Flag). Arrowhead indicates band of interest. (E) Representative blot of SUMOylated FGFR1 under hypoxic conditions. HMVECs were treated under normoxia (norm.) or hypoxia (hypo.; 1% oxygen) for 12 h and maintained at 37 °C. SUMOylation of FGFR1 was determined by IP with FGFR1 followed by immunoblotting for detection of SUMO1. Arrowhead indicates band of interest. (F) Representative blot of SUMOylated FGFR1 after VEGFA treatment for the indicated time points. Four dishes of HMVECs were starved in basic EBM-2 medium for 4 h. VEGFA165 was then added at indicated time points (30, 15, 5, and 0 min) into corresponding dishes before cell harvesting at the same time. SUMOylated FGFR1 was determined by IP with FGFR1 followed by immunoblotting for detection of SUMO1. (G) Representative blot of endogenous SUMOylated FGFR1 in HMVECs overexpressing GFP or SENP1. (H) Representative blot of endogenous SUMOylated FGFR1 in HMVECs transfected with control (CTL) siRNA or SENP1 siRNA. Arrowhead indicates band of interest. (I) Representative blot of SUMOylated FGFR1 in 293T cells transfected with Flag-FGFR1 plus SUMO1, SUMO1, and SENP1-WT or SUMO1 and the SENP1 mutant. Arrowhead indicates band of interest. (J) Representative blot showing the association between SENP1 and FGFR1 after FGF2 stimulation. HMVECs were starved in basic EBM-2 medium for 4 h followed by FGF2 stimulation at the different time points indicated. The association between SENP1 and FGFR1 was determined by IP with FGFR1 followed by immunoblotting for detection of SENP1. Arrowhead indicates band of interest. All the representative blots were from three independent experiments. (K) Model for SENP1-regulated endothelial FGFR1 SUMOylation in response to hypoxia.  : SUMO1;
: SUMO1;  : SENP1.
: SENP1.
SENP1 Is the Key deSUMOylase That Mediates FGFR1 deSUMOylation.
Dynamic conjugation/deconjugation is the central pattern of SUMOylation for regulating substrates, which is mediated by deSUMOylase SENP-family proteins. To study the roles of SENPs in regulating FGFR1 SUMOylation, we first verified the association of FGFR1 with SENPs. SENP1, SENP2, and SENP5, as they have been reported as major functional deSUMOylases in mammalian cells (36, 37). Co-IP experiments demonstrated FGFR1 was predominantly associated with SENP1 but only weakly associated with the other SENPs (SI Appendix, Fig. S1F). To determine whether SENP1 functions as an FGFR1 deSUMOylase in ECs, FGFR1 SUMOylation in HMVECs transfected with SENP1 adenovirus or SENP1 small interfering RNA (siRNA) was measured. Intriguingly, FGFR1 SUMOylation was significantly attenuated in the HMVECs transduced with Ad-Flag-SENP1 (Fig. 1G). In contrast, the amount of SUMOylated FGFR1 was significantly increased in SENP1-deficient ECs (Fig. 1H). Additionally, FGFR1 was coexpressed with wild-type (WT) SENP1 or a catalytic inactive form that inhibits deSUMOylase activity (SENP1 mutant) in an exogenous system. Co-IP results showed that the level of SUMOylated FGFR1 was robustly decreased by WT SENP1, whereas the SENP1 mutant increased SUMOylated FGFR1 levels (Fig. 1I). In vitro SUMOylation assay also shown that FGFR1 SUMOylation could be de-SUMOylated by recombinant SENP1 in vitro, which confirmed that SENP1 functions as FGFR1 deSUMOylase (SI Appendix, Fig. S1G). Interestingly, the endogenous dynamic association of FGFR1 with SENP1 in response to FGF2 treatment was observed in HMVECs (Fig. 1J). Taken together, these results indicate that SENP1 functions as the key deSUMOylase that mediates FGFR1 deSUMOylation in ECs responding to hypoxia and other proangiogenic stimuli (Fig. 1K).
SUMOylation Comprehensively Modulates the Tyrosine Kinase Activation of FGFR1.
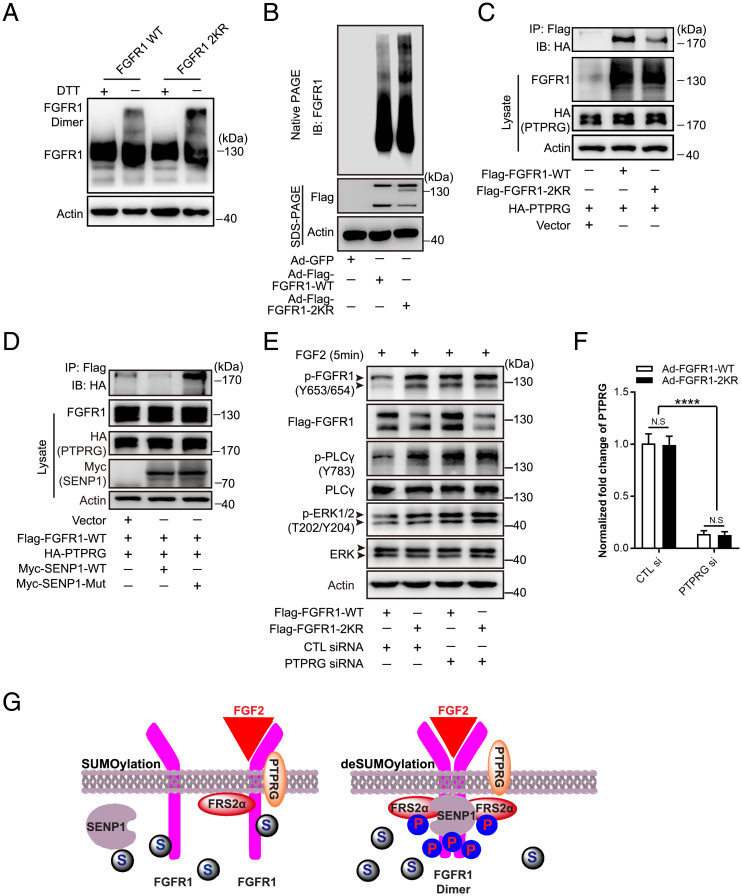
The manipulation of SENP1-regulated FGFR1 SUMOylation by angiogenic inducers enabled us to identify the biological effect of SUMOylation on endothelial FGFR1 during angiogenesis. The alteration regulated by SUMOylation/deSUMOylation may affect FGFR1 stability, cellular localization, and/or protein activity, as indicated by the multiple characteristic effects of SUMOylation on its substrates. In terms of FGFR1 stability, immunoblotting demonstrated that FGFR1-WT and the FGFR1-2KR mutant had similar basal expression levels and half-life in the presence of the protein synthesis inhibitor cycloheximide (SI Appendix, Fig. S2 A and B). Additionally, FGFR1-WT and FGFR1-2KR had a similar pattern of localization to the cell membrane and other subcellular compartments, as detected by immunoblotting after cell fractionation (SI Appendix, Fig. S2C) and immunofluorescence (SI Appendix, Fig. S2D). On the other hand, a molecular docking simulation revealed that both lysine 517 and lysine 714 residues are located at the core autophosphorylation region of the FGFR1 tyrosine kinase domain. Specifically, lysine 517 is located close to an ATP-binding pocket, providing FGFR1 with a phosphate group from ATP, and lysine 714 is located near the interface of the juxtaposed tyrosine kinase domains of dimerized FGFR1 molecules (SI Appendix, Fig. S3 A and B). Given that both ATP binding and receptor dimerization are indispensable for tyrosine kinase phosphorylation of FGFR1, we hypothesized that SUMO conjugation on FGFR1 might affect its tyrosine kinase activation. Accordingly, we examined the effect of SUMOylation on FGFR1 dimerization, which is essential for FGFR1 phosphorylation in response to ligand binding and for subsequent adaptor-protein docking for downstream signal activation. In the absence of the reducing agent dithiothreitol (DTT), immunoblotting demonstrated increased dimer formation in ECs bearing the FGFR1-2KR mutant compared with that of the FGFR1-WT group (Fig. 2A). Moreover, natural forms of FGFR1-WT or FGFR1-2KR in ECs were revealed by immunoblotting detection of FGFR1 in native polyacrylamide gel electrophoresis (PAGE) gels. Compared with that of the FGFR1-WT group, the signal of shifted bands was sharply augmented in the FGFR1-2KR mutant group, indicating an increase in the FGFR1 activation complex containing the FGFR1 dimer (Fig. 2B). In addition, we investigated the role of SUMO modification in FGFR1 dephosphorylation by examining the interaction between FGFR1 and its major phosphatase PTPRG (38) upon FGFR1 SUMOylation/deSUMOylation. SUMO deconjugation of FGFR1 (FGFR1-2KR) significantly attenuated the binding of FGFR1 to PTPRG, as demonstrated by co-IP assay (Fig. 2C), a finding confirmed by the molecular docking analysis (SI Appendix, Fig. S4 A and B). Consistently, the expression of WT SENP1, the major deSUMOylase of FGFR1, reduced the binding of FGFR1 to PTPRG, whereas the SENP1 mutant significantly enhanced the binding of FGFR1 to PTPRG (Fig. 2D). To further confirm whether FGFR1 SUMOylation regulates FGFR1 activity by mediating the association of FGFR1 with PTPRG, PTPRG was knocked down in FGFR1-WT or FGFR1-2KR expressing ECs by siRNA (the normalized mRNA level for verifying the efficiency of PTPRG siRNA is shown in Fig. 2F) followed by examining FGFR1 phosphorylation. Consistent with the FGFR1-PTPRG binding results, the FGFR1-2KR group had enhanced FGFR1 phosphorylation and signaling compared with FGFR1-WT group when PTPRG is normally expressed (Fig. 2E, lanes 1 and 2), while the reduction in PTPRG resulted in FGFR1 phosphorylation and signaling in FGFR1-WT–expressing ECs comparable to the level observed in the FGFR1-2KR group. (Fig. 2E, lanes 3 and 4). Taken together, these data showed that SUMOylation comprehensively modulated the tyrosine kinase activation of FGFR1 (Fig. 2G), a finding that was confirmed by a decrease in FGFR1 tyrosine phosphorylation concomitant with the gradient expression of SUMO1 in ECs (SI Appendix, Fig. S5).
