Significance
Immune checkpoint inhibitors (ICIs) are a revolutionary class of antitumor therapy that promote T cell responses by blocking immune checkpoint proteins. However, ICIs can cause inflammatory side effects known as immune-related adverse events (irAEs). These events impact virtually all tissues, predominantly those colonized by the microbiota. The etiology and mechanisms that drive the development of irAEs remain unclear. Here, we establish a preclinical model of irAEs and use it to establish that skin commensals can unleash aberrant commensal-specific T cell responses in the setting of ICI therapy, which leads to local inflammation and pathology. This work highlights how ICI therapy can disrupt the host–microbiota dialogue and provides groundwork for the design of novel therapies aimed at preventing and treating irAEs.
Keywords: immunotherapy, immunology, dermatology, T cells, microbiota
Abstract
Immune checkpoint inhibitors (ICIs) are essential components of the cancer therapeutic armamentarium. While ICIs have demonstrated remarkable clinical responses, they can be accompanied by immune-related adverse events (irAEs). These inflammatory side effects are of unclear etiology and impact virtually all organ systems, with the most common being sites colonized by the microbiota such as the skin and gastrointestinal tract. Here, we establish a mouse model of commensal bacteria–driven skin irAEs and demonstrate that immune checkpoint inhibition unleashes commensal-specific inflammatory T cell responses. These aberrant responses were dependent on production of IL-17 by commensal-specific T cells and induced pathology that recapitulated the cutaneous inflammation seen in patients treated with ICIs. Importantly, aberrant T cell responses unleashed by ICIs were sufficient to perpetuate inflammatory memory responses to the microbiota months following the cessation of treatment. Altogether, we have established a mouse model of skin irAEs and reveal that ICIs unleash aberrant immune responses against skin commensals, with long-lasting inflammatory consequences.
Immunotherapies have rapidly become one of the mainstays of cancer therapy, with Food and Drug Administration (FDA) approval for use in several cancer types, including melanoma, lung, genitourinary, and gastrointestinal malignancies (1–6). Among the most frequently used immunotherapies are immune checkpoint inhibitors (ICIs) that target the CTLA-4 and PD-1/PD-L1 pathways. These agents potentiate the antitumor T response by dampening inhibitory signals. In multiple clinical trials, CTLA-4 and PD-1 inhibitors have shown improved overall survival, response rates, and durable responses compared to standard cytotoxic therapies (7, 8). Although CTLA-4 and PD-1 inhibitors have been revolutionary in oncology, they can also cause severe, sometimes life-threatening systemic autoimmune-like events, termed immune-related adverse events (irAEs) (9). These events can involve virtually all organ systems but are particularly prevalent at sites colonized by the microbiota, including the skin and gastrointestinal tract (10–15). However, the role of the microbiota in eliciting these responses remains unclear. Dermatological toxicities are among the most common irAEs (16, 17). These events range substantially in form and severity, from mild pruritus to life-threatening epidermal necrolysis (18–21). While low-grade skin irAEs can be managed with emollients and oral antihistamines, more severe skin irAEs need to be treated with topical or systemic corticosteroids. In some instances, skin irAEs refractory to high-dose steroids have been reported (22–26), with treatment options for such cases not well established (27). Several hurdles have restricted the development of novel treatments for dermatological irAEs. In particular, the mechanisms underlying the pathogenesis of skin irAEs remain unclear and robust mouse systems that model skin irAEs have been challenging to develop. As more patients are treated with immune checkpoint inhibitors, a better understanding of the precise pathophysiology underlying skin irAEs will be essential to prevent and ameliorate skin irAEs.
Previous work from our group and others has revealed the complex and contextual relationship that exists between commensals and their hosts immune system (28–35). The diversity of the skin microbiome varies between individuals and can be influenced by both internal and external factors (36–38). New environmental exposures, antimicrobial treatment, dietary changes, and illness can all affect skin microbial communities, resulting in the acquisition of novel species and the loss of others (32, 39–41). In turn, these changes to the skin community can alter the hosts immune response. For instance, the acquisition of commensal bacteria that dominantly affect skin immunity, such as Staphylococci spp., induces a robust population of IL-17A–producing T cells that home to and persist within the skin (31). This response occurs in the complete absence of inflammation and is referred to as homeostatic immunity (29, 42). T cell responses directed toward the microbiota can provide long-term benefits, which include heterologous protection against diverse pathogens and accelerated wound healing (28, 43, 44). On the other hand, certain environmental contexts, such as inflammation and nutritional stress, can promote aberrant immune responses to this same bacterium, leading to tissue pathology (30, 33, 39, 45). However, to what extent aberrant immune responses to the microbiota cause or amplify disease remains poorly understood. Given the tumor-agnostic nature of skin irAEs and the fact that the skin is home to an abundance of diverse microbes (40), we hypothesized that the acquisition of a new commensal in the context of ICI treatment would impair the establishment of homeostatic immunity to the microbiota and promote an inflammatory response. Such lines of research are particularly important to consider for patients that frequently enter the hospital setting for treatment, where the chance of transiently acquiring novel commensals is relatively high (41). Here, we describe a mouse model of skin irAEs and use it to demonstrate that inhibiting immune checkpoint proteins at the time of exposure to a novel skin commensal induces commensal-specific T cell responses with pathogenic potential. The induction of these responses results in cutaneous inflammation that closely resembles the skin irAEs observed in patients treated with ICIs. Such findings provide a mechanistic explanation for the etiology of irAEs and lay the foundation for future clinical studies targeting the cross-talk between the immune system and microbiota to prevent and ameliorate irAEs.
Results
Acquisition of a Novel Skin Commensal in the Setting of CTLA-4 Blockade Promotes Pathology That Phenocopies Signatures of Human Cutaneous Inflammatory Disorders.
The relationship between commensal-specific T cell responses and immune checkpoint blockade in promoting skin disease has not been explored. To address this question, mice were treated systemically with an anti–CTLA-4 antibody concurrent with local skin colonization with the commensal bacterium Staphylococcus epidermidis (Fig. 1A) (32). Consistent with previous studies, application of S. epidermidis alone did not cause inflammation (31, 32, 46), nor did anti–CTLA-4 treatment alone (Fig. 1B). However, concurrent S. epidermidis colonization and CTLA-4 blockade resulted in striking skin inflammation on days 6 to 8 (Fig. 1 B and C). This was accompanied by an inflammatory infiltrate comprising proinflammatory cytokine-producing neutrophils and monocytes (which included Ly6Chigh monocytes, inflammatory monocytes, and monocyte-derived dendritic cells), as well as an increase in macrophages (Fig. 1 C and D, and SI Appendix, Fig. S1 A–C). Thus, immune checkpoint inhibition can unleash pathogenic immune responses against a newly acquired skin commensal.
