Significance
Mechanistic understanding of cellular defense mechanisms against ferroptosis can inform novel therapeutic strategies in disease treatment. This study reveals that glycerol-3-phosphate (G3P) dehydrogenase 2 (GPD2), an inner mitochondria membrane-bound enzyme that is involved in the G3P shuttle, has a previously unrecognized role in defending against ferroptosis in mitochondria and suggests that inhibiting GPD2-mediated ferroptosis defense can be explored in cancer therapy.
Keywords: GPD2, ferroptosis, mitochondria, lipid peroxidation, cell death
Abstract
Mechanisms of defense against ferroptosis (an iron-dependent form of cell death induced by lipid peroxidation) in cellular organelles remain poorly understood, hindering our ability to target ferroptosis in disease treatment. In this study, metabolomic analyses revealed that treatment of cancer cells with glutathione peroxidase 4 (GPX4) inhibitors results in intracellular glycerol-3-phosphate (G3P) depletion. We further showed that supplementation of cancer cells with G3P attenuates ferroptosis induced by GPX4 inhibitors in a G3P dehydrogenase 2 (GPD2)-dependent manner; GPD2 deletion sensitizes cancer cells to GPX4 inhibition-induced mitochondrial lipid peroxidation and ferroptosis, and combined deletion of GPX4 and GPD2 synergistically suppresses tumor growth by inducing ferroptosis in vivo. Mechanistically, inner mitochondrial membrane-localized GPD2 couples G3P oxidation with ubiquinone reduction to ubiquinol, which acts as a radical-trapping antioxidant to suppress ferroptosis in mitochondria. Taken together, these results reveal that GPD2 participates in ferroptosis defense in mitochondria by generating ubiquinol.
Ferroptosis is a form of iron-dependent cell death triggered by unchecked lipid peroxidation and subsequent oxidative damage in cellular membranes (1, 2). It is morphologically and mechanistically different from other forms of regulated cell death, such as apoptosis (1, 3, 4). Ferroptosis results from an imbalance between cellular metabolic activities and antioxidant activities that provoke and dampen lipid peroxidation, respectively (5). Polyunsaturated fatty acid (PUFA)-containing phospholipids are essential components of cellular membranes and have important roles in cellular signaling and homeostasis. However, PUFAs contain bis-allylic moieties with methylene groups flanked by C–C double bonds and the C–H bonds in such methylene groups are very weak; this structure nature renders PUFA-containing phospholipids susceptible to oxidation, particularly in an iron- and oxygen-rich cellular environment (6). If left unchecked, a lethal accumulation of lipid hydroperoxides in cellular membranes ultimately induces membrane rupture and ferroptotic cell death.
Ferroptosis appears to represent an inevitable adverse cost that animal cells must endure during evolution, particularly after PUFAs, oxygen, and iron metabolism were incorporated into cellular lives. Correspondingly, different cellular defense systems against ferroptosis have evolved to counteract these toxic effects induced by lipid peroxidation (3). The best-known ferroptosis defense mechanism is mediated by glutathione peroxidase 4 (GPX4), a glutathione-dependent peroxidase that reduces toxic lipid hydroperoxides to nontoxic lipid alcohols on cellular membranes, thereby mitigating ferroptosis (7, 8). In addition, ferroptosis suppressor protein 1 (FSP1, also known as AIFM2) suppresses ferroptosis independent of GPX4 by reducing ubiquinone (CoQ) to ubiquinol (CoQH2) at the plasma membrane; CoQH2 then acts as a lipophilic radical-trapping antioxidant to trap lipid peroxyl radicals (9, 10). Finally, GTP cyclohydroxylase-1 (GCH-1) mediates the synthesis of tetrahydrobiopterin, which is also a radical-trapping antioxidant capable of suppressing lipid peroxyl radicals and ferroptosis (11, 12).
Notably, GPX4 has both cytosolic and mitochondrial isoforms that are encoded from the same GPX4 gene but with different transcription start sites (5, 13, 14). Whereas cytosolic GPX4’s role in defending against ferroptosis is well-established, researchers initially believed that mitochondrial GPX4 does not have a role in suppressing ferroptosis (5). However, we recently showed that mitochondrial GPX4 acts in concert with dihydroorotate dehydrogenase (DHODH), an inner mitochondrial membrane-localized enzyme that converts CoQ to CoQH2, to suppress lipid peroxidation and ferroptosis in mitochondria. Consequently, inactivation of both mitochondrial GPX4 and DHODH induces mitochondrial lipid peroxidation and unleashes potent ferroptosis (15, 16). These findings suggest that ferroptosis defense systems can be compartmentalized into different subcellular locations (e.g., plasma membrane and mitochondria). However, whether additional players defend against ferroptosis in mitochondria remains to be determined.
Since ferroptosis has been linked to diverse diseases and pathological conditions, such as cancer, acute kidney injury, and neurodegenerative and cardiovascular diseases (3–5, 17), further studying mitochondria-localized ferroptosis defense mechanisms can identify additional therapeutic targets in disease treatment. In this study, we identified glycerol-3-phosphate (G3P) dehydrogenase 2 (GPD2) as another inner mitochondrial membrane-bound enzyme that suppresses mitochondrial lipid peroxidation and ferroptosis by producing CoQH2 and showed that GPD2 inactivation synergizes with GPX4 inactivation to suppress tumor formation.
Results
Metabolomic Analysis Links GPD2 to Ferroptosis Regulation.
We took a metabolomic approach to identify potential ferroptosis defense mechanisms that presumably are turned on following acute GPX4 inactivation by short-term (2-h) treatment with a GPX4 inhibitor (at which time point cells had not died of ferroptosis). In a recent study, we identified N-carbamoyl-l-aspartate (C-Asp), an intermediate of pyrimidine biosynthesis, as the most significantly depleted metabolite in cancer cells upon 2-h treatment with a GPX4 inhibitor (RSL3, ML210, or ML162); this observation led to the finding that DHODH, an enzyme involved in pyrimidine biosynthesis, acts as a key gatekeeper in ferroptosis defense in mitochondria (15). The same analyses uncovered G3P as another metabolite that was consistently depleted in cells upon treatment with all three GPX4 inhibitors (Fig. 1 A–D). G3P supplementation suppressed RSL3-induced ferroptosis in different cancer cell lines (Fig. 1E and SI Appendix, Fig. S1 A–D). We confirmed that G3P supplementation increased intracellular G3P levels (SI Appendix, Fig. S1 E–G). These findings suggested that G3P has a protective effect against GPX4 inactivation-induced ferroptosis.
Fig. 1.