Fig. 2.
SUMO conjugation to FGFR1 attenuates the formation of the FGFR1 functional complex and enhances its binding with phosphatase PTPRG. (A and B) Detection of the FGFR1 activation complex in HMVECs transfected with Ad-Flag-FGFR1-WT or the Ad-Flag-FGFR1-2KR mutant. (A) Representative blot showing the cell lysate mixed with or without DTT before boiling for Western blot analysis. The FGFR1 activation complex was detected with an anti-Flag antibody after SDS-PAGE gel separation. (B) Representative blot of the indicated FGFR1 activation complex. Samples were mixed with sample loading buffer without reducing reagent and loaded into native PAGE gel. (C and D) Representative blot showing the interaction between FGFR1 phosphatase PTPRG and FGFR1 in 293T cells. (C) Cells were cotransfected with HA-PTPRG plus vector plasmid, FGFR1-WT, or the FGFR1-2KR mutant. The association between FGFR1 and PTPRG was indicated by IP with Flag (FGFR1) followed by immunoblotting (IB) with HA (PTPRG). (D) Cells were cotransfected with Flag-FGFR1 plus PTPRG (HA), PTPRG (HA) and SENP1-WT, or PTPRG (HA) and the SENP1 mutant. FGFR1 (Flag) was immunoprecipitated followed by IB for detection of PTPRG (HA). (E) Representative blots showing FGF2-FGFR1 signaling in HMVECs transfected with Ad-FGFR1-WT plus control siRNA (lane 1), Ad-FGFR1-2KR plus control siRNA (lane 2), Ad-FGFR1-WT plus PTPRG siRNA (lane 3), and Ad-FGFR1-2KR plus PTPRG siRNA (lane 4). HMVECs were starved in basic EBM-2 medium for 4 h followed by FGF2 stimulation. Arrowhead indicates band of interest. (F) Normalized mRNA level of PTPRG after control (CTL) siRNA or PTPRG siRNA treatment. The knockdown of PTPRG was detected by real-time qPCR. The data were normalized to the those of the Ad-FGFR1-WT plus control siRNA–treated cells and are presented as the mean ± SEM from at least three independent experiments. ****P ≤ 0.0001. N.S., not significant. All the blots were from three independent experiments. (G) Model depicting the comprehensive modulation of SENP1-regulated SUMOylation in the tyrosine kinase activation of FGFR1.  : FRS2α;
: FRS2α;  : PTPRG;
: PTPRG;  : Phosphate;
: Phosphate;  : SUMO1;
: SUMO1;  : SENP1.
: SENP1.
SUMOylation of FGFR1 Controls the Competitive Recruitment of FRS2α to FGFR1 and VEGFR2.
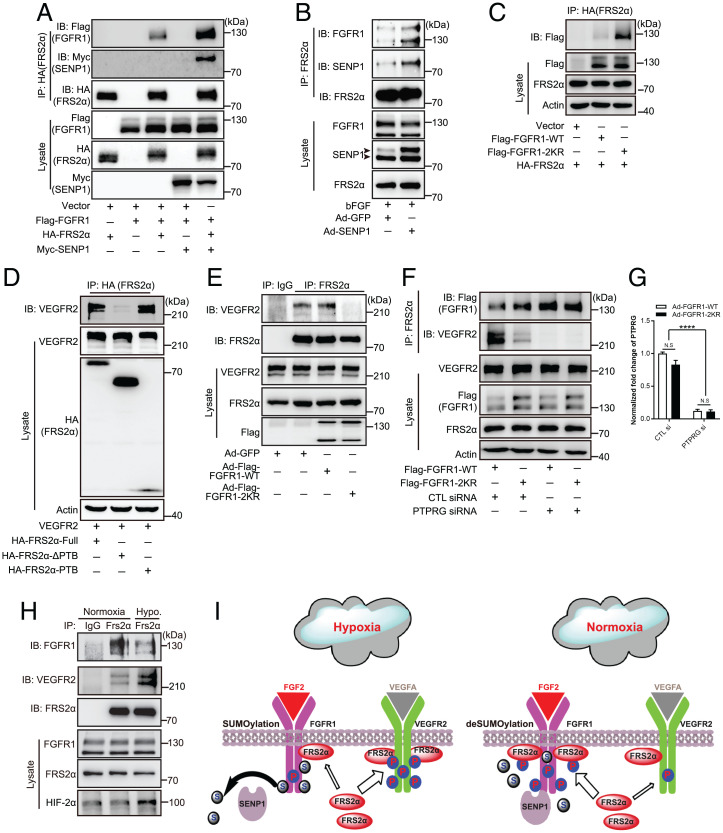
The negative regulation of SUMOylation in the tyrosine kinase activation of FGFR1 and the aforementioned interaction between SENP1 and FGFR1 led us to explore the regulatory mechanism of SUMOylation in FGFR1 active complex formation further, especially the recruitment of the key adaptor protein FRS2α. Notably, SENP1 overexpression significantly enhanced the binding between FGFR1 and FRS2α in 293T cells, as demonstrated by the co-IP assay (Fig. 3A). A similar result was also observed in the HMVECs overexpressing SENP1 (Fig. 3B), suggesting that SENP1-regulated deSUMOylation may facilitate further recruitment of FRS2α to FGFR1 in ECs. Indeed, we observed WT FGFR1 was moderately associated with FRS2α (Fig. 3C, lane 2), but the FGFR1 deSUMOylation mutant (FGFR1-2KR) remarkably promoted the level of FRS2α associated with FGFR1 (Fig. 3C, lane 3). On the other hand, our previous study identified FRS2α as the critical component and regulator of VEGFR2 signaling (26), which suggested to us that FGFR1 SUMOylation may be critical in the regulation of the VEGFR2-FRS2α interaction. Therefore, we first examined the exact association between VEGFR2 and FRS2α. IP assays demonstrated that FRS2α directly binds through its PTB domain to VEGFR2 (Fig. 3D), the same pattern as docking at FGFR1. Then, we asked whether the FRS2α-VEGFR2 association was affected by FGFR1 SUMOylation/deSUMOylation. To answer this question, FRS2α was immunoprecipitated from HMVECs overexpressing FGFR1-WT or the FGFR1-2KR mutant to determine the extent of VEGFR2-FRS2α complex enrichment. Immunoblotting for VEGFR2 showed that the FRS2α-VEGFR2 association was robustly detected in ECs overexpressing FGFR1-WT (Fig. 3E, lane 3) but was dramatically diminished in ECs bearing the FGFR1-2KR mutant (Fig. 3E, lane 4). Moreover, the reduction in the level of PTPRG (the normalized mRNA level for verifying the efficiency of PTPRG siRNA is shown in Fig. 3G), the phosphatase of FGFR1, augmented the binding between FGFR1 and FRS2α, which reached a level similar to that of FRS2α with FGFR1-2KR, while the FRS2α-VEGFR2 association was comparably reduced (Fig. 3F, lanes 3 and 4), corresponding to the enhanced FGFR1 phosphorylation in both groups (Fig. 2E, lanes 3 and 4). In contrast, SENP1 knockdown decreased the FRS2α binding with FGFR1 but increased the FRS2α binding with VEGFR2 (SI Appendix, Fig. S6A). More importantly, hypoxia, the central pathophysiological factor triggering angiogenesis, enhanced FGFR1 SUMOylation (Fig. 1E) and restrained the additional binding of FRS2α with FGFR1-WT, but enhanced FRS2α-VEGFR2 association in HMVECs, compared with those under normoxia (Fig. 3H and SI Appendix, Fig. S6B). Moreover, IP assays demonstrated that Y653/654 inactivation in FGFR1 by Y653F mutation eliminated the differences in the binding of FGFR1-WT or FGFR1-2KR with FRS2α and in the corresponding VEGFR2-FRS2α association (SI Appendix, Fig. S7). This indicates that both FGFR1-WT and FGFR1-2KR require FGFR1 phosphorylation at Y653/654 to recruit FRS2α. Therefore, SENP1-regulated FGFR1 SUMOylation restrains additional association of FGFR1-FRS2α but facilitates VEGFR2-FRS2α complex formation in ECs upon hypoxia (Fig. 3I), indicating the potential positive modulation of FGFR1 SUMOylation in proangiogenic signaling under hypoxia.
