Significance
Plants use photoperiod information to control flowering. Flowering in rice is accelerated by short days, while other plants, such as Arabidopsis, flower in response to long days. The molecular mechanisms controlling photoperiod-dependent flowering in rice are not fully understood. Previous experiments have demonstrated the presence of several floral repressors that confer relatively mild flowering phenotypes. Here we show that the rice evening complex of the circadian clock is essential for the activation of flowering. We find that the evening complex (EC) binds directly to the promoters of floral repressors, reducing their transcription. These findings are consistent with a key role for the EC in controlling flowering in many diverse species including soybean, barley, Brachypodium, and Arabidopsis.
Keywords: rice, flowering, ELF3, LUX, Evening Complex
Abstract
Plants use photoperiodism to activate flowering in response to a particular daylength. In rice, flowering is accelerated in short-day conditions, and even a brief exposure to light during the dark period (night-break) is sufficient to delay flowering. Although many of the genes involved in controlling flowering in rice have been uncovered, how the long- and short-day flowering pathways are integrated, and the mechanism of photoperiod perception is not understood. While many of the signaling components controlling photoperiod-activated flowering are conserved between Arabidopsis and rice, flowering in these two systems is activated by opposite photoperiods. Here we establish that photoperiodism in rice is controlled by the evening complex (EC). We show that mutants in the EC genes LUX ARRYTHMO (LUX) and EARLY FLOWERING3 (ELF3) paralogs abolish rice flowering. We also show that the EC directly binds and suppresses the expression of flowering repressors, including PRR37 and Ghd7. We further demonstrate that light acts via phyB to cause a rapid and sustained posttranslational modification of ELF3-1. Our results suggest a mechanism by which the EC is able to control both long- and short-day flowering pathways.
Photoperiod provides seasonal information to control flowering. Rice flowering is accelerated under short-day (SD) photoperiod while other cereals such as wheat and barley flower under long-day (LD) conditions. The activation of flowering in rice is dependent on the expression of florigens Heading date3a (Hd3a) and Rice FT1 (RFT1) (1). Inductive SD photoperiods lead to the up-regulation of Hd3a, which activates the floral transition (2). Under LD, flowering occurs later, via induction of RFT1 (1). In Arabidopsis (an LD plant), the circadian- and light-controlled CCT domain-containing protein CONSTANS (CO) activates flowering in LD. CO expression is circadianly controlled by GIGANTEA (GI), while CO protein stability is dependent upon light-active phytochromes to induce Arabidopsis florigen FLOWERING LOCUS T (FT) (3, 4). The GI-CO-FT pathway is conserved in other flowering plants. The Arabidopsis CO homolog in barley, CO1, was demonstrated to be similarly regulated to activate VRN3, an FT ortholog, in LD (5), as are the homologs CO1 and CO2 in wheat (6). In rice, Heading date1 (Hd1), a CO homolog, has been proposed to be bifunctional: promoting flowering under SD and inhibiting it under LD (7). Besides the conserved GI-CO-FT pathway, rice has another regulatory mechanism to activate flowering through the CCT-domain flowering repressor Grain number, Plant Height, and Heading date1 (Ghd7), which in LD inhibits the flowering activator Early heading date1 (Ehd1), a B-type response regulator capable of inducing the expression of Hd3a and RFT (8, 9).
Recently, it has been shown that Ghd7 directly interacts with Hd1 (10, 11) and the LD suppression of flowering by Hd1 depends on the floral repressors Ghd7. Similarly, the presence of another CCT domain-containing protein PSEUDO-RESPONSE REGULATOR37 (PRR37) is predicted to switch the activity of Hd1 from a flowering activator to a repressor (10–12) potentially via a heteromeric protein complex (13). The expression of Ghd7 and PRR37 under noninductive LD is therefore a key means to regulate flowering, but the mechanism by which they perceive photoperiod is unknown. Ghd7 expression is regulated by ELF3-1 and the phytochrome pathway (14) whereas PRR37 functions in the circadian clock (15). ELF3 and the transcription factor LUX are part of the evening complex (EC) in several plant species. In Arabidopsis, the EC acts as a transcriptional repressor, binding and reducing the expression of key target genes such as the flowering activator PIF4 (16, 17). LUX orthologs in wheat (18), barley (19), pea (20), and soybean (21) have been implicated in the control of photoperiodic flowering. ELF3 was also implicated in the same mechanism in barley (22, 23), pea, and lentil (24). A rice genome duplication event resulted in the establishment of two ELF3 paralogs, ELF3-1 and ELF3-2 (25). Recently, the existence of a ternary EC has been confirmed in rice (26). The role of LUX in rice flowering is not known but mutants in ELF3 paralogs or ELF4 have a mild late flowering response, consistent with a role for the EC in flowering (26–28).
Results
The EC is Essential for Flowering in Rice.