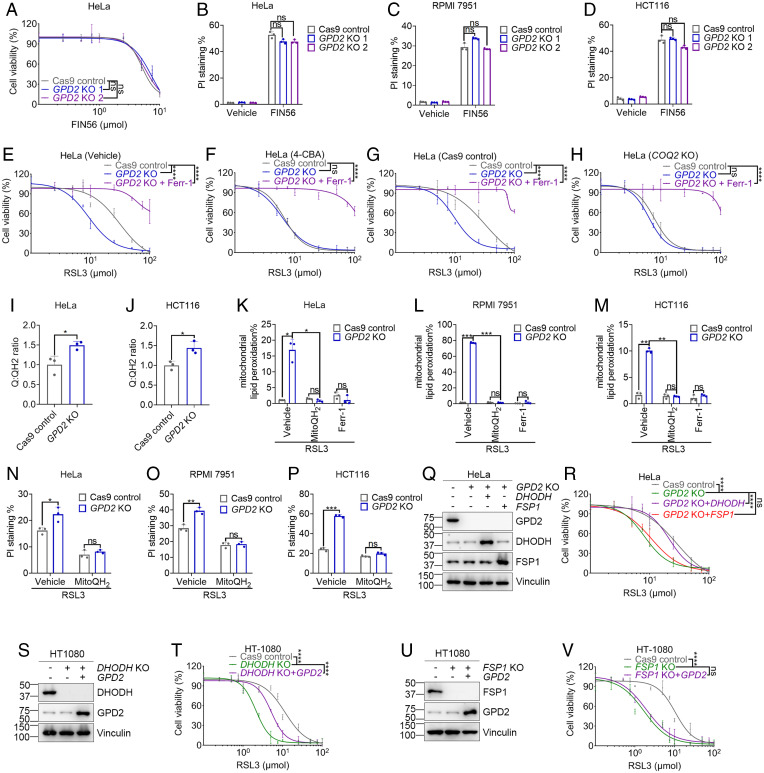
Metabolomic analysis links GPD2 to ferroptosis regulation. (A–C) Volcano plots comparing the metabolomic profiles of HT1080 cells treated with a vehicle, RSL3 (A; 10 μM), ML210 (B; 10 μM), or ML162 (C; 10 μM) for 2 h. (D) Fold changes in G3P levels in HT1080 cells upon treatment with RSL3 (10 μM), ML210 (10 μM), or ML162 (10 μM) for 2 h. (E) Cell viability measurement of HCT116 cells treated with RSL3 at different concentrations for 24 h following pretreatment with a vehicle or G3P (1 mM) for 24 h. (F) Schematic of G3P metabolism. (G and H) RSL3 resistance correlates with high GPD2 expression (G), and high GPD2 expression correlates with resistance to treatment with GPX4 inhibitors (RSL3, ML162, and ML210) in cancer cells (H). The plotted data were mined from the CTRP database. The plotted values are Pearson’s correlation coefficients. The scatter plots show the medians, 10th and 90th percentiles, and minima and maxima of the distributions. (I) Western blot of GPD2, ACSL4, FSP1, SLC7A11, DHODH, and GPX4 protein levels in Cas9 control and GPD2-KO cells. (J) Cell viability measurement of HeLa GPD2-KO and control cell after RSL3 treatment. (K–P) Results of PI staining of Cas9 control and GPD2-KO (numbers 1 and 2) HeLa (K and N), RPMI 7951 (L and O), and HCT116 (M and P) cells treated with RSL3 (10 μM), ML210 (10 μM), or ferrostatin-1 (Ferr-1; 5 μM) as indicated. HeLa and RPMI 7951 cells were treated for 6 h, and HCT116 cells were treated for 24 h. (Q–S) Lipid peroxidation measurement in Cas9 control, GPD2-KO HeLa (Q), and RPMI 7951 (R) cells treated with RSL3 (10 μM) for 3 h and HCT116 cells (S) treated with RSL3 (1.5 μM) for 24 h. (T and U) Cell viability measurement of HeLa control (T) and GPD2-KO (U) cells after G3P and RSL3 treatment. (V and W) Results of PI staining of Cas9 control and GPD2-KO HeLa (V) and RPMI 7951 (W) cells treated with RSL3 (10 μM) for 4 h following pretreatment with a vehicle or G3P (1 mM) for 24 h. Data are presented as means (± SD), n = 3 independent repeats. Unpaired, two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
These observations prompted us to further study the potential relevance of G3P-linked metabolic enzymes to ferroptosis regulation. Several enzymes are involved in G3P metabolism (Fig. 1F). Specifically, phosphoglycolate phosphatase (PGP) hydrolyzes G3P to glycerol, whereas glycerol kinase (GK) catalyzes the phosphorylation of glycerol to generate G3P. G3P acyltransferases (GPATs, including GPAT1–4) convert G3P to lysophosphatidic acid, which is subsequently used in triacylglycerol and phospholipid synthesis. In addition, cytosolic GPD1 (and possibly GPD1-like [GPD1L]) couples the oxidation of NADH to NAD+ with the reduction of dihydroxyacetone phosphate (DAP) to G3P; and inner mitochondrial membrane-bound GPD2 then converts G3P back to DAP and donates electrons to the electron transport chain in mitochondria (18). (This G3P shuttle serves to deliver NADH generated by glycolysis in the cytosol into mitochondria for oxidation because the inner mitochondrial membrane is impermeable to NADH; consistently, G3P supplementation increased the NAD+/NADH ratio [SI Appendix, Fig. S1H].) Analyses of data from the Cancer Therapeutics Response Portal (CTRP) (19) revealed that expression of GPD2, but not that of GPD1, GPD1L, GK, or PGP, correlated with cellular resistance to GPX4 inhibitors (Fig. 1 G and H and SI Appendix, Fig. S1 I–L; it should be noted that GPAT data were not available from the CTRP). Also, the correlation of GPD2 expression with GPX4 inhibitor resistance was particularly pronounced in large intestine, skin, and stomach cancer cell lines (SI Appendix, Fig. S1 M–O). We therefore focused on GPD2 in our subsequent studies.
GPD2 Deletion Promotes GPX4 Inactivation-Induced Ferroptosis.
To determine the role of GPD2 in ferroptosis regulation, we depleted GPD2 expression in HeLa (a cervical cancer cell line widely used in cancer research), RPMI 7951 (a skin cancer cell line), and HCT116 cells (a large intestine cancer cell line) using the CRISPR-Cas9 approach (we chose RPMI 7951 and HCT116 cells based on CTRP analyses) (Fig. 1I and SI Appendix, Fig. S1P). The results showed that GPD2 deletion did not affect basal cell viability but significantly sensitized these cell lines to cell death and lipid peroxidation induced by GPX4 inhibitors (Fig. 1 J–S). The cell death was completely suppressed by the ferroptosis inhibitor ferrostatin-1 or iron chelator deferoxamine (DFO) in HeLa and RPMI 7951 cells, confirming that these GPX4 inhibitors induced ferroptosis in these cell lines (Fig. 1 K, L, N, and O and SI Appendix, Fig. S1 Q and R); however, residual death of HCT116 cells remained after treatment with ferrostatin-1 or DFO (Fig. 1 M and P and SI Appendix, Fig. S1S), suggesting that these GPX4 inhibitors mostly trigger ferroptosis but can also induce other nonferroptotic cell death in HCT116 cells. GPD2 deletion did not appear to affect the expression of other ferroptosis regulators, including GPX4, FSP1, DHODH, SLC7A11 (an amino acid transporter that imports cystine for glutathione biosynthesis and ferroptosis protection) (20, 21), and ACSL4 (a lipid metabolism enzyme that mediates PUFA-containing phospholipid biosynthesis and is required for ferroptosis in many cell lines) (Fig. 1I) (22). As expected, GPD2 deletion decreased the NAD+/NADH ratio (SI Appendix, Fig. S1T). Finally, we showed that the protective effect of G3P against RSL3-induced cell death was abolished in GPD2-KO (knockout) cells (Fig. 1 T–W). Taken together, these data suggested that G3P protects cells against GPX4 inhibition–induced ferroptosis through GPD2 and that GPD2 deficiency sensitizes cells to ferroptosis triggered by GPX4 inhibition.
We also tested the impact of pharmacological inhibition of GPD2 on ferroptosis. iGP-1 was the only GPD2 inhibitor we could obtain from commercial sources (23). Whereas GPD2 deletion markedly increased intracellular G3P levels as expected (SI Appendix, Fig. S1U), we found that iGP-1 treatment across different doses and treatment durations did not have an effect on intracellular G3P levels (SI Appendix, Fig. S1 U–Y), indicating that, at least under our experimental conditions, iGP-1 did not appear to exert an inhibitory effect on GPD2. Consistent with this observation, treatment with iGP-1 did not have any sensitizing effect on RSL3-induced ferroptosis in HCT116 cells (SI Appendix, Fig. S1Z).
GPD2 converts G3P to DAP, which is then converted back to G3P by GPD1 (and potentially GPD1L), forming the G3P shuttle (Fig. 1F). By supplying G3P, GPD1 presumably should suppress ferroptosis. However, we found that GPD1 deletion did not affect RSL3-induced ferroptosis in HeLa cells (SI Appendix, Fig. S2 A and B), which is consistent with data from CTRP analyses (SI Appendix, Fig. S1J). We therefore reasoned that GPD1 and GPD1L may have redundant functions. To test this, we generated GPD1 and GPD1L (GPD1/1L) double-KO (DKO) HeLa cells (SI Appendix, Fig. S2C). Indeed, we found that GPD1/1L KO sensitized the cells to RSL3-induced ferroptosis (SI Appendix, Fig. S2D). We made similar observations with HCT116 cells (SI Appendix, Fig. S2 E and F). Analysis of data from the Cancer Cell Line Encyclopedia and Genotype-Tissue Expression (24, 25) revealed that whereas expression of GPD2 and GPD1L was relatively ubiquitous in different tissues and cancer cells, GPD1 expression was low in most cancer cell lines, and GPD1 was mainly expressed in adipose and mammary tissues (SI Appendix, Fig. S2 G–L), suggesting that GPD1L compensates for GPD1’s function in generating G3P in many cancer cells. This is in line with our observation that deleting both GPD1 and GPD1L is required to reveal ferroptosis sensitization phenotypes in cancer cells we have examined.
