Significance
Endogenous retroviruses (ERVs) are remnants of ancestral viral infections of cells and occupy substantial proportions of mammalian genomes. Most ERVs have numerous mutations, deletions, and insertions in the coding region; truncated envelope (env) genes are scattered throughout the genomes. The truncated Envelope (Env) proteins consist of a signal peptide and the N-terminal region of the surface unit, but they lack a transmembrane domain; these proteins are secreted from cells in soluble form and block viral infection via competition with entry receptors. This defense system could function to prevent both intraspecies and interspecies transmission of viral infections. Truncated Envs have been independently acquired by infection with ancient viruses, and they could prevent the spread of infectious diseases.
Keywords: antiviral, retrovirus, receptor, restriction factor, evolution
Abstract
Host genetic resistance to viral infection controls the pathogenicity and epidemic dynamics of infectious diseases. Refrex-1 is a restriction factor against feline leukemia virus subgroup D (FeLV-D) and an endogenous retrovirus (ERV) in domestic cats (ERV-DC). Refrex-1 is encoded by a subset of ERV-DC loci with truncated envelope genes and secreted from cells as a soluble protein. Here, we identified the copper transporter CTR1 as the entry receptor for FeLV-D and genotype I ERV-DCs. We also identified CTR1 as a receptor for primate ERVs from crab-eating macaques and rhesus macaques, which were found in a search of intact envelope genes capable of forming infectious viruses. Refrex-1 counteracted infection by FeLV-D and ERV-DCs via competition for the entry receptor CTR1; the antiviral effects extended to primate ERVs with CTR1-dependent entry. Furthermore, truncated ERV envelope genes found in chimpanzee, bonobo, gorilla, crab-eating macaque, and rhesus macaque genomes could also block infection by feline and primate retroviruses. Genetic analyses showed that these ERV envelope genes were acquired in a species- or genus-specific manner during host evolution. These results indicated that soluble envelope proteins could suppress retroviral infection across species boundaries, suggesting that they function to control retroviral spread. Our findings revealed that several mammalian species acquired antiviral machinery from various ancient retroviruses, leading to convergent evolution for host defense.
Endogenous retroviruses (ERVs) are the germline remnants of ancestral infections by exogenous retroviruses and are present in all vertebrate genomes (1). ERVs comprise a substantial proportion of the mammalian genome and are transmitted in a Mendelian fashion (2). When retroviruses are inserted into a host genome, they achieve increased copy numbers by repeated reinfection in germline cells (3). ERVs are gradually attenuated or inactivated by mutations that occur during viral replication, as well as by postinsertional mutations that occur during host genome replication. Nucleotide changes in viral genes disrupt or modify the functions of the encoded proteins (4–6). Hosts often control the gene expression of ERVs via epigenetic mechanisms, such as nucleotide CpG methylation or suppressive histone modification, to silence the ERVs (7). It is assumed that such events ensure that the ERVs do not become a threat to the host. Some ERVs are domesticated by their hosts and eventually gain physiological functions [e.g., in processes such as placentation (8), sex-specific muscle development (9), and viral resistance (10–13)], despite the loss of their ancestral viral properties. Domesticated ERVs are strictly regulated, and dysregulation can negatively impact their hosts (14).
Feline leukemia virus (FeLV) belongs to the genus Gammaretrovirus and is horizontally transmitted among cats. FeLV has been linked to various diseases, including lymphoma, myelodysplastic syndrome, anemia, and acute myelogenous leukemia, as well as immune suppression (15). Genetic changes resulting from substitutions, recombination, or both have generated viral subgroups defined on the basis of their capacities for viral interference. In particular, changes in FeLV pathogenicity and the occurrence of unexpected symptoms are associated with alterations of viral target cells (16).
ERV-DCs are endogenous gammaretroviruses of domestic cats (DCs) (17–19); they are classified as genotypes I, II, and III according to their phylogenetic relationships (SI Appendix, Fig. S1) and receptor interference groups. ERV-DCs were inserted into feline genomes several times within the past few million years. Some ERV-DCs have intact open reading frames (ORFs) that encode gag, pol, and envelope (env) genes; therefore, they are replication-competent in cultured cells and capable of infecting a broad range of cells, including human ones. Furthermore, FeLV-D was generated by recombination of genotype I ERV-DCs in the env region (17) and shows distinct tropism, compared with other FeLV subgroups. Importantly, ERV-DCs currently retain the ability to impact their hosts via their potential viral activity and their contribution to the emergence of recombinant viruses. Furthermore, it has been reported that an ERV-DC recombined with Baboon endogenous retrovirus (BaEV) and generated a distinct feline retrovirus, RD-114 (17–19). This past recombination event suggested interspecies retroviral transmission between cats and primates (17–19). Therefore, the interactions between exogenous retroviruses and ERVs have contributed to long-term retroviral diversification and evolution.
Refrex-1 is a feline soluble restriction factor against genotype I ERV-DCs and FeLV-D infection; it is encoded by truncated env genes of ERV-DC7 and ERV-DC16 loci, which are classified as genotype II ERV-DCs (20). Thus, Refrex-1 was presumably acquired through ERV domestication as part of an evolutionary arms race between cats and ERV-DCs. Indeed, resistance to viral infection associated with ERV has been discovered in chicken (21) and in mice (22–24), as an evolutionary arms race between hosts and viruses. Refrex-1 consists of a signal peptide and a surface unit (SU) N-terminal domain, also known as the receptor-binding domain (RBD); however, it lacks a transmembrane (TM) domain because of a premature stop codon and lethal mutations in the middle of the env ORF. Refrex-1 is efficiently secreted from cells and specifically blocks the infection of genotype I ERV-DCs and FeLV-D, suggesting that its restriction activity is mediated by a mechanism of receptor interference.
In this study, copper transport protein 1 (CTR1) was identified as the entry receptor for infection by FeLV-D and genotype I ERV-DCs. Functional Env proteins from novel primate ERVs were found through a mammalian genome database search, and receptor interference assays revealed that these primate ERVs also used CTR1 as the entry receptor for infection. The restriction capacity of Refrex-1 extended beyond feline retroviruses to primate ERVs, based on the involvement of CTR1. Furthermore, this study showed that endogenous retrovirus truncated envelopes with Refrex-1–like structures are present in primates; they also have antiviral activity toward feline and primate retroviruses. Because genetic analyses showed that these truncated env genes were independently endogenized during host evolution, antiviral machinery derived from truncated envelope genes was inferred to have evolved in a convergent manner in multiple mammalian species. This study provides a mechanistic finding that soluble Env proteins broadly suppress retroviruses from different host species through binding interactions with a common entry receptor.
Results
Identification of an Entry Receptor for FeLV-D and Genotype I ERV-DCs.