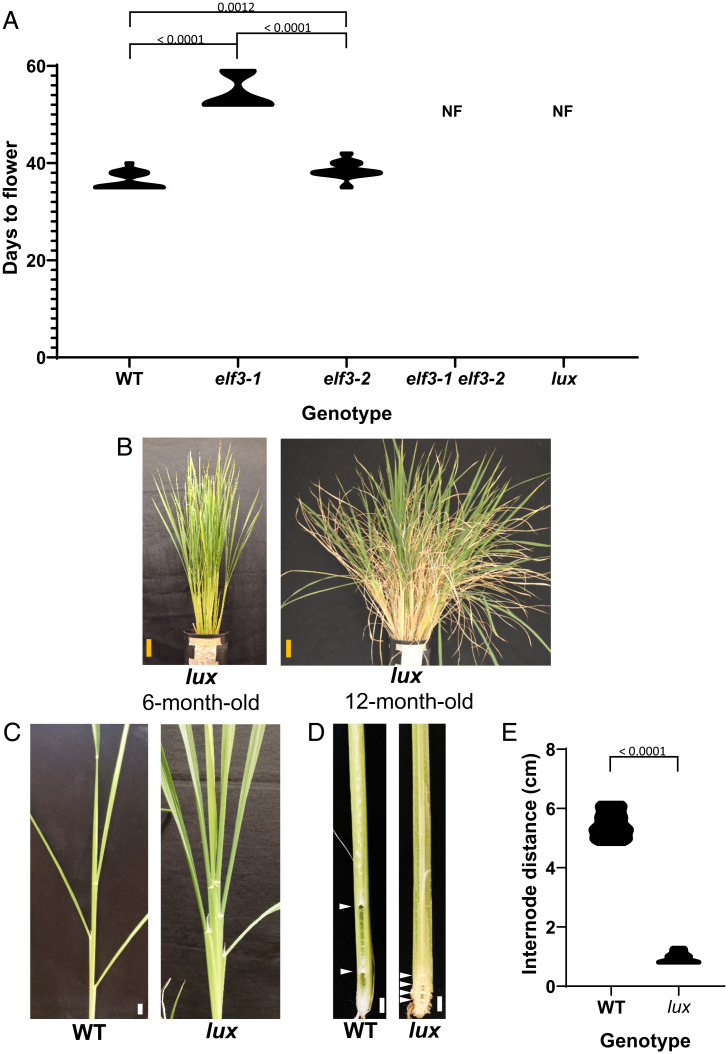
The EC is an important regulator of circadian programs in plants (16). To determine if it has a role in controlling photoperiodism in rice, we investigated mutants in LUX and two ELF3 paralogs generated by genome editing in the rice variety Nipponbare (hereafter referred as WT) (SI Appendix, Fig. S1A and Table S1). Rice contains two Arabidopsis ELF3 orthologs, ELF3-1 and ELF3-2. Individually, they have a mild effect on flowering (27, 28), however, the double elf3-1 elf3-2 mutant does not flower under our conditions (Fig. 1A). Similarly, we found that lux plants never flowered. This was despite growing plants under both long and short photoperiods for more than one year (Fig. 1B and SI Appendix, Fig. S1B). The EC mutants also exhibit short internode elongation (Fig. 1 C–E), a trait correlated with delayed flowering (29). ELF3-1 and ELF3-2 appear to both contribute to the formation of the EC, as only the double mutant shows the dramatic nonflowering phenotype of lux. Consistent with this, both ELF3-1 and ELF3-2 interact with LUX in yeast two-hybrid assays (SI Appendix, Fig. S1C) similar to previous reports (26). Taken together, these results indicate that the EC is essential for flowering in rice.
Fig. 1.
EC genes are essential for flowering in rice. (A) Flowering time for elf3-1, elf3-2, and lux in rice from the emergence of the second leaf. Plants were grown under ND conditions (12-h day, 12-h night). Nonflowering genotypes are marked with “NF”. P values are for two-tailed unpaired t test with n = 10. (B) Adult lux plants (line 120T7, 2 nucleotide insertion) at 6 and 12 mo growth in SD (10-h day, 14-h night). (Yellow scale bars represent 10 cm.) (C–E) lux (line 69S4b, insertion of one nucleotide in one allele and deletion of four nucleotides in the other allele) showing decreased distance between internodes. (White scale bars represent 1 cm.) P values are for two-tailed unpaired t test.
The EC Controls the Photoperiod Transcriptome.
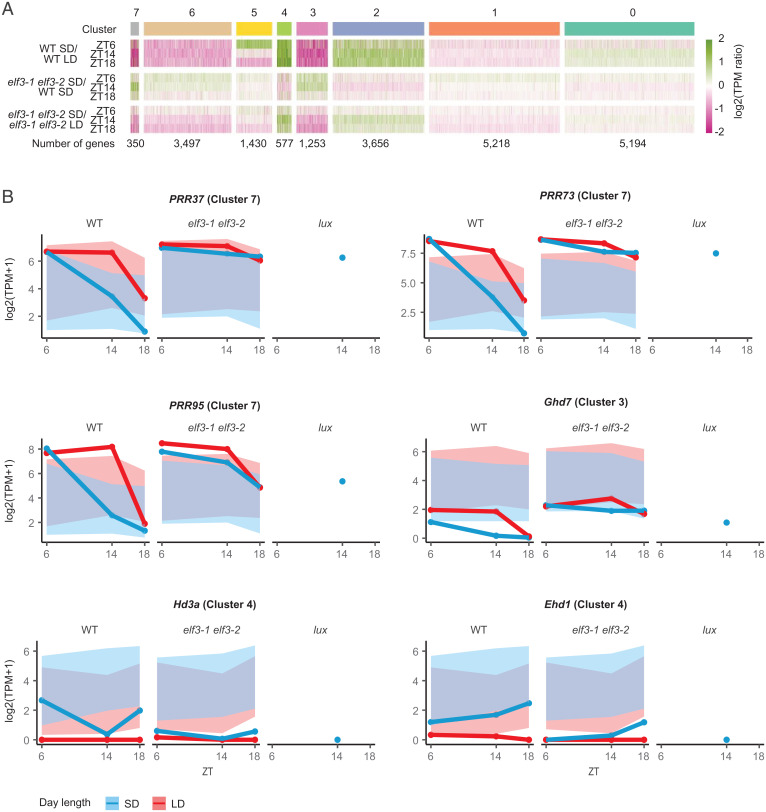
Flowering is a complex trait, and it is possible that disrupting the EC, an integral component of the circadian clock, may affect the floral transition indirectly. We therefore investigated the photoperiod-dependent transcriptome in rice by comparing inductive SD to noninductive LD conditions. We observe prominent clusters of photoperiod-dependent gene regulation at ZT6, ZT14, and ZT18, consistent with the major impact of photoperiod on rice development (Fig. 2A, SI Appendix, Fig. S2A, and Datasets S1 and S2). Particularly prominent are clusters 7 and 3, which show strong down-regulation in SD, and cluster 4, which is up-regulated in response to SD. The SD repressed cluster 7 contains key floral regulators, such as PRR37, PRR73, PRR95, GI, FKF1 and DOF12 (Fig. 2B) and is enriched for GO terms associated with circadian rhythm, rhythmic process, and photosynthesis (SI Appendix, Fig. S15) as has been observed for evening complex targets in Arabidopsis (17), while cluster 3 contains the flowering regulator Ghd7 (Fig. 2B) and is enriched for GO terms associated with salt stress (SI Appendix, Fig. S13) (26). The key outputs of flowering are found in the induced cluster 4, including Hd3a and RFT1. To investigate the potential role of the EC in this process, we examined the elf3-1 elf3-2 SD and LD transcriptome. By performing clustering using the same gene order as for the WT photoperiod response, we observe the transcriptome has a greatly reduced response to SD in elf3-1 elf3-2. Specifically, the strongly down-regulated SD clusters 7 and 3 become up-regulated in elf3-1 elf3-2, while cluster 4, which is up-regulated in SD in WT becomes down-regulated in elf3-1 elf3-2 (Fig. 2 A and B and SI Appendix, Fig. S2B). Indeed, similar patterns are observed across nearly all the clusters. Taken together, these results show that elf3-1 elf3-2 mutants in SD resemble WT plants grown in LD, at both the phenotype and transcriptome level. This behavior is also seen for lux at ZT14 (Fig. 2B and SI Appendix, Fig. S2 A and B), suggesting that the EC is required for the correct expression of the photoperiod transcriptome (SI Appendix, Fig. 2A). Consistent with the role of the EC as a transcriptional repressor in Arabidopsis, we find that nearly all the genes up-regulated in elf3-1 elf3-2 are also up-regulated in lux (SI Appendix, Fig. S3).