Fig. 3.
SENP1-regulated FGFR1 SUMOylation restrains the FGFR1/FRS2α association but facilitates VEGFR2/FRS2α complex formation. (A–C) Representative blots of FGFR1/FRS2α combination. (A) SENP1 increases the FGFR1/FRS2α association in 293T cells. The FGFR1/FRS2α complex was indicated by IP with HA (FRS2α) followed by immunoblotting (IB) with Myc (SENP1). (B) SENP1 increases the FGFR1/FRS2α association in HMVECs transduced by Ad-GFP or Ad-SENP1. The FGFR1/FRS2α complex was indicated by IP with FRS2α followed by IB with SENP1. Arrowhead indicates band of interest. (C) FGFR1-2KR enhances the FGFR1/FRS2α interaction in 293T cells. Cells were cotransfected with HA-FRS2α plus vector plasmid, FGFR1-WT, or the FGFR1-2KR mutant. HA (FRS2α) was immunoprecipitated followed by IB with Flag (FGFR1). (D) Representative blot showing the FRS2α/VEGFR2 association. 293T cells were cotransfected with VEGFR2 plus HA-FRS2α–full-length, HA-FRS2α-ΔPTB, or HA-PTB. HA was immunoprecipitated followed by IB using anti-VEGFR2 antibody. (E) Blots of the VEGFR2/FRS2α complex in HMVECs. HMVECs were infected with Ad-GFP, Ad-FGFR1-WT, or the Ad-FGFR1-2KR mutant, and endogenous FRS2α was immunoprecipitated followed by IB using an anti-VEGFR2 antibody. (F) Representative blots of additional FGFR1/FRS2α association with the VEGFR2/FRS2α association inhibited by knocking down FGFR1 phosphatase PTPRG. FRS2α was immunoprecipitated followed by IB using anti-FGFR1 and anti-VEGFR2. (G) Normalized mRNA level of PTPRG. The knockdown of PTPRG was indicated by real-time qPCR. The data were normalized to the those of the Ad-FGFR1-WT plus control siRNA-treated cells and are presented as the mean ± SEM from at least three independent experiments. ****P ≤ 0.0001. N.S., not significant. (H) Representative blots showing the FGFR1/FRS2α association and VEGFR2/FRS2α complex under hypoxic (hypo.) conditions. (I) Model for SENP1-regulated FGFR1 SUMOylation restrains additional association of FGFR1/FRS2α but facilitates VEGFR2/FRS2α complex formation in ECs under hypoxia.  : FRS2α;
: FRS2α;  : Phosphate;
: Phosphate;  : SUMO1;
: SUMO1;  : SENP1.
: SENP1.
SENP1-Regulated FGFR1 SUMOylation/deSUMOylation Maintains the Balance between FGFR1 Signaling and VEGFR2 Signaling in ECs.
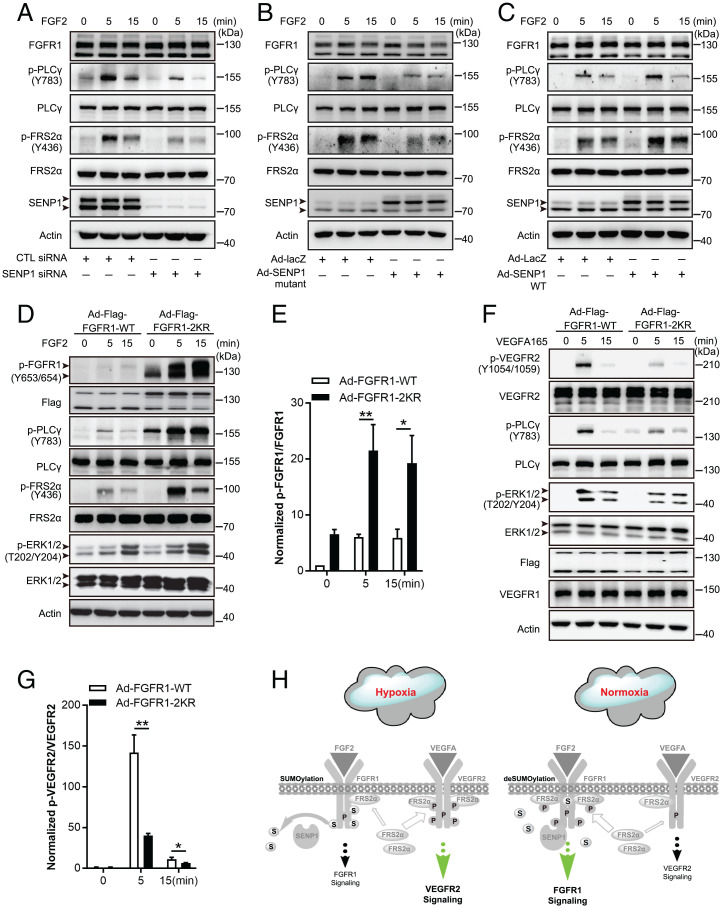
According to the important role of FGFR1 SUMOylation in FGFR1 and VEGFR2 signaling complex formation, we next examined the correlated angiogenic signaling in ECs to define the molecular regulation of this FGFR1 PTM. SENP1 knockdown in HMVECs resulted in a significant decrease in the phosphorylation of FRS2α (and also the reduced mobility shift) and in PLCγ following FGF2 treatment, as demonstrated by immunoblotting, indicating the attenuation of the signal transduction from FGF2/FGFR1 to the docking protein and downstream signaling (Fig. 4A, with quantification in SI Appendix, Fig. S8 A and B). A consistent result was obtained in the presence of the SENP1 C603A mutant (Fig. 4B, with quantification in SI Appendix, Fig. S8 C and D). In contrast, SENP1 overexpression by adenoviral delivery reinforced FGF2/FGFR1 signaling in HMVECs (Fig. 4C, with quantification in SI Appendix, Fig. S8 E and F). These results suggest that SENP1-regulated deSUMOylation may augment FGFR1 signaling in ECs. To verify this outcome, FGFR1-WT and FGFR1-2KR mutant were introduced into HMVECs by adenoviral delivery. In response to FGF2, the phosphorylation of FGFR1 and FRS2α, as well as downstream signaling, was sharply elevated in HMVECs expressing the FGFR1-2KR mutant compared with that in the FGFR1-WT transgene group (Fig. 4 D and E). However, FGFR1-2KR mutant overexpression resulted in significant inhibition of VEGFR2 phosphorylation and attenuated its downstream signaling following VEGFA treatment, while FGFR1-2KR had little effect on the expression of VEGFR2 or VEGFR1 in ECs (Fig. 4 F and G). The same trend was also verified in HUVECs with FGFR1-WT and FGFR1-2KR overexpression (SI Appendix, Fig. S9 A–D). To further appreciate the balance between endothelial FGF2/FGFR1 signaling and VEGFA/VEGFR2 signaling under the control of FGFR1 SUMOylation in response to angiogenic environment, we examined the effect of SKLB1002, a VEGFR2 signaling inhibitor, on downstream signaling upon FGF2 plus VEGFA stimulation under normoxia or hypoxia. Interestingly, the phosphorylation of PLCγ and ERK, the downstream signals shared by FGFR1 and VEGFR2 signaling, was mainly inhibited under hypoxia, while suppressed phosphorylation of VEGFR2 was detected both under normoxia and hypoxia (SI Appendix, Fig. S10 A–D). Thus, hypoxia-enhanced FGFR1 SUMOylation facilitates the recruitment of FRS2α to VEGFR2 and consequent VEGFR2 signaling, while FGFR1 deSUMOylation promotes additional recruitment of FRS2α to FGFR1 and consequent FGFR1 signaling (Fig. 4H).
Fig. 4.
SENP1-regulated FGFR1 SUMOylation/deSUMOylation maintains the balance between FGF2/FGFR1 signaling and VEGFA/VEGFR2 signaling in ECs. (A–C) Representative blots showing FGF/FGFR1 signaling in samples in HMVECs with SENP1 knockdown (A), inactive SENP1-mutant overexpression (B), and SENP1-WT overexpression (C). Arrowhead indicates band of interest. (D and E) FGF2/FGFR1 signaling in HMVECs infected with Ad-FGFR1-WT or Ad-FGFR1-2KR after FGF2 stimulation at the indicated time points. Representative blots are shown in D with quantification in E. Arrowhead indicates band of interest. The normalized value of p-FGFR1/FGFR1 is presented as mean ± SEM from three independent experiments. *P ≤ 0.05; **P ≤ 0.01. (F and G) VEGFA/VEGFR2 signaling in HMVECs infected with Ad-FGFR1-WT or Ad-FGFR1-2KR after VEGFA stimulation at the indicated time points. Representative blots are shown in F with quantification in G. Arrowhead indicates band of interest. The normalized value of p-VEGFR2/VEGFR2 is presented as mean ± SEM from three independent experiments. **P ≤ 0.01. (H) Model for SENP1-regulated FGFR1 SUMOylation/deSUMOylation maintains the balance between FGF2/FGFR1 signaling and VEGFA/VEGFR2 signaling in ECs; the consequent mechanism following SENP1-regulated FGFR1 SUMOylation restrains additional association of FGFR1/FRS2α but facilitates VEGFR2/FRS2α complex formation in ECs under hypoxia (gray part, the same procedures as shown in Fig. 3I).  : FRS2α;
: FRS2α;  : PTPRG;
: PTPRG;  : Phosphate;
: Phosphate;  : SUMO1;
: SUMO1;  : SENP1;
: SENP1;  : enhanced signaling;
: enhanced signaling;  : restrained signaling. CTL, control.