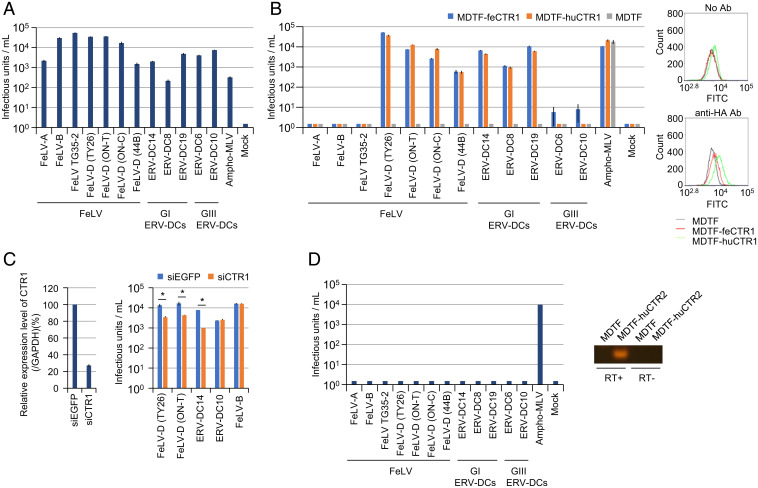
FeLV-D Env-pseudotyped virus could infect human cells (Fig. 1A). Therefore, we sought to screen an HEK293 cDNA library for genes encoding receptors used by FeLV-D for entry. Mus dunni tail fibroblast (MDTF) cells were resistant to FeLV-D infection (Fig. 1B), and cells transduced with the retroviral HEK293 cDNA library were challenged with FeLV-D (TY26)-enveloped virus carrying a puromycin-resistance gene. Fifty-one puromycin-resistant colonies were replated in culture plates. These cells were subsequently challenged with FeLV-D–enveloped viruses carrying a gene encoding green fluorescent protein (GFP), and 11 of 51 colonies were GFP+. Sequence analysis revealed that the cDNAs encoded human CTR1 (huCTR1, solute carrier 31A1 [SLC31A1]) in all GFP+ colonies.
Fig. 1.
CTR1 as the entry receptor for FeLV-D and Genotype I ERV-DCs. Viruses pseudotyped with the Envs shown in the x axis were tested in the following cell lines: (A) HEK293T cells, (B) MDTF cells, MDTF cells expressing human CTR1 (MDTF-huCTR1), and MDTF cells expressing feline CTR1 (MDTF-feCTR1) (Left). The expression of CTR1 was monitored by flow cytometry (Right). (C) Infection assay using HEK293T cells with endogenous CTR1 knockdown using siRNA. Expression of human CTR1 was analyzed by quantitative real-time RT-PCR and normalized by the expression level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Left). Infective titers of the indicated LacZ-coding Env-pseudotyped viruses in HEK293T cells with CTR1 siRNA (Right). Enhanced GFP (EGFP) siRNA was used as a negative control. Comparisons with siEGFP control were performed using Student’s t test (*P < 0.01). (D) Env-pseudotyped viruses shown in A were tested in MDTF cells expressing human CTR2 (MDTF-huCTR2) (Left). Expression of CTR2 was monitored by RT-PCR (Right). The infectious units were assessed via log10 β-galactosidase (LacZ)+ cells per milliliter of virus. Infective titers with SDs were means of three independent infection experiments. GI ERV-DCs, genotype I ERV-DCs; GIII ERV-DCs, genotype III ERV-DCs.
Next, to determine whether CTR1 might function as an entry receptor for FeLV-D, we obtained the coding regions of feline CTR1 (feCTR1) and huCTR1; both cDNAs were then introduced into MDTF cells, termed MDTF-feCTR1 and MDTF-huCTR1, respectively (Fig. 1B). MDTF-feCTR1 and MDTF-huCTR1 cells were permissive to FeLV-D Env-pseudotyped virus infection using four different FeLV-D Env proteins (TY26, ON-T, ON-C, and 44B) derived from leukemia and lymphoma in domestic cats (17) (Fig. 1B). Furthermore, the expression of feCTR1 or huCTR1 rendered MDTF cells permissive to infection by genotype I ERV-DC Env-pseudotyped viruses (loci ERV-DC8, DC14, and DC19) within the same interference group as FeLV-D (Fig. 1B). In contrast, other feline retroviruses (e.g., FeLV-A, FeLV-B, FeLV TG35-2, and genotype III ERV-DC [loci DC6 and DC10] Env-pseudotyped viruses) did not infect MDTF-feCTR1 or MDTF-huCTR1 cells (Fig. 1B). These results indicated that feCTR1 and huCTR1 conferred susceptibility to infection by FeLV-D and genotype I ERV-DC–pseudotyped viruses.
To further investigate whether the infection of HEK293T cells by FeLV-D and genotype I ERV-DCs was mediated by CTR1, we assessed the susceptibility of HEK293T cells transiently transfected with huCTR1-specific small-interfering RNA (siRNA). Silencing of endogenous CTR1 expression caused a 75% reduction in the infective titer of FeLV-D in HEK293T cells and an 88% reduction in the infective titer of ERV-DC14 (genotype I) in HEK293T cells, relative to HEK293T cells transfected with siEnhanced GFP (siEGFP) control (Fig. 1C). However, silencing of CTR1 expression did not affect the infective titers of ERV-DC10 and FeLV-B in HEK293T cells. These results provided further evidence that the infection of HEK293T cells by FeLV-D and genotype I ERV-DCs is CTR1-dependent.
Copper transporter 2 (CTR2) is a second member of the mammalian SLC31 copper transporter family and shares structural similarities with CTR1 (25). To determine the specificity of CTR1 as a viral entry receptor, MDTF cells expressing huCTR2 (MDTF-huCTR2) were generated. As shown in Fig. 1D, none of the viruses (except ampho-MLV [murine leukemia virus]) could infect MDTF-huCTR2 cells. These results indicated the specificity of CTR1 as the entry receptor for FeLV-D and genotype I ERV-DCs.
Expression of CTR1 in Feline and Primate Tissues.
FeCTR1, isolated based on the cat genome (Felis catus 9.0), was predicted to encode a protein of 180 amino acids (SI Appendix, Fig. S2). The amino acid sequence identity between feCTR1 and huCTR1 was 96.1%. The N-terminal region corresponding to the extracellular domain of CTR1 was most divergent. FeCTR1 mRNA expression was detected by qRT-PCR in total RNA extracted from various feline tissues and feline cell lines (SI Appendix, Fig. S3). Expression of feCTR1 was detected in all feline tissues and cell lines tested; expression levels were highest in the liver, small intestine, and lung.
Primate CTR1 expression levels were also investigated using public RNA-sequencing (RNA-seq) data collected from chimpanzee, crab-eating macaque, and rhesus macaque (26). Primate CTR1 expression was detected in all tissues tested; comparatively high expression levels were observed in kidney and liver (SI Appendix, Fig. S4A). We also detected CTR1 expression in testis and ovary from crab-eating macaque by qRT-PCR (SI Appendix, Fig. S4C).
Effect of Refrex-1 on CTR1.