Fig. 2.
EC activity is essential for the SD transcriptome. (A) Clustering of gene expression under SD (10-h day, 14-h night) and LD (14-h day, 10-h night) reveals groups of genes that are activated (e.g., clusters 4 and 2) and repressed (e.g., clusters 7, 6, and 3) by SD photoperiods. These clusters become largely daylength neutral in the elf3-1 elf3-2 background (line 246U1.6a2: 1 nucleotide insertion in ELF3-1 locus: two nucleotide deletion in ELF3-2 locus). Values for log2(TPM ratio) > 2 or < −2 are transformed to the range of ± 2. (B) Examples of circadian and flowering time genes that lose their photoperiod responsiveness in elf3-1 elf3-2 and lux (line 120T7, two nucleotide insertion). Blue and red ribbons denote the overall behavior of the relevant cluster in SD and LD, respectively.
The EC Binds to the Promoters of Key Floral Regulators.
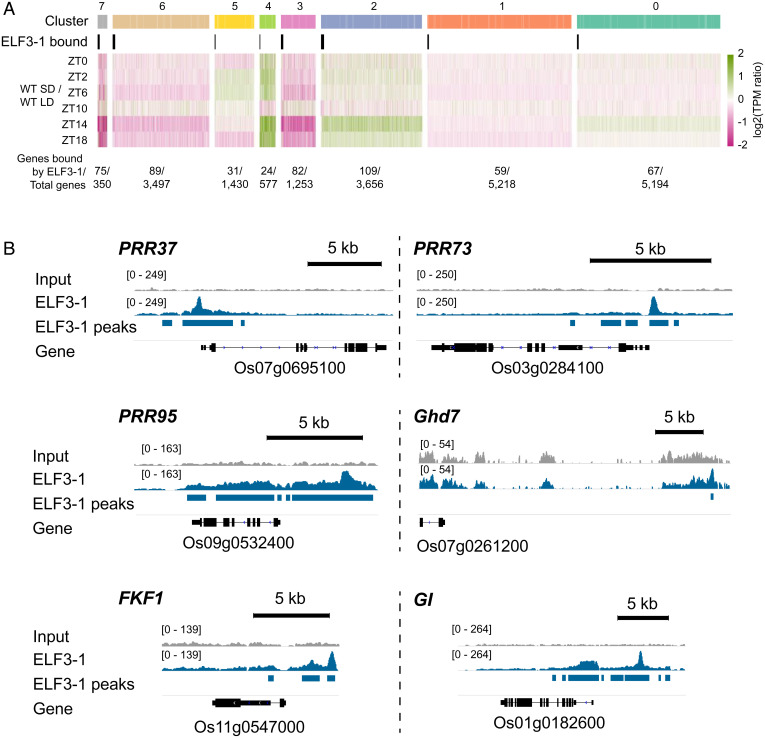
Since the clusters of genes repressed by SD become up-regulated in the elf3-1 elf3-2 background, we sought to determine if this is a direct effect of EC activity. We therefore investigated the genome-wide binding of ELF3-1 at ZT14. We observe enrichment for ELF3-1 binding in the promoters of cluster 7 genes (21% are bound by ELF3-1) and cluster 3 genes (7% bound) (Fig. 3A and Dataset S3). Cluster 7 is particularly prominent, with multiple floral regulator genes being directly bound by ELF3-1: PRR37, PRR73, PRR95 and GI (Fig. 3B). The major floral repressor present in cluster 3, Ghd7, is also directly bound by ELF3-1 (Fig. 3B and SI Appendix, Fig. S4). The association of ELF3 binding loci with gene repression is consistent with the EC role as a transcriptional repressor in Arabidopsis (17). By contrast, clusters of genes that are repressed in the elf3-1 elf3-2 background do not show an enrichment for ELF3-1 binding. For example, cluster 4 contains many key floral inducers, including Hd3a, RFT1, Ehd1, FTL10 and MADS14, but this cluster is not enriched for ELF3-1 binding events (Fig. 3A). This indicates that ELF3-1 controls flowering by binding to the promoter region of floral repressors, and not by repressing the expression of the florigen encoding genes directly. Since ELF3 does not have a DNA binding domain but is recruited to target promoters by LUX in Arabidopsis (30), we investigated if this is also the case in rice. We observe enrichment for sequences similar to LUX Binding Site (LBS) in the ELF3-1 ChIP-seq peaks, consistent with LUX recruiting ELF3-1 (SI Appendix, Fig. S3 A–C). ChIP-seq of a tagged version of LUX demonstrated an enrichment at the center of the peaks identified in the ELF3-1 ChIP-seq (SI Appendix, Figs. S3D and 4B). These results suggest that ELF3-1 is likely recruited by LUX to repress the expression of flowering-time regulators such as Ghd7 and PRR37. Since Ghd7 and PRR37 activity is linked to a delay in flowering in LD (9, 31), the photoperiodic-dependent expression of these genes suggests a direct mechanism to account for day-length–dependent flowering.