Fig. 1.
Acquisition of a novel skin commensal bacterium in the setting of CTLA-4 blockade promotes pathology that phenocopies signatures of human cutaneous disease. (A) S. epidermidis (S. epi) and anti–CTLA-4 treatment schematic. (B) Skin inflammation reported as fold change in ear-skin thickness (mean ± SEM) relative to baseline at day 0 (first day of S. epidermidis application) comparing mice associated with either S. epidermidis (white) or anti–CTLA-4 alone (gray), and mice treated with both S. epidermidis and anti–CTLA-4 (red). (C) Representative histopathological comparison of the ear pinnae of mice colonized with S. epidermidis alone or colonized with S. epidermidis receiving anti–CTLA-4 treatment at day 8. (Scale bars [white], 100 µm.) (D) Absolute number of neutrophils, macrophages, and total monocytes in the skin of mice treated with S. epidermidis alone (white) or with S. epidermidis and anti–CTLA-4 (red). Data are shown for day 8. (E) Volcano plot showing log2 fold change and −log10 (FDR) of gene expression measured by RNA-seq. Blue dots denote genes significantly down-regulated and yellow dots show genes significantly up-regulated in mice colonized with S. epidermidis and treated with anti–CTLA-4 compared to those colonized with S. epidermidis alone. Red dots show significantly up-regulated genes associated with immune responses. (F) Metascape pathway enrichment analysis for genes up-regulated in S. epi + anti–CTLA-4 in comparison to S. epidermidis + isotype. (G) IPA disease term analysis for genes significantly up-regulated in mice colonized with S. epidermidis and treated with anti–CTLA-4 compared to those colonized with S. epidermidis alone (Left). Zoom in heatmap view of genes associated with dermatitis and psoriasis pathway (Right). Each symbol represents the average of a pool of mice (B), or an individual mouse (D). Data are pooled from at least two experiments (B and D–G) or representative of three experiments (C). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, as calculated with two-way ANOVA with Holm–Šidák multiple-comparison test (B) or nonparametric Mann–Whitney test. (D). Data are represented as mean ± SEM.
To examine the impact of concurrent S. epidermidis colonization and CTLA-4 blockade on skin at the whole tissue level, we performed RNA sequencing (RNA-seq) on skin of mice at day 8 following S. epidermidis colonization alone or S. epidermidis colonization with anti–CTLA-4 treatment (Fig. 1A). Anti–CTLA-4 treatment in combination with S. epidermidis significantly altered the transcriptional program of the tissue compared to colonization with S. epidermidis alone, with 394 genes significantly up-regulated and 33 genes down-regulated (false discovery rate [FDR] < 0.05, fold change > 2) (Fig. 1E). Of note, many genes associated with cells of the innate and adaptive immune system, as well as proinflammatory responses, were up-regulated (Fig. 1E). Consistent with this, gene ontology (GO) enrichment analysis revealed that gene signatures associated with inflammatory responses, T cell activation, regulation of cytokine production, leukocyte migration, and myeloid cell activation were up-regulated in the skin of mice colonized with S. epidermidis receiving anti–CTLA-4 therapy compared to those colonized with S. epidermidis alone (Fig. 1F). This result was further confirmed by pathway analysis using ingenuity pathway analysis (IPA), which showed that pathways associated with T cell and innate cell activation were enriched, as were those linked to various forms of autoimmunity, and the cytokine IL-17 (SI Appendix, Fig. S1D). Importantly, IPA also revealed that the gene signature induced by CTLA-4 blockade and S. epidermidis colonization recapitulated several human cutaneous inflammatory conditions (Fig. 1G). These included psoriasis and dermatitis (Fig. 1G), both of which have noted similarities to the pathology observed in skin irAEs following ICI therapy (24, 47–49). Altogether, our results demonstrate that immune checkpoint inhibitors unleash a broad proinflammatory, pathogenic program against a newly acquired member of the skin microbiota, and that our mouse model may represent a unique tool to dissect the etiology and mechanism of action of skin irAEs.
Commensal Colonization of Skin in the Setting of CTLA-4 Blockade Promotes Aberrant Commensal-Specific T Cell Responses.
Based on the pathways enriched in our RNA-seq dataset, which reflected a broad proinflammatory program associated with T cell activation, we next assessed the magnitude and quality of the commensal-specific T cell response within skin at day 8 following S. epidermidis colonization, with or without anti–CTLA-4 treatment (Fig. 2A and SI Appendix, Fig. S2A). As previously reported, S. epidermidis colonization alone resulted in a significant increase in the number of CD4+ and CD8+ T cells producing IFN-γ or IL-17 in skin compared to unassociated controls (Fig. 2B), which occurred in the absence of inflammation (Fig. 1 B and C) (28, 31, 32, 44). Consistent with our RNA-seq data, anti–CTLA-4 treatment with S. epidermidis colonization induced a dramatic increase in the number of CD4+ and CD8+ T cells producing IFN-γ or IL-17 in skin when compared to mice colonized with S. epidermidis alone (Fig. 2B). Anti–CTLA-4 treatment alone had no impact on the number of T cells within the skin (Fig. 2B), supporting the idea that this treatment preferentially altered responses to a newly acquired microbe, as opposed to responses against preexisting microbes. The number of regulatory T cells (Treg) was also increased in the skin of mice colonized with S. epidermidis receiving anti–CTLA-4 treatment (Fig. 2C). However, the ratio of effector CD4+ T cells to Tregs was reduced in this context compared to mice colonized with S. epidermidis alone (Fig. 2C), indicating a preferential activation of effector T cells during anti–CTLA-4 treatment. T cell accumulation in skin in the context of S. epidermidis and CTLA-4 blockade was associated with increased proliferation (SI Appendix, Fig. S2B). While the number of cytokine-producing T cells was greatly increased in the skin of mice colonized with S. epidermidis receiving anti–CTLA-4 treatment compared to the skin of mice colonized with S. epidermidis alone (Fig. 2B), the frequency of cytokine-positive cells between these groups was similar (SI Appendix, Fig. S2C). Thus, commensal colonization in the context of anti–CTLA-4-treatment broadly promoted inflammatory T cell responses.
Fig. 2.