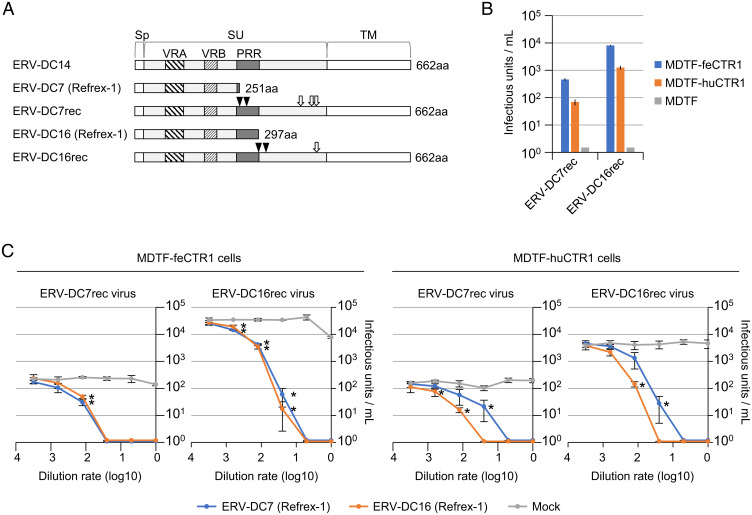
Refrex-1, encoded by the ERV-DC7 and ERV-DC16 envelope genes, is a soluble restriction factor that blocks infection by FeLV-D and genotype I ERV-DCs (20). The ERV-DC7 and ERV-DC16 env genes are defective, but their reconstructed sequences (ERV-DC7rec and ERV-DC16rec) have been restored to their infectious origin belonging to the same interference group as genotype I (27) (Fig. 2A). We used ERV-DC7rec and ERV-DC16rec to determine whether the ancestor of Refrex-1 infects cells via the CTR1 entry receptor. ERV-DC7rec and ERV-DC16rec Env-pseudotyped viruses infected MDTF-feCTR1 and MDTF-huCTR1 cells, but they did not infect MDTF cells (Fig. 2B). These results suggest that CTR1 was used as an entry receptor for infection by the ancient retrovirus from which Refrex-1 originated.
Fig. 2.
Refrex-1 inhibition of ancestral virus infection via CTR1. (A) Schematic representation of the structures of Refrex-1 and reconstructed ancestral Env proteins (ERV-DC7rec and ERV-DC16rec). Env genes of ERV-DC7rec and ERV-DC16rec were reconstructed by the removal of stop codons (triangles) and amino acid substitutions (arrows) based on ERV-DC7 and ERV-DC16 Env sequences, respectively. The amino acid substitutions were R407G, I427N, and T429A in ERV-DC7rec, and D431Y in ERV-DC16rec. PRR, proline-rich region; Sp, signal peptide. The number of amino acids (aa) is indicated on the right. (B) Infection of MDTF-feCTR1, MDTF-huCTR1, and MDTF cells by LacZ-coding Env-pseudotyped ERV-DC7rec and ERV-DC16rec viruses. The y axis indicates infectious units per milliliter of supernatant. (C) Dose-dependent inhibitory effect of Refrex-1 on viral infection. Supernatants of HEK293T cells transfected with expression vectors encoding ERV-DC7 Refrex-1 (blue), ERV-DC16 Refrex-1 (orange), or empty vector (Mock; gray) were diluted with medium; 250 µL of diluted supernatant was added to the culture when MDTF-feCTR1 or MDTF-huCTR1 cells were infected with ERV-DC7rec or ERV-DC16rec pseudotyped viruses. Infection titers with SDs are means of three independent infection experiments. Comparisons with Mock control were performed using Student’s t test (*P < 0.01).
Because Refrex-1 is secreted into the supernatant by feline cells and specifically suppressed FeLV-D and genotype I ERV-DCs, its antiviral mechanism was expected to involve receptor interference (20). Therefore, we investigated whether the antiviral effect of Refrex-1 depended on the viral entry receptor, CTR1. Refrex-1 was prepared from the supernatants of HEK293T cells that had been transfected with expression vectors encoding ERV-DC7 Env or ERV-DC16 Env. The supernatants were incubated with MDTF-feCTR1 and MDTF-huCTR1 cells; viral challenge experiments were then conducted. Refrex-1 blocked infection of MDTF-feCTR1 and MDTF-huCTR1 cells by ERV-DC7rec and ERV-DC16rec Env-pseudotyped viruses in a concentration-dependent manner (Fig. 2C). In contrast, the supernatants of HEK293T cells transfected with an empty vector could not block infection by these viruses. These results suggested that Refrex-1 restricts viral infection via competitive binding with the CTR1 entry receptor.
Identification of Functional Envs in Primates and FeLV-D Interference Subgroup.
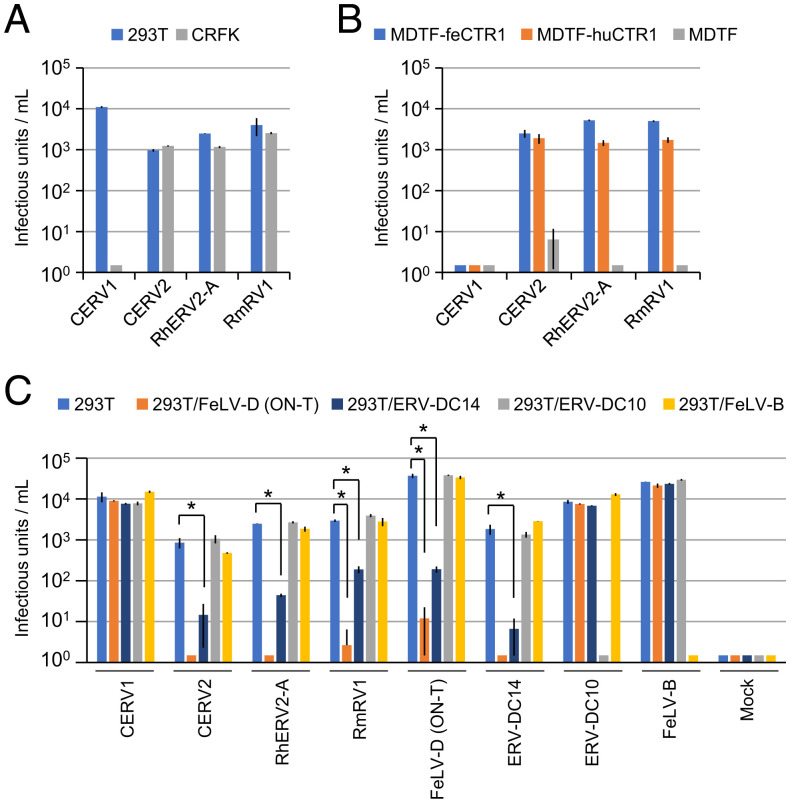
Human CTR1 is reportedly a receptor for extinct chimpanzee and rhesus macaque ERVs (i.e., CERV2 and RhERV2-A, respectively) (28). CERV1 was identified as a phylogenetically distinct virus from CERV2 and did not use CTR1 as an entry receptor. Although a previous study reported that these primate ERVs were extinct and described artificial reconstruction of their Envs (28), we sought to identify intact env genes related to CERV2 and RhERV2-A. Our genomic database search identified six env sequences encoding intact Env protein related to RhERV2-A in rhesus macaques and crab-eating macaques (i.e., RmRV1, and CmRV1, CmRV1_1, and CmRV1_2, respectively) (SI Appendix, Fig. S5 and Table S1), but the CERV-related ERV Env sequences were not found in the human genome. The amino acid sequences of Env proteins were identical in RmRV1 and CmRV1, but not in CmRV1_1 and CmRV1_2; as a result, three different amino acid sequences were revealed (SI Appendix, Fig. S5). The infectivity of RmRV1 (CmRV1), CmRV1_1, and CmRV1_2 Env-pseudotyped viruses with MLV Gag-Pol was found in human and feline cells (Fig. 3A and SI Appendix, Fig. S6A), and these pseudotyped viruses infected MDTF-feCTR1 and MDTF-huCTR1 cells, but not MDTF cells (Fig. 3B and SI Appendix, Fig. S6B). RmRV1 (CmRV1) Env-pseudotyped virus broadly infected cell lines from various mammalian species (SI Appendix, Table S2). These results indicated that CERV2- and RhERV2-A–related functional Env genes are present in primates and the pseudotyped viruses infect cells using feCTR1 and huCTR1 as entry receptors.