: restrained signaling. CTL, control.
FGFR1 deSUMOylation-Induced Inhibition of EC Angiogenic Capabilities Is Reversed by FRS2α through the Restoration of VEGFR2 Signaling.
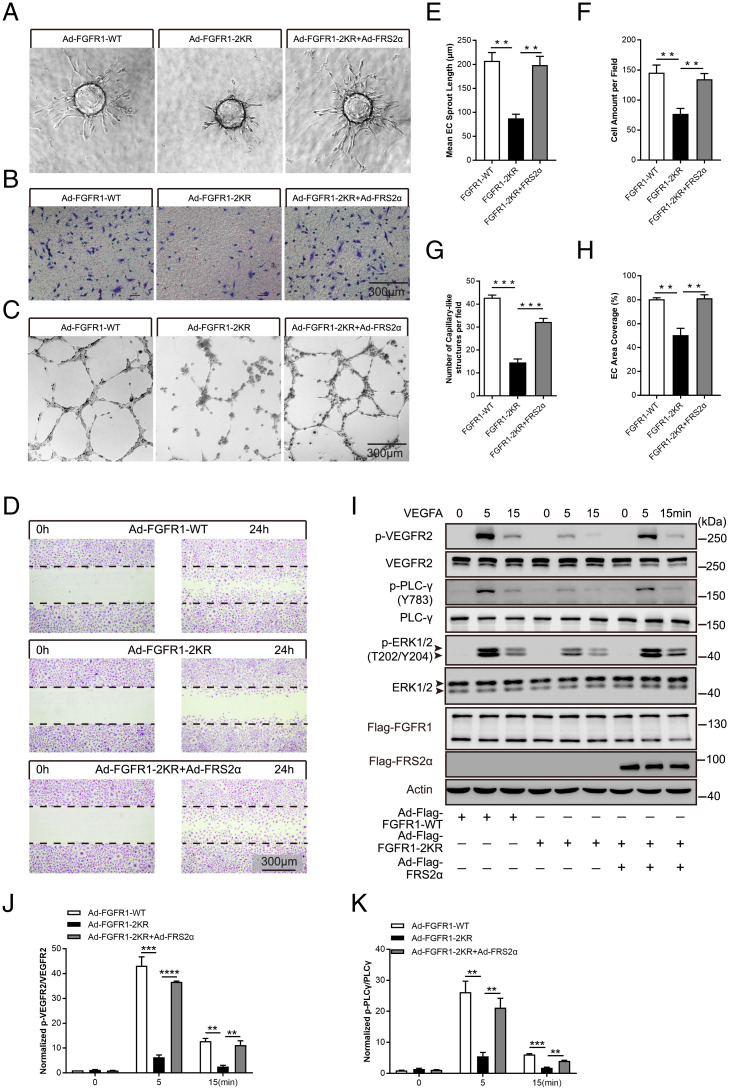
Considering the findings in angiogenic signaling coordination, we further determined the biological function of FGFR1 SUMOylation in ECs. Specifically, the angiogenic capabilities of primary ECs were investigated in HUVECs transduced with adenoviral vectors containing WT FGFR1 or the FGFR1-2KR mutant. First, a spheroid sprouting assay was employed to determine the role of FGFR1 deSUMOylation in controlling the sprouting capability of ECs in vitro. The results showed that the sprouting length formed by the HUVECs transduced with the FGFR1-2KR mutant was significantly shorter than that of the HUVECs expressing FGFR1-WT (SI Appendix, Fig. S11A with quantification in SI Appendix, Fig. S11E). Next, transmigration and tube-like structure analyses were performed to examine the migration and tube-formation capability of the ECs. Compared with the FGFR1-WT group, the migration and tube formation capabilities were moderately decreased in the ECs harboring the FGFR1-2KR mutant (SI Appendix, Fig. S11 B and C with quantification shown in SI Appendix, Fig. S11 F and G). Moreover, a similar trend was observed in the wound-healing assay (SI Appendix, Fig. S11D with quantification shown in SI Appendix, Fig. S11H).
These data and the role of FGFR1 SUMOylation in receptor signaling activation enabled us to infer that FGFR1 deSUMOylation suppresses the angiogenic capabilities of ECs by inhibiting VEGFR2 signaling through the prevention of VEGFR2 association with FRS2α. To verify this mechanism, FRS2α (Ad-Flag-FRS2α) was transduced by adenoviral delivery into HMVECs overexpressing WT FGFR1 or the FGFR1-2KR mutant. The angiogenic capabilities (i.e., vessel sprouting, migration, capillary-like structure formation, and wound healing) of the ECs bearing FGFR1-2KR were reversed to a level comparable with that in ECs bearing WT FGFR1 by FRS2α overexpression (Fig. 5 A–D, with quantification in Fig. 5 E–H). Concomitantly, VEGFA-induced VEGFR2 phosphorylation and the signaling cascade were successfully restored by overexpressing FRS2α in ECs (Fig. 5I, with quantification in Fig. 5 J and K). Corresponding to the modulatory mechanism of FGFR1 SUMOylation on the interplay of FGFR1 and VEGFR2 signaling complexes, these data proved the critical role of FGFR1 SUMOylation in maintaining the angiogenic capabilities of ECs via facilitating VEGFR2/FRS2α complex formation for VEGFR2 signaling activation.
Fig. 5.
The inhibited angiogenic capabilities of ECs bearing the FGFR1-2KR mutant can be restored by overexpressing FRS2α. HMVECs infected with Ad-FGFR1-WT, Ad-FGFR1-2KR mutant, or the Ad-FGFR1-2KR mutant plus Ad-FRS2α were evaluated for proangiogenic capabilities. Representative images of sprout outgrowth, migration capability, tube formation capability, and wound-healing assay are shown in A–D, with quantification shown in E–H. (B–D) Scale bar, 300 μm. (I–K) VEGFA/VEGFR2 signaling was restored in HMVECs carrying the FGFR1-2KR mutant by overexpressing FRS2α. Representative blots are shown in I with normalized quantification in J and K. Arrowhead indicates band of interest. All experiments were performed in triplicate. The quantification results are presented as the mean ± SEM from at least three independent experiments. **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
FGFR1 deSUMOylation in ECs Compromises Developmental Retina Angiogenesis.