Commensal colonization in the setting of CTLA-4 inhibition promotes aberrant commensal-specific T cell responses in skin. (A) S. epidermidis and anti–CTLA-4 treatment schematic. (B) Absolute number of IL-17A+ (Left) and IFN-γ+ (Right) CD4+ and CD8+ T cells in the treatment groups. Data are shown for day 8. (C) Absolute number of Tregs (Left). Representative flow cytometry plot showing the frequency of Foxp3+ CD4+ Treg cells in skin (Middle). Ratio of Treg/effector CD4+ T cells (Right). (D) S. epidermidis-specific TCR-transgenic CD8+ T cells (BowieTg) were adoptively transferred to WT mice 1 d prior to colonization with S. epidermidis (Left). Time course showing BowieTg cells in draining lymph nodes (Middle). Absolute number and frequency of BowieTg cells (of total CD8+ T cells) in skin at day 8 treated with S. epidermidis alone (white) or with S. epidermidis and anti–CTLA-4 (red) (Right). (E) Absolute number of IL-17A+ (Left) and IFN-γ+ (Right) BowieTg cells in skin at day 8 treated with S. epidermidis alone (white) or with S. epidermidis and anti–CTLA-4 (red). Each symbol represents the average of a pool of mice (D) or an individual mouse (B–E). Data shown are pooled from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant, as calculated with one-way ANOVA with Holm–Šidák multiple-comparison test (B and C) or two-way ANOVA with Holm–Šidák multiple-comparison test (D) or nonparametric Mann–Whitney test (D and E). Data are represented as mean ± SEM.
S. epidermidis-specific CD8+ T cell responses depend on the nonclassical major histocompatibility complex class I (MHC-I)–like molecule H2-M3 (44). To begin investigating the restriction and specificity of the amplified T cell responses described above, we associated wild-type (WT) and H2-M3–deficient mice with S. epidermidis with or without CTLA-4 blockade. Regardless of anti–CTLA-4 treatment, CD8+ T cell responses were completely abolished in H2-M3–deficient mice, supporting the idea that CTLA-4 blockade was amplifying nonclassical immunity to S. epidermidis (SI Appendix, Fig. S2D). To formally link such responses to commensal-specific T cells, we utilized our previously described T cell receptor (TCR)–transgenic mice (BowieTg) (28) to track the S. epidermidis-specific CD8+ T cell response. Naive S. epidermidis-specific CD8+ T cells (BowieTg) were adoptively transferred into WT mice 1 d prior to colonization with S. epidermidis with or without anti–CTLA-4 treatment (Fig. 2D). A significant increase in the number of S. epidermidis-specific CD8+ T cells was detected in the draining lymph nodes as early as day 5 in mice colonized with S. epidermidis receiving anti–CTLA-4 treatment compared to mice colonized with S. epidermidis alone (Fig. 2D), suggesting that this regiment was aberrantly amplifying the early stages of T cell activation. Within the lymph nodes, the number of S. epidermidis-specific CD8+ T cells continued to drastically increase until day 8 postassociation (Fig. 2D). At that time, the number and frequency of S. epidermidis-specific CD8+ T cells was significantly higher in the skin of mice colonized with S. epidermidis receiving anti–CTLA-4 treatment compared to those colonized with S. epidermidis alone (Fig. 2D). Furthermore, the number of these cells producing IL-17 or IFN-γ was significantly increased in the context of CTLA-4 blockade (Fig. 2E). Altogether, these data indicate that anti–CTLA-4 treatment aberrantly amplifies the induction of commensal-specific T cell responses.
Skin Pathology Induced by Commensal Colonization and CTLA-4 Blockade Is Driven by Production of IL-17 by Commensal-Specific T Cells.
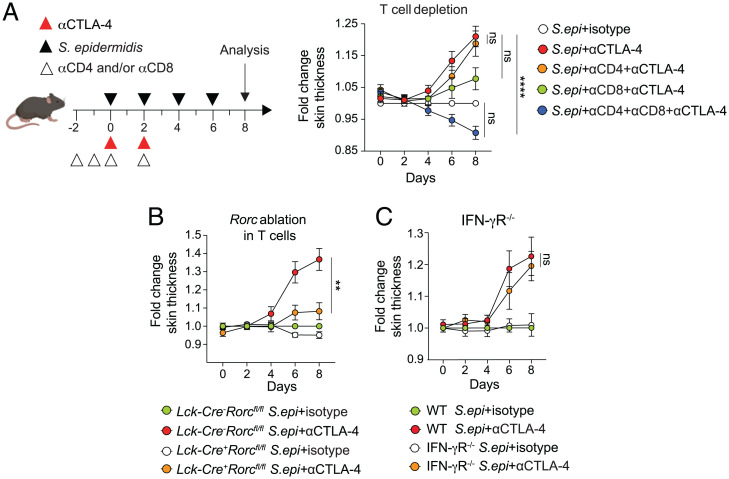
Our results supported the idea that cutaneous inflammation in the context of commensal colonization and CTLA-4 blockade depended on aberrantly amplified commensal-specific T cell responses. To directly address this, we depleted T cells in vivo using anti-CD4 and anti-CD8 antibodies in mice colonized with S. epidermidis alone and those colonized with S. epidermidis receiving anti–CTLA-4 treatment (Fig. 3A and SI Appendix, Fig. S3A). Depletion of either T cell subset alone did not significantly reduce skin inflammation in mice colonized with S. epidermidis receiving anti–CTLA-4 treatment (Fig. 3A). However, simultaneous depletion of both T cell subsets completely abolished skin inflammation (Fig. 3A and SI Appendix, Fig. S3B). Innate cell infiltrates were also significantly reduced when T cells were depleted in mice colonized with S. epidermidis receiving anti–CTLA-4 treatment compared to those colonized with S. epidermidis alone (SI Appendix, Fig. S3C), indicating that T cells are the primary source of inflammation in this context. As such, aberrant commensal-specific T cell responses in the context of CTLA-4 blockade required the coordinated action of both CD4+ T and CD8+ T cells.
Fig. 3.
Skin pathology in the setting of commensal colonization and CTLA-4 blockade depends on T cell-derived IL-17. (A) WT mice were associated with S. epidermidis (S. epi) alone (white), S. epidermidis and treated with anti–CTLA-4 alone (red) or combined with anti-CD4 (orange), anti-CD8 (green), or both anti-CD4 and anti-CD8 depleting antibodies (blue). Ear thickness measurement is reported as fold change in ear-skin thickness (mean ± SEM) relative to baseline at day 0 (first day of S. epidermidis application). (B) Lckcre+ Rorcflox and Lckcre- Rorcflox mice were associated with S. epidermidis and treated with anti–CTLA-4 as indicated. Ear thickness measurement is reported as fold change in ear-skin thickness (mean ± SEM) relative to baseline at day 0 (first day of S. epidermidis application). (C) WT and IFN-γR−/− mice were associated with S. epidermidis and treated with anti–CTLA-4 as indicated. Ear thickness measurement is reported as fold change in ear-skin thickness (mean ± SEM) relative to baseline at day 0 (first day of S. epidermidis application). Each symbol represents the average of a pool of mice. (A–C) Data are pooled from two independent experiments. **P < 0.01, ****P < 0.0001, ns, not significant, as calculated with one-way ANOVA with Holm–Šidák multiple-comparison test (A–C). Data are represented as mean ± SEM.