Fig. 3.
Infection of primate Env-pseudotyped viruses belonging to FeLV-D interference groups. Infection of LacZ-coding primate Env-pseudotyped viruses in (A) HEK293T and CRFK cells and (B) MDTF-feCTR1, MDTF-huCTR1, and MDTF cells. (C) Infection of HEK293T cells and HEK293T cells preinfected with FeLV-D (ON-T), ERV-DC14, ERV-DC10, or FeLV-B by Env-pseudotyped viruses indicated in the x axis. Comparisons with 293T control were performed using Student’s t test (*P < 0.01). The y axis indicates infectious units per milliliter of virus. Infective titers with SDs are means of three independent infection experiments.
Next, we performed viral interference assays using Env-pseudotyped viruses to determine whether these feline and primate retroviruses belong to the same interference group. RmRV1, CERV2, and RhERV2-A interfered with FeLV-D and a genotype I ERV-DC (ERV-DC14), but they did not interfere with a genotype III ERV-DC (ERV-DC10) (Fig. 3C). These results indicated that CERV2, RhERV2-A, RmRV1, FeLV-D, and genotype I ERV-DCs belong to the same viral interference group. In contrast, CERV1, ERV-DC10, and FeLV-B belong to distinct interference groups. Taken together, these experiments showed that intact ERV env sequences capable of generating infectious viral proteins, which belong to the FeLV-D interference group, are present in the genomes of crab-eating macaques and rhesus macaques.
Antiviral Effect of Refrex-1 against Primate ERV Infection.
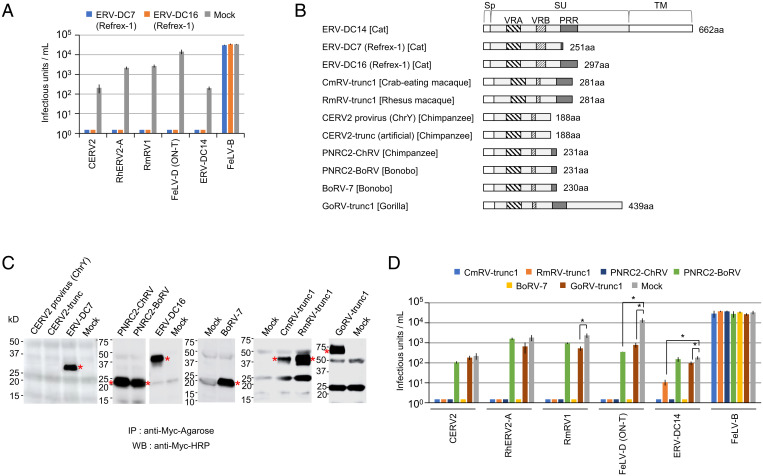
The common usage of CTR1 for infections by FeLV-D, feline ERVs, and primate ERVs implied that Refrex-1 could block these primate retroviral infections. To test this hypothesis, we challenged feline and primate ERV Env-pseudotyped viruses with HEK293T cells that had been treated with Refrex-1 (Fig. 4A). Refrex-1 corresponding to ERV-DC7 Env and ERV-DC16 Env, blocked infection by CERV2, RhERV2-A, RmRV1, FeLV-D, and ERV-DC14; it did not block infection by FeLV-B (Fig. 4A). These results suggested that Refrex-1 has broad inhibitory activity toward feline and primate retroviral infections.
Fig. 4.
Effects of Refrex-1 and primate soluble Env proteins on retroviral infection. (A) Supernatants were collected from HEK293T cells transfected with expression vectors encoding ERV-DC7 Refrex-1, ERV-DC16 Refrex-1, or empty vector (Mock); HEK293T cells preincubated with the supernatant were then infected with pseudotyped viruses indicated in the x axis. (B) Schematic representation of Env structures. Number of amino acids (aa) is indicated on the right side. (C) Immunoprecipitation (IP) and Western blotting (WB) with anti–c-Myc antibody of supernatants from HEK293T cells transfected with indicated plasmids. Asterisks indicate truncated Env proteins. (D) Supernatants were collected from HEK293T cells that had been transfected with expression vectors encoding primate truncated Env proteins or empty vector (Mock); HEK293T cells preincubated with the supernatant were then infected with Env-pseudotyped viruses indicated in the x axis. The bar colors correspond to the truncated Env protein contained in the supernatant. The infectious units were assessed via log10 LacZ+ cells per milliliter of virus. Infection titers with SDs are means of three infection experiments. Comparisons with Mock control were performed using Student’s t test (*P < 0.01).
Antiviral Effect of Truncated Envs in Primate Genomes against Retroviral Infection.
Next, we hypothesized that the antiviral mechanism derived from truncated env genes is also present in primates (i.e., it is not limited to Refrex-1 in cats). To test this hypothesis, we performed a sequence search for genes encoding only the SU domain of RmRV1-related Envs lacking TM regions. Consequently, we found seven truncated Envs (CmRV-trunc1, RmRV-trunc1, PNRC2-ChRV, CERV2 provirus [ChrY], PNRC2-BoRV, BoRV-7, and GoRV-trunc1) in crab-eating macaque, rhesus macaque, chimpanzee, bonobo, and gorilla genomes (Fig. 4B and SI Appendix, Fig. S7 and Table S1). The expression plasmids of primate truncated Envs were transfected into HEK293T cells. Following immunoprecipitation of the supernatants, Western blot analysis detected Myc-tagged Env proteins and Refrex-1. These results indicated that the primate truncated Env proteins except for CERV2 provirus (ChrY) were proteins secreted in a manner similar to Refrex-1 (Fig. 4C). The supernatants containing soluble Env proteins were incubated with HEK293T cells prior to viral challenge experiments to evaluate their inhibitory effects against viral infection. CmRV-trunc1, RmRV-trunc1, PNRC2-ChRV, and BoRV-7 specifically blocked infection by feline and primate retroviruses within the same interference group: FeLV-D, ERV-DC14, CERV2, RhERV2-A, and RmRV1 (Fig. 4D). Furthermore, GoRV-trunc1, which is larger than the other truncated Envs in terms of molecular weight, showed a reduced ability to block infection by RmRV1, FeLV-D, and ERV-DC14 (Fig. 4D); similar results were observed after viral infection of cells transfected with GoRV-trunc1 (SI Appendix, Fig. S8A).