GPD2 Suppresses Mitochondrial Lipid Peroxidation.
GPD2 is an enzyme localized on the inner mitochondrial membrane (18). Fractionation analyses confirmed mitochondrial localization of endogenous GPD2 in several cancer cell lines (Fig. 2A). Consistent with previous reports (7, 15), staining of MitoPerOx [mitochondria-targeted C11-BODIPY (26)] showed that RSL3 treatment did not induce strong mitochondrial lipid peroxidation; importantly, GPD2 deletion markedly enhanced mitochondrial lipid peroxidation under RSL3 treatment (Fig. 2 B–E). Restoration of wild-type GPD2, but not its mutant defective in mitochondrial localization (42AA) (27), in GPD2-KO cells rescued the effect of GPD2 deficiency on RSL3-induced cell death and mitochondrial lipid peroxidation (Fig. 2 A, F–L and SI Appendix, Fig. S3 A–C). Further analyses confirmed that GPD2-KO cells exhibited decreased GPD2 activity and basal oxygen consumption rate, and these defects were rescued by wild-type GPD2 restoration (SI Appendix, Fig. S3 D and E); GPD2 42AA mutant restoration also rescued GPD2 activity (as this mutant has the intact enzymatic domain) but failed to rescue oxygen consumption rate defect in GPD2 KO cells (SI Appendix, Fig. S3 D and E), suggesting that mitochondrial localization is required for GPD2’s function in regulating ferroptosis and oxidative phosphorylation. Furthermore, in wild-type cells for GPD2, whereas treatment with the radical-trapping antioxidant TEMPO or ferrostatin-1 completely (or largely) suppressed RSL3-induced cell death, treatment with MitoTEMPO (28) did not provide a protective effect (Fig. 2 M–P and SI Appendix, Fig. S3 F–H). In contrast, in GPD2-KO cells, MitoTEMPO, similar to TEMPO and ferrostatin-1, completely suppressed RSL3-induced mitochondrial lipid peroxidation (Fig. 2 Q–S) and significantly suppressed RSL3-induced cell death (Fig. 2 T–W). Collectively, these data suggested that GPD2 suppresses mitochondrial lipid peroxidation and that ferroptosis induced by RSL3 in GPD2-deficient cells is at least partly triggered by mitochondrial lipid peroxidation.
Fig. 2.
GPD2 suppresses mitochondrial lipid peroxidation. (A) Western blot showing GPD2 protein levels in cytosolic and mitochondrial fractions from GPD2-KO cells expressing the indicated GPD2 constructs. mito, mitochondrial; cyto, cytosolic. (B–E) Mitochondrial lipid peroxidation measurement by flow cytometry or immunofluorescence in Cas9 control and GPD2-KO cells upon treatment with RSL3 (1 μM) for 2 h (B–D) or 24 h (E). (F–H) Results of PI staining of GPD2-KO HeLa (F), RPMI 7951 (G), and HCT116 (H) cells expressing the indicated GPD2 constructs upon treatment with RSL3 (10 μM). (I–L) Mitochondrial lipid peroxidation measurement by flow cytometry or immunofluorescence in GPD2-KO HeLa (I and J), RPMI 7951 (K), and HCT116 (L) cells expressing the indicated GPD2 constructs upon treatment with RSL3 (1 μM). (M) Cell viability measurement of HeLa cas9 control cells with RSL3, TEMPO, or MitoTEMPO treatment as indicated. (N–P) Results of PI staining of Cas9 control HeLa (N), RPMI 7951 (O), and HCT116 (P) cells treated with RSL3 (10 μM) for 6 h (N and O) or 24 h (P) following pretreatment with a vehicle, TEMPO (10 μM), MitoTEMPO (10 μM), or ferrostatin-1 (Ferr-1; 5 μM) for 24 h. (Q–S) Mitochondrial lipid peroxidation measurement in GPD2-KO HeLa (Q), RPMI 7951 (R), and HCT116 (S) cells treated with RSL3 (1 μM) for 2 h (Q and R) or 24 h (S) following pretreatment with a vehicle, TEMPO (10 μM), MitoTEMPO (10 μM), or Ferr-1 (5 μM) for 24 h. (T) Cell viability measurement of HeLa GPD2-KO cell with RSL3, TEMPO, or MitoTEMPO treatment as indicated. (U–W) Results of PI staining of GPD2-KO HeLa (U), RPMI 7951 (V), and HCT116 (W) cells treated with RSL3 (10 μM) for 6 h (U and V) or 24 h (W) following pretreatment with a vehicle, TEMPO (10 μM), MitoTEMPO (10 μM), or Ferr-1 (5 μM) for 24 h. Data are presented as means (± SD) for three independent repeats. Unpaired, two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
GPD2 Suppresses Ferroptosis through Reducing CoQ to CoQH2 in Mitochondria.
Next, we sought to understand the mechanism by which GPD2 suppresses mitochondrial lipid peroxidation and ferroptosis. We found that GPD2 deletion sensitized cells to GPX4 inhibitors (RSL3 or ML210) (Fig. 1 J–P) but not FIN56, another ferroptosis inducer that depletes CoQ and GPX4 protein (Fig. 3 A–D) (29). Because the major difference between these GPX4 inhibitors and FIN56 lies in their different effects on CoQ depletion, these data suggested that GPD2 suppresses ferroptosis in a CoQ-dependent manner (i.e., when CoQ is depleted by FIN56, the effect of GPD2 deficiency on ferroptosis is blunted). Consistent with this, we found that the ferroptosis-sensitizing effect of GPD2 deficiency was ablated under CoQ synthesis blockade conditions by treatment with 4-chlorobenzoic acid (4CBA) (Fig. 3 E and F, SI Appendix, Fig. S4 A–C). COQ2 catalyzes a key step in CoQ biosynthesis (30). Likewise, COQ2 deletion abolished the sensitizing effect of GPD2 deficiency on RSL3-induced ferroptosis (Fig. 3 G and H and SI Appendix, Fig. S4 D and E). Of note, these data also revealed that 4CBA treatment or COQ2 deletion markedly sensitized cells to RSL3-induced ferroptosis (Fig. 3 E–H and SI Appendix, Fig. S4 A–E), which is consistent with the known role of CoQ in suppressing lipid peroxidation and ferroptosis (3).
Fig. 3.
GPD2 suppresses ferroptosis through reduction of CoQ to CoQH2 in mitochondria. (A) Cell viability measurement of HeLa Cas9 control and GPD2-KO cells with FIN56 treatment. (B–D) Results of PI staining of Cas9 control and GPD2-KO HeLa (B), RPMI 7951 (C), and HCT116 (D) cells treated with FIN56 (5 μM) for 24 h. (E–H) Cell viability measurement of HeLa cells under RSL3 treatment with 4-CBA (E and F) or COQ2 KO (G and H). (I and J) CoQ/CoQH2 ratios in GPD2-KO HeLa (I) and HCT116 (J) cells. (K–M) Mitochondrial lipid peroxidation measurement in Cas9 control and GPD2-KO HeLa (K), RPMI 7951 (L), and HCT116 (M) cells upon treatment with RSL3 (1 μM) for 2 h (K and L) or 24 h (M) following pretreatment with a vehicle, MitoQH2 (10 μM), or Ferr-1 (5 μM) for 24 h. (N–P) Results of PI staining of Cas9 control and GPD2-KO HeLa (N), RPMI 7951 (O), and HCT116 (P) cells upon treatment with RSL3 (10 μM) for 6 h (N and O) or 24 h (P) following pretreatment with a vehicle, MitoQH2 (10 μM), or Ferr-1 (5 μM) for 24 h. (Q and R) The effect of DHODH or FSP1 overexpression on RSL3-induced ferroptosis in GPD2 KO cells. (S–V) The effect of GPD2 overexpression on RSL3-induced ferroptosis in DHODH KO cells (S and T) or FSP1 KO cells (U and V). Data are presented as means (± SD) for three independent repeats. Unpaired, two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
GPD2 couples the oxidation of G3P to DAP with the reduction of CoQ to CoQH2 in the inner mitochondrial membrane (SI Appendix, Fig. S4F). We confirmed that GPD2 deletion increased the CoQ/CoQH2 ratio in HeLa and HCT116 cells (Fig. 3 I and J and SI Appendix, Fig. S4 G–J); GPD2-deficient RPMI 7951 cells also exhibited a trend toward greater CoQ/CoQH2 ratios than did their GPD2 wild-type counterparts, although the difference was not statistically significant (SI Appendix, Fig. S4K). Furthermore, supplementation with MitoQH2 (mitochondria-targeted analog of CoQH2) completely suppressed mitochondrial lipid peroxidation in RSL3-treated GPD2-KO cells (Fig. 3 K–M) and abolished the ferroptosis-sensitizing effect caused by GPD2 deletion (Fig. 3 N–P). Finally, we found that GPD2 deletion, but not GPD1/1L deletion, abolished the protective effect of G3P against RSL3-induced cell death (Fig. 1 T and U and SI Appendix, Fig. S4L), which is in line with the model that CoQH2 generation from G3P depends on GPD2 but not GPD1/1L. Taken together, these data suggested that GPD2 suppresses mitochondrial lipid peroxidation and ferroptosis by reducing CoQ to CoQH2 in mitochondria.