We next sought to establish how commensal-specific T cells were eliciting inflammatory skin responses in the context of S. epidermidis colonization and CTLA-4 blockade. IL-17 plays a key role in host immunity and homeostasis by promoting epithelial cell integrity, as well as recruiting and activating cells of the innate immune system (50, 51). However, IL-17 can also be pathogenic and has been implicated in several autoimmune disorders, including psoriasis, inflammatory bowel disease, multiple sclerosis, and rheumatoid arthritis (52–55). Based on these studies and our findings of a pronounced increase in IL-17–producing T cells after immune checkpoint inhibition and commensal colonization (Fig. 2 B and E), we next assessed to what extent T cell–derived IL-17 plays a role in driving skin inflammation in this context. To this end, we utilized LckCre Rorcflox mice, a strain with a floxed orphan receptor Rorc gene, which abrogates differentiation of IL-17–producing T cells (56, 57). LckCre+ Rorcflox mice colonized with S. epidermidis and treated with anti–CTLA-4 antibodies showed reduced skin inflammation, as well as reduced numbers of IL-17+ T cells and several populations of innate cells in skin when compared to wild-type LckCre-Rorcflox littermate control mice colonized with S. epidermidis and treated with anti–CTLA-4 antibodies (Fig. 3B and SI Appendix, Fig. S3 D and E). In contrast, mice deficient for the IFN-γ receptor showed a similar response to wild-type mice, indicating that IFN-γ does not play a role in this context (Fig. 3C and SI Appendix, Fig. S3F). Thus, these results suggest that excessive commensal-specific T cell–derived IL-17 drives skin pathology in the context of CTLA-4 blockade and S. epidermidis skin colonization.
CTLA-4 Blockade Aberrantly Promotes Inflammatory Memory T Cell Responses against the Microbiota.
Clinical observations indicate that although skin irAEs frequently occur early following ICI treatment, they can also occur as late as 3 mo after the cessation of treatment (58, 59). In light of these clinical observations, we used our model of commensal bacteria–driven skin irAEs to determine whether there were long-term consequences of encountering a novel commensal while receiving CTLA-4 blockade. As expected, S. epidermidis colonization followed by another S. epidermidis colonization 1 mo later did not result in an inflammatory response (Fig. 4A). However, an initial S. epidermidis colonization with concurrent CTLA-4 blockade followed by recolonization with S. epidermidis alone 1 mo later resulted in a significantly more rapid and potent inflammatory response compared to the initial colonization (Fig. 4A). Such findings suggested increased generation of commensal-specific memory T cells in the context of S. epidermidis association and CTLA-4 blockade. This was indeed the case, with a significant increase in number of S. epidermidis-specific CD8+ T cells (BowieTg) found in skin 1 mo following association in mice that had undergone anti–CTLA-4 treatment, with the vast majority of these cells being CD69+ CD103+ resident memory T cells (TRM) (Fig. 4B and SI Appendix, Fig. S4A) (60, 61). Furthermore, an increase in circulating S. epidermidis-specific CD8+ memory T cells was observed in the draining lymph node, whereas the lymph node resident memory T cell population was unaltered in mice previously associated with S. epidermidis receiving anti–CTLA-4 treatment compared to those associated with S. epidermidis alone (Fig. 4C). As such, encountering a novel commensal bacterium in the setting of CTLA-4 blockade may break tolerance to that commensal. Our data suggest that this results in chronically aberrant memory T cell responses upon subsequent encounters with that same commensal, even when CTLA-4 blockade is not actively occurring.
Fig. 4.
CTLA-4 blockade primes inflammatory memory responses to the microbiota. (A) WT mice were initially colonized with S. epidermidis (S. epi) alone (white) or with S. epidermidis and anti–CTLA-4 treatment (red) (1° colonization). Thirty days later, they were recolonized with S. epidermidis (2° encounter) (Left). Ear thickness measurement is reported as fold change in ear-skin thickness (mean ± SEM) relative to baseline at day 0 (first day of S. epidermidis application) (Right). (B) Absolute number of resident memory (CD44+ CD69+ CD103+ BowieTg cells in skin of mice treated with S. epidermidis alone (white) or with S. epidermidis and anti–CTLA-4 (red). (C) Absolute number of circulating (CD69−) and resident (CD69+) memory BowieTg cells in draining lymph nodes of mice treated with S. epidermidis alone (white) or with S. epidermidis and anti–CTLA-4 (red). (D) Homeostatic interactions with the microbiota induce commensal-specific T cells that produce IL-17 and benefit the host by promoting heterologous protection and wound healing. However, immune checkpoint blockade at the time of exposure to a novel commensal induces unrestrained inflammatory responses, with excessive numbers of IL-17–producing commensal-specific T cells infiltrating skin. These cells drive proinflammatory cytokine-producing innate cell populations that altogether promote skin pathology. Each symbol represents the average of a pool of mice (A) or an individual mouse (B and C). *P < 0.05, ns, not significant, as calculated with one-way ANOVA with Holm–Šidák multiple-comparison test, the peak of ear-skin thickness in the primary colonization (day 6) was compared to the peak in the secondary encounter (day 43) (A) or nonparametric Mann–Whitney test (B and C).
Discussion
Commensal microbes and the immune system are constantly engaged in a complex and dynamic dialogue, which oscillates between being beneficial or detrimental to the host depending on the environmental setting (62, 63). Previous studies from our group have demonstrated that the induction of IL-17–producing commensal-specific T cells is beneficial by promoting wound repair (28, 43) and heterologous immunity against invasive pathogens (31). However, environmental shifts such as consumption of a high-fat diet induces excessive, pathogenic responses to that same commensal, resulting in tissue inflammation (45). Our current study on immune checkpoint inhibitors expands the known repertoire of environmental stimuli that disrupt the homeostatic host–microbiota dialogue. Based on our data, we propose that the acquisition of a novel commensal in the context of immune checkpoint blockade results in excessive, dysregulated activation of IL-17–producing commensal-specific T cells, which in turn induce a hyperactive, pathogenic innate immune response (Fig. 4B). Therefore, it would be of great interest for future studies to assess the composition of the skin microbiota in patients before and during treatment of ICIs to determine how alterations to the skin microbiome influence the severity and development of skin irAEs.