Furthermore, PNRC2-ChRV inhibited FeLV-D, ERV-DC14, CERV2, RhERV2-A, and RmRV1 (Fig. 4D), while the PNRC2-ChRV ortholog, PNRC2-BoRV, inhibited only FeLV-D infection; PNRC2-BoRV did not block infection by CERV2, RhERV2-A, RmRV1, ERV-DC14, or FeLV-B, despite abundant expression of this Env in HEK293T cell supernatants (Fig. 4C). Only one amino acid at position 82 differed between PNRC2-ChRV (isoleucine) and PNRC2-BoRV (threonine) (SI Appendix, Fig. S7), indicating that this isoleucine in PNRC2-ChRV exerts an inhibitory effect on viral infection. There are seven amino acid differences between PNRC2-BoRV and BoRV-7 (isoleucine at position 82) (SI Appendix, Fig. S7); BoRV-7 Env blocked infection by FeLV-D, ERV-DC14, CERV2, RhERV2-A, and RmRV1 (Fig. 4D).
Notably, the truncated Env protein (188 amino acids in length) encoded by the CERV2 provirus (ChrY) was not secreted extracellularly (Fig. 4C), but expressed intracellularly (SI Appendix, Fig. S8B). Likewise, artificial CERV2-trunc (188 amino acids), which has a sequence identical to amino acid positions 1 to 188 of full-length CERV2 Env, was not secreted extracellularly (Fig. 4C), but expressed intracellularly (SI Appendix, Fig. S8B). There are five amino acid differences between CERV2 provirus (ChrY) Env and CERV2-trunc Env (SI Appendix, Fig. S7). The expression of CERV2 provirus (ChrY) Env and CERV2-trunc Env in 293T cells did not result in an inhibitory effect against viral infection (SI Appendix, Fig. S8B).
These results indicated that the antiviral mechanism derived from truncated env genes is also present in primates, and primate truncated Envs characterized as secreted proteins have inhibitory activity toward feline and primate retroviral infections. Truncated Env proteins with lengths of 230 to 439 amino acids are efficiently secreted from cells and exhibited antiviral activity. A subtle amino acid change in SU variable region A (VRA), which could be the RBD (SI Appendix, Fig. S7) (29), affected antiviral activity.
A previous study reported that Refrex-1 was expressed in various feline tissues, with greatest expression in the blood (30). To determine whether the truncated env genes are currently expressed in primate tissues, we analyzed the expression levels of primate env genes using publicly available RNA-seq data collected from chimpanzee, crab-eating macaque, and rhesus macaque (26). Notably, we identified abundant expression of truncated env genes (i.e., PNRC2-ChRV, CmRV-trunc1, and RmRV-trunc1) in various tissues (SI Appendix, Fig. S4B), including thymus and bone marrow; their expression levels were much higher than the expression levels of full-length env genes (i.e., CmRV1_1, CmRV1_2, and RmRV1) (SI Appendix, Fig. S9) in each species. We also detected the CmRV-trunc1 expression in testis and ovary from crab-eating macaque, as determined by qRT-PCR (SI Appendix, Fig. S4C). Expression of CERV2 provirus (ChrY) env gene was not clearly detected in tissues (SI Appendix, Fig. S4B). These results suggested that RNA expression of truncated env genes in reproductive tissues and immune-related tissues may contribute to prevent endogenous and exogenous retroviral infection and endogenization of ERVs. Additionally, the mapping pattern of RNA-seq reads to CmRV-trunc1, RmRV-trunc1, and PNRC2-ChRV suggested that the spliced transcripts encode Env proteins (SI Appendix, Fig. S9), although we do not know the protein levels in vivo. These results suggest that truncated env genes intrinsically function as antiviral factors in primates. Expression of truncated env genes, but not full-length env genes, may have evolutionarily important implications for the host.
Genetic Analyses of Feline and Primate ERVs.
The evolutionary relationships of feline and primate ERVs were investigated by genetic analyses. Phylogenetic trees constructed using gag and pol genes indicated that ERV-DC and CERV-related viruses were located in the same clade (SI Appendix, Fig. S10 A and B). However, env genes of ERV-DC–related viruses and CERV-related viruses were distinct (SI Appendix, Figs. S7 and S10C). As an example, comparison of amino acid sequences between ERV-DC14 Env and RmRV1 Env showed 43.8% identity. These results suggest that the backbones of feline and primate ERVs are derived from a common ancestor, while the env genes have a different origin.
A previous study reported that the timing of integration of ERV-DCs was 2.8 million y ago (Mya) (17). The timing of integration of CERV-related viruses has been calculated using the identification of ortholog env genes in sister species (31). For example, the integration timings of CmRV-trunc1 and RmRV-trunc1 were estimated to be ∼3.69 to 4.53 Mya; the integration timings of PNRC2-ChRV and PNRC2-BoRV were estimated to be ∼2.82 to 6.55 Mya, corresponding to the diversification of crab-eating macaque (Macaca fascicularis) and rhesus macaque (Macaca mulatta), and of chimpanzee (Pan troglodytes) and bonobo (Pan paniscus), respectively (SI Appendix, Fig. S11). Each locus was identified as the true ortholog (SI Appendix, Figs. S11–S14). These results suggested that ancestral primate retroviruses might have spread their infections in host lineages several million years ago; moreover, the primate ERV env genes were independently acquired in the ancestral genome of macaque monkeys, that of chimpanzee and bonobo, or in a species-specific manner.
Discussion
Refrex-1, a soluble restriction factor for FeLV-D and genotype I ERV-DCs, is secreted from feline cells (20). This antiviral machinery is driven by truncated Env proteins encoded by ERV-DC7 and ERV-DC16, which are classified as genotype II ERV-DCs (SI Appendix, Fig. S1). In this study, we found that CTR1 was the entry receptor for FeLV-D and genotype I ERV-DCs; moreover, the antiviral effect of Refrex-1 was found to depend on CTR1 (Figs. 1 and 2). These results imply that receptor interference is the mechanism underlying Refrex-1 activity. Furthermore, we determined that Refrex-1 could inhibit primate ERV infections using CTR1 as an entry receptor; likewise, we also found the antiviral machinery derived from truncated Envs in primate species (Fig. 4), suggesting that they could function as a barrier to cross-species transmission of ancient retroviruses. These results suggest that truncated Envs might constitute a convergently evolving antiviral system in feline and primate species. Our findings provide insight into the coevolutionary history between retroviruses and their hosts.
Evolutionary Arms Race between ERV-DC–Related Viruses and Refrex-1.