We and others recently showed that FSP1 and DHODH suppress ferroptosis by generating CoQH2 on the plasma membrane and in mitochondria, respectively (9, 10, 15). Importantly, FSP1 overexpression drives ferroptosis resistance in wild-type cells but not in DHODH KO cells (15), suggesting that FSP1 and DHODH operate in separate cellular compartments to defend against ferroptosis. We further studied the genetic interactions between GPD2 and DHODH (or FSP1) in regulating ferroptosis. Notably, overexpression of DHODH, but not FSP1, rescued the ferroptosis sensitization phenotype in GPD2-KO cells (Fig. 3 Q and R). In addition, GPD2 overexpression partially rescued ferroptosis sensitization phenotype in DHODH KO cells (Fig. 3 S and T) but not that in FSP1-KO cells (Fig. 3 U and V). These results therefore support a model that GPD2 operates in parallel to DHODH in mitochondria, but not to FSP1 on the plasma membrane, to defend against ferroptosis.
GPD2 Acts in Parallel with Mitochondrial GPX4 to Suppress Ferroptosis.
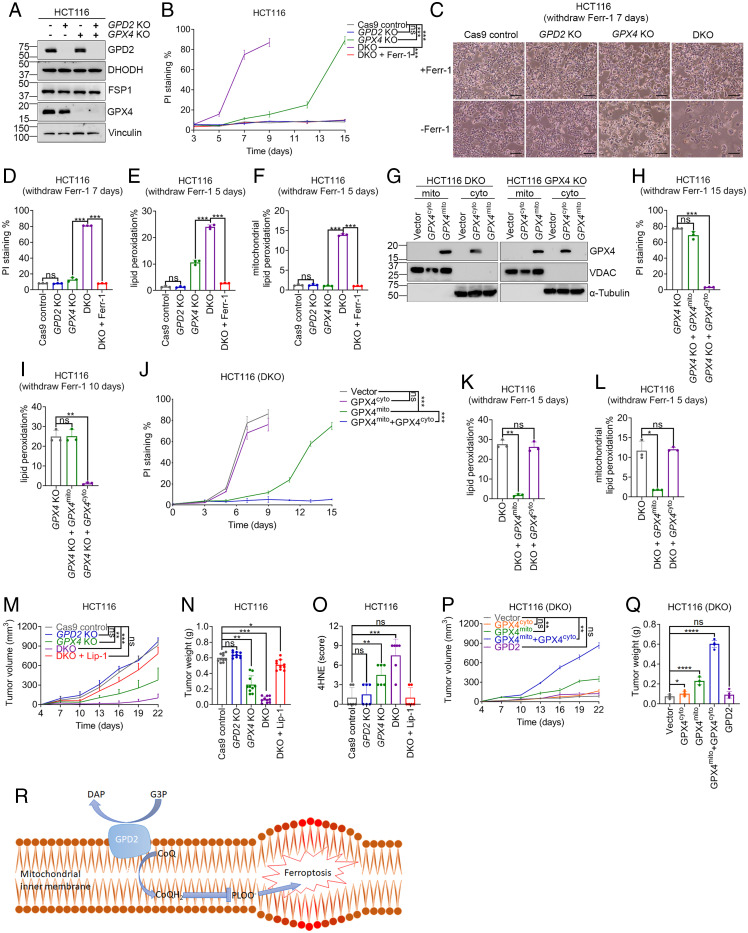
The data described above prompted us to further study the genetic interactions between GPD2 and GPX4 in suppressing ferroptosis. To this end, we generated GPX4 and GPD2 single-KO and DKO HCT116 cells using the CRISPR-Cas9 approach (Fig. 4A). These cell lines did not exhibit obvious difference in cell proliferation rate under the medium supplemented with the ferroptosis inhibitor ferrostatin-1 or liproxstatin-1 (SI Appendix, Fig. S5A). The ferroptosis inhibitor was subsequently removed from the medium to unleash lipid peroxidation and ferroptosis in GPX4-KO cells. Analysis of the data from the CTRP (19) showed that HCT116 is an RSL3-resistant cell line (SI Appendix, Fig. S5 B and C). Consistently, we found that long periods of culture time in ferrostatin-1–free medium (14 to 15 d) were required to induce complete death of GPX4-KO HCT116 cells (Fig. 4B). GPD2 deletion did not affect the viability of GPX4 wild-type cells but dramatically accelerated the onset of death of GPX4-KO cells such that most of the DKO cells died after 7 d of culture in ferrostatin-1–free medium (Fig. 4 B–D). Of note, ferrostatin-1 completely suppressed HCT116 cell death induced by genetic ablation of GPX4 but not by RSL3 treatment, demonstrating that RSL3 has off-target effects, resulting in the induction of nonferroptotic cell death of HCT116 cells (Fig. 1M versus Fig. 4 B and D). Consistently, lipid peroxidation levels were markedly higher in DKO cells than in GPX4-KO cells (Fig. 4E). Furthermore, DKO cells but not the single-KO cells exhibited strong mitochondrial lipid peroxidation after ferrostatin-1 removal (Fig. 4F).
Fig. 4.