While our findings remain to be confirmed in a clinical setting, an association with IL-17 and skin irAEs has been demonstrated. For example, a case report has shown psoriasiform dermatologic toxicity induced by immune checkpoint inhibition that was successfully resolved with IL-17A blockade (26). These observations, together with our data, support the idea that cytokine blockade could be a future treatment option for steroid-refractory skin irAEs. In future studies it will be important to determine whether all commensal-specific immune responses are pathogenic in the context of immune checkpoint inhibition or if only defined classes of commensals elicit these types of responses. Importantly, numerous commensals elicit a type 17 immune response (30, 64, 65), suggesting that the observations in our study could reflect a broader phenomenon. Considering our demonstration that a single commensal microbe can drive skin irAEs, it would be of interest in future studies to use wild mice that have been exposed to a diverse array of commensals to gain further unique insights into the pathogenesis of irAEs (34, 35, 66, 67).
We focused on the CTLA-4 pathway because of the high frequency of irAEs that occur with anti–CTLA-4 therapy, with up to 35% of patients developing a grade I-II skin irAE after treatment (68). Blocking anti–CTLA-4 is well established to enhance T cell priming (69, 70). Consistent with this, our data suggested that commensal colonization and concurrent CTLA-4 blockade resulted in unrestrained expansion of commensal-specific T cells in draining lymph nodes. These T cells then infiltrated skin and triggered a broad inflammatory response. Such results indicate that the CTLA-4 pathway provides an essential layer of regulation that promotes beneficial homeostatic interactions between the host and its microbiota by restricting excessive activation of commensal-specific T cells. CTLA-4 blockade has been shown to promote effector T cell responses directly and also indirectly by reducing Treg-mediated immunosuppression (71). Therefore, the inflammatory response seen in our model could be due to altered CTLA-4 function in both commensal-specific effector T cells and Tregs. In line with this, skin Treg cells play a crucial role in mediating tolerance to commensal organisms (72). As such, future studies delineating the distinct impact of CTLA-4 blockade on effector T cells and Treg cells in the context of commensal colonization could provide additional therapeutic targets to prevent and treat irAEs.
While we focused on effector T cell responses to a newly acquired commensal in the context of CTLA-4 blockade, development of skin irAEs can also arise through other pathways. For example, blocking the PD-1/PD-L1 pathway can promote the function of established memory T cells within tissues (69, 73). As such, it is possible that therapeutic inhibition of the PD-1/PD-L1 pathway in patients could have the side effect of aberrantly unleashing established commensal-specific memory T cell responses (35, 74). Consistent with this, blocking both the anti–CTLA-4 and PD-1 pathways has been reported to increase the frequency of irAEs compared to monotherapy (75). Given the increase in use of ICIs to treat cancer, it will be critical in future clinical studies to establish the relationship between inhibition of multiple checkpoint pathways, commensals, and irAEs.
Based on clinical studies on the chronic effects of ICI-induced skin irAEs, we also used our model to explore the long-term consequences of ICI treatment on the skin. We found that encountering a novel commensal bacterium in the setting of immune checkpoint blockade may permanently break tolerance to that commensal, resulting in chronically aberrant memory T cell responses upon subsequent encounters, even when checkpoint blockade was not actively occurring. These data are in line with clinical observations reporting that although skin irAEs frequently occur early after ICI treatment, skin irAEs can also occur as late as 3 mo after the cessation of treatment (58, 59). Previous studies from our group and others have found that acute gastrointestinal infection can induce long-lived commensal-specific memory T cells and that tissue resident memory T cells are the dominantly activated in ICI-mediated colitis (76, 77). It will be important for future clinical studies examining these late developing irAEs to analyze the skin microbiota of patients to determine whether defined commensals are involved in driving inflammatory responses. Furthermore, while we describe here how skin commensals can affect the development of irAEs, it would be expected that commensal-specific T cell responses within other tissues such as the gut, lung, or liver are impacted similarly. As more data are emerging from long-term patient followup, it may become clear that chronic irAEs associated with checkpoint blockade are more common than previously recognized (78, 79).
Altogether, our mouse model provides an accessible, translational approach to exploring and treating the acute and chronic dermatological adverse events that arise from ICIs in the clinic. In 2019, over a third of cancer patients in the United States were eligible to receive ICIs (80). The cumulated number of cases per year of irAEs has increased exponentially since 2002, with nearly 13,000 irAEs cases reported in 2018 (81). Given the increasing use of ICIs and rising number of irAEs, further understanding of the underlying pathogenesis of irAEs will be critical in managing the increasing incidence of irAEs in the future. In addition, patients with active autoimmune disease are frequently excluded from clinical trials out of concern that ICIs can lead to autoimmune flares (82–85). Therefore, future research into modulating the microbiota to reduce irAE incidence and severity may allow this patient population to be included in clinical trials using novel immunotherapy treatments. Overall, we have generated an animal model that recapitulates irAEs induced by ICIs. We then utilized this model to demonstrate the importance of aberrant T cell responses directed toward the commensal microbiota in the etiology of irAEs. Such findings lay the groundwork for future clinical studies targeting the cross-talk between the immune system and microbiota to prevent and ameliorate irAEs.
Materials and Methods
Mice.
C57BL/6 mice specific pathogen free (SPF) positive for segmented filamentous bacteria (SFB) were purchased from Taconic Farms. B6.SJL Cd45a(Ly5a)/Nai (B6.SJL) and IFNγR−/− mice were obtained through the National Institute of Allergy and Infectious Diseases (NIAID)–Taconic exchange program. LckCre+ x Rorcfl/fl mice, a kind gift from Remy Bosselut, NIH, National Cancer Institute, Bethesda, MD, were generated by breeding B6.Cg-Tg(Lck-icre)3779Nik/J (Lckcre+) mice with B6(Cg)- Rorctm3Litt/J (Rorcfl/fl) mice (both strains from The Jackson Laboratory). H2-M3−/− mice were a gift from Chyung-Ru Wang, Northwestern University, Evanston, IL. All mice were bred and maintained under SPF conditions at an American Association for the Accreditation of Laboratory Animal Care–accredited animal facility and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (86). All experiments were performed at the NIAID under an animal study proposal approved by the NIAID Animal Care and Use Committee. Gender- and age-matched mice between 6 and 16 wk of age were used for each experiment. When possible, preliminary experiments were performed to determine requirements for sample size, taking into account resources available and ethical, reductionist animal use. Animals were assigned randomly to experimental groups.
Topical Association with Commensal Bacteria.
S. epidermis strain LM087 (87) was cultured in tryptic soy broth at 37 °C for 18 h. For topical association, mice were associated with S. epidermidis by applying bacterial suspension (109 colony-forming units [CFU]/mL) using a sterile cotton swab. Application of bacterial suspension was repeated every other day for a total of four associations. For secondary challenge, mice were associated once.