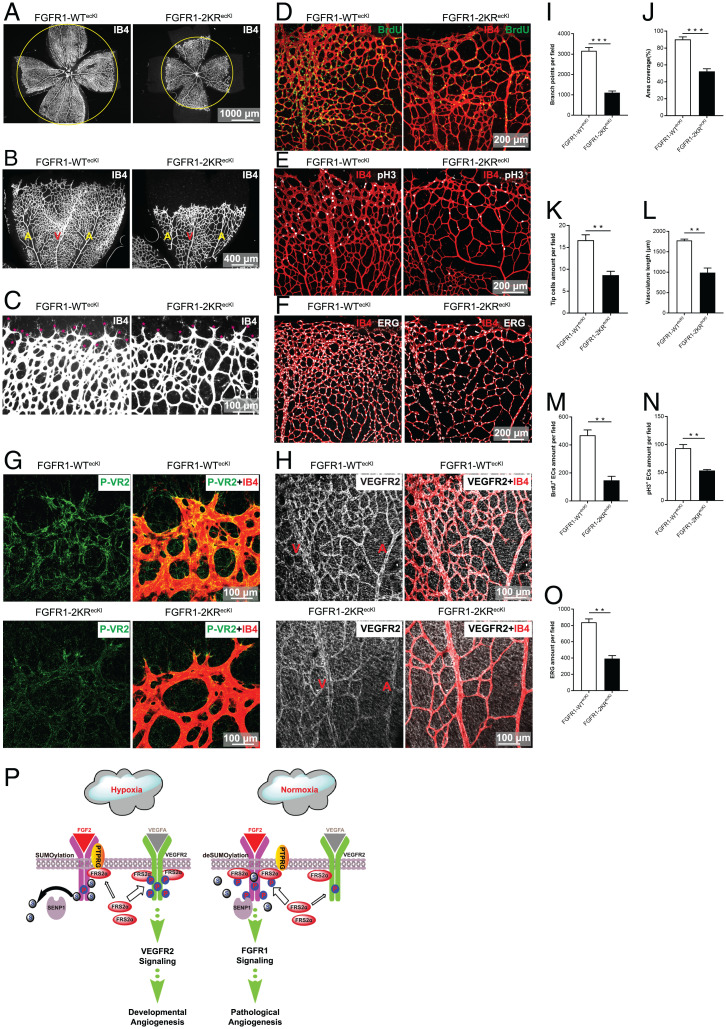
The function of endothelial FGFR1 SUMOylation in vivo was studied by constructing an EC-specific FGFR1-2KR mutant knock-in (KI) mouse employing a ROSA26 KI strategy (Rosa26-flox-stop codon-flox-FGFR1-2KR) under the control of an inducible Cdh5 Cre deleter (Cdh5-CreERT2) on the stop codon. As a control, an EC-specific FGFR1-WT KI mouse (Rosa26-flox-stop codon-flox-FGFR1-WT) was also constructed using the same strategy. Endothelial-specific FGFR1-WT KI mice (FGFR1-WTecKI) and FGFR1-2KR KI mice (FGFR1-2KRecKI) were obtained by administering tamoxifen to FGFR1-WTflox/flox-Cdh5-CreERT2 mice and FGFR1-2KRflox/flox-Cdh5-CreERT2 mice, respectively. Both mouse strains were viable and fertile. The construction strategy of these genetic mice is shown in SI Appendix, Fig. S12A, and the endothelial expression of FGFR1-WT and FGFR1-2KR is shown in SI Appendix, Fig. S12 B and C. Postnatal retina angiogenesis, classical hypoxia-induced angiogenesis, was assessed in P5 and P7 pups. Retinas from FGFR1-2KRecKI mice demonstrated a delayed expansion of the vascular plexus to the periphery compared with retinal formation in FGFR1-WTecKI mice; this finding was evident by the decrease in vascular branching and reduced vessel coverage at P5 (SI Appendix, Fig. S13 A–C, with quantification in SI Appendix, Fig. S13 H and I) and P7 (Fig. 6 A and B, with quantification in Fig. 6 I and J). Further analysis revealed that the tip-cell number and vascular length were significantly reduced in the sprouting region in the FGFR1-2KRecKI pups at P5 (SI Appendix, Fig. S13D, with quantification in SI Appendix, Fig. S13 J and K) and P7 (Fig. 6C, with quantification in Fig. 6 K and L). Pulse BrdU labeling and immunostaining of phosphorylated Histone-3 (p-H3) revealed a significant decrease in the proliferation of retinal ECs in the FGFR1-2KRecKI pups, and a similar trend was observed by the nuclear EC marker ERG (ETS transcription factor) stained in the retina at P5 (SI Appendix, Fig. S13 E–G with quantification in SI Appendix, Fig. S13 L–N) and P7 (Fig. 6 D–F, with quantification in Fig. 6 M–O). Moreover, the FGFR1 deSUMOylation-induced VEGFR2 phosphorylation blockade was examined in vivo. The immunofluorescence signal of phosphorylated VEGFR2 was observed in the angiogenic front, especially in the tip cells of the FGFR1-WTecKI retinas but was obscure and limited to only some tip cells in the vasculature of the FGFR1-2KRecKI retinas, a finding that is consistent with the results obtained in vitro (Fig. 6G). In addition, the expression of total VEGFR2 was similar, as evident throughout the retinal endothelium of the FGFR1-WTecKI and FGFR1-2KRecKI pups (Fig. 6H). Taken together, these data confirmed the functional effect of FGFR1 SUMOylation on angiogenesis in vivo.
Fig. 6.
Endothelial expression of the FGFR1-2KR mutant delays neonatal retinal angiogenesis by inhibiting VEGFR2 phosphorylation. (A–C) Representative images showing P7 vascular outgrowth in FGFR1-WTecKI control (n = 10) and FGFR1-2KRecKI retinas (n = 12) stained with IB4. The FGFR1-2KR mutant delays neonatal retinal angiogenesis. Red asterisks in C indicate tip cells. (D–F) EC proliferation test of P7 FGFR1-WTecKI (n ≥ 4 for all assays) and P7 FGFR1-2KRecKI (n ≥ 4 for all assays) for retinal vasculature. BrdU labeling (D), anti–phospho-histone H3 (E), and anti-ERG (F) were employed to determine the proliferation of retinal vasculature. (G) Representative images showing p-VEGFR2 in the P7 FGFR1-WTecKI and FGFR1-2KRecKI retinal vasculature. (H) Representative images showing total VEGFR2 in the P7 FGFR1-WTecKI and FGFR1-2KRecKI retinal vasculature. The quantification data of the branch points, area coverage, tip cell amounts, and vasculature length are presented as the mean ± SEM (n ≥ 4) in I–L. The quantification data of BrdU+ cells, p-H3+ cells, and ERG are presented as the mean ± SEM in M–O. All experiments were performed in triplicate, and significance was determined by an unpaired t test. **P ≤ 0.01; ***P ≤ 0.001. (P) Working model of FGFR1 SUMOylation coordinates endothelial angiogenic signaling in angiogenesis. In ECs, hypoxia-enhanced FGFR1 SUMOylation restricts the tyrosine kinase activation of FGFR1 by modulating the dimerization of FGFR1 and FGFR1 binding with its phosphatase PTPRG, which, in turn, facilitates the recruitment of FRS2α to VEGFR2 but limits additional recruitment of FRS2α to FGFR1, supporting the developmental angiogenesis by activating VEGFA/VEGFR2 signaling. On the other hand, pathological release of FGF2 induces FGFR1 deSUMOylation and promotes additional recruitment of FRS2α to FGFR1, which highly activates FGFR1 signaling to support pathological angiogenesis under normoxia.  : FRS2α;
: FRS2α;  : PTPRG;
: PTPRG;  : Phosphate;
: Phosphate;  : SUMO1;
: SUMO1;  : SENP1.
: SENP1.
Discussion
The FGF2/FGFR1 pathway in ECs has been implied to be extensively involved in vascular growth and vascular homeostasis maintenance; however, the exact role and the complete regulatory mechanism of this signaling axis are largely unclear. The key issues are the equivocal vascular phenotypes by gene deficiency in vivo and the complex interplay of the growth factor systems. The present study revealed the constitutive posttranslational SUMOylation of endothelial FGFR1 in response to proangiogenic stimulation, for which the reverse is mediated by sentrin-specific protease SENP1. The SENP1-regulated SUMOylation of FGFR1 functions as an intrinsic regulatory machinery of FGFR1 activation and the balance between FGF2/FGFR1 signaling and VEGFA/VEGFR2 signaling for angiogenic capacity of ECs in response to an angiogenic environment. The key link between the two pathways is the competition of FRS2α, the adaptor protein critically required for initiating both signaling pathways. SENP1-regulated FGFR1 SUMOylation/deSUMOylation controls the competitive recruitment of FRS2α by FGFR1 and VEGFR2 to switch receptor complex formation as required, thus ensuring the downstream signal transduction for adapting angiogenic environment. Hypoxia enhances SUMOylation of FGFR1 to frame its tyrosine kinase activation that is essential for the optimal binding of FGFR1 with FRS2α (39). Consequently, it facilitates the recruitment of FRS2α to the VEGFR2 complex but restrains additional recruitment of FRS2α to the FGFR1 side, resulting in high activation of VEGFR2 signaling in ECs to support developmental angiogenesis. On the other hand, pathological release of FGF2 induces FGFR1 deSUMOylation and further promotes additional recruitment of FRS2α to FGFR1, which, in turn, highly activates FGFR1 signaling to support pathological angiogenesis under normoxia (Fig. 6P).
Posttranslational SUMOylation Tightly Regulates the FGFR1 Signaling Complex in ECs.
FGFR1 undergoes glycosylation, ubiquitination, and phosphorylation, but whether FGFR1 undergoes other types of PTMs remains unknown. In this study, we discovered that SUMOylation is a PTM of FGFR1. FGFR1 is partially SUMOylated in quiescent ECs, and the modification responds to hypoxia and other proangiogenic stimuli throughout FGFR1 cellular functions. Due to its high dynamics and reversibility, SUMOylation may alter a plethora of protein properties. Our investigation revealed that SUMOylation does not affect FGFR1 stability or subcellular localization. However, SUMOylation directly participates in FGFR1 tyrosine kinase complex formation in ECs. The critical finding in this respect is that the FGFR1 SUMOylation inhibits, while FGFR1 deSUMOylation promotes, the binding of FGFR1 with FRS2α, the major docking protein in the FGFR1 signaling complex. As a substrate of the FGFR1 tyrosine kinase, FRS2α is tyrosine phosphorylated at its C-terminus downstream of the PTB domain upon FGF-induced FGFR1 activation (40, 41). In addition to initiating downstream signaling, the tyrosine phosphorylation eliminates the negative impact on FGFR1/FRS2α binding imposed by the C-terminal sequence (39). Interestingly, the two SUMOylation sites of FGFR1 are located in the tyrosine kinase domain, and molecular simulation analysis conducted as part of this study indicates that SUMO conjugation may induce conformational changes in the tyrosine kinase domain and thereby affect the phosphorylation of FGFR1. Indeed, our further investigations demonstrate that FGFR1 deSUMOylation promotes the dimerization of FGFR1 but inhibits the binding of FGFR1 to PTPRG, a phosphatase that directly dephosphorylates activated FGFR1 at the cell membrane (38). Consequently, deSUMOylated FGFR1 is highly phosphorylated, enabling it to recruit and activate FRS2α and other downstream signal components. Therefore, SUMOylation/deSUMOylation plays an important role in regulating FGFR1 tyrosine autophosphorylation and signaling complex across the cell membrane and subsequent signal pathway activation. Whether SUMO modification has an effect on FGF2 binding to FGFR1 is not determined and is worthy of further investigation.