GPD2 cooperates with mitochondrial GPX4 to suppress ferroptosis both in vitro and in tumors. (A) Western blot of GPD2, DHODH, FSP1, and GPX4 protein levels in HCT116 cells with the indicated genotypes. (B) Results of PI staining of Cas9 control, GPD2-KO, GPX4-KO, and DKO HCT116 cells with or without treatment with ferrostatin-1 (Ferr-1; 5 μM) at different time points (days). (C and D) Results of microscopic analysis (C) and PI staining (D) of Cas9 control, GPD2-KO, GPX4-KO, and DKO HCT116 cells with or without treatment with ferrostatin-1 (5 μM) at 7 d. (Scale bars, 20 μM.) (E and F) Total (E) and mitochondrial (F) lipid peroxidation in Cas9 control, GPD2-KO, GPX4-KO, and DKO HCT116 cells with or without treatment with ferrostatin-1 (5 μM) at 5 d. (G) Western blot showing GPX4 protein levels in cytosolic and mitochondrial fractions of GPX4-KO and DKO HCT116 cells expressing the indicated GPX4 constructs. (H and I) Results of PI staining (H) and lipid peroxidation levels of (I) GPX4-KO HCT116 cells expressing the indicated GPX4 constructs at 15 d (H) or 10 d (I) after ferrostatin-1 removal. (J) Result of PI staining of DKO HCT116 cells expressing the indicated GPX4 constructs in 15 d after ferrostatin-1 removal. (K and L) Total (K) and mitochondrial lipid peroxidation (L) of DKO HCT116 cells expressing the indicated GPX4 constructs at 5 d after ferrostatin-1 removal. (M) Volumes of Cas9 control, GPX4-KO, GPD2-KO, and DKO HCT116 xenografts treated with or without liproxstatin-1 (Lip-1) at different time points (days). (N) Weights of Cas9 control, GPX4-KO, GPD2-KO, and DKO HCT116 xenograft tumors with and without treatment with liproxstatin-1 at the experimental endpoints. (O) Immunohistochemical scores for 4-HNE staining of Cas9 control, GPX4-KO, GPD2-KO, and DKO HCT116 xenograft tumors with the indicated treatments. (P and Q) The effect of various GPX4 or GPD2 restoration as indicated on the growth of GPX4/GPD2 DKO tumors. (R) Working model depicting how GPD2 suppresses ferroptosis in mitochondria. Data are presented as means (± SD), n = 3 independent repeats. Unpaired, two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
There are several GPX4 isoforms with different subcellular localizations, including cytosolic and mitochondrial GPX4. We restored cytosolic or mitochondrial GPX4 in GPX4-KO and DKO cells. Fractionation analysis confirmed that cytosolic and mitochondrial GPX4 were correctly localized to their respective compartments (Fig. 4G). As expected, overexpression of cytosolic GPX4 but not mitochondrial GPX4 potently suppressed lipid peroxidation and ferroptosis in GPX4-KO cells (Fig. 4 H and I). Of note, removal of ferrostatin-1 did not induce obvious mitochondrial lipid peroxidation in GPX4-KO cells (SI Appendix, Fig. S5D). In stark contrast, we observed that cytosolic GPX4 failed to suppress ferroptotic cell death in DKO cells (Fig. 4J), whereas overexpression of mitochondrial GPX4 significantly delayed cell death in DKO cells, such that the cell death kinetics from DKO cells with mitochondrial GPX4 restoration appeared to be similar to that from GPX4-KO cells (Fig. 4B versus Fig. 4J). At 5 d after ferrostatin-1 removal (before DKO cells started to undergo ferroptosis), the induction of total or mitochondrial lipid peroxidation in DKO cells was markedly suppressed by restoration of mitochondrial GPX4 but not cytosolic GPX4 (Fig. 4 K and L). DKO cells restored with mitochondrial GPX4 eventually still died of ferroptosis, ostensibly because these cells lacked cytosolic GPX4 and therefore were incapable of detoxifying lipid peroxides accumulated on nonmitochondrial compartments at later time points; correspondingly, restoration of both cytosolic and mitochondrial GPX4 completely suppressed ferroptosis in DKO cells (Fig. 4J). Taken together, these data suggested that GPD2 operates in parallel with mitochondrial GPX4 but not cytosolic GPX4 to suppress lipid peroxidation and ferroptosis in mitochondria.
GPD2 Cooperates with GPX4 to Suppress Tumor Ferroptosis.
We further studied whether GPD2 and/or GPX4 inactivation affects tumor growth through induction of ferroptosis. Because the available GPD2 inhibitor (iGP-1) did not work in our experimental systems (SI Appendix, Fig. S1 U–Z) and because no existing GPX4 inhibitor is suitable for animal studies, we took a genetic approach to study this question. We injected control, GPX4-KO, GPD2-KO, or DKO HCT116 cells (which were cultured in ferrostatin-1–containing medium) into nude mice and monitored their tumor growth. We found that GPX4 deficiency, but not GPD2 deficiency, suppressed the growth of HCT116 xenograft tumors, whereas DKO tumors exhibited even more pronounced growth reduction than did GPX4-KO tumors; importantly, treatment with the ferroptosis inhibitor liproxstatin-1 almost completely restored the growth of DKO tumors to that of control tumors (Fig. 4M and SI Appendix, Fig. S5E). Similar observations were made in end-point tumor weight analyses (Fig. 4N). We found no differences in animal weights in these studies (SI Appendix, Fig. S5F). Further analyses revealed that, although cleaved caspase-3 and Ki67 staining did not differ significantly among the different genotypes or treatments (SI Appendix, Fig. S5 G–I), lipid peroxidation marker 4-HNE exhibited increased staining in GPX4-KO tumors and even higher staining in DKO tumors compared with control and GPD2-KO tumors, and treatment with liproxstatin-1 completely normalized the level of 4-HNE staining to that in control tumors (Fig. 4O and SI Appendix, Fig. S5G). Finally, we showed that, in the DKO background, restoration of mitochondrial GPX4 partially restored tumor growth, and restoration of both cytosolic and mitochondrial GPX4 completely restored tumor growth to a level similar to that in control tumors, whereas restoration of cytosolic GPX4 or GPD2 had no effect on tumor growth (Fig. 4 P and Q and SI Appendix, Fig. S5J). These in vivo observations mirrored in vitro cell death data in corresponding cell lines (Fig. 4J). These data demonstrated that in HCT116 xenograft models, GPD2 deletion synergizes with GPX4 deletion to suppress tumor growth through inducing ferroptosis in vivo.
Discussion
Mitochondria are multifunctional organelles that are surrounded by two phospholipid bilayer membranes and have vital roles in energy production and cellular metabolism (31). However, enormous bioenergetic, biosynthetic, and redox activities within mitochondria render mitochondrial membranes particularly susceptible to lipid peroxidation. For example, mitochondria are major organelles that produce reactive oxygen species, which can promote lipid peroxidation through Fenton reactions (5, 32). Mitochondria also generate ATP, which has been demonstrated to promote lipid peroxidation and ferroptosis by inactivation of energy sensor AMP-activated protein kinase (AMPK) (33, 34). Other metabolic activities within mitochondria, such as glutaminolysis and the citric acid cycle, also drive ferroptosis (1, 35, 36). To combat these adverse effects of diverse metabolic activities in mitochondria, cells have evolved to equip mitochondria with antioxidant systems to detoxify lipid peroxides that accumulate in mitochondrial membranes and to defend against ferroptosis. The study described herein, together with our recent study (15), identified at least three such surveillance mechanisms in mitochondria: mitochondrial GPX4, DHODH, and GPD2. Our study further demonstrated that GPD2 suppresses mitochondrial lipid peroxidation by generation of CoQH2 in the inner mitochondrial membrane (Fig. 4R).
Importantly, GPX4 restoration experiments demonstrated that cytosolic GPX4 exerts rescuing effects on GPX4-KO cells but not GPX4/GPD2 DKO cells, whereas mitochondrial GPX4 does not have any rescuing effect on GPX4-KO cells but can substantially delay ferroptosis onset in GPX4/GPD2 DKO cells (Fig. 4 G–L). Our data suggest a model that GPX4/GPD2 DKO cells die of ferroptosis mainly triggered by mitochondrial lipid peroxidation; how ferroptosis in this context can bypass the surveillance of cytosolic GPX4 remains unknown and will be an interesting topic for future investigations. Mitochondrial GPX4 restoration suppresses ferroptosis induced by mitochondrial lipid peroxidation in GPX4/GPD2 DKO cells; however, these cells (much like GPX4-KO cells) eventually undergo ferroptosis triggered by nonmitochondrial lipid peroxidation.
Together, we propose that mitochondrial GPX4, DHODH, and GPD2 operate in parallel to suppress mitochondrial lipid peroxidation within mitochondria. As long as cells maintain high expression of GPX4, deletion of DHODH or GPD2 alone typically is not sufficient to induce mitochondrial lipid peroxidation and ferroptosis. On the other hand, deletion of mitochondrial GPX4 alone (as occurred in cytosolic GPX4-restored GPX4-KO cells) is not sufficient to induce mitochondrial lipid peroxidation and ferroptosis either. It appears that inactivation of both arms (mitochondrial GPX4 and DHODH/GPD2-mediated CoQH2 generation) is required to trigger potent mitochondrial lipid peroxidation and ferroptosis.
In this study, we also showed that GPD2 overexpression is capable of suppressing ferroptosis in DHODH KO (but not FSP1 KO) cells, whereas DHODH (but not FSP1) overexpression could suppress ferroptosis in GPD2 KO cells. Together with results of our recent study (15), these findings suggest that cytosolic GPX4/FSP1 and mitochondrial GPX4/DHODH/GPD2 operate in two separate systems to repair lipid peroxides and defend against ferroptosis, highlighting the importance of compartmentalization in ferroptosis defense. We reason that the compartmentalization of cytosolic versus mitochondrial ferroptosis defense systems probably relates to the double-membrane structure of mitochondria. Cytosolic GPX4 or FSP1 (which mainly localizes on the plasma membrane) cannot enter into mitochondria to repair lipid peroxides that accumulate in the inner mitochondrial membrane, forcing cells to build up separate ferroptosis surveillance systems in mitochondria. Potential ferroptosis defense systems localized within other organelles remain to be identified.