Tissue Processing.
Mice were euthanized 8 d after the first topical association with S. epidermidis. Single-cell suspension from ear pinnae were isolated as previously described (32) and digested in RPMI 1640 containing 2 mM L-glutamine, 1 mM sodium pyruvate and nonessential amino acids, 55 mM β-mercaptoethanol, 20 mM Hepes, 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 mg/mL Liberase TL purified enzyme blend; Roche). Ears were incubated for 1 h 45 min at 37 °C in 5% CO2. Digested skin sheets were homogenized using the Medicon/Medimachine tissue homogenizer system (Becton, Dickinson and Company).
Ex Vivo Restimulation.
To analyze cytokine potential, single-cell suspensions from tissues were cultured and restimulated ex vivo in a 96-well U-bottom plate in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1 mM sodium pyruvate and nonessential amnio acids, 20 mM HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin, 50 mM β-mercaptoethanol, and stimulated with 50 ng/mL phorbol myristate acetate (PMA) (Sigma-Aldrich) and 5 μg/mL ionomycin (Sigma-Aldrich) in the presence of a 1:1,000 dilution of brefeldin A (GolgiPlug, BD Biosciences), diluted per manufacturer’s instructions, for 2.5 h at 37 °C in 5% CO2. Following stimulation, cells were analyzed for intracellular cytokine production as described below.
Adoptive Transfer of BowieTg CD8+ T Cells.
BowieTg mice were backcrossed to a CD45.2 Rag1−/− background to exclude the possibility of dual TCR expression and facilitate identification of transferred cells. Recipient mice (CD45.1) received 1 × 105 BowieTg CD45.2 Rag1−/− CD8+ T cells by intravenous injection 24 h prior to the first application of S. epidermidis.
Flow Cytometry.
Single-cell suspensions were first stained with fluorochrome-conjugated antibodies against surface markers for 20 to 30 min at 4 °C in phosphate-buffered saline (PBS) and then washed twice with cold PBS. LIVE/DEAD Fixable Blue Dead Cell Stain Kit (Invitrogen Life Technologies) was used to exclude dead cells. For intracellular staining, cells were fixed/permeabilized for 30 min at 4 °C, washed twice, and then stained intracellularly for at least 1 h at 4 °C using the Foxp3/transcription factor staining buffer set (Life Technologies, eBioscience). The following fluorochrome-conjugated antibodies from eBiosciences, BD, or R&D were used: CCR2 (FAB5538P), CD4 (RM4-5), CD8β (H35–17.2), CD11b (M1/70), CD44 (IM7), CD45 (30-F11), CD24 (M1/69), CD45.1 (A20), CD45.2 (104), CD45R (RA3-6B2), CD64 (FA11), CD90.2 (30-H12), CD103 (2E7), (MAR-1), Ly6C (HK1.4), Ly6G (1A8), MHC-II (M5/114.15.2), TCRb (H57-597), gdTCR (eBioGL3), IFN-γ (XMG1.2), FOXP3 (FJK-16s), IL-17A (TC11-18H10.1), Ki-67 (SolA15), and TNF-a (MP6-XT22). Flow cytometric data were acquired on an LSR Fortessa (BD Biosciences) and analyzed using FlowJo software (Treestar).
Histology.
Mice were euthanized 8 d after the first topical application of S. epidermidis. The ear pinnae from each mouse were removed and fixed in PBS containing 10% formalin. Paraffin-embedded sections were cut at 0.5 mm, stained with hemotoxylin and eosin, and examined histologically.
Microscopy of Ear Pinnae.
Ear pinnae images were captured on a Leica M205 stereomicroscope and analyzed using Aperio Imagescope software.
In Vivo Antibody Administration.
Mice were treated intraperitoneally (i.p.) with anti–CTLA-4 antibody (clone UC10-4F10-11, BioXCell) or Armenian hamster IgG isotype control (BioXcell). Treatment schedule was as follows: 300 μg of either antibody was injected i.p. in each mouse on days 0 and 2 after the first topical application of S. epidermidis. For CD4+ and CD8+ depletion, mice were treated i.p. with 200 μg of anti-CD4 antibody (GK1.5, BioXCell), 200 μg of anti-CD8 antibody (clone 2.43, BioXCell) or both antibodies on days −2, −1, 0, and 3 after first topical application of S. epidermidis.
Total Tissue RNA-Seq Library Preparation.
A ∼1-mm ear skin region was microdissected at day 8, submerged in RNAlater (Sigma-Aldrich), and stored at −20 °C. Total tissue RNA was isolated from ear skin tissue using the RNeasy Fibrous Tissue Mini kit (Qiagen), as per manufacturer’s instructions. Total mRNA library was prepared using 200 to 500 ng of input RNA with the Universal Plus# RNA-seq with NuQuant Library Prep Kit (Tecan) as per manufacturer’s instructions. Libraries were quantified using an Agilent Tapestation (High Sensitivity D1000 ScreenTape) and Qubit (Thermo Fisher Scientific). Libraries were pooled at equimolar concentrations and sequenced on an Illumina NextSeq 550 using the High Output v2 kit (75 cycles). Data were demultiplexed with bcl2fastq.
RNA-Seq Analysis.
RNA-seq samples were mapped to the mm10 mouse genome with STAR. Samples that clustered together by treatment using Spearman correlation were kept, resulting in five replicates for S. epidermidis treatment and four replicates for S. epidermidis + anti–CTLA-4 treatment. Gene counts were calculated with HOMER’s analyzeRepeats with parameters -condenseGenes. For plots, reads were normalized to transcripts per million (TPM) (parameter -tpm), and for differential gene expression reads were not normalized (parameter -noadj). Differential gene expression was calculated using DESeq2 (with HOMER’s getDiffExpression.pl) and genes with a fold change greater than 2 and FDR smaller than 0.05 were considered differentially expressed. Pathway analysis was performed with metascape for up- and down-regulated genes separately, as well as IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) for all differentially expressed genes (88). Furthermore, IPA’s disease term enrichment analysis was performed for all differentially expressed genes.
Statistics.
Groups were compared with Prism V7.0 software (GraphPad) using the two-tailed unpaired Student’s t test, nonparametric Mann–Whitney test, one-way analysis of variance (ANOVA) with Holm–Šidák multiple-comparison test, or two-way ANOVA with Holm–Šidák multiple-comparison test where appropriate. Differences were considered to be statistically significant when P ≤0.05.