Fig. 3.
ELF3-1 directly binds and represses the expression of LD responsive genes in rice. (A) 21.4% of the genes in cluster 7 and 6.5% of genes in cluster 3 are directly repressed by ELF3-1 at ZT14. Values for log2(TPM ratio) > 2 or < −2 are transformed to the range of ± 2. (B) Representative views showing ELF3-1 binding peaks at key circadian and floral regulators.
ELF3-1 Activity is Controlled by phyB in Response to Light.
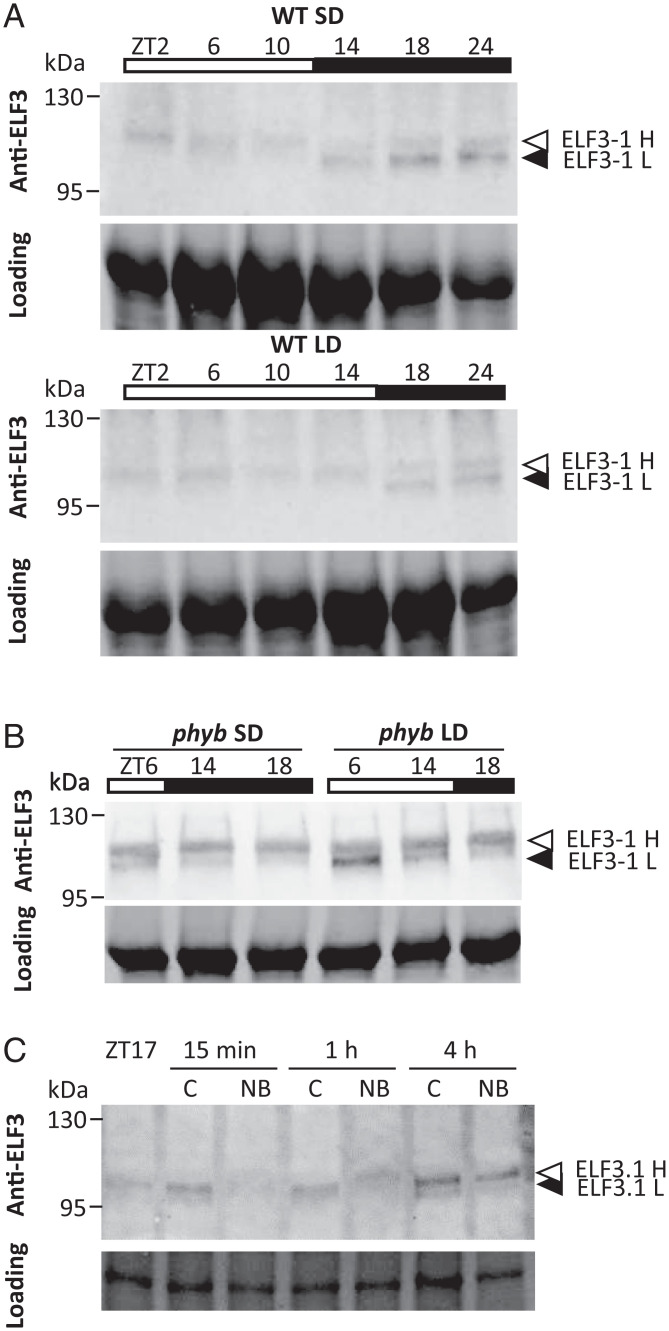
Since the EC is able to control the photoperiod transcriptome, we sought to determine how light signals are integrated by the EC. LUX protein levels rise at dusk and remain high for the next 12 h (SI Appendix, Fig. 6A), suggesting that LUX is likely not the limiting factor for EC activity. In Arabidopsis, ELF3 activity is controlled by temperature through a reversible phase change mediated by a prion-related domain (32). In the case of rice ELF3 homologs, we do not observe in silico evidence for such a prion. We therefore investigated if rice ELF3 protein levels are influenced by photoperiod. We observe that there is a clear difference between light and dark conditions for ELF3-1 protein behavior (Fig. 4A). In the light, a higher migrating protein band is present. By contrast, an additional lower sharp ELF3-1 band is visible in darkness. These are specific ELF3-1 protein bands since neither is observed in the elf3-1 background (SI Appendix, Fig. 6B). We therefore hypothesized that under LD conditions ELF3-1 is largely in the higher, likely posttranslationally modified form, rendering it inactive. By contrast, in SD the plants experience longer nights, enabling the lower, likely active, form to accumulate during darkness. By assaying ELF3-1 in plants grown under SD and LD, we observe this is the case with the lower ELF3-1 accumulating to a higher level in SD conditions (Fig. 4A).
Fig. 4.
ELF3-1 protein stability responds to light. (A) Anti-ELF3 specifically recognizes ELF3-1, which occurs in two bands during darkness. During light periods, only the higher (H) band is present. During SD (10-h day, 14-h night) the low (L) band is able to accumulate during the long night. (B) In the phyb background ELF3-1 loses photoperiod responsiveness, and both H and L bands are present in both light and dark periods. (C) The reduction in ELF3-1 levels in response to light is rapid, with only 15 min of NB being sufficient.