Furthermore, the crosstalk between SUMOylation and other PTMs of FGFR1 should also be nonnegligible. Glycosylation is implemented in extracellular and transmembrane domains of FGFR1; however, the potential ubiquitination sites were ascertained in the tyrosine kinase domain of FGFR1 (42) in the same modification domain of SUMOylation. Collectively considering the mapped putative modification sites and comparable ubiquitination level between the FGFR1 WT and SUMOylation-deficient mutant in our pilot study, our findings indicate SUMOylation and ubiquitination may not share or compete for the same site-specific lysine residues in FGFR1. However, we cannot rule out the regulation of SUMOylation on the recruitment of de-ubiquitinases and ligases and recognition of putative ubiquitylated residues by different linkages of ubiquitin chains on FGFR1, based on possible SUMOylation-dependent substrate conformational alternation, ATP binding, and ubiquitin transfer (43, 44). In the present study, expressing mutant FGFR1 with lysine-to-arginine (K-to-R) mutations of SUMOylation sites was employed as the strategy in investigating the molecular and physiological role of FGFR1 SUMOylation. So far, it may be the only way to study protein modifications like SUMOylation, and is widely used and accepted, by changing the original K into R that has the most similar biochemical character, but cannot be modified. Our data validated that the overexpressed FGFR1-2KR mutant demonstrated the consistent SUMO deconjugation/deSUMOylation state of FGFR1 and consequent receptor complex and signaling changes as those that occur under the action of the deSUMOylase SENP1 in physiological conditions, which could preserve the effectiveness of FGFR1-2KR mutant expression (two- to threefold for both FGFR1-2KR and -WT in mice) for understanding the physiological role of FGFR1 deSUMOylation in vivo. Nevertheless, the K-to-R mutation may alter stability and folding of the SUMOylation target peptides in the tyrosine kinase domain of FGFR1, owing to the residues’ different geometric structures and capabilities in electrostatic interactions and ionic interactions. Correlatively, this may interfere with the ubiquitination code on FGFR1 and the absence or presence of other potential PTMs of FGFR1, especially the ones having mutual interactions with SUMOylation, in ECs. Also, because this strategy mimics deSUMOylation without interaction with SUMO enzymes, it may limit the precise physiological understanding of the spatiotemporal modulation of SUMOylation during angiogenesis, considering that SUMOylation is dynamically balanced by SUMO conjugases/ligases and SENPs in vivo. Thus, our further studies would focus on the mechanism of inter- and counteraction between SUMOylation and ubiquitination or other potential PTMs of FGFR1 and correlated physiological and pathological functions in vasculature.
Dual Roles of FGFR1 in the Modulation of VEGFR2 Signaling.
Several lines of evidence have indicated the dependency of FGFR on VEGFR2 signaling during the angiogenic process (45–47). Mechanistically, our previous study showed that FGFR1 acts as a positive regulator in endothelial VEGFR2 expression via the FGFR1–ERK1/2–ETS axis. Activated ERK1/2 translocates to the nucleus and promotes ETS transcription-factor binding to the ETS site of the VEGFR2 enhancer (23). In the present study, we revealed the reversible FGFR1 SUMOylation as the other FGFR regulatory mechanism over VEGFR signaling in angiogenesis. FGFR1 deSUMOylation dampened VEGFA/VEGFR2 signaling in ECs, which resulted in retarded angiogenic responses in vitro and in vivo. Interestingly, deSUMOylation of FGFR1 did not disturb the expression of VEGFR2 and VEGFR1, although it enhanced ERK activation upon FGF2 treatment. The indirect upregulation of the NOTCH1-HEY2 cascade and saturated availability of activated ERK may provide elegant explanations for the unaffected VEGFR level. Nevertheless, reversible SUMOylation of FGFR1 aims at FRS2α in regulating VEGFR2 signaling. FRS2α binds, through its PTB domain, to FGFR1 and acts as key adaptor protein in the FGF signaling axis that links FGFR kinase to various downstream signaling pathways (18, 48, 49). On the other hand, FRS2α is also considered critical for VEGFR2 activation, and FRS2α ablation in ECs results in serious angiogenic defects by suppressing VEGFR2 signaling (26, 50). Our findings uncovered FRS2α as a major component of the VEGFR2 signaling complex by binding to VEGFR2 through its PTB domain. Under the regulation of SUMOylation, FGFR1 competes for FRS2α with VEGFR2 based on FGFR1 activation at specific tyrosine residues but not the FGFR1 downstream signaling. More importantly, upon hypoxia, the enhanced SUMOylation of FGFR1 restrains the recruitment of FRS2α to FGFR1 and thereby guarantees the VEGFR2-FRS2α association that activates downstream signaling for the angiogenic capacity of ECs. Collectively, we speculate there is a fine-tuned mechanism by which FGFR1 modulates VEGFR2 signaling responsiveness in angiogenesis via competing for the recruitment of FRS2α upon SUMOylation/deSUMOylation.
The Potential Coordinating Functionalities of FGFR1 SUMOylation in Controlling the Homeostasis of the Vascular Endothelium.
The role of the FGF/FGFR system in the vasculature is still controversial, although their functional phenotypes in vascular growth and maintenance were observed. The findings in our present and previous studies showed that FGFR1 signaling controls the angiogenic process upstream of VEGFR2 signaling. In contrast to VEGFR2/R3 directly driving the biological activity of ECs in angiogenesis, FGFR1 may act mainly through signal integration of more specialized growth-factor systems in ECs and vascular smooth muscle cells (51–53). Intriguingly, the present study revealed that SUMOylation of FGFR1 alone could modulate the adaptor protein distribution between FGFR1 and VEGFR2 complexes and, consequently, control the balance of angiogenic signaling to support angiogenesis in response to different angiogenic environments. At the pathophysiological level, FGFR1/FRS2α–driven FGFR1 signaling is more required in pathological angiogenesis under normoxia, while VEGFR2/FRS2α–driven VEGFR2 signaling is dominant in hypoxic angiogenesis, by the regulatory mechanism of SUMOylation (SI Appendix, Fig. S14). This may explain the confusion regarding physiological angiogenesis in FGF1 and FGF2 double-knockout mice and some FGFR-deficient mice, in which the FGFR1 SUMOylation maintains the angiogenic signaling. Furthermore, the FGFR1 SUMOylation-regulated signaling switch of VEGFR2/FGFR1 pathways may also be part of the molecular basis of vasculogenesis, based on the spatiotemporal availability of VEGFA and FGF2 (54, 55). The coordinating role of FGFR1 may apply to its function in vascular pathology. FGF2/FGFR1 signaling has been identified as a critical antagonistic system against endothelial activation and endothelial-to-mesenchymal transition during arteriosclerosis and pulmonary hypertension by inhibiting transforming growth factor β signaling (56, 57). Together, these findings indicate FGFR1 is important in maintaining the homeostasis of the vascular endothelium by orchestrating multiple vascular biological processes. It will be valuable to investigate the roles of FGFR1 SUMOylation in controlling EC fate and endothelial function in different pathological milieus, especially during vascular inflammation, which substantially impairs FGFR1 stability. Defects in FGF/FGFR and VEGF-VEGFR signaling closely correlate with several cardiovascular diseases, such as angiogenesis-related disease, atherosclerosis, metabolic vascular disorders, as well as tumors and more (58). These pathways are conspicuous as potential therapeutic targets, and the complex molecular circuitry governing them is highly desirable for use in drug design. Therefore, FGFR SUMOylation, which integrates angiogenic signaling spatiotemporally, may be an important piece of the puzzle in this perspective.
Materials and Methods
Information about reagents and antibodies, cell lines and culture conditions, plasmids and transfection, siRNA transfection, adenovirus infection, cell lysates and cell fractionation, immunoblotting, protein stability assay, real-time qPCR, immunofluorescence staining, and mouse lung EC isolation can be found in SI Appendix, SI Materials and Methods.
Animals.
Both endothelial-specific FGFR1-WTecKI mice and endothelial-specific FGFR1-2KRecKI mice were generated by employing a ROSA26 KI strategy (Rosa26-flox-stop codon-flox-FGFR1-WT or Rosa26-flox-stop codon-flox-FGFR1-2KR) based on the Cre-loxP system. EC-specific FGFR1-WTecKI mice and FGFR1-2KRecKI mice were obtained by administering tamoxifen to FGFR1-WTflox/flox-Cdh5-CreERT2 mice and FGFR1-2KRflox/flox-Cdh5-CreERT2 mice, respectively. Cre activity and gene deletion in the postnatal pups were induced by daily intraperitoneal injections of 50 μL (P1) or 100 μL (P2-P5) of tamoxifen solution (Sigma-Aldrich, T5648; 1 mg/mL in corn oil).