Supplementary Material
Acknowledgments
We thank the members of the Y.B. laboratory for their suggestions, support, and critical reading of the manuscript. We thank the NIAID animal facility staff and P. J. P. Chaparro, K. Beacht, and E. Lewis for technical assistance. Y.B. is supported by the Division of Intramural Research of the NIAID, NIH (1ZIA-AI001115 and 1ZIA-AI001132). Z.I.H. was supported by the NCI Medical Oncology Fellowship program. N.C. is supported by a K99/R00 Pathway to Independence Award.
Footnotes
Reviewers: D.M., University of Minnesota Medical School; and M.D.R., University of California San Francisco Medical Center
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200348119/-/DCSupplemental.
Data Availability
RNA sequencing datasets generated during this study are available at the NCBI GEO: GSE176030.
References
- 1.Paz-Ares L., et al. ; KEYNOTE-407 Investigators, Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N. Engl. J. Med. 379, 2040–2051 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Motzer R. J., et al. ; CheckMate 214 Investigators, Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 378, 1277–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le D. T., et al. , PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larkin J., et al. , Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi L., et al. ; KEYNOTE-189 Investigators, Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Balar A. V., et al. , First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): A multicentre, single-arm, phase 2 study. Lancet Oncol. 18, 1483–1492 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Pons-Tostivint E., et al. , Comparative analysis of durable responses on immune checkpoint inhibitors versus other systemic therapies: A pooled analysis of phase III trials. JCO Precis. Oncol. 3, 1–10 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Topalian S. L., et al. , Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postow M. A., Hellmann M. D., Adverse events associated with immune checkpoint blockade. N. Engl. J. Med. 378, 1165 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Di Giacomo A. M., et al. , Therapeutic efficacy of ipilimumab, an anti-CTLA-4 monoclonal antibody, in patients with metastatic melanoma unresponsive to prior systemic treatments: Clinical and immunological evidence from three patient cases. Cancer Immunol. Immunother. 58, 1297–1306 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishino M., Giobbie-Hurder A., Hatabu H., Ramaiya N. H., Hodi F. S., Incidence of programmed cell death 1 inhibitor-related pneumonitis in patients with advanced cancer: A systematic review and meta-analysis. JAMA Oncol. 2, 1607–1616 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Barroso-Sousa R., et al. , Incidence of endocrine dysfunction following the use of different immune checkpoint inhibitor regimens: A systematic review and meta-analysis. JAMA Oncol. 4, 173–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naidoo J., et al. , Toxicities of the anti-PD-1 and anti-PD-L1 immune checkpoint antibodies. Ann. Oncol. 26, 2375–2391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allenbach Y., et al. , Immune checkpoint inhibitor-induced myositis, the earliest and most lethal complication among rheumatic and musculoskeletal toxicities. Autoimmun. Rev. 19, 102586 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Yamauchi I., et al. , Incidence, features, and prognosis of immune-related adverse events involving the thyroid gland induced by nivolumab. PLoS One 14, e0216954 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sibaud V., Dermatologic reactions to immune checkpoint inhibitors: Skin toxicities and immunotherapy. Am. J. Clin. Dermatol. 19, 345–361 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., et al. , Treatment-related adverse events of PD-1 and PD-L1 inhibitors in clinical trials: A systematic review and meta-analysis. JAMA Oncol. 5, 1008–1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirza S., Hill E., Ludlow S. P., Nanjappa S., Checkpoint inhibitor-associated drug reaction with eosinophilia and systemic symptom syndrome. Melanoma Res. 27, 271–273 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Griffin L. L., et al. , Toxic epidermal necrolysis (TEN) associated with the use of nivolumab (PD-1 inhibitor) for lymphoma. JAAD Case Rep. 4, 229–231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomura H., et al. , Unexpected recalcitrant course of drug-induced erythema multiforme-like eruption and interstitial pneumonia sequentially occurring after nivolumab therapy. J. Dermatol. 44, 818–821 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Maloney N. J., Ravi V., Cheng K., Bach D. Q., Worswick S., Stevens-Johnson syndrome and toxic epidermal necrolysis-like reactions to checkpoint inhibitors: A systematic review. Int. J. Dermatol. 59, e183–e188 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Fixsen E., Patel J., Selim M. A., Kheterpal M., Resolution of pembrolizumab-associated steroid-refractory lichenoid dermatitis with cyclosporine. Oncologist 24, e103–e105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hines J., et al. , Steroid-refractory dermatologic and pulmonary toxicity in a patient on rituximab treated with pembrolizumab for progressive urothelial carcinoma: A case report. J. Med. Case Reports 15, 124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seervai R. N. H., et al. , Severe de novo pustular psoriasiform immune-related adverse event associated with nivolumab treatment for metastatic esophageal adenocarcinoma. J. Cutan. Pathol. 49, 472–481 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Randhawa M., et al. , Combined immune therapy grade IV dermatitis in metastatic melanoma. Asia Pac. J. Clin. Oncol. 15, 262–265 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Johnson D., et al. , IL17A blockade successfully treated psoriasiform dermatologic toxicity from immunotherapy. Cancer Immunol. Res. 7, 860–865 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Phillips G. S., et al. , Treatment outcomes of immune-related cutaneous adverse events. J. Clin. Oncol. 37, 2746–2758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison O. J., et al. , Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363, eaat6280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belkaid Y., Harrison O. J., Homeostatic immunity and the microbiota. Immunity 46, 562–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurabielle C., et al. , Immunity to commensal skin fungi promotes psoriasiform skin inflammation. Proc. Natl. Acad. Sci. U.S.A. 117, 16465–16474 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naik S., et al. , Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naik S., et al. , Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridaura V. K., et al. , Contextual control of skin immunity and inflammation by Corynebacterium. J. Exp. Med. 215, 785–799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beura L. K., et al. , Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wijeyesinghe S., et al. , Expansible residence decentralizes immune homeostasis. Nature 592, 457–462 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costello E. K., et al. , Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh J., Byrd A. L., Park M., Kong H. H., Segre J. A.; NISC Comparative Sequencing Program, Temporal stability of the human skin microbiome. Cell 165, 854–866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flores G. E., et al. , Temporal variability is a personalized feature of the human microbiome. Genome Biol. 15, 531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang D., Shi B., Erfe M. C., Craft N., Li H., Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci. Transl. Med. 7, 293ra103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh J., et al. ; NISC Comparative Sequencing Program, Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lax S., et al. , Bacterial colonization and succession in a newly opened hospital. Sci. Transl. Med. 9, eaah6500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooper L. V., Macpherson A. J., Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Constantinides M. G., et al. , MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linehan J. L., et al. , Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lima-Junior D. S., et al. , Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell 184, 3794–3811.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byrd A. L., et al. ; NISC Comparative Sequencing Program, Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 9, eaal4651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Curry J. L., et al. , Diverse types of dermatologic toxicities from immune checkpoint blockade therapy. J. Cutan. Pathol. 44, 158–176 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Totonchy M. B., Ezaldein H. H., Ko C. J., Choi J. N., Inverse psoriasiform eruption during pembrolizumab therapy for metastatic melanoma. JAMA Dermatol. 152, 590–592 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Elosua-González M., et al. , A case of de novo palmoplantar psoriasis with psoriatic arthritis and autoimmune hypothyroidism after receiving nivolumab therapy. Dermatol. Online J. 23, 9–12 (2017). [PubMed] [Google Scholar]