A key contributor to photoperiodism in rice is the photoreceptor phyB, with phyb mutants flowering earlier in LD, and being less responsive to night-break (NB) experiments (33, 34). Examining the transcriptome of the previously described phyb mutant (35) shows that under noninductive LD conditions, it resembles a WT transcriptome in SD, with repression of clusters 7 and 3 and up-regulation of cluster 4 (SI Appendix, Fig. S7 A and B). We therefore investigated if phyB may be necessary for the light-dependent behavior of ELF3-1. We observe a complete loss of responsiveness of ELF3-1 to light in phyb (Fig. 4B), with both the higher and lower forms of ELF3-1 present in day and night. This loss of responsiveness is independent of the photoperiod (Fig. 4B). Even short NB experiments are sufficient to greatly delay flowering in rice, a response that is dependent upon phyB (36). To determine if ELF3-1 levels respond sufficiently rapidly to account for this behavior, we assayed ELF3-1 in response to NB over a short time course. We observe loss of ELF3-1 signal within 15 min of NB, indicating this regulation is sufficiently rapid to account for the observed sensitivity of rice to even brief exposure to light during the night (Fig. 4C). This indicates that phyB is the photoreceptor that mediates the perception of photoperiod to control ELF3-1 activity.
Discussion
Photoperiodism enables plants to flower in the appropriate season. For temperate plants this is often spring, when daylength is increasing, while tropical plants such as rice often flower in subjective winter in response to short days. While LD and SD plants have opposite photoperiod requirements, it is noticeable that the EC has been found to regulate flowering responses in a variety of plants in both cases.
For example, the EC represses flowering in Arabidopsis (37–41) and other LD plants (18–23, 42). In contrast, genetic evidence from SD plants suggests the EC induces flowering (14, 21, 27, 43, 44). Here we find that LUX and ELF3-1 ELF3-2 are essential for flowering to occur in rice. As seen by others (14, 27, 43, 44), elf3-1 and elf3-2 single mutants have mild late flowering, suggesting that ELF3-1 and ELF3-2 are partially redundant in the formation of the EC. In soybean, two LUX homologs also act redundantly to activate flowering (21).
In Arabidopsis, the EC is recruited to the promoter regions of temperature, stress, and circadian clock signaling genes (17, 45, 46). We observed that the rice EC can also directly regulate gene expression. Direct targets of rice EC already implicated in the regulation of flowering include PRR37, Ghd7, GI, PRR73, and PRR95. It has been demonstrated that Arabidopsis homologs to PRR37, PRR73, and PRR95 coordinately activate flowering in Arabidopsis (47), while overexpressing either rice PRR37 or Arabidopsis PRR5 in rice delays flowering (48, 49). Our observations are consistent with the EC having a conserved role as a transcriptional repressor (16) and explain the nonflowering phenotype of lux and elf3-1 elf3-2. We observed that EC mutants have lower levels of Hd3a and RFT transcripts, as well as Ehd1 and FTL10, another FT ortholog in rice. We propose that the EC mutants are nonflowering due to the low levels of these flowering activators (1). Interestingly, neither Ghd7 nor PRR37 have been shown to directly interact with florigen promoters. However, Hd1, which can interact with Ghd7 and PRR37, is proven to bind and repress florigens under LD conditions (10–13). In the double mutant ghd7 prr37 however, Hd1 acts as a flowering activator in both SD and LD conditions (12). Hence, Hd1 can act as a flowering activator in SD and as a repressor in LD, since Hd1 by itself activates gene expression, while when in a complex with Ghd7 or PRR37 it has the opposite role (10–13) (Fig. 5).
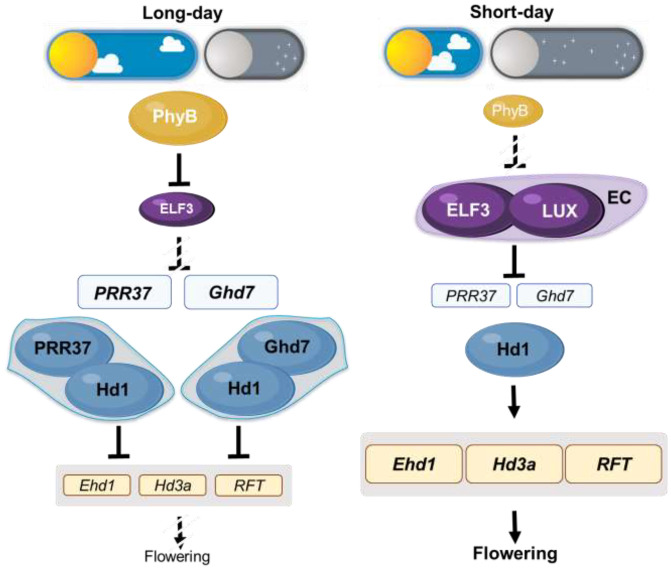
Fig. 5.
The EC integrates photoperiod information in rice. ELF3 activity increases during darkness, enabling the EC to reduce expression of key floral repressors such as PRR37 and Ghd7 in SD conditions. Activation of PhyB under LD conditions enables repression of ELF3-1, resulting in the enhanced expression of the transcriptional regulators that, in concert with Hd1, repress florigen genes Hd3a and RFT, therefore delaying flowering.
Phytochromes and ELF3 have antagonistic roles in flowering in many plants. For example, in the LD grass Brachypodium distachyon, phyC is essential for flowering, while elf3 mutants flower early and are photoperiod insensitive (50). In rice, mutants in the light receptor phyb are early flowering (36). While NB delays flowering in rice due to increased expression of Ghd7 (14), this effect is suppressed in phyb (34). Light signaling through PHYB and PHYC is also necessary for the NB response in wheat, which accelerates flowering through the up-regulation of PPD1 (50, 51). We find that ELF3-1 protein accumulates similarly in day and night in phyb, indicating a role for phyB in modifying ELF3-1 activity in response to light. Since phyB has been shown to bind and regulate target proteins by posttranslation modification (52–54), we suggest that light-controlled phyB is responsible for the conversion of the lower migrating ELF3-1 form to a higher form during the day, thus decreasing the accumulation of the EC complex in LD (Fig. 5). Previous results have shown that phyB can interact with ELF3-1 and ELF3-2 in vitro (14). The two distinctly migrating ELF3-1 bands are characteristic of posttranslation modification regulation. Indeed, ubiquitination of ELF3-1 by an E3 ubiquitin ligase (55) has been demonstrated, while a similar mechanism was described for ELF3-2 (56). Such posttranslation modifications independent of phyB may be responsible for the presence of the higher migrating ELF3-1 band even in phyb mutants. Consistent with this model, photoperiod dependent flowering in Arabidopsis is influenced by the ubiquitination of ELF3 and GI by the E3 ubiquitin ligase COP1 (57). Thus, while the key inputs and outputs of the photoperiod pathway are conserved (phytochrome signaling and florigens, respectively), the targets of EC repression determine the photoperiodic specificity of the response. In this way plants have been able to evolve dramatically different responses to photoperiod, facilitating adaptation to new environments.