Mice were housed in the animal facilities at Zhejiang University and exposed to light on a 12-h cycle in a humidity- and temperature-controlled environment with no pathogenic microorganisms. All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Zhejiang University. Mice were grouped with no blinding but operated randomly and blindly during the experiments. Samples from every animal allocated to the indicated groups were fully blindly analyzed by different persons. No criteria for inclusions and exclusions were set before the study. No samples or animals were excluded from analysis.
Mass Spectrometry Analysis.
LC-MS/MS analysis was performed by the PTM-Bio Company according to established protocols. Briefly, peptides were obtained by in-gel digestion using trypsin; the peptides were extracted with 50% acetonitrile/5% formic acid and 100% acetonitrile. Then, the peptides were completely freeze-dried and resuspended in water containing 2% acetonitrile/0.1% formic acid. For the LC-MS/MS analysis, peptides were dissolved in 0.1% formic acid and directly loaded onto a homemade, reversed-phase analytical column (15-cm long with a 75-μm inner diameter). Highly effective separation of the peptide mixture was obtained by employing an EASY-nLC 1000 UPLC system. The peptides separated by ultra-performance liquid chromatography were subjected to an nanospray ionization source followed by MS in a Q Exactive Plus MS (Thermo). The electrospray voltage was applied at 2.0 kV. The m/z scan range was 350 to 1,800 for full scan, and intact peptides were detected in an Orbitrap at a resolution of 70,000. The peptides were then selected for MS/MS using the Normalized Collision Energy setting of 28, and the fragments were detected in the Orbitrap at a resolution of 17,500. The automatic gain control was set at 5E4. The MS data were processed using Proteome Discoverer 1.3.
Immunoprecipitation.
For IP using anti-Flag M2 magnetic beads to characterize SUMOylated lysine residues in the MS and other analyses, cell pellets were resuspended and lysed with 100 μL 1.25× sodium dodecylsulfate (SDS) loading buffer (75 mM Tris⋅HCl, pH 6.8; 12.5% glycerol; 2.5% SDS; 0.01% bromophenol blue and 1.25× protease inhibitor mixture; 20 mM N-ethylmaleimide [NEM]) and added protease inhibitors and DTT (150 mM) on ice, then cell lysates were boiled for 5 min at 100 °C to denature protein complexes. Then denatured cell lysates were mixed with 10 volumes of 0.1% Triton X-100 lysis buffer (50 mM Tris⋅HCl, pH 7.5; 150 mM NaCl; 1 mM EGTA; 1 mM EDTA; 0.1% Triton X-100; and 1× protease inhibitor mixture; 20 mM NEM). The cell lysates were centrifuged at 12,000g for 20 min at 4 °C to remove insoluble material. The supernatant was collected into 15-mL tubes on ice followed by incubation with equilibrated anti-Flag M2 magnetic beads overnight at 4 °C with gentle rotation. The beads were collected by a magnetic separator and washed seven times with 0.1% Triton X-100 lysis buffer to remove nonspecifically bound proteins. The bound Flag fusion proteins were eluted by competitive elution buffer containing the Flag peptide (100 μg/mL; Millipore Sigma, F3290) in Tris-buffered saline and resuspended in SDS buffer. After heating for 5 min at 100 °C, samples were placed in ice and cooled for use.
For IP using protein-specific antibodies, cells were washed twice with cold PBS after various treatments and harvested in SUMOylation lysis buffer (50 mM Tris⋅HCl, pH 7.5; 150 mM NaCl; 1 mM EGTA; 1 mM EDTA; 0.1% Triton X-100; 1× protease inhibitor mixture, and 20 mM NEM), followed by boiling for 5 min at 100 °C to denature protein complexes for detecting SUMOylation or harvested in lysis buffer (50 mM Tris⋅HCl, pH 7.5; 150 mM NaCl; 1 mM EGTA; 1 mM EDTA; 1.0% Triton X-100; and 1× protease inhibitor mixture) to assay proteins. Cell lysates were then centrifuged at 12,000g for 20 min at 4 °C. The supernatants were immediately prepared for IP by incubating with protein A/G Sepharose (GE Health Care; 10 μL per test) for 1 h on ice to preclear the solution before incubating it with the first protein-specific antisera (e.g., anti-FGFR1 or anti-HA). Then, 20 μL of protein A/G Sepharose was added to the antibody–lysate mixture and incubated for 3 to 5 h or overnight in a cold room. Immune complexes were collected by centrifugation at 2,500 rpm for 2 min followed by five to seven washes with SUMOylation or regular lysis buffer. The immune complexes were resuspended in 2× SDS buffer. After heating for 5 min at 100 °C, the samples were placed in ice and cooled for use.
In Vitro SUMOylation Assay.
The in vitro SUMOylation assay was performed according to the in vitro SUMOylation kit (Enzo, BML-UW8955). Briefly, an immune-purified FGFR1 active peptide (aa 458–765), containing SUMO binding motifs, was used as a substrate in the presence of SUMO E1 (AOS1/UBA2), SUMO E2 (UBC9), ATP, and recombinant SENP1 (catalytic domain). The reaction mixture was then subjected to immunoblotting with anti-SUMO1 antibody.
Retinal Dissection and Whole-Mount Staining.
The procedures for retina dissection, tamoxifen injection, and whole-mount staining were performed as previously described. In brief, for isolectin B4 (IB4) staining, retinas were fixed in freshly prepared 4% paraformaldehyde (PFA) at room temperature (RT) for 0.5 h, and the dissected retinas were blocked in TNTB blocking buffer (100 mM Tris⋅HCl, pH 7.4; 150 mM NaCl; 0.4% Triton X-100; and 0.5% tyramide signal amplification blocking reagent (weight per volume; PerkinElmer, FP1020) at RT for 1 h followed by IB4 staining at RT for 1 h. For antibody immunostaining, retinas were fixed in 4% PFA for 1 to 2 h on ice, and the dissected retinas were incubated with phosphate-buffered solution (PBS) containing 0.5% Triton X-100, 2% bovine serum albumin, and 2% horse serum (PBST blocking buffer) for permeabilization and blocked for 1 h at RT. Then, the retinas were successively incubated with first antibodies and second antibodies in PBST blocking buffer followed by IB4 staining. All retinal staining procedures were performed on a platform shaker. EC proliferation was first measured by a pulse-BrdU labeling assay following daily intraperitoneal injections of 200 μg of BrdU into P3 to P5 pups. In addition, both phospho-histone H3 and ERG were measured to determine the proliferation capability of the retinal vasculature.
Native Gel for FGFR1 WT/2KR Proteins.
HUVECs cultured in 6-cm dishes were washed with cold PBS and drained on ice. Then, 300 μL of lysis buffer (20 mM Tris⋅HCl, pH 7.5; 150 mM NaCl; 1.0% Triton X-100; 10% glycerol; 1 mM Na3VO4; and 1× protease inhibitor mixture) was added to each dish, and the cells were scraped and transferred to a 1.5-mL tube. Cell lysates were then centrifuged at 12,000g for 10 min at 4 °C to remove any insoluble material. The supernatant was collected into a sterile 1.5-mL tube. A 20-μL aliquot of lysate, 1 μL of 10% deoxycholic acid sodium salt (DOC), and 5 μL of 5× native sample buffer were mixed to prepare the samples. Gels were prepared as regular gels for Western blotting without SDS, and the gels were prerun at 200 V for 60 min in a cold room. Then, loaded samples were run in the gel in running buffers (cathode buffer [inside]: 25 mM Tris, 192 mM glycine, 0.4% DOC-Na2 [final pH 8.3 to ∼8.8] and anode buffer [outside]: 25 mM Tris, 192 mM glycine). The transfer process and antibody incubation times were the same as those for regular Western blotting.
Spheroid-Based Angiogenesis, EC Transmigration, Capillary-like Structure Formation, and Wound-Healing Assays.
ECs were infected with Ad-FGFR1-WT or Ad-FGFR1-2KR with or without Ad-FRS2α and then assayed. Forty-eight hours after initial infection, spheroid-based angiogenesis assays, Transwell migration assays, capillary-like structure formation assays, and monolayer EC wound-healing assays were performed as described in previous studies (35).
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism 7 (GraphPad Software, Inc.). The data are presented as the mean ± SEM from at least three independent experiments. Single comparisons between two groups were performed by unpaired t test. Multiple comparisons were performed by two-way ANOVA followed by Tukey’s test or Bonferroni’s multiple comparisons test. P < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Ralf Adams (Max Planck Institute, Munster) for the Cdh5-CreERT2 mice, Dr. Pei-Yu Chen (Yale University School of Medicine) for helpful discussions of the manuscript, and Dr. Shelong Zhang and Dr. Fangliang Huang (Equipment and Technology Service Platform, College of Life Sciences, Zhejiang University) for their excellent technical support with microscopy and cell transfection.