Both DHODH and GPD2 are inner mitochondrial membrane-bound enzymes that reduce CoQ to CoQH2, which is capable of quenching lipid peroxyl radicals in mitochondria as a radical-trapping antioxidant. We propose that, in response to acute inactivation of GPX4 (which forms the major cellular defense arm against ferroptosis), much of the CoQH2 pool is oxidized to CoQ via quenching of mitochondrial lipid peroxyl radicals, resulting in an increased CoQ/CoQH2 ratio, which further drives DHODH and GPD2 reactions to generate more CoQH2 for ferroptosis defense in mitochondria. This explains why C-Asp and G3P, the upstream metabolites of DHODH and GPD2 reactions, respectively, were acutely depleted in cancer cells treated with RSL3 (Fig. 1 A–D).
DHODH is a key enzyme involved in pyrimidine biosynthesis, which is required for rapid proliferation of cancer cells. Consistently, in most of the cancer cell lines we examined, DHODH deletion or treatment with the DHODH inhibitor brequinar significantly sensitized cells to RSL3-induced ferroptosis (15), suggesting that DHODH has a general role in suppressing ferroptosis in cancer cells. GPD2, on the other hand, is involved in the G3P shuttle to transfer NADH from the cytosol to mitochondria for oxidative phosphorylation. However, cells can also employ other shuttle systems, such as the malate–aspartate shuttle, to transfer NADH from the cytosol into mitochondria. Considering potential redundancies of the G3P shuttle and other NADH shuttle systems, we speculate that GPD2 has a more restricted role than DHODH does in regulating ferroptosis in cancer cells. In support of this, we found that whereas DHODH deletion in GPX4low cancer cells is sufficient to trigger ferroptosis (15), GPD2 deficiency does not appear to impair the basal viability of GPX4low RPMI 7951 cells (but significantly sensitizes such cells to RSL3-induced ferroptosis; see Fig. 1L and SI Appendix, Fig. S1R). Further studies are needed to determine the relative importance of DHODH and GPD2 in generating CoQH2 for ferroptosis defense in different cellular contexts. In addition, several other mitochondrial enzymes, including complexes I and II in the electron transport chain, also produce CoQH2. Their exact roles in ferroptosis regulation remain to be defined.
While our data strongly suggest GPD2 suppresses ferroptosis through producing mitochondrial CoQH2, we cannot rule out the possibility of other potential mechanisms. For example, recent studies have linked NAD+ recycling to PUFA desaturation (37), which is known to affect ferroptosis sensitivity (17). Therefore, it will be interesting to determine whether GPD2 regulates PUFA desaturation and ferroptosis through affecting cytosolic NAD+/NADH ratio in future studies.
Finally, from a therapeutic perspective, our study identified GPD2 as a potential therapeutic target in cancer cells and suggests that combining GPD2 inhibitors with other ferroptosis inducers can be explored in cancer therapy through induction of tumoral ferroptosis. Although a lack of suitable GPD2 inhibitors prevented us from formally testing this idea in the present study, we hope that our study inspires other investigators to develop potent, effective GPD2 inhibitors and to further test the impact of such inhibitors in cancer treatment.
Materials and Methods
Cell Culture Studies.
All cell lines used in this study were purchased from the American Type Culture Collection and were free of mycoplasma contamination (tested by the vendor). No cell line used in this study has been found in the International Cell Line Authentication Committee database of commonly misidentified cell lines, based on short tandem-repeat profiling performed by the vendor. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (vol/vol) fetal bovine serum (FBS) and 1% (vol/vol) penicillin/streptomycin in a humidified atmosphere of 5% CO2 at 37 °C as described previously (38, 39). All cell lines were seeded into a 12-well plate for cell death and lipid peroxidation measurements. Cells were treated with ferroptosis inducers, including RSL3 (Selleck Chemicals), ML210 (Cayman Chemical), ML162 (Cayman Chemical), and FIN56 (Cayman Chemical); ferroptosis inhibitors, including liproxstatin-1 (Cayman Chemical) and ferrostatin-1 (Sigma-Aldrich); deferoxamine (Sigma-Aldrich); glycerol (Sigma-Aldrich); sn-G3P (Cayman Chemical); the GPD2 inhibitor iGP-1 - Calbiochem (Sigma-Aldrich); and antioxidants, including 2,2,6,6-tetramethylpiperidinyl-1-oxy (TEMPO; Sigma-Aldrich), mitochondria-targeted TEMPO (MitoTEMPO; Sigma-Aldrich), MitoQ (Cayman Chemical), and MitoQH2 (Cayman Chemical).
Plasmid Constructs.
GPX4, DHODH, and FSP1 expression plasmids were described in our recent publication (15). GPD2 complementary DNA (cDNA) was purchased from the Functional Genomics Core of The University of Texas MD Anderson Cancer Center and subsequently cloned into the lentiviral vector pLVX-Puro as described previously (40, 41). All constructs were confirmed via DNA sequencing.
CRISPR-Cas9–Mediated Gene KO.
Single-guide RNAs (sgRNAs) and a CRISPR-Cas9 expression system were used to generate GPD2-, GPX4-, and COQ2-KO cell lines as described previously (33, 42). sgRNAs were cloned into a lentiviral lentiGuide vector. The sequences of the sgRNAs used in this study are listed in SI Appendix, Table S1. LentiGuide clones were transfected into HEK293T cells with a psPAX2 packaging plasmid and pMD2.G-expressing plasmid. Lentiviruses were collected and used to infect cells with 0.8 μg/mL Polybrene for 24 h. Cells were selected with puromycin (1 μg/mL; InvivoGen), blasticidin (2 μg/mL; InvivoGen), or hygromycin B (2 μg/mL; InvivoGen) for 3 d, and single cells were sorted into 96-well plates. Single cells were cultured in DMEM with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin in 5% CO2 at 37 °C in an incubator for 3 to 4 wk, and Western blotting was used to verify the target gene deletion efficiency of each clone.
Generation of Cell Lines Overexpressing Target Genes.
Cell lines with stable overexpression of target genes were generated as described previously (43, 44). In brief, HEK293T cells were transfected with either a target gene construct or pLVX-empty vector together with a psPAX.2 and pMD2.G third-generation lentiviral packaging system and 0.8 μg/mL Polybrene. Seventy-two hours later, media with lentiviral particles were collected and filtered to infect the target cell lines. Forty-eight hours afterward, puromycin was added to select stable cell lines with successful expression of target genes.
Metabolomic Analysis.
Metabolomic analysis was performed as described previously (45, 46). Cells were seeded in 35-mm culture plates. When the cell density reached 70 to 80%, cells were treated with RSL3, ML210, or ML162 for 2 h. Metabolites were then extracted from cells by rapidly aspirating the culture medium and incubating the plates with 0.6 mL of an 80% methanol:20% water mixture on a cold block for 15 min. Next, the cell material was scraped into Eppendorf tubes prechilled on ice. After centrifugation at 13,000 rcf for 5 min at 4 °C, the supernatant was collected into a fresh tube and stored on dry ice until analysis. Just before analysis, 500 μL of extract was dried under a nitrogen gas flow and then resuspended in 100 μL of water.
The complete metabolomic platform consisted of an Accela 1250 high-performance liquid chromatography system, Accela Open Autosampler, MayLab MistraSwitch column oven, and Exactive Orbitrap mass spectrometer controlled by the Xcalibur 3.0.63 software package. Chromatography was performed with a Phenomenex Synergi Hydro-Rp column (100 × 2 mm, 2.5-μm particle size). Solvent A contained 10 mM tributylamine and 15 mm acetic acid in water, and solvent B was methanol. The gradient was 0 min, 0% B; 2.5 min, 0% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 0% B; and 25 min, 0% B. The injection volume was 10 μL. The column temperature was set to 40 °C, and the flow speed was 200 μL/min. The Exactive was operated in negative ionization mode with an electrospray ionization interface. The instrument parameters were as follows: sheath gas flow rate, 30 (arbitrary units); sweep gas flow rate, 3 (arbitrary units); aux gas flow rate, 10 (arbitrary units); capillary temperature, 325 °C; spray voltage, 3 kV; tube lens voltage, −50 V; and capillary voltage, −25 V. The scan range was 80 to 1,000 m/z, with a maximum injection time of 250 ms, resolution of 100,000 at 1 Hz, and AGC (automatic gain control) target 1 × 106. The data were analyzed using the Maven software suite, with signal intensity determined as the peak area (top). The metabolomic data have been deposited into Metabolomics Workbench (Study ID: ST002115). The annotation of detected common metabolites is listed in SI Appendix, Table S2.