ERV-DCs are classified into three genotypes (SI Appendix, Fig. S1). Genotype II ERV-DCs comprise the most ancient ERV-DC group, and the integration time was estimated to be ∼2.8 Mya (17); this is consistent with the observation that genotype II ERV-DCs exhibit early phylogenetic divergence from the common ancestor of genotype I and genotype III ERV-DCs (SI Appendix, Fig. S1). Genotype I and II ERV-DCs belong to the same interference group, but genotype III ERV-DCs do not (27). This study revealed that genotype I and II ERV-DCs can use CTR1 as the entry receptor, but ERV-DC6 and ERV-DC10 (genotype III) cannot (Fig. 1B). These results suggest that genotype III ERV-DCs may have been generated by changes of entry receptor usage to escape host pressure (e.g., host immunity and antiviral effect of Refrex-1). Similarly, the occurrence of RD-114 might have resulted from selective pressure by the host because RD-114 is a recombinant virus that consists of an ERV-DC–like backbone and a BaEV-related env gene (17–19, 32). In fact, host resistance genes provide important defenses against viral infection (33, 34), but also produce selective pressures that favor the evolution of virus variants that subvert those blocks (35–37). Therefore, we assume that the antiviral machinery, such as truncated Envs, has driven an evolutionary arms race against viral infection.
Antiviral Activity by Feline and Primate Truncated Envelopes.
The truncated Env proteins with antiviral activity comprise the 5′ region of the envelope, which contains the signal sequence, VRA, and variable region B (VRB) with or without the proline-rich region. These proteins are extracellularly released because they lack the TM region (20). VRA and VRB correspond to the RBD; thus, they mediate the first step of the interaction between the viral envelope and its receptors for viral entry (29). The mechanism by which truncated Envs exhibit antiviral activity was shown by analyzing Refrex-1 and its ancestral viruses, ERV-DC7rec and ERV-DC16rec, both of which use CTR1 as the entry receptor. Here, we suggested that these truncated Envs compete for viral binding with the entry receptor, CTR1 (Fig. 2C).
Refrex-1 has been shown to protect against FeLV-D and genotype I ERV-DCs (20); it also effectively restricts infections by primate-derived ERVs (e.g., RmRV1/CmRV1, CERV2, and RhERV2-A) (Fig. 4A), despite their divergence from feline viruses (SI Appendix, Fig. S10). Truncated Envs with structures similar to Refrex-1 were identified in crab-eating macaque, rhesus macaque, chimpanzee, bonobo, and gorilla genomes (Fig. 4B); some of these Envs also effectively prevented infections by primate and feline retroviruses. Ten truncated Env proteins analyzed in this study exhibited lengths ranging from 188 to 439 amino acids; eight truncated Env proteins with lengths of 230 to 439 amino acids were efficiently secreted from cells (Fig. 4C) and exhibited antiviral activity (Fig. 4 A and D).
It was revealed that threonine at position 82 of the VRA amino acid sequence in PNRC2-BoRV influenced protection against infection in comparison with its ortholog PNRC2-ChRV (isoleucine at position 82) (Fig. 4D and SI Appendix, Figs. S7 and S11). In total, five amino acid differences (isoleucine, valine, threonine, glutamic acid, and leucine) at position 82 in ERV Envs were found in primate genomes, and three amino acid differences (isoleucine, valine, and threonine) at this position were found in primate truncated Envs analyzed in this study (SI Appendix, Fig. S7). These findings suggest that this position may be subject to selection pressure for viral entry of exogenous as well as endogenous viruses. Alternatively, these changes may target distinct viruses because a subtle mutation within the VRA of Env SU alters receptor usage (38).
Overall, characteristic soluble Env protein structure, lengths, or specific amino acid sequences may be necessary for truncated Envs to efficiently suppress viral infection. The precise mechanism behind the antiviral activity is unknown, but it is inferred to be due to the difference in the binding affinity, strength, or stability between CTR1 and truncated Env.
Two truncated Envs, BoRV-7 and PNRC2-BoRV, were identified in the bonobo genome; BoRV-7 showed broad antiviral activity, while PNRC2-BoRV showed limited inhibitory potential (Fig. 4D). Interestingly, the PNRC2-BoRV ortholog, PNRC2-ChRV, showed broad antiviral activity. These findings suggest that functional evolution has occurred between PNRC2-ChRV and PNRC2-BoRV after their speciation. It is speculated that the functional limitation of PNRC2-BoRV may have been replaced or complemented by BoRV-7, as it is suggested that the captured syncytins can be replaced in the divergent lineage by the env gene, which has a selective advantage in placental formation (39).
The finding that the mRNA expression of truncated env genes was detected in many tissues from primates (SI Appendix, Figs. S4 B and C and S9) and cats (20, 30), and the differences of those expression levels in each tissue may suggest the role of truncated Env in particular tissues (e.g., hematopoietic tissues, immune-related tissues). It should be noted that protein and RNA expression levels are not always correlated, and protein expression could not be investigated in this study.
Further elucidation of the precise mechanism behind the antiviral activity of the truncated Env proteins in vitro and in vivo is needed, which should provide insights for controlling various viral diseases.
Functional Full-Length Env in Primate ERVs.
To our knowledge, this study is unique in showing that two primates (crab-eating macaque and rhesus macaque) possess intact Envs capable of viral infection through viral receptor CTR1 (Fig. 3 and SI Appendix, Fig. S6). The existence of intact full-length env genes in the genome suggests the possibility of a viral threat, such as the formation of infectious particles due to the generation of recombinant viruses [e.g., FeLV-B and FeLV-D (17, 40, 41)], endogenous MLV (42, 43), or activation of infectious ERV provirus [e.g., ERV-DC (17, 30)].
In addition to making an infectious virus with the consequent risks of retroviral insertional mutagenesis, the TM domain of the Env protein has immunosuppressive effects with negative influences on the host (44), and ERV Env, which is known to have cell fusion potential in the placenta (8), can also pose a risk to the host (45).
Reconstruction of full-length env genes, DC7rec and DC16rec from Refrex-1, revealed that there are additional gene mutations with some stop codons that can result in an inability to make infectious particles (Fig. 2A) (27). Furthermore, expression analysis of the primate ERV env genes (SI Appendix, Figs. S4 B and C and S9) and ERV-DC env genes (20, 30) showed the suppression of intact full-length ERV env genes in primates and cats. In contrast, the truncated env genes were abundantly expressed in various tissues in primates (SI Appendix, Figs. S4 B and C and S9) and cats (20, 30). Combined with our previous studies (27, 30), it can be speculated that truncated env genes pose less risk to the host than full-length env genes.
Trade-off for Truncated Env between Antiviral Effect and Copper Metabolism.
There may be a functional trade-off of truncated Envs regarding their benefits to cats and primates; for example, CTR1 is characterized as a high-affinity copper uptake transporter (25), while the truncated Envs have antiviral activity against viral infection depending on CTR1. Therefore, it cannot be ruled out that the interaction between truncated Env and CTR1 perturbs copper metabolism. The effects of full-length and truncated Envs on copper metabolism are currently unknown. Elucidation of the effects of ERV Env–receptor interactions on copper metabolism would be a worthwhile topic for future study.