Materials and Methods
Cloning of CRISPR/Cas9 Transformation Cassettes.
Binary vectors containing a hygromycin resistance gene driven by the cauliflower mosaic virus 35S promoter were used to generate Cas9-mediated genomic insertion and/or deletion events at LUX, ELF3-1, and ELF3-2 loci (SI Appendix, Fig. S1A and Table S1). The final vectors were introduced into Agrobacterium tumefaciens EHA105 for rice transformation (58, 59).
Cloning of LUX Genomic Tagged Transformation Cassette.
For generating a genomic tagged line of LUX, a 2,935 bp sequence comprising 2,000 bp of the promoter sequence, 217 bp of 5′UTR, and 717 bp of LUX coding sequence was fused to a 5xMYC tag followed by a 500 bp fragment downstream of LUX stop codon (3′UTR) by PCR. The final vector was introduced into Agrobacterium tumefaciens EHA105 for rice transformation.
Rice calli Culture and Transformation.
Stable transformations were generated using wild-type rice cv. Nipponbare following established methods (60).
Identification of Tagged LUX, lux, elf3-1, elf3-2, and elf3-1 elf3-2 Mutants.
Plant lines obtained from tissue culture harboring the hygromycin cassette were further tested. The LUX tagged lines were validated by testing the expression of LUX::MYC by RT-qPCR. Cas9 transformed lines were then tested for the presence of insertions or deletions (InDels) in the target loci using PCR and Sanger sequencing (61).
Rice Growth Conditions.
For the measurement of flowering-time, wild-type, elf3-1 (line 246U2.7a1), elf3-2 (line 246U2.7a1), and elf3-1 elf3-2 (line 246U2.7a2). Germinated seedlings were planted in containers with soil (soil:peat:vermiculite in 2:2:1 volume ratio) in controlled conditions at 28 °C under 12 h/12 h light/dark photoperiod (ND conditions) at 70% humidity. lux mutants were grown under similar conditions to compare flowering time, while initial plant material used was prepared from calli, so time to flower was counted from the emergence of the second leaf. Additionally, WT and lux rice (from calli culture) were grown under LD conditions (14-h day, 10-h night) and SD conditions (10-h day, 14-h night).
For gene expression or protein accumulation, wild-type, phyb-1 [generated elsewhere (36)], elf3-1 (line 246U2.7a1), elf3-2 (line 246U2.7a1) and elf3-1 elf3-2 (line 246U2.7a2) rice seeds were surface sterilized and germinated under dark for 3 d at 28 °C. Rice lux mutants (line 75Z3a) tillers were obtained by a donor plant and grown under similar conditions in order to compare results. Seedlings were grown on 1/2 MS solid medium. Pools of four to five plants were collected 21 d after germination at specific time-points: wild-type plants were collected at indicated times.
For the identification of EC direct targets, wild-type and LUX genomic 5xMYC tagged line seeds were surface sterilized and germinated under dark for 3 d at 28 °C. Germinated seedlings were then transferred to tubes containing sterile 1/2 MS solid medium and placed in a growth chamber in SD conditions for 14 d.
For the NB experiment, wild-type and phyb-1 rice seeds were germinated as described above but placed in a growth chamber at SD conditions for 10 d. Part of the plants was exposed to a NB of 300 μmol photons m−2s−1 of photoactive radiation for 15 min in the middle of the night (ZT17) while others remained under dark.
Extraction of Total RNA.
Total RNA was extracted from WT, phyb-1, lux (line 75Z3a), and elf3.1 elf3.2 (line 246U2.7a2), sampled at multiple points in the day under SD or LD conditions, by homogenizing samples to powder using a freeze cold mortar and pestle followed by total RNA extraction using the RNeasy plant kit (Qiagen), followed by DNase I treatment (Ambion). Total RNA was quantified using Qubit4 (Thermo Fisher Scientific) and tested for integrity using an RNA screentape in an Agilent2200 tape station (Agilent).
Chromatin Immunoprecipitation.
Three grams of plant material for each genotype was fixed in 1× phosphate buffered saline (10 mM PO43−, 137 mM NaCl, and 2.7 mM KCl) containing 0.5% formaldehyde (Sigma). The reaction was quenched with glycine to a final concentration of 62 mM. Chromatin immunoprecipitation (ChIP) was performed as described (62), with the exception that 100 μL of anti-cMyc agarose affinity gel (A7470-1ML) was used per sample or custom anti-ELF3 (Agrisera, AS184168, lot# 1808). Three microliters of anti-ELF3 was preincubated 1 h with Novex DYNAL Dynabeads Protein A and G (50 μL each. Sequencing libraries were prepared using TruSeq ChIP Sample Preparation Kit (Illumina). Libraries were sequenced on a NextSeq-500 (Illumina; single end, 75 bp reads).
Messenger RNA and DNA Library Preparation and Sequencing.
Ultrapure total RNA from wild-type, phyb-1, lux (line 75Z3a), and elf3-1 elf3-2 (line 246U2.7a2) rice genotypes was used to prepare high-throughput mRNA sequencing libraries using NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs). ChIP DNA libraries were performed using wild-type and LUX tagged line via TruSeq ChIP Sample Preparation Kit (Illumina). Nucleotide sequencing was performed in a NEXTSEQ550 (Illumina) using a high-throughput flow cell.
RNA-seq and ChIP-seq Data Processing.
HISAT2 with parameters ‘–no-mixed, –rna-strandness RF –dta –fr’ was used for aligning the raw RNA-seq reads to the rice genome assemblies (63). Transcripts per million (TPM) values for genes were calculated by StringTie with default settings directed by gene annotation file IRGSP-1.0 (64). Mean TPM values were calculated from replicates.