This work was supported by the National Natural Science Foundation of China (Grants 91839104, 81770444, 81600354, 81970372, 11932017, and 31800972), the National Key R&D Program of China (Grants 2018YFA0800504 and 2021YFA1101101), the Zhejiang Provincial Natural Science Foundation of China (Grant LZ20H020002), the Fundamental Research Funds for the Central Universities of China and the Medical and Health Science and Technology Program of Health Commission of Zhejiang Province, China (Grant 2021KY633).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202631119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Carmeliet P., Jain R. K., Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shalaby F., et al. , Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Shibuya M., VEGFR and type-V RTK activation and signaling. Cold Spring Harb. Perspect. Biol. 5, a009092 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons M., Eichmann A., Molecular controls of arterial morphogenesis. Circ. Res. 116, 1712–1724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami M., Simons M., Fibroblast growth factor regulation of neovascularization. Curr. Opin. Hematol. 15, 215–220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner N., Grose R., Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 10, 116–129 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Dow J. K., deVere White R. W., Fibroblast growth factor 2: Its structure and property, paracrine function, tumor angiogenesis, and prostate-related mitogenic and oncogenic functions. Urology 55, 800–806 (2000). [DOI] [PubMed] [Google Scholar]
- 8.De Smet F., et al. , Fibroblast growth factor signaling affects vascular outgrowth and is required for the maintenance of blood vessel integrity. Chem. Biol. 21, 1310–1317 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Deng C. X., et al. , Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 8, 3045–3057 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Yanagisawa-Miwa A., et al. , Salvage of infarcted myocardium by angiogenic action of basic fibroblast growth factor. Science 257, 1401–1403 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Tsuboi R., Shi C. M., Rifkin D. B., Ogawa H., A wound healing model using healing-impaired diabetic mice. J. Dermatol. 19, 673–675 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Compagni A., Wilgenbus P., Impagnatiello M. A., Cotten M., Christofori G., Fibroblast growth factors are required for efficient tumor angiogenesis. Cancer Res. 60, 7163–7169 (2000). [PubMed] [Google Scholar]
- 13.Karsan A., et al. , Fibroblast growth factor-2 inhibits endothelial cell apoptosis by Bcl-2-dependent and independent mechanisms. Am. J. Pathol. 151, 1775–1784 (1997). [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto T., Turesson I., Book M., Gerwins P., Claesson-Welsh L., p38 MAP kinase negatively regulates endothelial cell survival, proliferation, and differentiation in FGF-2-stimulated angiogenesis. J. Cell Biol. 156, 149–160 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami M., Simons M., Regulation of vascular integrity. J. Mol. Med. (Berl.) 87, 571–582 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortega S., Ittmann M., Tsang S. H., Ehrlich M., Basilico C., Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc. Natl. Acad. Sci. U.S.A. 95, 5672–5677 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou M., et al. , Fibroblast growth factor 2 control of vascular tone. Nat. Med. 4, 201–207 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong S. H., et al. , FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol. Cell. Biol. 20, 979–989 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller D. L., Ortega S., Bashayan O., Basch R., Basilico C., Compensation by fibroblast growth factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice. Mol. Cell. Biol. 20, 2260–2268 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oladipupo S. S., et al. , Endothelial cell FGF signaling is required for injury response but not for vascular homeostasis. Proc. Natl. Acad. Sci. U.S.A. 111, 13379–13384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu P., et al. , FGF-dependent metabolic control of vascular development. Nature 545, 224–228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross M. J., Claesson-Welsh L., FGF and VEGF function in angiogenesis: Signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol. Sci. 22, 201–207 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Murakami M., et al. , FGF-dependent regulation of VEGF receptor 2 expression in mice. J. Clin. Invest. 121, 2668–2678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molin D., Post M. J., Therapeutic angiogenesis in the heart: Protect and serve. Curr. Opin. Pharmacol. 7, 158–163 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Jeltsch M., Leppänen V. M., Saharinen P., Alitalo K., Receptor tyrosine kinase-mediated angiogenesis. Cold Spring Harb. Perspect. Biol. 5, a00913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen P. Y., et al. , The docking protein FRS2α is a critical regulator of VEGF receptors signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 5514–5519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickey C. M., Wilson N. R., Hochstrasser M., Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Celen A. B., Sahin U., Sumoylation on its 25th anniversary: Mechanisms, pathology, and emerging concepts. FEBS J. 287, 3110–3140 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Xie M., Yu J., Ge S., Huang J., Fan X., SUMOylation homeostasis in tumorigenesis. Cancer Lett. 469, 301–309 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Flotho A., Melchior F., Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Qiu C., et al. , The critical role of SENP1-mediated GATA2 deSUMOylation in promoting endothelial activation in graft arteriosclerosis. Nat. Commun. 8, 15426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao L., Zhong X., Jin J., Li J., Meng X. M., Potential targeted therapy and diagnosis based on novel insight into growth factors, receptors, and downstream effectors in acute kidney injury and acute kidney injury-chronic kidney disease progression. Signal Transduct. Target. Ther. 5, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson K. A., Henley J. M., Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428, 133–145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu L., et al. , SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J. Exp. Med. 207, 1183–1195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X., et al. , SUMOylation negatively regulates angiogenesis by targeting endothelial NOTCH signaling. Circ. Res. 121, 636–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hay R. T., SUMO-specific proteases: A twist in the tail. Trends Cell Biol. 17, 370–376 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Mikolajczyk J., et al. , Small ubiquitin-related modifier (SUMO)-specific proteases: Profiling the specificities and activities of human SENPs. J. Biol. Chem. 282, 26217–26224 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Kostas M., et al. , Protein tyrosine phosphatase receptor type G (PTPRG) controls fibroblast growth factor receptor (FGFR) 1 activity and influences sensitivity to FGFR kinase inhibitors. Mol. Cell. Proteomics 17, 850–870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., McKeehan K., Lin Y., Zhang J., Wang F., Fibroblast growth factor receptor 1 (FGFR1) tyrosine phosphorylation regulates binding of FGFR substrate 2α (FRS2α) but not FRS2 to the receptor. Mol. Endocrinol. 22, 167–175 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotoh N., et al. , The docking protein FRS2α is an essential component of multiple fibroblast growth factor responses during early mouse development. Mol. Cell. Biol. 25, 4105–4116 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lax I., et al. , The docking protein FRS2α controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol. Cell 10, 709–719 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Haugsten E. M., Malecki J., Bjørklund S. M., Olsnes S., Wesche J., Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol. Biol. Cell 19, 3390–3403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yau R., Rape M., The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Chang Y. C., Oram M. K., Bielinsky A. K., SUMO-targeted ubiquitin ligases and their functions in maintaining genome stability. Int. J. Mol. Sci. 22, 5391 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rousseau B., Larrieu-Lahargue F., Bikfalvi A., Javerzat S., Involvement of fibroblast growth factors in choroidal angiogenesis and retinal vascularization. Exp. Eye Res. 77, 147–156 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Magnusson P., et al. , Deregulation of Flk-1/vascular endothelial growth factor receptor-2 in fibroblast growth factor receptor-1-deficient vascular stem cell development. J. Cell Sci. 117, 1513–1523 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Huang X., et al. , Ectopic activity of fibroblast growth factor receptor 1 in hepatocytes accelerates hepatocarcinogenesis by driving proliferation and vascular endothelial growth factor-induced angiogenesis. Cancer Res. 66, 1481–1490 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Kouhara H., et al. , A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89, 693–702 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Gotoh N., Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 99, 1319–1325 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoletov K. V., Ratcliffe K. E., Terman B. I., Fibroblast growth factor receptor substrate 2 participates in vascular endothelial growth factor-induced signaling. FASEB J. 16, 1283–1285 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Cao R., et al. , Angiogenic synergism, vascular stability and improvement of hind-limb ischemia by a combination of PDGF-BB and FGF-2. Nat. Med. 9, 604–613 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Kano M. R., et al. , VEGF-A and FGF-2 synergistically promote neoangiogenesis through enhancement of endogenous PDGF-B-PDGFRβ signaling. J. Cell Sci. 118, 3759–3768 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Pickering J. G., et al. , Fibroblast growth factor-2 potentiates vascular smooth muscle cell migration to platelet-derived growth factor: Upregulation of α2β1 integrin and disassembly of actin filaments. Circ. Res. 80, 627–637 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Poole T. J., Finkelstein E. B., Cox C. M., The role of FGF and VEGF in angioblast induction and migration during vascular development. Dev. Dyn. 220, 1–17 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Chung A. S., Ferrara N., Developmental and pathological angiogenesis. Annu. Rev. Cell Dev. Biol. 27, 563–584 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Chen P. Y., et al. , FGF regulates TGF-β signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2, 1684–1696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sánchez-Duffhues G., García de Vinuesa A., Ten Dijke P., Endothelial-to-mesenchymal transition in cardiovascular diseases: Developmental signaling pathways gone awry. Dev. Dyn. 247, 492–508 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Tas S. W., Maracle C. X., Balogh E., Szekanecz Z., Targeting of proangiogenic signalling pathways in chronic inflammation. Nat. Rev. Rheumatol. 12, 111–122 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.