Receptors for FeLV.
A viral interference experiment is a powerful way of identifying viral receptors and interference groups (46). Cells persistently infected by retroviruses display resistance to the same or related retroviruses due to the interaction of viral envelope protein with the receptor by masking or down-regulating the receptor in the cell (46). We have revealed the FeLV-D interference group and its receptor CTR1 via a viral interference experiment. Likewise, thiamine transporter 1 (THTR1), sodium-dependent phosphate transporters (Pit-1 and Pit-2), feline leukemia virus subgroup C receptor (FLVCR), FeLV infectivity X-essory protein (FeLIX), and reduced folate carrier have been identified to be involved in FeLV entry (38, 47–54). In other words, the substitutions of the nucleotide sequence as well as recombination in Env genes have led to a change in the usage of the virus receptor, and variants of FeLV accompanied by changes in viral entry receptors have frequently emerged in domestic cats (16).
The expression of viral receptors in germline cells may contribute to the formation of ERVs (55). Although CTR1 appears to be ubiquitously expressed (SI Appendix, Figs. S3 and S4 A and C), it is unknown whether it is expressed in germline cells. However, the expression of CTR1 in the ovary and testis (SI Appendix, Figs. S3 and S4 A and C) suggests a risk of viral transmission (e.g., exogenous or endogenous virus) to offspring, while the presence of viral interference, including that due to truncated Env or Refrex-1, may contribute to prevention of viral infection or additional viral endogenization (46).
Viruses that can infect a wide range of species, such as the FeLV-D interference group, can cause epizootics in multiple species. Against such viruses, hosts may acquire truncated Env proteins as a common mechanism for preventing interspecies and intraspecies viral infection. Even under epizootics of infectious diseases, the acquisition of such resistant molecules appears to be effective. Convergent acquisition of these antiviral systems by multiple species could prevent epizootics of infectious diseases and greatly contribute to viral extinction. This study provides mechanistic insights into antiretroviral factors derived from truncated Envs and broad suppression of retroviruses across different species by targeting a common viral entry receptor.
Materials and Methods
Receptor Screening.
MDTF cells were transduced with a retroviral HEK293 cDNA library constructed in the pMX retroviral vector. FeLV-D (TY26) Env-pseudotyped viruses were used to challenge cDNA-transduced MDTF cells in eight 10-cm culture dishes. Cells were selected with 6 µg/mL puromycin. Chromosomal DNA from puromycin-resistant cells was extracted and served as a template for PCR amplification using primers annealing in the pMX vector. The PCR products were directly sequenced (SI Appendix, Supplementary Materials and Methods).
cDNA Cloning and Establishment of Cell Lines.
FeCTR1 and huCTR1 were PCR-amplified using cDNA from FT-1 cells and HEK293 cells as templates; they were cloned into the pFUΔss and pMSCVneo vectors, respectively. A DNA construct encoding huCTR2 was synthesized and cloned into the pMSCVneo vector. MDTF cells expressing these genes were termed MDTF-feCTR1, MDTF-huCTR1, and MDTF-huCTR2 (SI Appendix, Supplementary Materials and Methods).
Env-Pseudotyped Virus Preparation.
GPLac cells, an Env− packaging cell line containing a LacZ-coding retroviral vector, were transfected with Env expression plasmids to produce LacZ-carrying Env-pseudotyped viruses. Cell culture supernatants were collected, filtered, and stored at −80 °C (SI Appendix, Supplementary Materials and Methods).
Infection Assay.
Cells inoculated with viruses were cultured for 2 d postinfection. The LacZ assay was conducted by counting blue-stained nuclei (SI Appendix, Supplementary Materials and Methods).
RNA Interference.
MISSION esiRNA was used to knock down huCTR1 expression. Cells transfected with siRNA were infected with the pseudotyped viruses and CTR1 expression was monitored by qRT-PCR (SI Appendix, Supplementary Materials and Methods).
Genetic Analyses.
Primate ERV sequences were searched in the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/) and in the University of California, Santa Cruz, genome browser (https://genome.ucsc.edu/). Phylogenetic trees were constructed based on the maximum-likelihood method and the neighbor-joining method (SI Appendix, Supplementary Materials and Methods).
Supplementary Material
Acknowledgments
We thank Dr. Alan Rein (National Cancer Institute-Frederick) for his help editing and writing the manuscript; Dr. Tanya Jelacic (US Army Medical Research Institute of Infectious Diseases) for her help editing the manuscript; Hajime Tsujimoto (Department of Veterinary Internal Medicine, University of Tokyo) for providing the FeLV-B/pFGB plasmid; Toshio Kitamura (The Institute of Medical Science, University of Tokyo) for providing the pMXs and pMX retroviral vectors, as well as PLAT-E and PLAT-GP cells; and Edanz for editing a draft of this manuscript. This study was supported by Japan Society for the Promotion of Science KAKENHI Grants 20H03152 (to K.N.) and 20K06775 (to S.N.).
Footnotes
The authors declare no competing interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114441119/-/DCSupplemental.
Data Availability
The nucleotide sequence reported in this study was deposited in the DNA Data Bank of Japan database (accession no. LC705724)(56). The relevant codes and data are available in GitHub at https://github.com/Junna-Kawasaki/Refrex_2021 (57). All other study data are included in the main text and supporting information.