For processing ChIP-seq fastq files, BWA was used to map raw reads to rice genome IRGSP-1.0. Unmapped reads, mate unmapped reads, nonprimary alignment and duplicate reads were removed. Peaks were identified using MACS2 and filtered by q value < 0.05. BigWig files for IGV tracks were generated using deepTools function bamCoverage and normalized using RPKM.
The accession number for the raw and processed data from RNA-seq and ChIP-seq in this paper is Gene Expression Omnibus (GEO): GSE181836.
RNA-seq and ChIP-seq Data Analysis.
TPM values were transformed into log2(TPM+1). Genes with the maximum log2(TPM+1) > 2 were kept. Of 37,849 reference genes, 21,175 were kept. To perform clustering of transcriptomic data, a time-course perturbation matrices was constructed between SD and LD in WT, for example, . The selected perturbation matrices are described in the SI Appendix.
ELF3 and LUX bound genes were determined if ChIP-seq peaks overlap with the genomic regions of gene body extended by 2 kb upstream and downstream, respectively.
Motifs were predicted using HOMER2 (de novo and known motifs), using the genomic regions of 100 bp upstream and downstream of peak summits as target sequences and permuted sequences (excluding target sequences) as background. R package motifStack was used for generating SI Appendix, Fig. S5 and motifs were filtered using the following criteria: P value < 1e-5 for known motifs, P value < 1e-10 for de novo motifs.
Software Used for Analysis.
Graphpad Prism 8.0.2; Geneious Prime 2020.2.2; HISAT2 version 2.2.1; StringTie version 2.1.1; bwa version: 0.7.17-r1188; macs2 version 2.2.7.1; deeptools version 3.5.0; homer version 4.11; samtools version 1.11; bedtools version 2.30.0; R version 4.1.0;
Custom code for using R packages are deposited at https://github.com/yl-lu/Rice_EC.
Validation of Sequencing Data Using RT-PCR.
To validate the RNA-seq results, the same samples were used for RNA extraction using RNeasy Plant Mini Kit (Qiagen). cDNA was made from 1 µg of RNA in the Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics). cDNA was diluted to 1:100 to obtain a working concentration. The PCR mix was performed using 3 µL of diluted cDNA, gene-specific primers (available in SI Appendix, Table S2), and LightCycler 480 SYBR Green I Master (Roche Diagnostics) set up according to the manufacturer’s instructions. The cDNA was then amplified using a LightCycler 480 machine (Roche Diagnostics). Ubiquitin-conjugating enzyme E2 (UBC2) was used as an internal normalization. The experiment was done in triplicate for each sample. The results are given as a normalization of the target of interest/UBC2 amplification using double delta Ct analysis (65).
To validate ChIP-seq results, ChIP DNA and input DNA were de-crosslinked by submitting the samples to 65 °C for 8 h, followed by a cleaning step using Ampure beads according to manual. The obtained DNA was diluted to 1:100 to obtain a working concentration. The PCR mix was performed using 2 µL of diluted DNA, region-specific primers (available in SI Appendix, Table S2), and LightCycler 480 SYBR Green I Master (Roche Diagnostics) set up according to the manufacturer’s instructions. The DNA was then amplified using a LightCycler 480 machine (Roche Diagnostics). A region showing no particular enrichment was used as an internal control. The experiment was done in triplicate for each sample. Results are given as fold change enrichment relative to the internal control region using double delta Ct analysis (65).
Protein Extraction and Analyses.
Protein extracts were prepared from wild-type, elf3-1 (line 246U2.7a1), elf3-2 (line 246U2.7a1), elf3-1 elf3-2 (line 246U2.7a2), phyb-1, and LUX::MYC. Whole plants were homogenized to powder using a freeze cold mortar. For each 50 mg of plant homogenized material was added 150 µL of freshly made 2 × Laemmli buffer (Bio-Rad) supplemented with 50 mM dithiothreitol. Samples were subjected to gel electrophoresis and immunoblotted as described in the SI Appendix.
Yeast Two-Hybrid.
To test in vitro interaction, the coding sequence of ELF3-1 and ELF3-2 were cloned into pGADT7 vector (Clontech) whereas the coding sequence of LUX was cloned in pGBKT7 (Clontech). LUX in pGBKT7 and either ELF3 in pGADT7 vectors were used to transform Saccharomyces cerevisiae Y2HGold (Clontech). Positively transformed yeast grew in a synthetic defined medium lacking the amino acids leucine and tryptophan (SD). Interaction screening was performed in SD without adenine (-Ade) and histidine (-His).
Supplementary Material
Acknowledgments
We thank Makato Takano for providing the phyb mutant in rice. We thank Emmanuel Guiderdoni and Wendy Harwood for providing the CRISPR/Cas9 backbone vectors used in this study. Work in the lab of N.J.M.S. was supported by Fundação para a Ciência e Tecnologia (FCT) through the projects PTDC/BIA-FBT/31070/2017 and GREEN-IT Bioresources for Sustainability R&D Unit (UIDB/04551/2020, UIDP/04551/2020) and the fellowship PD/BD/114416/2016 (L.A.). K.E.J. and P.A.W. are supported by startup funds from the Leibniz IGZ and the Leibniz Association.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122582119/-/DCSupplemental.
Data Availability
All the sequencing data in this study is publicly available in GEO with the reference GSE181836.
All code used in this study for data analysis is publicly available on GitHub (https://github.com/yl-lu/Rice_EC).