Cell Viability Assay.
Viable cells were measured using a crystal violet assay. Ten thousand cells per well were seeded in a 96-well plate; 50 µL of 0.5% crystal violet staining solution was added to each well after aspirating the medium and incubating for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute. After the plate was washed with phosphate-buffered saline (PBS) and air-dried at room temperature overnight. Two hundred microliters of methanol was added to each well and incubated for 10 min at room temperature. The optical density of each well was measured at 570 nm (OD570) with a plate reader. The percentage of viable cells was determined by comparing the OD570 values.
Cell Death Assay.
Cell death was measured using a flow cytometer after propidium iodide (PI) (Roche) staining as described previously (47, 48). For the staining, cells were seeded at a density of 50% confluence into 12-well plates. The next day, cells were treated with different reagents. To measure cell death, the cells (including floating dead cells) were harvested and stained with 5 μg/mL PI, and the percentage of PI-positive dead cells was determined using a BD Accuri C6 flow cytometer (BD Biosciences) and an FL2 detector. At least 10,000 single cells per well were analyzed, and all experiments were carried out at least three times.
Lipid Peroxidation Measurement.
Total or mitochondrial lipid peroxidation was measured as described previously (15, 49, 50). Briefly, cells were seeded at a density of 50% confluence on 12-well plates and incubated overnight. Cells were then treated with the indicated compounds for the indicated times, collected via trypsinization, and resuspended in 200 μL PBS with 5 μM C11-BODIPY 581/591 (Invitrogen) to measure lipid peroxidation or 5 μM MitoPerOx (Abcam) to measure mitochondrial lipid peroxidation. Next, cells were incubated for 30 min at 37 °C. Lipid peroxidation was assessed using the flow cytometer BD Accuri C6 with a 488-nm laser and an FL1 detector (in which at least 10,000 single cells per well were analyzed) or was analyzed by confocal imaging by an inverted Zeiss LSM880 laser scanning confocal microscope with a 63× oil immersion objective lens, 488- and 543-nm laser line.
GPD2 Enzyme Activity Assay.
GPD2 activity was measured spectrophotometrically at 610 nm. The assay medium included 10 mM Tris⋅HCl, 50 mM KCl, 1 mg/mL bovine serum albumin, 1 mM KCN, and 1 mM ethylenediaminetetraacetic acid, pH 7.4. The reaction was started by adding 100 μM DCPIP and 25 mM G3P at 30 °C. Absorbance at 610 nm was monitored after 1-min incubation. Enzyme activity was shown as the ratio of absorbance.
NAD+/NADH Ratio Measurement.
NAD+/NADH ratios were determined by using the NAD+/NADH Quantitation Colorimetric Kit (BioVision, K337). Briefly, cells were seeded at a density of 50% confluence on 24-well plates and incubated overnight, washed with PBS, and spun at 2,000 rpm for 5 min and the supernatant was discarded. Cells were extracted with 400 µL of extraction buffer by two freeze/thaw cycles and centrifuged for 5 min at 15,000 rpm in a cold microcentrifuge to remove insoluble material. Supernatants were divided for measurement of NADH+NAD+ and NADH (after NAD+ decomposition at 60 °C for 30 min), respectively.
Basal Oxygen Consumption Rate Measurement.
Basal oxygen consumption rate levels were determined by using the Oxygen Consumption Rate Assay Kit (Cayman Chemical, 600800). Briefly, 40,000 cells per well were seeded in a black, clear-bottom 96-well tissue culture plate. Twenty microliters of culture medium, 10 µL glucose oxidase stock solution, 10 µL antimycin A stock solution, and 10 µL phosphorescent oxygen probe solution were added to each well. The plate was read immediately (380 nm for excitation and 650 nm for emission).
CoQ and CoQH2 Analysis.
Intracellular CoQ and CoQH2 levels were measured as we described previously (15). Cells growing on 35-mm plates to about 70% confluence were quickly washed with 1 mL of room-temperature, serum-free culture medium to remove serum-derived CoQ and CoQH2 with minimal metabolic perturbation to the cells. Next, 600 μL of freshly prepared ice-cold isopropanol containing 100 μM tert-butyl-hydroquinone was added to the plates, and plates were then placed on ice. The cells were then scraped with cell lifters and transferred to Eppendorf tubes on ice. After brief vortexing, the tubes were clarified using centrifugation at 13,000 rcf at 4 °C for 5 min, and the supernatants was transferred to fresh tubes. These extracts were stored in the dark on dry ice until analysis by liquid chromatography with mass spectrometry. The relative levels of CoQ and CoQH2 under these conditions were stable for at least 48 h.
CoQ and CoQH2 levels were determined via liquid chromatography with mass spectrometry using an analytical method derived from a previously described lipidomic method (51). The extracts were transferred to amber glass sample vials in an autosampler maintained at 4 °C. Separation was performed using a Thermo Fisher Scientific UltiMate 3000 HPLC system with an Acquity UPLC HSS T3 column (2.1 × 100 mm, 1.8-μm particle size). Mobile phase A consisted of acetonitrile:water (60:40, vol/vol) with 10 mM ammonium acetate and 0.1% acetic acid, whereas mobile phase B consisted of isopropanol:acetonitrile:water (85:10:5, vol/vol/vol) with 10 mM ammonium acetate and 0.1% acetic acid. The gradient was 0 min, 40% B; 1.5 min, 40% B; 12 min, 100% B; 15 min, 100% B; 16 min, 40% B; and 17 min, 40% B. The injection volume was 10 μL. The column temperature was set to 55 °C, and the flow rate was 400 μL/min.
Samples were analyzed using the Exactive Orbitrap mass spectrometer in positive ionization mode with a heated electrospray ion source. The instrument parameters were as follows: sheath gas flow rate 30 (arbitrary units), aux gas flow rate 10 (arbitrary units), sweep gas flow rate 3 (arbitrary units), spray voltage 4 kV, capillary temperature 120 °C, heater temperature 500 °C, capillary voltage 65 V, tube lens voltage 100 V. The scan range was set to 200 to 1,000 m/z, with a maximum inject time of 100 ms, resolution of 100,000 at 1 Hz, and AGC (automatic gain control) target 1 × 106. The data were analyzed using the Maven software suite 57 with signal intensity determined as the peak area (top). Both CoQ and CoQH2 were detected as their ammonium adducts ([M+NH4]+).
Immunoblotting.
Western blotting was used to analyze protein expression in cells as described previously (52, 53). In brief, cell pellets were lysed using IP lysis buffer (Thermo Fisher Scientific), and the protein concentrations were measured using a bicinchoninic acid protein assay (Thermo Fisher Scientific) with a FLUOstar Omega microplate reader (BMG LABTECH). Mitochondrial protein was extracted by using a mitochondrial fractionation kit (K256; BioVision). Thirty micrograms of protein was used for immunoblot analysis with antibodies against GPD1 (1:1,000,13451-1-AP; Proteintech), GPD1L (1:1,000, 17263-1-AP; Proteintech), GPD2 (1:1,000, 17219-1-AP; Proteintech), GPX4 (1:1,000, MAB5457; R&D Systems), DHODH (1:1,000, 14877-1-AP; Proteintech), SLC7A11 (1:1,000, 12691; Cell Signaling Technology), ACSL4 (1:1,000, sc-271800; Santa Cruz Biotechnology), COQ2 (1:1,000, sc-517107; Santa Cruz Biotechnology), FSP1 (1:1,000, sc-377120; Santa Cruz Biotechnology), and vinculin (1:3,000, V4505; Sigma-Aldrich).
Histology and Immunohistochemistry.
Fresh tumor samples were fixed in 10% neutral buffered formalin for 16 h, washed with PBS, and stored in 70% ethanol at 4 °C. The samples were dehydrated and embedded in paraffin at the MD Anderson Research Histology Core Laboratory according to standard protocols. Embedded tissues were sliced at a thickness of 5 μm for hematoxylin and eosin or immunohistochemical (IHC) staining. IHC staining was performed as described previously (54, 55). Briefly, primary antibodies, including those against cleaved caspase-3 (1:500, 9661s; Cell Signaling Technology), Ki-67 (1:500, 9027s; Cell Signaling Technology), and 4-HNE (1:400, ab46545; Abcam), were incubated overnight at 4 °C. Staining was performed using a VECTASTAIN Elite ABC Kit and DAB peroxidase substrate kit (Vector Laboratories). Images were randomly taken (five images per tumor) at 200× magnification using an Olympus BX43 microscope.