References
- 1.Herniou E., et al. , Retroviral diversity and distribution in vertebrates. J. Virol. 72, 5955–5966 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pontius J. U., et al. ; Agencourt Sequencing Team; NISC Comparative Sequencing Program, Initial sequence and comparative analysis of the cat genome. Genome Res. 17, 1675–1689 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belshaw R., Katzourakis A., Paces J., Burt A., Tristem M., High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol. Biol. Evol. 22, 814–817 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Spodick D. A., Ghosh A. K., Parimoo S., Roy-Burman P., The long terminal repeat of feline endogenous RD-114 retroviral DNAs: Analysis of transcription regulatory activity and nucleotide sequence. Virus Res. 9, 263–283 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Oliveira N. M., Satija H., Kouwenhoven I. A., Eiden M. V., Changes in viral protein function that accompany retroviral endogenization. Proc. Natl. Acad. Sci. U.S.A. 104, 17506–17511 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y. N., Bieniasz P. D., Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3, e10 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slotkin R. K., Martienssen R., Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Mi S., et al. , Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Redelsperger F., et al. , Genetic evidence that captured retroviral envelope syncytins contribute to myoblast fusion and muscle sexual dimorphism in mice. PLoS Genet. 12, e1006289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasachar Badarinarayan S., Sauter D., Switching sides: How endogenous retroviruses protect us from viral infections. J. Virol. 95, e02299–e02220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco-Melo D., Gifford R. J., Bieniasz P. D., Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. eLife 6, e22519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mura M., et al. , Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc. Natl. Acad. Sci. U.S.A. 101, 11117–11122 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujino K., Horie M., Honda T., Merriman D. K., Tomonaga K., Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc. Natl. Acad. Sci. U.S.A. 111, 13175–13180 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Treger R. S., et al. , The lupus susceptibility locus Sgp3 encodes the suppressor of endogenous retrovirus expression SNERV. Immunity 50, 334–347.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann K., Hofmann-Lehmann R., What’s new in feline leukemia virus infection. Vet. Clin. North Am. Small Anim. Pract. 50, 1013–1036 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Chiu E. S., Hoover E. A., VandeWoude S., A retrospective examination of feline leukemia subgroup characterization: Viral interference assays to deep sequencing. Viruses 10, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anai Y., et al. , Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J. Virol. 86, 8634–8644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beyer W., Möhring R., Drescher B., Nötzel U., Rosenthal S., Molecular cloning of an endogenous cat retroviral element (ECE 1)—A recombinant between RD-114 and FeLV-related sequences. Brief report. Arch. Virol. 96, 297–301 (1987). [DOI] [PubMed] [Google Scholar]
- 19.van der Kuyl A. C., Dekker J. T., Goudsmit J., Discovery of a new endogenous type C retrovirus (FcEV) in cats: Evidence for RD-114 being an FcEV(Gag-Pol)/baboon endogenous virus BaEV(Env) recombinant. J. Virol. 73, 7994–8002 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito J., et al. , Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J. Virol. 87, 12029–12040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payne L. N., Pani P. K., Weiss R. A., A dominant epistatic gene which inhibits cellular susceptibility to RSV(RAV-O). J. Gen. Virol. 13, 455–462 (1971). [DOI] [PubMed] [Google Scholar]
- 22.Odaka T., Yamamoto T., Inheritance of susceptibility to Friend mouse leukemia virus. 11. Spleen foci method applied to test the susceptibility of crossbred progeny between a sensitive and a resistant strain. Jpn. J. Exp. Med. 35, 311–314 (1965). [PubMed] [Google Scholar]
- 23.Lilly F., Susceptibility to two strains of Friend leukemia virus in mice. Science 155, 461–462 (1967). [DOI] [PubMed] [Google Scholar]
- 24.Suzuki S., FV-4: A new gene affecting the splenomegaly induction by Friend leukemia virus. Jpn. J. Exp. Med. 45, 473–478 (1975). [PubMed] [Google Scholar]
- 25.Zhou B., Gitschier J., hCTR1: A human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. U.S.A. 94, 7481–7486 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng X., et al. , Tissue-specific transcriptome sequencing analysis expands the non-human primate reference transcriptome resource (NHPRTR). Nucleic Acids Res. 43, D737–D742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito J., Baba T., Kawasaki J., Nishigaki K., Ancestral mutations acquired in Refrex-1, a restriction factor against feline retroviruses, during its cooption and domestication. J. Virol. 90, 1470–1485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soll S. J., Neil S. J. D., Bieniasz P. D., Identification of a receptor for an extinct virus. Proc. Natl. Acad. Sci. U.S.A. 107, 19496–19501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenwood A. D., Ishida Y., O’Brien S. P., Roca A. L., Eiden M. V., Transmission, evolution, and endogenization: Lessons learned from recent retroviral invasions. Microbiol. Mol. Biol. Rev. 82, e00044–e00017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuse K., et al. , Existence of two distinct infectious endogenous retroviruses in domestic cats and their different strategies for adaptation to transcriptional regulation. J. Virol. 90, 9029–9045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiewsakun P., Katzourakis A., Endogenous viruses: Connecting recent and ancient viral evolution. Virology 479-480, 26–37 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Shimode S., Nakagawa S., Miyazawa T., Multiple invasions of an infectious retrovirus in cat genomes. Sci. Rep. 5, 8164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda H., Laigret F., Martin M. A., Repaske R., Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J. Virol. 55, 768–777 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Best S., Le Tissier P., Towers G., Stoye J. P., Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382, 826–829 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Stevens A., et al. , Retroviral capsid determinants of Fv1 NB and NR tropism. J. Virol. 78, 9592–9598 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Y., Buckler-White A., Wollenberg K., Kozak C. A., Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc. Natl. Acad. Sci. U.S.A. 106, 3259–3263 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Y., Liu Q., Kozak C. A., Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 6, 87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyake A., et al. , Reduced folate carrier: An entry receptor for a novel feline leukemia virus variant. J. Virol. 93, e00269–e00219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esnault C., Cornelis G., Heidmann O., Heidmann T., Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet. 9, e1003400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart M. A., et al. , Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J. Virol. 58, 825–834 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overbaugh J., Riedel N., Hoover E. A., Mullins J. I., Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature 332, 731–734 (1988). [DOI] [PubMed] [Google Scholar]
- 42.Young G. R., et al. , Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491, 774–778 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu P., et al. , Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity 37, 867–879 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Mathes L. E., et al. , Immunosuppressive properties of a virion polypeptide, a 15,000-dalton protein, from feline leukemia virus. Cancer Res. 39, 950–955 (1979). [PubMed] [Google Scholar]
- 45.Stoye J. P., Proviral protein provides placental function. Proc. Natl. Acad. Sci. U.S.A. 106, 11827–11828 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter E., “Viral entry and receptors” in Retroviruses, Coffin J. M., Hughes S. H., Varmus H. E., Eds. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1997), pp. 71–119. [PubMed] [Google Scholar]
- 47.Anderson M. M., Lauring A. S., Burns C. C., Overbaugh J., Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287, 1828–1830 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Mendoza R., Anderson M. M., Overbaugh J., A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J. Virol. 80, 3378–3385 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Hara B., et al. , Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1, 119–127 (1990). [PubMed] [Google Scholar]
- 50.Takeuchi Y., et al. , Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66, 1219–1222 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson M. M., Lauring A. S., Robertson S., Dirks C., Overbaugh J., Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J. Virol. 75, 10563–10572 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tailor C. S., Willett B. J., Kabat D., A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J. Virol. 73, 6500–6505 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quigley J. G., et al. , Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95, 1093–1099 (2000). [PubMed] [Google Scholar]
- 54.Shalev Z., et al. , Identification of a feline leukemia virus variant that can use THTR1, FLVCR1, and FLVCR2 for infection. J. Virol. 83, 6706–6716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarlinton R. E., Meers J., Young P. R., Retroviral invasion of the koala genome. Nature 442, 79–81 (2006). [DOI] [PubMed] [Google Scholar]
- 56.M. H. Ngo, A. Miyake, K. Nishigaki, Convergent evolution of antiviral machinery derived from endogenous retrovirus truncated envelope genes in multiple species. DNA Data Bank of Japan. http://getentry.ddbj.nig.ac.jp/getentry/na/LC705724. Deposited 21 June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. Kawasaki, Refrex_2021. GitHub. https://github.com/Junna-Kawasaki/Refrex_2021. Deposited 8 June 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequence reported in this study was deposited in the DNA Data Bank of Japan database (accession no. LC705724)(56). The relevant codes and data are available in GitHub at https://github.com/Junna-Kawasaki/Refrex_2021 (57). All other study data are included in the main text and supporting information.