References
- 1.Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K., Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Kojima S., et al. , Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Suárez-López P., et al. , CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Valverde F., et al. , Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Campoli C., Shtaya M., Davis S. J., von Korff M., Expression conservation within the circadian clock of a monocot: Natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 12, 97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw L. M., Turner A. S., Herry L., Griffiths S., Laurie D. A., Mutant alleles of Photoperiod-1 in wheat (Triticum aestivum L.) that confer a late flowering phenotype in long days. PLoS One 8, e79459 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yano M., et al. , Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doi K., et al. , Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue W., et al. , Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Nemoto Y., Nonoue Y., Yano M., Izawa T., Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86, 221–233 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., et al. , Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci. Rep. 7, 5388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujino K., Yamanouchi U., Nonoue Y., Obara M., Yano M., Switching genetic effects of the flowering time gene Hd1 in LD conditions by Ghd7 and OsPRR37 in rice. Breed. Sci. 69, 127–132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goretti D., et al. , Transcriptional and post-transcriptional mechanisms limit heading date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13, e1006530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh H., Tanaka Y., Izawa T., Genetic relationship between phytochromes and OsELF3-1 reveals the mode of regulation for the suppression of phytochrome signaling in rice. Plant Cell Physiol. 60, 549–561 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Murakami M., Tago Y., Yamashino T., Mizuno T., Characterization of the rice circadian clock-associated pseudo-response regulators in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71, 1107–1110 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Nusinow D. A., et al. , The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezer D., et al. , The Evening Complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 3, 17087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno N., Nitta M., Sato K., Nasuda S., A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotype to einkorn wheat. Genes Genet Syst. 87, 357–367 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Campoli C., et al. , HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: Phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 199, 1045–1059 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liew L. C., Hecht V., Sussmilch F. C., Weller J. L., The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO 1. Plant Physiol. 165, 648–657 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bu T., et al. , A critical role of the soybean Evening Complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. U.S.A. 118, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faure S., et al. , Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. U.S.A. 109, 8328–8333 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zakhrabekova S., et al. , Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. U.S.A. 109, 4326–4331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weller J. L., et al. , A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. U.S.A. 109, 21158–21163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami M., Tago Y., Yamashino T., Mizuno T., Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 110–121 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Wang X., He Y., Wei H., Wang L., A clock regulatory module is required for salt tolerance and control of heading date in rice.Plant. Cell Environ. 44, 3283–3301 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Fu C., et al. , OsEF3, a homologous gene of Arabidopsis ELF3, has pleiotropic effects in rice. Plant Biol. 11, 751–757 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Zhao J., et al. , OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One 7, e43705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gómez-Ariza J., et al. , A transcription factor coordinating internode elongation and photoperiodic signals in rice. Nat. Plants 5, 358–362 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Silva C. S., et al. , Molecular mechanisms of Evening Complex activity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 6901–6909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao H., et al. , Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 16337–16342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung J.-H., et al. , A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa R., et al. , Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell 17, 3326–3336 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishikawa R., Shinomura T., Takano M., Shimamoto K., Phytochrome dependent quantitative control of Hd3a transcription is the basis of the night break effect in rice flowering. Genes Genet. Syst. 84, 179–184 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Takano M., et al. , Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17, 3311–3325 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa R., et al. , Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genomics 285, 461–470 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Hicks K. A., et al. , Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274, 790–791 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Zagotta M. T., et al. , The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10, 691–702 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Doyle M. R., et al. , The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Hazen S. P., et al. , LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 102, 10387–10392 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onai K., Ishiura M., PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10, 963–972 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Boden S. A., et al. , EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 26, 1557–1569 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito H., et al. , Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol. 53, 717–728 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Yang Y., Peng Q., Chen G. X., Li X. H., Wu C. Y., OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol. Plant 6, 202–215 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Chow B. Y., Helfer A., Nusinow D. A., Kay S. A., ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal. Behav. 7, 170–173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C., et al. , LUX ARRHYTHMO mediates crosstalk between the circadian clock and defense in Arabidopsis. Nat. Commun. 10, 2543 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamichi N., et al. , Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 48, 822–832 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Liu C., et al. , OsPRR37 confers an expanded regulation of the diurnal rhythms of the transcriptome and photoperiodic flowering pathways in rice. Plant Cell Environ. 41, 630–645 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Nakamichi N., et al. , Flowering time control in rice by introducing Arabidopsis clock-associated PSEUDO-RESPONSE REGULATOR 5. Biosci. Biotechnol. Biochem. 84, 970–979 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Gao M., et al. , Phytochromes measure photoperiod in Brachypodium. bioRxiv [Preprint] (2019). 10.1101/697169 (Accessed 10 December 2021). [DOI]
- 51.Pearce S., et al. , Night-break experiments shed light on the Photoperiod1-mediated flowering. Plant Physiol. 174, 1139–1150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P. H., Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Park E., et al. , Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45, 968–975 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Shen Y., Khanna R., Carle C. M., Quail P. H., Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu C., et al. , The E3 ubiquitin ligase HAF1 modulates circadian accumulation of EARLY FLOWERING3 to control heading date in rice under long-day conditions. Plant Cell 30, 2352–2367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ning Y., et al. , OsELF3-2, an ortholog of Arabidopsis ELF3, interacts with the E3 Ligase APIP6 and negatively regulates immunity against Magnaporthe oryzae in rice. Mol. Plant 8, 1679–1682 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Yu J. W., et al. , COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32, 617–630 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miao J., et al. , Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawrenson T., Hundleby P., Harwood W., Creating targeted gene knockouts in Brassica oleracea using CRISPR/Cas9. Methods Mol. Biol. 1917, 155–170 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Hiei Y., Komari T., Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Brinkman E. K., Chen T., Amendola M., van Steensel B., Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaeger K. E., Pullen N., Lamzin S., Morris R. J., Wigge P. A., Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25, 820–833 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.European Bioinformatics Institute, EnsemblePlant release 47, Oryza sativa Japonica Group (IRGSP-1.0): fasta. http://ftp.ensemblgenomes.org/pub/plants/release-47/fasta/oryza_sativa/dna/. Accessed 3 June 2022. [Google Scholar]
- 64.European Bioinformatics Institute, EnsemblePlant release 47, Oryza sativa Japonica Group (IRGSP-1.0): gtf. http://ftp.ensemblgenomes.org/pub/plants/release-47/gtf/oryza_sativa/. Accessed 3 June 2022. [Google Scholar]
- 65.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the sequencing data in this study is publicly available in GEO with the reference GSE181836.
All code used in this study for data analysis is publicly available on GitHub (https://github.com/yl-lu/Rice_EC).