Cell-Line–Derived Xenograft Model.
Xenograft experiments were performed as described previously (56) and in accordance with a protocol approved by the MD Anderson Institutional Animal Care and Use Committee and Institutional Review Board. The xenograft study was compliant with all relevant ethical regulations regarding animal research. Rodent housing conditions used in this study were a temperature set point of 72 °F (high limit, 74 °F; low limit, 70 °F), humidity set point of 45% (high limit, 55%; low limit, 40%), and light cycle of 12 h light–dark. Animals were killed when their xenograft tumor volumes reached 1,500 mm3. No mice exhibited severe loss of body weight (>15%) or evidence of infections or wounds. Female 4- to 6-wk-old athymic nude mice (Foxn1nu/Foxn1nu) were purchased from the Experimental Radiation Oncology Breeding Core Facility at MD Anderson and housed in the animal care facility at the MD Anderson Department of Veterinary Medicine and Surgery. To generate cell-line–derived xenograft models, cancer cell lines were suspended and counted in cold PBS, and 3 × 106 cells were injected into the mice subcutaneously. Liproxstatin-1 diluted in PBS was intraperitoneally injected daily at a dose of 10 mg/kg. The daily injections were continued until the endpoint as indicated in the corresponding figures. Tumor length and width were measured every 3 d until the endpoint and tumor volumes were calculated according to the equation volume = length × width2 × 1/2.
Statistics and Reproducibility.
The results of cell culture experiments were collected from at least three independent replicates. Volumes or weights from at least 10 tumors in each group of mice were plotted. Data were presented as means ± SD. Statistical significance (P values) was calculated using an unpaired Student’s t test or a log-rank test by GraphPad Prism software (version 8.0; GraphPad Software) or SPSS software (version 25.0; IBM). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. No statistical methods were used to predetermine sample size. The investigators were not blinded to allocation during the experiments or outcome assessment.
Supplementary Material
Acknowledgments
We thank Donald Norwood from the Research Medical Library at MD Anderson for editing the manuscript. This research was supported by Institutional Research Fund and Bridge Fund from The University of Texas MD Anderson Cancer Center, Emerson Collective Cancer Research Fund, Cancer Prevention & Research Institute of Texas Grant RP220258, and R01CA181196, R01CA244144, and R01CA247992 from the National Institutes of Health (to B.G.). B.G. was an Andrew Sabin Family Fellow. This research has also been supported by the National Institutes of Health Cancer Center Support Grant P30CA016672 to The University of Texas MD Anderson Cancer Center.
Footnotes
Competing interest statement: K.O. is a full-time employee of Barer Institute and a former full-time employee of Kadmon Corporation. L.K. and M.V.P. are full-time employees of Kadmon Corporation. B.G. is an inventor on patent applications involving targeting ferroptosis in cancer therapy.
This article is a PNAS Direct Submission. B.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121987119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Dixon S. J., et al. , Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stockwell B. R., et al. , Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang X., Stockwell B. R., Conrad M., Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang D., Chen X., Kang R., Kroemer G., Ferroptosis: Molecular mechanisms and health implications. Cell Res. 31, 107–125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng J., Conrad M., The metabolic underpinnings of ferroptosis. Cell Metab. 32, 920–937 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Conrad M., Pratt D. A., The chemical basis of ferroptosis. Nat. Chem. Biol. 15, 1137–1147 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Friedmann Angeli J. P., et al. , Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W. S., et al. , Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bersuker K., et al. , The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doll S., et al. , FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Kraft V. A. N., et al. , GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 6, 41–53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soula M., et al. , Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat. Chem. Biol. 16, 1351–1360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pushpa-Rekha T. R., Burdsall A. L., Oleksa L. M., Chisolm G. M., Driscoll D. M., Rat phospholipid-hydroperoxide glutathione peroxidase. cDNA cloning and identification of multiple transcription and translation start sites. J. Biol. Chem. 270, 26993–26999 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Maiorino M., et al. , Distinct promoters determine alternative transcription of gpx-4 into phospholipid-hydroperoxide glutathione peroxidase variants. J. Biol. Chem. 278, 34286–34290 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Mao C., et al. , DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 593, 586–590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan B., Mitochondrial regulation of ferroptosis. J. Cell Biol. 220, 202105043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei G., Zhuang L., Gan B., Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 10.1038/s41568-022-00459-0. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mráček T., Drahota Z., Houštěk J., The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim. Biophys. Acta 1827, 401–410 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Basu A., et al. , An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koppula P., Zhang Y., Zhuang L., Gan B., Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. (Lond.) 38, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koppula P., Zhuang L., Gan B., Cytochrome P450 reductase (POR) as a ferroptosis fuel. Protein Cell 12, 675–679 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doll S., et al. , ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 13, 91–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr A. L., et al. , Novel inhibitors of mitochondrial sn-glycerol 3-phosphate dehydrogenase. PLoS One 9, e89938 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghandi M., et al. , Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium G. T.; GTEx Consortium, The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prime T. A., et al. , A ratiometric fluorescent probe for assessing mitochondrial phospholipid peroxidation within living cells. Free Radic. Biol. Med. 53, 544–553 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Pecinová A., et al. , Role of mitochondrial glycerol-3-phosphate dehydrogenase in metabolic adaptations of prostate cancer. Cells 9, 1764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trnka J., Blaikie F. H., Smith R. A., Murphy M. P., A mitochondria-targeted nitroxide is reduced to its hydroxylamine by ubiquinol in mitochondria. Free Radic. Biol. Med. 44, 1406–1419 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Shimada K., et al. , Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 12, 497–503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefely J. A., Pagliarini D. J., Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem. Sci. 42, 824–843 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman J. R., Nunnari J., Mitochondrial form and function. Nature 505, 335–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy M. P., How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H., et al. , Energy-stress-mediated AMPK activation inhibits ferroptosis. Nat. Cell Biol. 22, 225–234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C., et al. , LKB1-AMPK axis negatively regulates ferroptosis by inhibiting fatty acid synthesis. Signal Transduct. Target. Ther. 5, 187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X., Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell 59, 298–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao M., et al. , Role of mitochondria in ferroptosis. Mol. Cell 73, 354–363.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim W., et al. , Polyunsaturated fatty acid desaturation is a mechanism for glycolytic NAD+ recycling. Cell Metab. 29, 856–870.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X., et al. , LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat. Cell Biol. 18, 431–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X., Gan B., lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle 15, 3471–3481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., et al. , H2A monoubiquitination links glucose availability to epigenetic regulation of the endoplasmic reticulum stress response and cancer cell death. Cancer Res. 80, 2243–2256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koppula P., Zhang Y., Shi J., Li W., Gan B., The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 292, 14240–14249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei G., et al. , The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 30, 146–162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Koppula P., Gan B., Regulation of H2A ubiquitination and SLC7A11 expression by BAP1 and PRC1. Cell Cycle 18, 773–783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chauhan A. S., et al. , STIM2 interacts with AMPK and regulates calcium-induced AMPK activation. FASEB J. 33, 2957–2970 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X., et al. , Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 22, 476–486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koppula P., et al. , KEAP1 deficiency drives glucose dependency and sensitizes lung cancer cells and tumors to GLUT inhibition. iScience 24, 102649 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai F., et al. , BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc. Natl. Acad. Sci. U.S.A. 114, 3192–3197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H., et al. , BAF180 regulates cellular senescence and hematopoietic stem cell homeostasis through p21. Oncotarget 7, 19134–19146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y., et al. , BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 20, 1181–1192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., et al. , mTORC1 couples cyst(e)ine availability with GPX4 protein synthesis and ferroptosis regulation. Nat. Commun. 12, 1589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., et al. , Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 26, 623–633 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin A., et al. , FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacologic inhibition of the PI3K-AKT pathway. Cancer Res. 74, 1682–1693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin A., et al. , The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene 33, 3183–3194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lei G., et al. , Ferroptosis as a mechanism to mediate p53 function in tumor radiosensitivity. Oncogene 40, 3533–3547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gan B., et al. , FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18, 472–484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao Z. D., et al. , Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 8, 783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.