- 50.Lee J. S., et al. , Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 43, 727–738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jovanovic D. V., et al. , IL-17 stimulates the production and expression of proinflammatory cytokines, IL-beta and TNF-alpha, by human macrophages. J. Immunol. 160, 3513–3521 (1998). [PubMed] [Google Scholar]
- 52.Fujino S., et al. , Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziolkowska M., et al. , High levels of IL-17 in rheumatoid arthritis patients: IL-15 triggers in vitro IL-17 production via cyclosporin A-sensitive mechanism. J. Immunol. 164, 2832–2838 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Luchtman D. W., Ellwardt E., Larochelle C., Zipp F., IL-17 and related cytokines involved in the pathology and immunotherapy of multiple sclerosis: Current and future developments. Cytokine Growth Factor Rev. 25, 403–413 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Martin D. A., et al. , The emerging role of IL-17 in the pathogenesis of psoriasis: Preclinical and clinical findings. J. Invest. Dermatol. 133, 17–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi G. B., et al. , The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Q., Strong J., Killeen N., Homeostatic competition among T cells revealed by conditional inactivation of the mouse Cd4 gene. J. Exp. Med. 194, 1721–1730 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang S.-Q., et al. , The pattern of time to onset and resolution of immune-related adverse events caused by immune checkpoint inhibitors in cancer: A pooled analysis of 23 clinical trials and 8,436 patients. J. Clin. Oncol. 38, e15110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghisoni E., et al. , Late-onset and long-lasting immune-related adverse events from immune checkpoint-inhibitors: An overlooked aspect in immunotherapy. Eur. J. Cancer 149, 153–164 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Jameson S. C., Masopust D., Understanding subset diversity in T cell memory. Immunity 48, 214–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mueller S. N., Mackay L. K., Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 16, 79–89 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Stacy A., Belkaid Y., Microbial guardians of skin health. Science 363, 227–228 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Gallo R. L., Nakatsuji T., Microbial symbiosis with the innate immune defense system of the skin. J. Invest. Dermatol. 131, 1974–1980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ivanov I. I., et al. , Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duan J., Chung H., Troy E., Kasper D. L., Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe 7, 140–150 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosshart S. P., et al. , Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, eaaw4361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosshart S. P., et al. , Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015–1028.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michot J. M., et al. , Immune-related adverse events with immune checkpoint blockade: A comprehensive review. Eur. J. Cancer 54, 139–148 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Krummel M. F., Sullivan T. J., Allison J. P., Superantigen responses and co-stimulation: CD28 and CTLA-4 have opposing effects on T cell expansion in vitro and in vivo. Int. Immunol. 8, 519–523 (1996). [DOI] [PubMed] [Google Scholar]
- 70.Sotomayor E. M., Borrello I., Tubb E., Allison J. P., Levitsky H. I., In vivo blockade of CTLA-4 enhances the priming of responsive T cells but fails to prevent the induction of tumor antigen-specific tolerance. Proc. Natl. Acad. Sci. U.S.A. 96, 11476–11481 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain N., Nguyen H., Chambers C., Kang J., Dual function of CTLA-4 in regulatory T cells and conventional T cells to prevent multiorgan autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 107, 1524–1528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scharschmidt T. C., et al. , A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pardoll D. M., The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosato P. C., et al. , Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat. Commun. 10, 567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xing P., et al. , Incidence rates of immune-related adverse events and their correlation with response in advanced solid tumours treated with NIVO or NIVO+IPI: A systematic review and meta-analysis. J. Immunother. Cancer 7, 341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hand T. W., et al. , Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasson S. C., et al. ; Oxford Inflammatory Bowel Disease Cohort Investigators, Interferon-gamma-producing CD8+ tissue resident memory T cells are a targetable hallmark of immune checkpoint inhibitor-colitis. Gastroenterology 161, 1229–1244.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.J. R. Patrinely, Jr, et al. , Chronic immune-related adverse events following adjuvant anti-PD-1 therapy for high-risk resected melanoma. JAMA Oncol. 7, 744–748 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghisoni E., et al. , Long-lasting, irreversible and late-onset immune-related adverse events (irAEs) from immune checkpoint inhibitors (ICIs): A real-world data analysis. J. Clin. Oncol. 38, e15095 (2020). [Google Scholar]
- 80.Haslam A., Gill J., Prasad V., Estimation of the percentage of US patients with cancer who are eligible for immune checkpoint inhibitor drugs. JAMA Netw. Open 3, e200423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramos-Casals M., et al. , THU0628 immune-related adverse events induced by cancer immunotherapies. Big Data analysis of 13,051 cases (Immunocancer International Registry). Ann. Rheum. Dis. 78, 607–608 (2019). [Google Scholar]
- 82.Hellmann M. D., et al. , Nivolumab plus ipilimumab in advanced non-small-cell lung cancer. N. Engl. J. Med. 381, 2020–2031 (2019). [DOI] [PubMed] [Google Scholar]
- 83.André T., et al. ; KEYNOTE-177 Investigators, Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 383, 2207–2218 (2020). [DOI] [PubMed] [Google Scholar]
- 84.O’Hara M. H., et al. , CD40 agonistic monoclonal antibody APX005M (sotigalimab) and chemotherapy, with or without nivolumab, for the treatment of metastatic pancreatic adenocarcinoma: An open-label, multicentre, phase 1b study. Lancet Oncol. 22, 118–131 (2021). [DOI] [PubMed] [Google Scholar]
- 85.Shitara K., et al. , Efficacy and safety of pembrolizumab or pembrolizumab plus chemotherapy vs chemotherapy alone for patients with first-line, advanced gastric cancer: The KEYNOTE-062 phase 3 randomized clinical trial. JAMA Oncol. 6, 1571–1580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.National Research Council, Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [PubMed] [Google Scholar]
- 87.Conlan S., et al. ; NISC Comparative Sequencing Program, Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 13, R64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou Y., et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing datasets generated during this study are available at the NCBI GEO: GSE176030.